Abstract
Hepatitis C virus (HCV) replication in infected patients produces large and diverse viral populations, which give rise to drug-resistant and immune escape variants. Here, we analyzed HCV populations during transmission and diversification in longitudinal and cross-sectional samples using 454/Roche pyrosequencing, in total analyzing 174,185 sequence reads. To sample diversity, four locations in the HCV genome were analyzed, ranging from high diversity (the envelope hypervariable region 1 [HVR1]) to almost no diversity (the 5′ untranslated region [UTR]). For three longitudinal samples for which early time points were available, we found that only 1 to 4 viral variants were present, suggesting that productive infection was initiated by a very small number of HCV particles. Sequence diversity accumulated subsequently, with the 5′ UTR showing almost no diversification while the envelope HVR1 showed >100 variants in some subjects. Calculation of the transmission probability for only a single variant, taking into account the measured population structure within patients, confirmed initial infection by one or a few viral particles. These findings provide the most detailed sequence-based analysis of HCV transmission bottlenecks to date. The analytical methods described here are broadly applicable to studies of viral diversity using deep sequencing.
Hepatitis C virus (HCV) is a positive-strand enveloped RNA virus of the flavivirus family. HCV infects ∼170 million people worldwide with a high rate of persistence (1, 2) and is a major etiological agent of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. The current standard of therapy is the combined use of pegylated alpha interferon (IFN-α) and ribavirin (9), although there are substantial limitations due to toxicity and resistance profiles (47). Recent development of various small-molecule inhibitors that specifically target HCV offer some promise (13), but challenges still remain because the size and diversity of viral populations promote rapid development of drug resistance (28, 42). In an infected individual, serum HCV RNA levels can reach 10 to 100 million IU/ml (40). The viral RNA polymerase is estimated to make 1 error per 10,000 to 100,000 bp copied (22), but the viral genome is only 9,600 bases, resulting in diversification of the viral population, so that most viral genomes differ in sequence from the population consensus (16, 20, 21). Thus, when antiviral pressure is exerted on a viral population, sequence variants with reduced sensitivity may expand in the presence of the selective pressure (30, 41) and cause resistance (37). Consistent with this, differential sequence diversity in HCV populations has been linked to clinical outcome (7, 8).
The size and complexity of HCV populations has made their analysis challenging. However, new deep-sequencing and bioinformatics methods are well suited to analyzing this problem. Using the 454/Roche technology, it is possible to generate more than 108 bases of DNA sequence in a single 1-day run, albeit in fragments 200 to 500 bases in length (24). In addition, many samples can be multiplexed in single experiments using DNA barcodes introduced in amplification primers to tag each sample (3, 12, 45, 46), allowing many viral sequences to be characterized in a single experiment.
Here, we analyze HCV diversity by pyrosequencing a series of representative viral regions contained within PCR amplicons, and we use methods from the environmental microbiology field for data processing and analysis. In both virology and environmental microbiology, populations of interest commonly consist of many related but nonidentical sequences (e.g., viral lineages with related sequences or bacteria harboring related 16S rRNA gene sequences). Assembly of short pyrosequence reads into longer scaffolds is quite difficult in such a setting, because the related sequences present in the population can be assembled in many different ways. Complicated data-processing methods yield at best complex probabilistic models of variants likely to be present in the population (6). For this reason, in studies of bacterial 16S DNA from uncultured communities, many groups have used simplified analysis of single 16S amplicons that query short regions of the 16S rRNA gene (5, 11, 14, 23, 25, 38, 44). Extensive simulations and practical applications show that analysis of such “sequence tags” can disclose biologically meaningful clusters and gradients in collections of samples. Here, we apply a similar approach, using sequence tags for several regions of the HCV genome. This approach has the disadvantage of losing linkage information between amplicons, but it does allow the efficient analysis of large numbers of viral variants over many samples.
A major challenge, however, is distinguishing variations authentically present in viral populations from artifactual mutations introduced as a result of the isolation procedure or sequencing error. Sequence recovery involves PCR steps that can result in base pair substitutions or artifactual chimera formation. The 454/Roche method, like any sequencing method, has a characteristic error rate and particularly elevated error rates at homopolymer runs (24). In this study, we took advantage of improved methods for error control using the PyroNoise program of Quince and colleagues, which was first used for analysis of 16S rRNA gene sequences (29). The PyroNoise program preclusters the raw light intensity data generated during pyrosequencing by the 454/Roche method, which removes most homopolymer errors. In reconstruction experiments, Quince and colleagues showed that 454/Roche sequence analysis of artificially constructed mock 16S rRNA gene communities yielded greatly inflated numbers of sequence types due to error, but preclustering using PyroNoise reduced the diversity to values much closer to the correct value. Here, we used a two-stage clustering method to remove noise. In the first stage, raw light intensity data were preclustered with PyroNoise (29); then, in the second stage, after interpretation of the sequence as base calls, sequences were clustered at 98.5% identity. The second step allowed us to take advantage of redundancy in the reads to improve sequence quality, though distinguishing genuine low-level variations in the viral populations from error is a challenge.
We determined 174,185 high-quality HCV sequence reads to characterize (i) longitudinal variation in HCV populations following transmission, (ii) differences in HCV variation between HCV-monoinfected and HIV-HCV-coinfected subjects, and (iii) variation in a control HCV genome cloned in a bacterial plasmid to quantify variation arising during the isolation and analytical procedure. We developed amplicons to characterize four regions of the HCV genome (Fig. 1A) and found that HCV sequence diversity ranged from almost nonexistent to extreme depending on the region of the viral genome studied. Using the deep-sequencing data, we estimate that only one or a few viral variants seeded initial infection, but after that, viral variants could expand to >100 in a single individual. Thus, these data specify the numbers of particles seeding productive infection and provide a general framework for the use of deep-sequencing data to characterize the structures of viral populations.
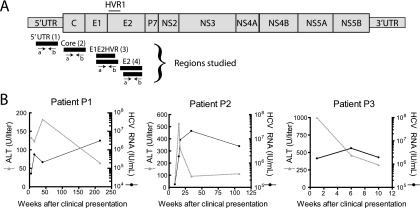
FIG. 1.
HCV genome and characteristics of three subjects studied longitudinally during acute HCV infection. (A) The HCV genome and the positions of amplicons studied. The amplicons are numbered 1 to 4 from left to right, and a letter is used to indicate the direction of sequence determination. For the E1E2 HVR1 (3) and E2 (4) amplicons, two slightly different primers were used in each direction in an effort to maximize the diversity of recovered sequence variants, and these are indicated by the two bars. (B) HCV load and ALT levels for patients 1 to 3 during acute HCV infection. The x axis shows the number of weeks after clinical presentation, which for these patients was close in time to initial infection. Further patient characteristics were as follows: patient 1, injecting drug user, anti-HCV negative on 18 June 2001, first ALT flare (ALT, 677) on 6 July 2001, anti-HCV positive on 11 October 2001; patient 2, possible medical exposure, anti-HCV negative on 16 May 2001, initial ALT flare (ALT, 467) on 9 January 2004, anti-HCV positive on 22 April 2004; patient 3, injecting drug user, anti-HCV negative on 31 January 2006 (slightly abnormal ALT, 73), initial ALT flare (ALT, 640) on 10 April 2006, anti-HCV positive on 11 April 2006.
