Abstract
Influenza A virus buds through the apical plasma membrane, forming enveloped virus particles that can take the shape of pleomorphic spheres or vastly elongated filaments. For either type of virion, the factors responsible for separation of viral and cell membranes are not known. We find that cellular Rab11 (a small GTP-binding protein involved in endocytic recycling) and Rab11-family interacting protein 3 ([FIP3] which plays a role in membrane trafficking and regulation of actin dynamics) are both required to support the formation of filamentous virions, while Rab11 is additionally involved in the final budding step of spherical particles. Cells transfected with Rab11 GTP-cycling mutants or depleted of Rab11 or FIP3 content by small interfering RNA treatment lost the ability to form virus filaments. Depletion of Rab11 resulted in up to a 100-fold decrease in titer of spherical virus released from cells. Scanning electron microscopy of Rab11-depleted cells showed high densities of virus particles apparently stalled in the process of budding. Transmission electron microscopy of thin sections confirmed that Rab11 depletion resulted in significant numbers of abnormally formed virus particles that had failed to pinch off from the plasma membrane. Based on these findings, we see a clear role for a Rab11-mediated pathway in influenza virus morphogenesis and budding.
Influenza A virus is a highly infectious respiratory pathogen, causing 3 to 5 million severe cases yearly while the recent H1N1 pandemic has spread to over 200 countries and resulted in over 15,000 WHO-confirmed deaths since its emergence in March 2009 (57). Influenza virus particles are enveloped structures that contain nine identified viral polypeptides. The lipid envelope is derived by budding from the apical plasma membrane and contains the viral integral membrane proteins hemagglutinin (HA) and neuraminidase (NA) as well as the M2 ion channel. Internally, virus particles contain a matrix protein (M1), small quantities of the NS2/NEP polypeptide, and eight genomic segments of negative-sense RNA that are separately encapsidated into ribonucleoprotein (RNP) particles by the viral nucleoprotein (NP) and tripartite polymerase complex (PB1, PB2, and PA). M1 is thought to form a link between the RNPs and the cytoplasmic tails of the viral membrane proteins though M2 may also play a role (39). The minimal viral protein requirements for budding are disputed; while initial studies suggested that M1 was the main driver of budding (21, 34), more recent work proposes that the glycoproteins HA and NA are responsible (8).
Further complicating the analysis of influenza A virus budding is the observation that most strains of the virus form two distinct types of virions: spherical particles approximately 100 nm in diameter and much longer filamentous particles up to 30 μm in length (38). Of the viral proteins, M1 is the primary determinant of particle shape (3, 17) although other virus genes also play a role. It is also likely that host factors are involved in the process as cells with fully differentiated apical and basolateral membranes produce more filaments than nonpolarized cell types (42). While it is tempting to speculate that virus morphology and budding are regulated by the same cellular process, the fact that spherical budding occurs in the absence of an intact actin cytoskeleton while filament formation does not (42, 48) indicates some level of divergence in the mechanisms responsible for spherical and filamentous virion morphogenesis.
The means by which viral and cellular membranes are separated are also unclear. Unlike many other enveloped viruses, including retroviruses (19, 36, 52) and herpes simplex virus (12), influenza A virus does not utilize the cellular endosomal sorting complex required for transport (ESCRT) pathway (5, 8). However, recent reports indicate that some viruses, including human cytomegalovirus (HCMV) (32), the hantavirus Andes virus (44), and respiratory syncytial virus (RSV) may employ a Rab11-mediated pathway during assembly and/or budding (4, 51). The Rab family of small GTPases is involved in targeting vesicle trafficking, mediating a wide range of downstream processes including endosomal trafficking and membrane fusion/fission events (reviewed in references 53 and 58). Rab11 is involved in trafficking proteins and vesicles between the trans-Golgi network (TGN), recycling endosome, and the plasma membrane (9, 49, 50) as well as playing a role in actin remodeling, cytokinesis, and abscission (27, 41, 55). Apical recycling endosome (ARE) trafficking is of particular interest in the context of viral infection as other negative-sense RNA viruses have been shown to assemble and/or traffic virion components through the ARE prior to final assembly and budding at the plasma membrane (4, 44, 51). Rab11 function is modulated and targeted through interactions with Rab11 family interacting proteins (Rab11-FIPs) that direct it to specific subcellular locations (23, 25, 26) by binding to actin or microtubule-based motor proteins (24, 26, 47). While Rab11-FIPs recognize both isoforms of Rab11 (a and b [Rab11a/b]) through a conserved amphipathic α-helical motif, they differ in their ability to bind either the GTP-bound form of Rab11 (FIP1, FIP3, FIP4, and Rip11) or both the GTP and GDP-bound forms (FIP2) (23, 30). FIP1 and FIP2 have been implicated in RSV budding (4, 51) while FIP4 is important for trafficking of HCMV components (32). FIP3 has not previously been linked with virus budding but plays an important role in both cell motility and cytokinesis, regulating actin dynamics and endosomal membrane trafficking (29, 55).