MATERIALS AND METHODS
HCV RNA isolation and multiplex PCR amplification.
HCV RNA from patient plasma samples was purified using the QIAamp viral-RNA mini kit (Qiagen, Valencia, CA). All samples were amplified using the OneStep RT-PCR kit (Qiagen, Valencia, CA) following the manufacturer's recommendations. The RNase inhibitor RNasin (Promega, Madison, WI) was added to each reaction mixture to a final concentration of 0.2 U/μl. The reverse transcription (RT)-PCR cycling conditions were as follows: 1 cycle at 50°C for 60 min, followed by 15 min at 95°C; 40 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and then extension at 72°C for 1 min; and final extension at 72°C for 10 min. The kit was chosen in part because we needed the more robust polymerase to amplify low-abundance samples. In a previous study (12), the RT step was found to be a negligible source of error compared to other sources. In order to multiplex samples in the downstream pyrosequencing procedure, barcoded primers, each containing a unique 8-base sequence tag, were used during the RT-PCR (Fig. 2A), as previously described (12, 46). For primer design, representative genotype 1 and 2 HCV sequences from the Los Alamos HCV sequence database (19) were aligned, and primers were designed to specifically amplify the 5′ untranslated region (UTR), the core, or the E1 and E2 envelope regions.
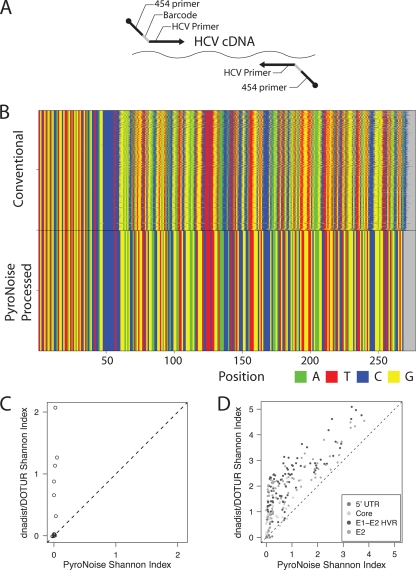
FIG. 2.
Methods for pyrosequence acquisition: DNA barcoding and controlling error using PyroNoise. (A) The DNA barcoding strategy. For each primer pair, an 8-base sequence (barcode) that indexed the patient and time point was included. (B) Controlling 454/Roche pyrosequencing error using PyroNoise. Sequences from a control plasmid amplicon (5′ UTR 1A amplicon) were aligned using the left-hand primer sequence. The x axis shows the base number in the read, and the y axis shows the cumulative number of sequence reads. The top sequence block shows the unprocessed raw pyrosequence reads, which contain considerable homopolymer error. The bottom block shows the improved alignments after application of the PyroNoise algorithm. (C) Comparison of the Shannon Diversity Index for OTUs from plasmid controls made by PyroNoise (x axis) versus DNADist and Dotur (y axis). The correct answer for all amplicons is 0. Default parameters were used for each program. (D) Comparison of the Shannon Diversity Index for OTUs from HCV samples made by PyroNoise (x axis) versus DNADist and Dotur (y axis). The correct answer for all amplicons varies from 0 (ultraconserved HCV regions) to higher values (hypervariable HCV regions). Default parameters were used for each program. Dotur OTUs were generated at 97% identity.
Sequence determination.
PCR products were separated by agarose gel electrophoresis, the bands of interest were excised, and DNA was extracted from the gel slices using the Qiagen gel extraction kit (Qiagen, Valencia, CA). Gel-purified PCR products were pooled and subjected to pyrosequencing using the 454/Roche GS FLX platform (24). The initial sequence reads were processed using the following criteria: (i) a perfect match to the barcode and primer sequences, (ii) >200 bases in length, and (iii) no undetermined bases (Ns). The barcodes and primer sequences were trimmed from the sequences, ultimately yielding a total of 174,185 high-quality sequences for downstream analysis. For analysis, the reads were trimmed to remove primers, yielding a final length of 201 nucleotides (nt).
Bioinformatics analysis.
All statistical analysis was carried out using R version 2.9.0 (http://www.R-project.org). All sequences were processed to remove reads containing Ns, as called by the 454/Roche software, because sequence errors are known to be autocorrelated in 454/Roche pyrosequencing data.
To control errors in the 454/Roche method, the data were further processed using a two-stage implementation of PyroNoise (29). In the first stage, the raw light intensity values generated from the 454/Roche instrument were clustered to remove homopolymer errors; in the second stage, the sequences themselves were clustered to remove base pair substitutions. The PyroNoise parameters were a cluster size of 1/20 and a 0.05 cutoff for initial hierarchical clustering. As a control, a plasmid carrying the HCV genome, pFL-H77/JFH (26), was amplified and processed in parallel.
After interpretation of flowgrams as base calls, alignments were generated using Mosaik. Distances between sequences were quantified as the Levenshtein distance. Indels were treated as mismatches (that is, a 1-base indel would be scored as a 1-base mismatch). For a new operational taxonomic unit (OTU) to be called under our analytical conditions, it needed to be represented by >2 reads and to differ by >5 positions out of ∼210 nt. Homopolymers greater than 2 nt were not counted in the distance analysis. For the furthest neighbor-clustering method used, reads with fewer than 3 nucleotide differences were grouped together. We chose 3 nt because, at an error rate of 1/1,000, if errors are randomly distributed, one would expect 2 errors in 1.6% of the reads and 3 or more errors in 0.1%. Thus, 3 nt represents a reasonable compromise for trying to maintain some of the rare authentic variability while minimizing error-induced variability.
We used a 3% OTU from Dotur (33) based on distances calculated by DNADIST (default parameters) (http://www.med.nyu.edu/rcr/rcr/phylip/faqs.html#citation) in the PHYLIP package for the analysis shown in Fig. 2. The Chao1 method uses counts of rare species (represented by one or two individuals) to determine species richness. For the conservative analysis reported here, in which OTUs supported by only one or two sequence reads were excluded, we redefined the categories containing three or four reads as the one- or two-read sets.
Nucleotide sequence accession numbers.
All sequence reads have been deposited in NCBI under accession numbers SRA012580.
RESULTS
HCV samples studied.
Table S1 in the supplemental material summarizes the HCV samples studied and the numbers of sequence reads determined for each. Three patients (P1 to P3) diagnosed close to the time of initial infection were studied longitudinally. Longitudinal HCV viral-load data are shown in Fig. 1B, together with serum alanine aminotransferase (ALT) levels, a marker for hepatic inflammation. These patients were tracked from the time of clinical presentation, with newly diagnosed positive HCV RNA (all genotype 1) and elevated ALT levels consistent with acute hepatitis C virus infection, as previously defined (15). All had negative HCV tests preceding diagnosis. Patients P1 and P2 developed apparently chronic infection over 6 and 2 years of follow-up, respectively. The long-term outcome is not known for patient P3, who was lost to follow-up 10 weeks after clinical presentation.
For the cross-sectional study, eight HIV-HCV-coinfected patients were studied. With the exception of one patient, all patients had CD4 counts below 300 cells/μl, enabling us to investigate the consequences of decreased immune function during HIV coinfection. As controls, the HCV populations in two additional monoinfected patients were characterized.
We also analyzed plasmid DNA carrying the HCV genome (pFL-H77/JFH1) (26), which provided a homogeneous control template of known sequence. Any diversity detected in sequence reads from the plasmid template were due to either mutation during PCR amplification or errors in the sequence determination steps, providing data for optimization of our error control procedures.
Pyrosequence data acquisition.
HCV RNA was purified from plasma, reverse transcribed, and then PCR amplified using primers complementary to HCV sequences. Amplicons were targeted to four regions (Fig. 1A): the 5′ UTR, the core nucleocapsid protein-coding region, and two regions of the envelope genes. Amplicons were chosen based on previous literature to sample both highly conserved (5′ UTR and core) and diverse (envelope) regions. The amplicons were numbered 1 to 4. For the two envelope amplicons, two primer pairs were used in an effort to minimize recovery biases possibly arising due to mismatches in the primer binding sites (indicated as A and B, with a and b indicating the direction of sequence determination). The primer sequences are shown in Table S2 in the supplemental material. All primers were composites containing the required primer binding sites for the 454/Roche sequencing procedure linked to the HCV-specific primers (Fig. 2A). An 8-base DNA barcode, which indexed the patient and time point from which the sample was derived, was positioned between the two (3, 12). This allowed PCR products to be pooled for sequencing, and then reads were parsed into individual amplicons after sequence determination.
We analyzed 288 patient-time-point-amplicon combinations (see Table S1 in the supplemental material). A total of 174,185 pyrosequence reads passed quality control (∼40 million bases of DNA sequence). The quality control criteria (14) included (i) a perfect match to the barcode and primer sequences, (ii) >200 bases in length, and (iii) no Ns. For HCV monoinfection, 69,825 sequences were recovered; for HIV-HCV coinfection, 73,656 sequences were recovered; and for the plasmid DNA, 30,704 sequences were recovered.
Data processing using PyroNoise.
Sequencing error can artificially inflate the numbers of sequence variants. This is particularly severe at homopolymeric runs, where the 454/Roche method can have difficulty calling the correct number of identical bases. Quince et al. have shown that improved accuracy can be obtained by first clustering the quantitative data from the initial light intensities generated during pyrosequencing using a distance measure that models pyrosequencing noise. The method has been implemented in the PyroNoise program (29). In addition, in this study, we incorporated a second stage of sequence clustering to remove PCR base pair substitutions, in which denoised sequences are clustered into OTUs with 98.5% identity (accepting three differences over 200 nt). PyroNoise preclustering greatly suppressed inflation of OTUs due to errors in published model studies (29), and similar results were obtained here.
Figure 2B presents an example, using the data from amplification of the HCV-bearing control plasmid pFL-H77/JFH1. At the top are the sequences after conventional processing and alignment by the left PCR primer sequence. As can be seen, many reads go out of alignment as the sequence extends from left to right due to errors at homopolymers. At the bottom is the alignment after error control using PyroNoise, which removed homopolymer errors.
The use of PyroNoise processing prior to formation of OTUs was then compared to a widely used program for making OTUs (Dotur), which uses conventional percent identity of the base calls as the basis for clustering sequence reads (33). In this analysis, OTUs were (i) clustered by Dotur using a 97% identity threshold or (ii) first preclustered using PyroNoise and then clustered at 97% identity in a second step. To allow comparison, the number of OTUs and the distribution of reads within OTUs were summarized using the Shannon Diversity Index for output from Dotur and PyroNoise. Values for the Shannon Index are increased by either adding more species (in this case, OTUs) to a community or distributing individuals (in this case, sequence reads) more evenly among the species (OTUs) present. Figure 2C shows that Shannon values for control plasmid sequence reads were much higher for the OTUs made using Dotur than for OTUs made using PyroNoise. We also compared the two methods over the HCV sequence data from patient samples, though unlike the case of the plasmid control, we did not know the true number of OTUs in the patient samples. Figure 2D shows that values were higher with Dotur than with PyroNoise, particularly for low Shannon values, which we infer to be erroneous inflation of the diversity values in the OTUs generated with Dotur. This general conclusion was not sensitive to variations in the parameters used in each program for data processing (data not shown). Thus, subsequent analyses were carried out on the data after PyroNoise processing.
Diversity along the HCV genome.
Variation in the HCV sequences is summarized in Fig. 3. Each base is color coded, and all 174,185 sequences are shown as “stacks” aligned on the HCV genome. The x axis indicates the position on the HCV genome, corresponding to the indicated positions on the genetic map. The y axis indicates the number of sequence reads, showing a cumulative sum over all samples and amplicons. The samples were analyzed separately for reads from the plasmid control (top), HCV-monoinfected (bottom), and HCV/HIV-dual-infected (middle) subjects.
FIG. 3.
Sequence variation across the HCV genome. Shown is a summary of sequence data. The x axis shows the position on the HCV genome, and the y axis indicates the cumulative numbers of sequences analyzed. Each DNA base is colored as indicated at the bottom right. Amplicons are numbered as in Fig. 1.
For the plasmid control, each of the four amplicons showed almost exclusively continuous vertical colored bands. This indicates that the same base was found in each position in almost every read after PyroNoise processing.
For the two sets of HCV samples, corresponding to HCV monoinfection and HIV-HCV coinfection, the results varied similarly by amplicon. For the leftmost 5′-UTR amplicon, little diversity in sequence was seen. Previous literature has established that this region is extremely highly conserved. For the core amplicon, the sequence composition was generally similar, though some variation could be discerned. For the two amplicons in the envelope region (rightmost in Fig. 3A), much more variation was observed. The variation was higher throughout the read but was particularly high around base position 1500, in HVR1 of the E2 envelope. Both the HCV monoinfection and HIV-HCV coinfection samples showed a high degree of variation in this region (discussed further below).