In light of the normal cellular functions of Rab11 and its effectors and of their reported involvement in the budding of other viruses, we examined the role of this cellular pathway in influenza virus budding. We find that Rab11-FIP3 is essential for filamentous but not spherical virion formation while Rab11 is required for both forms of virus budding.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
Human embryonic kidney 293T cells, CaCo2 colon carcinoma cells, and Madin-Darby canine kidney (MDCK) cells were cultured as previously described (6). Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and incubated overnight before further analysis. For transfection of small interfering RNAs (siRNAs), 293T cells were transfected approximately 18 h after seeding using DharmaFECT transfection reagent (1 μl/well; Dharmacon) to a final siRNA concentration of 0.1 μM and allowed to incubate for 72 h before further analysis or infection.
An MDCK-adapted version of A/PR/8/34 virus (PR8) rescued from plasmids (14) and grown in MDCK cells was used. A filamentous variant of PR8 virus containing segment 7 from A/Udorn/72 (PR8 MUd) has been previously described (40). A/Udorn/72 (Udorn) was the gift of Richard Compans (42, 48). Herpes simplex virus (HSV) strain SC16 was used as previously described (5). For influenza virus infections, virus was adsorbed to cells for 1 h in serum-free medium at 37°C and overlaid in serum-free medium supplemented with 1 μg/ml trypsin and 0.14% bovine serum albumin (BSA) (for viral growth assays) or medium containing serum (for microscopy). Titers of influenza viruses were determined by plaque assay in MDCK cells (28, 37). Plasmids encoding green fluorescent protein (GFP)-tagged dominant negative (DN) Rab11a (with the mutation S25N) and constitutively active (CA) Rab11a (Q70L) (13), wild-type (WT) Rab4, CA Rab4 (Q67L), and DN Rab4 (N121I) (46) proteins were generously supplied by Stephen Ferguson while WT Rab8, CA Rab8 (Q67L), and DN Rab8 (T22N) (45) were provided by Folma Buss.
Protein analysis, immunofluorescence, antisera, and microscopy.
For Western blotting, samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes according to standard procedures. Blots were developed using secondary antibodies conjugated to IR Dye 680 or 800 and imaged and quantified on a LiCor Biosciences Odyssey near-infrared imaging platform.
For fluorescence microscopy, cells were fixed with 4% formaldehyde and processed for immunohistochemistry as previously described (18, 48). Rabbit polyclonal antiserum to whole PR8 virus has been described previously (1). Rabbit polyclonal antiserum to Rab11-FIP3 used in Western blotting was a gift from Rytis Prekeris (55). Other reagents used in immunofluorescence were as follows: rabbit polyclonal Rab11 (71-5300; Invitrogen), mouse monoclonal NP (AA5H; Abcam ab20343), rabbit polyclonal NP (2915) (40), and Alexa Fluor 568-conjugated wheat germ agglutinin (WGA) (W11262; Invitrogen). Other antibodies used in Western blotting were as follows; mouse monoclonal Rab11a (47; Abcam ab78337), mouse polyclonal Rab11b (H00009230-B02F; Abnova), and rat monoclonal α-tubulin (YL1/2; MCA77G; AbD-Serotec). Rabbit polyclonal antisera to PR8 PB1 (V19), M1 (A2917), and whole PR8 virus have been previously described (1, 15, 40).