Longitudinal sequence variation in early HCV infection.
We next investigated longitudinal trends in sequence diversity for patients 1 to 3. Samples from each time point were condensed into OTUs after PyroNoise processing, and the diversity was summarized using the Shannon Index. Figure 4 shows longitudinal plots for each of the amplicons for patients 1 to 3. Each subject is shown by a single color. The raw numbers of sequence reads for each sample, and the Shannon values for each, are presented in Table S1 in the supplemental material.
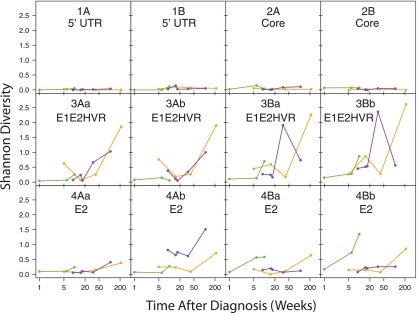
FIG. 4.
Longitudinal trends in sequence diversity analyzed using the Shannon Index. Data for each amplicon and read direction are shown separately. Each patient is indicated by a color. Times after diagnosis for the samples are shown on the x axis, and Shannon Diversity Index values are shown on the y axis. The data are shown in tabular form in Table S1 in the supplemental material. Data are shown for PyroNoise-processed amplicons (OTUs formed from one or two sequence reads were retained in this analysis). Patient 1, yellow; patient 2, purple; patient 3, green. All pairwise comparisons between pooled data from each amplicon achieved P values of <0.02 (t test). Analysis using a generalized linear model, comparing the Shannon diversity to amplicon and time data while controlling for the patient, showed that diversity significantly increased with time.
The Shannon values for the 5′-UTR (1) and core (2) regions were low (<0.3) and did not increase significantly over time. These findings provide a critical control for other amplicons with greater diversity. Mutation during RT-PCR or errors in sequence determination could falsely inflate diversity, but results with the 5′-UTR and core amplicons show that the magnitude of such errors was comparatively small.
Shannon Index values for the envelope amplicons were much higher than for the 5′ UTR and core (P < 0.02 for pairwise comparisons) and tended to increase over time (P < 0.05 for comparison of the first and last points of any amplicon). Patient 1 showed the most extreme behavior, with a high Shannon Index value (up to 2.5) for the envelope HVR1 amplicon at 215 weeks, indicating either diversification of the envelope sequences or a second infection. Patient 2 also showed a notable increase in diversity in HVR1 over time, reaching values up to 2.0. Patient 3, who was sampled over the shortest time (from 1 to 10 weeks following the initial clinical presentation), showed a longitudinal increase in diversity for some but not all the envelope amplicons.
Changes in sequence diversity were not strictly linked between amplicons. While the envelope-coding regions varied, the 5′-UTR and core regions did not. Even within the envelope-coding region, patterns of diversity varied. For example, in the HVR1 amplicon (3Aa) for patient 1, there was a sharp increase in diversity over time, whereas in the 4Aa amplicon, the diversity increased much less. Evidently different parts of the genome, or even different parts of the envelope region, diversified at different rates after infection.
The longitudinal patterns of PyroNoise-processed OTUs were analyzed further using phylogenetic trees. The analysis of diversity using the Shannon Index presented above is not sensitive to rare sequence variants, such as those due to mutation from PCR error or chimera formation. However, the phylogenetic survey, which emphasizes rare variants, would be. For that reason, in this and subsequent studies, we excluded variants supported by only one or two sequence reads. Control experiments showed that these reads were enriched in sequences with low sequence similarity to a collection of all available HCV sequences. Inspection of individual sequences indicated that several were apparent chimeras of HCV sequences with unidentified sequences. After being processed by this approach, 11 of 12 plasmid control samples showed single OTUs, with the 12th showing two (see Table S1, column K, in the supplemental material).
For the 5′-UTR amplicon (Fig. 5A), all the sequences clustered in a single OTU. In the figure, each rectangle indicates a single sequence variant, and the extent of the black bar indicates the percentage of the total population contributed by that variant. The plasmid control also formed a single OTU. Longitudinal analysis of the core amplicon showed similar highly conserved structure with only rare variation (data not shown).
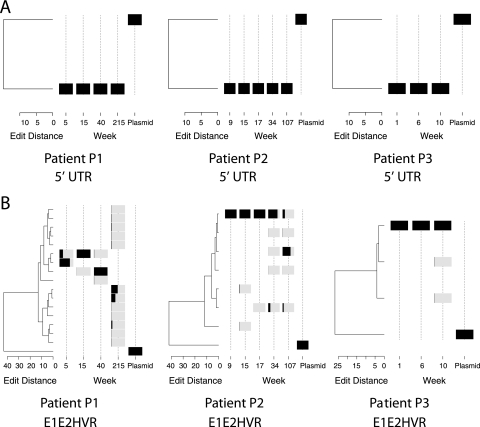
FIG. 5.
Analysis of HCV sequence data on phylogenetic trees. (A) Diversification of 5′ UTR amplicons (1A) during HCV replication. The relative abundance of a type is shown by the black shading, where the extent of the black region from left to right within the gray bar indicates the proportion in the population. Trees were generated using neighbor joining. (B) Diversification of HVR1 sequences (3B) during HCV replication. The plasmid control is shown for comparison on each tree.
In contrast, the amplicons for the envelope region showed much greater variation (Fig. 5B). For patient 1, three related OTUs predominated for the first three time points sampled (5 to 40 weeks after clinical presentation). Over this time period, the principal OTUs differed by about 5 base substitutions out of ∼200 total. By 215 weeks after infection, a different branch of the lineage predominated, separated from the earlier OTUs by an edit distance of at least 12 substitutions. For patient 2, the population was relatively stable for the first 17 weeks, dominated by a single OTU. Diversification was seen at 34 weeks, and by 107 weeks, the population had changed over to a divergent lineage. Patient 3 was sampled over only 10 weeks, and relatively little diversification was seen. Similar patterns were seen for the three patients for the other envelope amplicons analyzed (data not shown).
HCV diversity during HIV-HCV coinfection.
Approximately one-third of HIV-infected patients are coinfected with HCV (35), and HIV coinfection increases HCV-related liver diseases (10), raising the question of how HIV coinfection affects the structure and evolution of HCV populations. One possibility is that immune impairment due to advanced HIV infection results in reduced immune pressure on HCV populations (4, 18) and thus in diminished HCV diversity. To investigate this issue, we compared HCV sequence diversities in HIV-HCV-coinfected and HCV-monoinfected patients. We analyzed plasma HCV RNAs from eight HIV-HCV-coinfected subjects. For HCV monoinfection, we analyzed the last time points for patients 1 to 3 described above and two additional chronically infected HCV subjects, providing five HCV-monoinfected patients for comparison.
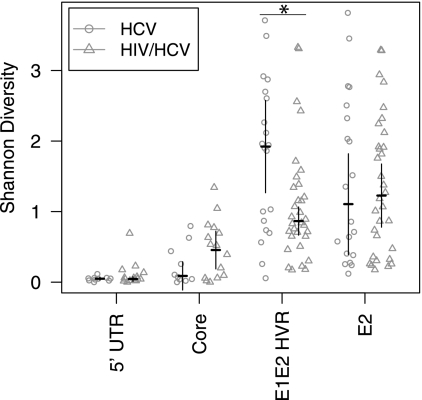
The monoinfected and coinfected individuals were compared over each genomic region to identify reproducible differences between the two patient groups. The Shannon Index value was calculated for each subject individually, and then the mean Shannon Index values for the two groups were compared (Fig. 6). For the 5′-UTR amplicon, almost no diversity was detected, and there was no difference between monoinfected and coinfected subjects. Higher diversity levels were seen for the core and the E2 envelope (excluding HVR1) amplicons for both groups, but there also were no significant differences in the mean diversity values.
FIG. 6.
Comparison of HCV diversity in samples from HCV-monoinfected and HIV-HCV-coinfected subjects. Shannon Index values were calculated for each subject, and then mean Shannon Index values were compared between monoinfected and coinfected subjects. The error bars show the 95% confidence interval of the difference (so that a lack of overlap of the error bars indicates a statistically significant difference in the means). The asterisk denotes a significant difference (P = 0.02616, Mann-Whitney comparison of means) between the two groups indicated by the horizontal line.
In contrast, significantly higher diversity was seen for the monoinfected subjects in the HVR1 envelope region than for coinfected subjects (P = 0.02616; Mann-Whitney comparison of means for pooled 3Aa, 3Ab, 3Ba, and 3Bb). This is consistent with the idea that loss of immune function due to advanced HIV infection may diminish selective pressure on HCV populations, so that the HCV population diversifies to a lesser extent. However, the magnitude of the effect was modest, and we note that there are possible complications in this analysis that could influence the outcome. We return to these issues in the Discussion below.
Estimating the number of OTUs in an HCV population.
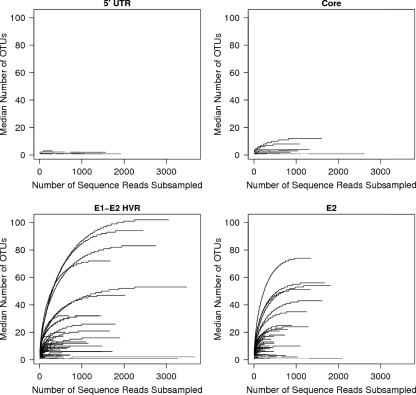
We investigated how close we were to completely sampling the HCV sequence populations using rarefaction analysis. In this method, random subsamples of sequences are drawn from the sequence pool for each amplicon from each individual, and the numbers of OTUs found in the subsample are plotted (Fig. 7A to D). The numbers of OTUs are then plotted for many different subsample sizes. If the curve reaches a plateau at values below the total number of sequences in the sample, it indicates that further sampling would yield few or no new OTUs. For this analysis, we chose the conservative approach of excluding variants supported by only one or two sequence reads.
FIG. 7.
Collector's curve (rarefaction) analysis of HCV sequences from each subject. Repeated sampling of sequence subsets was used to investigate whether additional sampling would likely yield additional OTUs. The numbers of sequences in sequence subsets are shown on the x axis, and the numbers of OTUs in the subset are shown on the y axis. OTUs supported by only one or two sequence reads were removed prior to rarefaction analysis. The amplicons studied were as follows: UTR (1A) (A), core (2A) (B), E1E2HVR1 (3A) (C), and E2 (4A) (D).
For the 5′-UTR sequences (Fig. 7A), no samples from any subject showed more than four OTUs, even at a sampling depth of 1,500 sequences per sample. For most of the samples, the values were near saturation at low sampling depths. We thus concluded that most or all of the variation in the 5′-UTR sequence pool had been sampled. For the core region (Fig. 7B), all of the samples appeared to be near plateau, with the highest OTU value <20.
For the envelope region, many of the OTU values were much higher (Fig. 7C and D), with a maximum value of ∼100. Nevertheless, the curves all reached clear plateaus, indicating that most or all of the variants in the sample were detected.
How many undetected OTUs are there in the HVR1 sequence populations? We investigated this using the Chao1 estimator for species richness, which allows the total number of species in a population to be inferred from counts of individuals from that population. If many species (OTUs) are represented by low counts of individuals (sequences), then it is likely that further sampling will yield new species (OTUs). Conversely, if most OTUs are represented by many sequences, then few additional OTUs will be recovered by further sampling. The Chao1 method uses such frequency of isolation information to estimate of the number of unseen species.
The Chao1 values for all HCV amplicons studied are shown in Table S1 in the supplemental material. When Chao1 was used to analyze the data in Fig. 7 (after PyroNoise clustering and removal of OTUs supported by 1 or 2 sequences), there were only slightly higher estimates. The highest value for any of the HVR1 samples was 124, only slightly higher than the measured number of OTUs.
The number of virions transmitted early during infection.
For HIV, exhaustive sequencing effort has suggested that typically only one or a few virions initiate infections during initial transmission (17, 32). Our data allow us to address this for HCV. Although we do not know the precise time of transmission for the patients studied longitudinally here, ALT values indicated that infection was probably close to the time of diagnosis. We thus assessed the diversity, and hence the number of virions transmitted, at the earliest available time points for patients 1 to 3.
For this, we used the HVR1 amplicons (3Ba and 3Bb), since they are the most diverse and so are most suited to reporting the number of HCV variants. We used the conservative Chao1 analysis described above to estimate population sizes (see Table S1 in the supplemental material). For the two HVR amplicons, the numbers of sequence reads ranged from 148 to 551, but the numbers of OTUs ranged from only 1 to 4 for the first time points. The higher numbers of OTUs were from patient 2, who was first sampled the longest time after diagnosis (9 weeks). For all three patients, OTU numbers were higher for the HVR1 amplicons at the last time point than for the first time point (see Table S1 in the supplemental material). Thus, these data indicate that during transmission or shortly after infection, the HCV sequence population was comprised of only one or a few OTUs.
The viral population structure strongly affects estimation of the size of the transmitted inoculum.