Samples were imaged using a Leica TCS SP confocal microscope. Postcapture processing of images to allow daylight visualization and reproduction was applied equally to each experiment using Adobe Photoshop.
For scanning electron microscopy (SEM), cells were washed in 0.1 M HEPES (pH 7.4)-2 mM CaCl2 and fixed overnight in the same buffer containing 0.3% H2O2 and 4% electron microscopy (EM)-grade glutaraldehyde. Samples were further processed by the Cambridge University Multi-Imaging Centre, Department of Anatomy. For transmission electron microscopy (TEM), monolayers of cells grown in 6-cm dishes were washed in 0.1 M HEPES, pH 6.4. The monolayer was then covered in HEPES-glutaraldehyde-H2O2 and left at 4°C for 2 h. The monolayer was washed with HEPES buffer, and cells were scraped into buffer and harvested by centrifugation at 14,000 × g for 10 min. The supernatant was replaced with fresh HEPES buffer and sent to Cambridge University Multi-Imaging Centre for further fixation and screening.
RESULTS
Role of Rab11 in influenza virus budding.
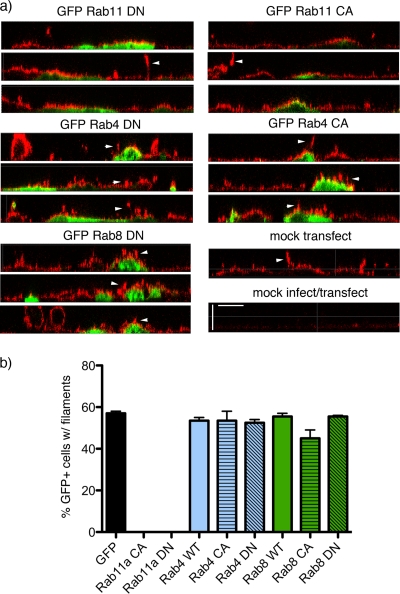
To explore the possibility that influenza virus buds via a Rab11-associated mechanism, we first tested whether mutant Rab proteins with alterations to their GTP-binding sites that resulted in either a permanently GTP-bound, constitutively active (CA) protein or a permanently GDP-bound, dominant negative (DN) protein affected viral filament formation. CaCo2 cells were transfected with plasmids that expressed various members of the Rab family fused to GFP (or GFP only) and then infected with a reassortant influenza virus (PR8 MUd) that produces large numbers of filamentous virions (40). Cells were fixed at 16 h postinfection (p.i.), stained for surface HA protein, and imaged in z-plane slices by confocal microscopy. Only background levels of antibody reactivity were observed in uninfected cells while the apical plasma membrane of infected but untransfected cells stained strongly for HA, some of which could be seen in characteristic micrometer-length protrusions away from the plasma membrane (Fig. 1a, arrowheads), indicative of bundles of viral filaments (40, 42, 48). Similar filamentous structures were also seen protruding from cells transfected with WT, CA, or DN forms of Rab4 or Rab8 or with GFP only (Fig. 1a and data not shown). However, no filaments were visible on the surface of cells expressing either the CA or DN forms of Rab11 even though abundant cell surface HA was still visible (Fig. 1a, top panels). When GFP-positive cells were examined and scored for the presence or absence of filamentous projections, around 60% of cells transfected with GFP alone were found to have at least one large bundle of filaments (Fig. 1b), in line with previous observations (40, 42, 48). Cells expressing the WT, CA, or DN forms of Rab4 or Rab8 formed filaments at a similar frequency. In contrast, there was a complete absence of large filamentous structures protruding from the surface of over 400 individual transfected cells expressing either Rab11 CA or Rab11 DN (Fig. 1b).
FIG. 1.
Effect of overexpressing GFP-tagged Rab polypeptides on influenza virus filament formation. CaCo2 cells were transfected (or mock transfected) with plasmids expressing the indicated GFP-tagged polypeptides and incubated overnight before infection at a multiplicity of infection of 10 with PR8 MUd. Cells were fixed at 16 h p.i., and cell surface HA (stained in red by anti-PR8 serum) and GFP (green) were imaged by confocal microscopy. (a) Representative z-plane reconstructions are shown. Scale bar, 5 μm. (b) A minimum of 200 cells from two independent experiments were scored for the presence or absence of filamentous projections. The mean and range are plotted.