In previous literature, detection of low viral sequence diversity immediately after transmission has been taken as evidence that only one or a few viral particles seeded the initial infection. However, this conclusion is highly dependent on the structure of the viral population in the individual virus donor. For example, in the extreme case in which the viral population in the donor is homogeneous, finding a single variant in the recipient does not establish transmission of only a single virion. Conversely, if the donor harbors a highly diverse viral population and only a single viral variant is observed in the recipient, then a single particle probably initiated the productive infection. Here, we developed a quantitative framework to analyze the probability of transmitting one viral sequence variant given known structure of the donor population and used the pyrosequence data to model the size of the initial HCV inoculum transmitted in the patients studied here.
The probability of receiving only one OTU from the donor is as follows: Prtotal = p1n + p2n + … + pin, where p1, p2, … pi are the proportions of OTU1, OTU2, … OTUi in the donor, Prtotal is the total probability of transmission of a single variant, and n is the number of virions transmitted initially. For example, if OTU1 comprises 90% of the population in the donor, then the first virion transmitted to the recipient has a 90% chance of being OTU1. The probability of transmission of two virions belonging to OTU1 is thus 0.9 × 0.9, or 0.81. The squares of each probability are then summed over all OTUs in the sample to yield the total probability of transmission of a single variant given infection by two particles. If three viral particles started the infection, the probability would be the proportion of each OTU cubed, and so forth for larger numbers of virions transmitted initially. Thus, either increasing the diversity of the viral population in the donor or increasing the numbers of viral particles transmitted decreases the probability of obtaining a single variant in the recipient.
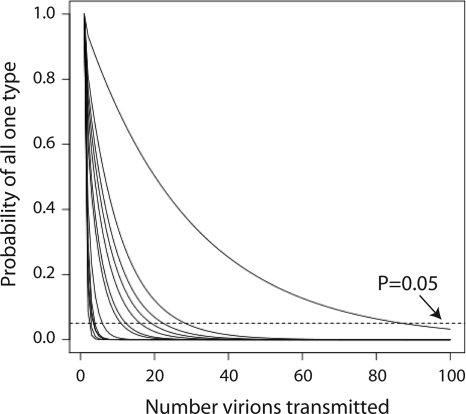
We used the HVR1 amplicon (3Ba) to represent proportions of OTUs in experimentally measured HCV populations (see Table S1 in the supplemental material). For subjects for whom multiple time points were available, the last time point was studied. Figure 8 shows the calculated probability of obtaining a single OTU from a single transmission of n virions from the viral populations in the subjects studied. The x axis shows the number (n) of virions in hypothetical transmission events, comparing n values of 1 to 100 virions. The y axis shows the probability of obtaining only one OTU given the numbers of virions transmitted. Data for each HCV donor (modeled as individuals from our study) are shown by a separate curve on the graph.
FIG. 8.
Calculation of the probability of transmission of a single viral variant given the measured HCV population structures. The x axis shows the number of viruses modeled to be involved in functional HCV transmission. The y axis shows the probability of detecting only one type of viral particle in the recipient in the donor-recipient transmission pair. Each of the subjects studied here was modeled individually as a potential donor and is shown as a single curve. The E1E2 HVR1 amplicon was used for this analysis. For subjects for whom multiple time points were available, the last time point was analyzed. Probabilities were calculated as described in the text. The dashed line indicates a P value of 0.05.
Inspection of the graph shows that most curves fall quickly, indicating that transmission of even small numbers of virions would result in the appearance of multiple sequence variants in the recipient. For most subjects, transmission of 20 functional viral particles would likely result in accumulation of more than one HVR1 variant. However, there is clearly also considerable variation among the subjects studied. The most extreme case (the rightmost curve in Fig. 8) is patient 8, 96% of whose viral population was made up of a single sequence variant. Thus, in general, the measured structures of the HCV populations in the subjects studied were sufficiently complex that transmission of >20 particles would be unlikely to result in a single OTU appearing in the recipient, but the variation among subjects was considerable.
DISCUSSION
Analyzing the structure and dynamics of HCV populations in patients presents a considerable challenge. HCV population sizes can be very large (>107/ml), and HCV replication is quite error prone, so that many different variants accumulate during viral replication. A deeper understanding of population genetics is critical to effective HCV treatment and prevention. Here, we used the 454/Roche pyrosequencing technology to quantify HCV population structure and dynamics, allowing a quantitative analysis of transmission bottlenecks.
A major challenge in studies of viral populations using massively parallel sequencing is the potential artifactual inflation of variants due to mutation during sample preparation and errors in sequence determination. Without careful filtering, these errors lead to an erroneous proliferation of sequence variants. However, comparison to control data sets showed that denoising with PyroNoise followed by clustering in OTUs removed most pyrosequencing errors. Another source of error was mutation accumulation during sample preparation. Judging by the results with the plasmid controls and the 5′-UTR and core amplicons, sporadic base changes introduced during reverse transcription or PCR were effectively suppressed as well, since mostly single OTUs were recovered in these samples. For control plasmid amplicons, most were reduced to single OTUs, while HVR1 samples ranged from 3 to 118 OTUs, indicating good retention of biological sequence variation while controlling false proliferation of variants due to error.
We developed a method for modeling the number of virions initiating productive HCV infection and applied it to the filtered HVR1 pyrosequence data. We first devised a method for calculating the probability of transmitting a single viral variant given any population structure in the individual virus donor. Using this method, and given the observed population structures in the donor and single variants in the recipient, we then calculated the probabilities for transmitting different numbers of particles initially. In this approach, each infected subject studied is treated as a candidate infection donor. We found that the observed HCV populations are sufficiently diverse that it would be unlikely for an infection to be founded by a single variant after transmission of even a modest number of infectious particles (>20). Thus, we conclude that the low diversity seen early after HCV infection is a result of transmission of one or a few virions and is not due to viral populations in the donors having low diversity at the time of transmission. The conclusion that one or a few virions were transmitted applies only to the virions that seeded productive infection—nothing in our data rules out the possibility that many additional particles were in the initial inoculum but did not replicate in the new host. We also note that the numbers of virions initiating HCV infection could well differ with different routes of infection, and it would be of interest to use this method to analyze samples early after transmission by various means.
In the HIV field, diversity estimates have been generated by using single-genome amplification, in which PCR templates are serially diluted until only a minority of dilution wells show amplification, allowing single variants to be collected and sequenced one by one (17, 32). The reason for this laborious approach is that falsely low diversity can be seen if PCR mixtures contain only small numbers of initial templates, so that amplification results in many products with relatively homogeneous sequence composition. This is a particular challenge for studies of HIV latency, where relatively few template molecules are typically available. However, for the HCV samples studied here, the viral-load values were >50,000 copies per ml, suggesting that low diversity at early time points was not simply a result of low template abundance.
HIV-HCV coinfection was associated with reduced HVR1 diversity compared to monoinfection, and our data help us to understand why this point has been controversial in previous literature (4, 27, 31, 34, 36, 39, 43). There was considerable variation in HCV diversity among subjects within each group, which is particularly evident thanks to the depth of the pyrosequencing data, suggesting that factors in addition to HIV coinfection also affect diversity. Analysis of Shannon diversity in the context of a generalized linear model, including the HCV load, the HIV load, and time after infection, showed that all these variables influence diversity significantly (P < 0.05; data not shown). In addition, methods for quantifying diversity in viral populations are still under development, and the Shannon Index used here may not be ideal for this application. We found in the longitudinal analysis of viral envelope sequences that diversification was not a homogenous process across the full sequence tree. Over time, particular branches tended to bloom with multiple variants, while others withered away. This is what would be expected if an immune response to particular HVR1 variants eliminates those members of the population, allowing other HVR1 sequences to grow out. The timing of sampling relative to this process may affect the diversity ultimately measured. Alternative approaches to quantifying sequence variation may better capture this underlying biology, and such methods are under ongoing development.
Intense efforts are presently focused on developing therapeutics for HCV, and it is already clear that viral drug evasion mutations are a severe problem. Similarly, there is considerable interest in studying how HCV evades host immune responses. The methods described here should be useful for studying the dynamics and nature of HCV responses to these selective pressures.
Supplementary Material
Acknowledgments
We are grateful to members of the Bushman laboratory for help and suggestions. We are grateful to Charles Rice at Rockefeller University for the pFL-H77-JFH1 plasmid. We thank Ian Frank at the University of Pennsylvania Center for AIDS Research for the HIV-HCV-coinfected plasma samples used in the study.
This work was supported by a Human Microbiome Demonstration Project Grant to F.D.B., James Lewis, and Gary Wu (UH2DK083981). G.P.W. was supported by NIH K08 AI077713 and a University of Pennsylvania Department of Medicine Measey Basic Science Fellowship Award.
Footnotes
Published ahead of print on 7 April 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. [PubMed] [Google Scholar]
- 3.Binladen, J., M. T. Gilbert, J. P. Bollback, F. Panitz, C. Bendixen, R. Nielsen, and E. Willerslev. 2007. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackard, J. T., Y. Yang, P. Bordoni, K. E. Sherman, R. T. Chung, and the AIDS Clinical Trials Group 383 Study Team. 2004. Hepatitis C virus (HCV) diversity in HIV-HCV-coinfected subjects initiating highly active antiretroviral therapy. J. Infect. Dis. 189:1472-1481. [DOI] [PubMed] [Google Scholar]
- 5.Costello, E. K., C. L. Lauber, M. Hamady, N. Fierer, J. I. Gordon, and R. Knight. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eriksson, N., L. Pachter, Y. Mitsuya, S. Y. Rhee, C. Wang, B. Gharizadeh, M. Ronaghi, R. W. Shafer, and N. Beerenwinkel. 2008. Viral population estimation using pyrosequencing. PLoS Comput. Biol. 4:e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 8.Farci, P., R. Strazzera, H. J. Alter, S. Farci, D. Degioannis, A. Coiana, G. Peddis, F. Usai, G. Serra, L. Chessa, G. Diaz, A. Balestrieri, and R. H. Purcell. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc. Natl. Acad. Sci. U. S. A. 99:3081-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghany, M. G., D. B. Strader, D. L. Thomas, L. B. Seeff, and the American Association for the Study of Liver Diseases. 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49:1335-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, C. S., L. R. Baden, E. Yu, J. M. Mrus, J. Carnie, T. Heeren, and M. J. Koziel. 2001. Influence of human immunodeficiency virus infection on the course of hepatitis C virus infection: a meta-analysis. Clin. Infect. Dis. 33:562-569. [DOI] [PubMed] [Google Scholar]
- 11.Hildebrandt, M. A., C. Hoffmann, S. A. Sherrill-Mix, S. A. Keilbaugh, M. Hamady, Y. Y. Chen, R. Knight, R. S. Ahima, F. Bushman, and G. D. Wu. 2009. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137:1716-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann, C., N. Minkah, J. Leipzig, G. Wang, M. Q. Arens, P. Tebas, and F. D. Bushman. 2007. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 35:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoofnagle, J. H. 2009. A step forward in therapy for hepatitis C. N. Engl. J. Med. 360:1899-1901. [DOI] [PubMed] [Google Scholar]
- 14.Huse, S. M., J. A. Huber, H. G. Morrison, M. L. Sogin, and D. M. Welch. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan, D. E., K. Sugimoto, K. Newton, M. E. Valiga, F. Ikeda, A. Aytaman, F. A. Nunes, M. R. Lucey, B. A. Vance, R. H. Vonderheide, K. R. Reddy, J. A. McKeating, and K. M. Chang. 2007. Discordant role of CD4 T-cell response relative to neutralizing antibody and CD8 T-cell responses in acute hepatitis C. Gastroenterology 132:654-666. [DOI] [PubMed] [Google Scholar]
- 16.Kasprowicz, V., Y. H. Kang, M. Lucas, J. Schulze zur Wiesch, T. Kuntzen, V. Fleming, B. E. Nolan, S. Longworth, A. Berical, B. Bengsch, R. Thimme, L. Lewis-Ximenez, T. M. Allen, A. Y. Kim, P. Klenerman, and G. M. Lauer. 2010. Hepatitis C virus (HCV) sequence variation induces an HCV-specific T-cell phenotype analogous to spontaneous resolution. J. Virol. 84:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearney, M., F. Maldarelli, W. Shao, J. B. Margolick, E. S. Daar, J. W. Mellors, V. Rao, J. M. Coffin, and S. Palmer. 2009. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 83:2715-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, A. Y., J. Schulze zur Wiesch, T. Kuntzen, J. Timm, D. E. Kaufmann, J. E. Duncan, A. M. Jones, A. G. Wurcel, B. T. Davis, R. T. Gandhi, G. K. Robbins, T. M. Allen, R. T. Chung, G. M. Lauer, and B. D. Walker. 2006. Impaired hepatitis C virus-specific T cell responses and recurrent hepatitis C virus in HIV coinfection. PLoS Med. 3:e492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21:379-384. [DOI] [PubMed] [Google Scholar]