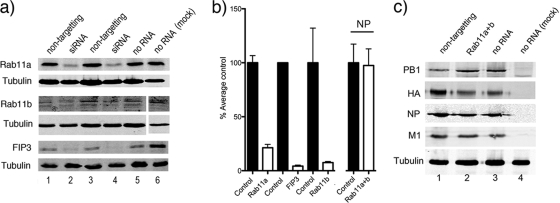
Given the greatly decreased filamentous budding we observed in the presence of Rab11 mutants unable to exchange GTP, we tested whether the endogenous Rab11 protein or two of its effectors were required for the formation of filamentous virions. 293T cells were transfected with siRNAs targeted against Rab11a, Rab11b, Rab11-FIP2, or Rab11-FIP3 or, as a control, with nontargeting siRNAs to deplete the endogenous pools of the respective proteins. First, we validated the effectiveness of our siRNA treatments by Western blotting of cell lysates at 72 h posttransfection. Comparison of samples from cells treated with specific siRNAs with those of cells treated with nontargeting siRNAs or nontreated cells showed reductions in the amounts of Rab11a, Rab11b, and FIP3 (Fig. 2a, compare lanes 2 and 4 with lanes 1, 3, and 5). Quantification of protein levels from replicate blots by densitometry confirmed substantial depletion of all the targeted proteins: on average Rab11a was depleted to 21% of normal levels, Rab11b was depleted to 8%, and Rab11-FIP3 was depleted to 4% (Fig. 2b). We were unable to acquire a sample of an antibody to FIP2, but reverse transcription-PCR (RT-PCR) analysis indicated knockdown of mRNA levels by around 80% (data not shown).
FIG. 2.
Cellular and viral protein accumulation in siRNA-treated cells. 293T cells were transfected (or mock transfected) with siRNA sequences targeting the indicated cellular polypeptides. At 72 h post transfection the cells were infected (or mock infected where indicated) with PR8 virus at a multiplicity of infection of 5 for 24 h before further analysis. (a and c) Cells were lysed and analyzed by Western blotting for the indicated proteins. (b) Levels of the indicated proteins were quantified by densitometry, normalized to tubulin levels, and plotted as percentages of the average amount contained in cells treated with nontargeting control or no RNA. The means and standard deviations (Rab11a, n = 16; Rab 11b and FIP3, n = 4; NP, n = 4) are plotted. Note that the gel conditions used in the upper blot of panel c did not resolve PB1 and PB1-N40 (an N-terminally truncated variant of PB1) (56).
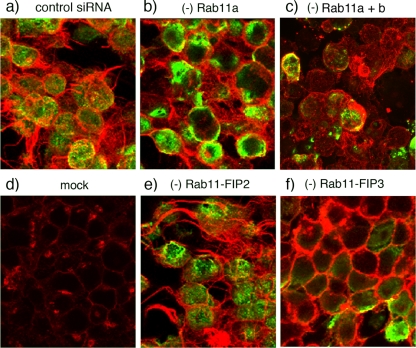
Next, to assess the effect of Rab depletion on virus filament formation, the morphology of the plasma membrane in siRNA-treated, PR8 MUd-infected cells was investigated by staining with the lectin WGA while virus infection was detected by staining for intracellular NP. Mock-infected cells displayed prominent WGA staining of a perinuclear compartment as well as faint staining of the lateral plasma membrane (Fig. 3d). In contrast, the apical surfaces of cells treated with a nontargeting siRNA and infected with PR8 MUd were decorated with numerous filamentous projections, often protruding 10 to 20 μm from the cell while, as expected, the cytoplasm stained strongly for NP (Fig. 3a). Treatment of cells with siRNAs against Rab11-FIP2 did not reduce the numbers of virus-induced filamentous structures (Fig. 3e). However, depletion of Rab 11a or of both Rab11a and Rab11b or of Rab11-FIP3 resulted in the almost complete absence of large membrane protrusions produced by infected cells (Fig. 3b, c, and f).
FIG. 3.