- 20.Kuntzen, T., J. Timm, A. Berical, N. Lennon, A. M. Berlin, S. K. Young, B. Lee, D. Heckerman, J. Carlson, L. L. Reyor, M. Kleyman, C. M. McMahon, C. Birch, J. Schulze Zur Wiesch, T. Ledlie, M. Koehrsen, C. Kodira, A. D. Roberts, G. M. Lauer, H. R. Rosen, F. Bihl, A. Cerny, U. Spengler, Z. Liu, A. Y. Kim, Y. Xing, A. Schneidewind, M. A. Madey, J. F. Fleckenstein, V. M. Park, J. E. Galagan, C. Nusbaum, B. D. Walker, G. V. Lake-Bakaar, E. S. Daar, I. M. Jacobson, E. D. Gomperts, B. R. Edlin, S. M. Donfield, R. T. Chung, A. H. Talal, T. Marion, B. W. Birren, M. R. Henn, and T. M. Allen. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology. 48:1769-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuntzen, T., J. Timm, A. Berical, L. L. Lewis-Ximenez, A. Jones, B. Nolan, J. Schulze zur Wiesch, B. Li, A. Schneidewind, A. Y. Kim, R. T. Chung, G. M. Lauer, and T. M. Allen. 2007. Viral sequence evolution in acute hepatitis C virus infection. J. Virol. 81:11658-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., and C. M. Rice. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933-938. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Z., C. Lozupone, M. Hamady, F. D. Bushman, and R. Knight. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna, P., C. Hoffmann, N. Minkah, P. P. Aye, A. Lackner, Z. Liu, C. A. Lozupone, M. Hamady, R. Knight, and F. D. Bushman. 2008. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 4:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMullan, L. K., A. Grakoui, M. J. Evans, K. Mihalik, M. Puig, A. D. Branch, S. M. Feinstone, and C. M. Rice. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. U. S. A. 104:2879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Netski, D. M., Q. Mao, S. C. Ray, and R. S. Klein. 2008. Genetic divergence of hepatitis C virus: the role of HIV-related immunosuppression. J. Acquir. Immune Defic. Syndr. 49:136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh, T. S., and C. M. Rice. 2009. Predicting response to hepatitis C therapy. J. Clin. Invest. 119:5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quince, C., A. Lanzen, T. P. Curtis, R. J. Davenport, N. Hall, I. M. Head, L. F. Read, and W. T. Sloan. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639-641. [DOI] [PubMed] [Google Scholar]
- 30.Ray, S. C., L. Fanning, X. H. Wang, D. M. Netski, E. Kenny-Walsh, and D. L. Thomas. 2005. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J. Exp. Med. 201:1753-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roque-Afonso, A. M., M. Robain, D. Simoneau, P. Rodriguez-Mathieu, M. Gigou, L. Meyer, and E. Dussaix. 2002. Influence of CD4 cell counts on the genetic heterogeneity of hepatitis C virus in patients coinfected with human immunodeficiency virus. J. Infect. Dis. 185:728-733. [DOI] [PubMed] [Google Scholar]
- 32.Salazar-Gonzalez, J. F., M. G. Salazar, B. F. Keele, G. H. Learn, E. E. Giorgi, H. Li, J. M. Decker, S. Wang, J. Baalwa, M. H. Kraus, N. F. Parrish, K. S. Shaw, M. B. Guffey, K. J. Bar, K. L. Davis, C. Ochsenbauer-Jambor, J. C. Kappes, M. S. Saag, M. S. Cohen, J. Mulenga, C. A. Derdeyn, S. Allen, E. Hunter, M. Markowitz, P. Hraber, A. S. Perelson, T. Bhattacharya, B. F. Haynes, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman, K. E., C. Andreatta, J. O'Brien, A. Gutierrez, and R. Harris. 1996. Hepatitis C in human immunodeficiency virus-coinfected patients: increased variability in the hypervariable envelope coding domain. Hepatology 23:688-694. [DOI] [PubMed] [Google Scholar]
- 35.Sherman, K. E., S. D. Rouster, R. T. Chung, and N. Rajicic. 2002. Hepatitis C virus prevalence among patients infected with human immunodeficiency virus: a cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin. Infect. Dis. 34:831-837. [DOI] [PubMed] [Google Scholar]
- 36.Shuhart, M. C., D. G. Sullivan, K. Bekele, R. D. Harrington, M. M. Kitahata, T. L. Mathisen, L. V. Thomassen, S. S. Emerson, and D. R. Gretch. 2006. HIV infection and antiretroviral therapy: effect on hepatitis C virus quasispecies variability. J. Infect. Dis. 193:1211-1218. [DOI] [PubMed] [Google Scholar]
- 37.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 38.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka, Y., K. Hanada, H. Hanabusa, F. Kurbanov, T. Gojobori, and M. Mizokami. 2007. Increasing genetic diversity of hepatitis C virus in haemophiliacs with human immunodeficiency virus coinfection. J. Gen. Virol. 88:2513-2519. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, D. L., J. Astemborski, D. Vlahov, S. A. Strathdee, S. C. Ray, K. E. Nelson, N. Galai, K. R. Nolt, O. Laeyendecker, and J. A. Todd. 2000. Determinants of the quantity of hepatitis C virus RNA. J. Infect. Dis. 181:844-851. [DOI] [PubMed] [Google Scholar]
- 41.Timm, J., G. M. Lauer, D. G. Kavanagh, I. Sheridan, A. Y. Kim, M. Lucas, T. Pillay, K. Ouchi, L. L. Reyor, J. Schulze zur Wiesch, R. T. Gandhi, R. T. Chung, N. Bhardwaj, P. Klenerman, B. D. Walker, and T. M. Allen. 2004. CD8 epitope escape and reversion in acute HCV infection. J. Exp. Med. 200:1593-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timm, J., and M. Roggendorf. 2007. Sequence diversity of hepatitis C virus: implications for immune control and therapy. World J. Gastroenterol. 13:4808-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyoda, H., Y. Fukuda, Y. Koyama, J. Takamatsu, H. Saito, and T. Hayakawa. 1997. Effect of immunosuppression on composition of quasispecies population of hepatitis C virus in patients with chronic hepatitis C coinfected with human immunodeficiency virus. J. Hepatol. 26:975-982. [DOI] [PubMed] [Google Scholar]
- 44.Turnbaugh, P. J., M. Hamady, T. Yatsunenko, B. L. Cantarel, A. Duncan, R. E. Ley, M. L. Sogin, W. J. Jones, B. A. Roe, J. P. Affourtit, M. Egholm, B. Henrissat, A. C. Heath, R. Knight, and J. I. Gordon. 2009. A core gut microbiome in obese and lean twins. Nature 457:480-484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, G. P., A. Garrigue, A. Ciuffi, K. Ronen, J. Leipzig, C. Berry, C. Lagresle-Peyrou, F. Benjelloun, S. Hacein-Bey-Abina, A. Fischer, M. Cavazzana-Calvo, and F. D. Bushman. 2008. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 36:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, G. P., B. L. Levine, G. K. Binder, C. C. Berry, N. Malani, G. McGarrity, P. Tebas, C. H. June, and F. D. Bushman. 2009. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol. Ther. 17:844-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wohnsland, A., W. P. Hofmann, and C. Sarrazin. 2007. Viral determinants of resistance to treatment in patients with hepatitis C. Clin. Microbiol. Rev. 20:23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.