Influenza virus filament formation in siRNA-treated cells. 293T cells were transfected with siRNA sequences targeting the indicated proteins or with a nontargeting control (a and d). At 72 h posttransfection cells were infected (or mock infected) with PR8 MUd at a multiplicity of infection of 5, fixed at 16 h p.i., and stained with fluorescently tagged WGA to visualize the cell surface (red) as well as with anti-NP serum to validate infection (green). Images are maximum intensity projections of confocal z-stacks taken through the cell at approximately 0.5-μm steps.
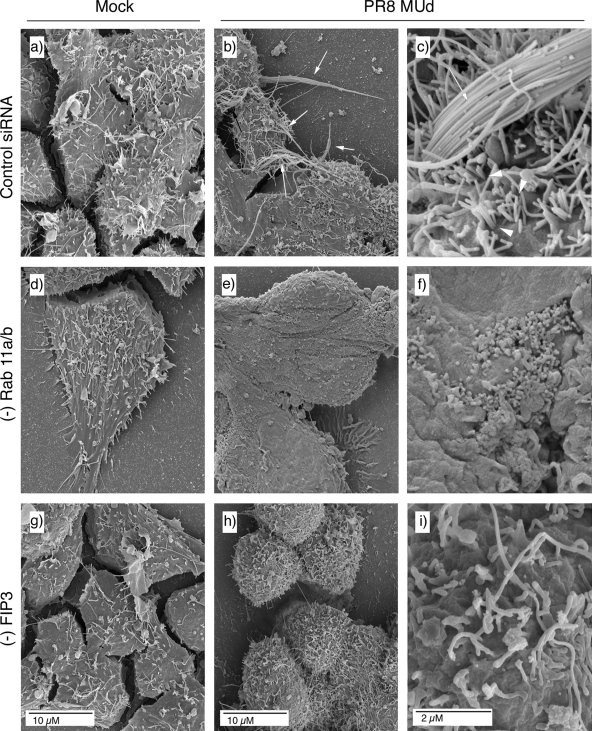
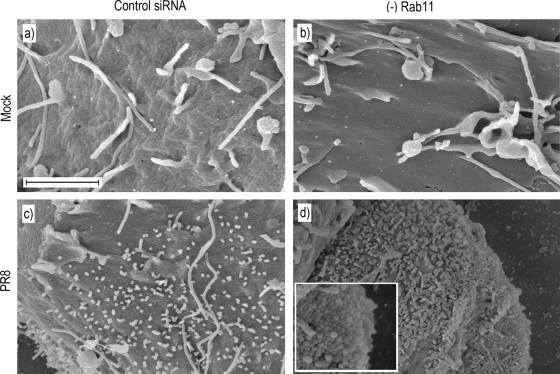
Light microscopy readily images bundles of large filaments but does not reveal shorter virions of <1-μm length (42, 48). Accordingly, using SEM we imaged the surface of cells doubly treated with siRNAs against Rab11a and Rab11b, singly treated with siRNA against FIP3, or treated with control siRNAs. At low magnification, the surfaces of mock-infected cells treated with control or specific siRNA sequences appeared similar, containing a number of microvilli or filopodia-like structures as well as more lamellar protrusions (Fig. 4a, d, and g), consistent with previous observations (20, 33). Low-magnification imaging of the surfaces of PR8 MUd-infected cells treated with control siRNA sequences showed similar features but with the addition of striking 10- to 20-μm length spikes (Fig. 4b, arrows). At higher magnification, these were seen to consist of bundles of multiple filamentous virions in parallel array (Fig. 4c). Other patches of the plasma membrane were profusely decorated with individual filamentous virus particles around 0.5 μm in length, distinguishable from normal cellular structures by their numbers and consistent diameter of just under 100 nm (Fig. 4c arrowheads; see also Fig. 9a for similar magnification images of uninfected cells). The surfaces of PR8 MUd-infected cells depleted of Rab11 appeared much smoother than those of the control siRNA-treated cells (whether infected or uninfected), lacking obvious long bundles of filamentous particles (Fig. 4e). At higher magnification, it was evident that short filamentous virions as well as cellular filopodia were also absent from the surfaces of infected Rab11 siRNA-treated cells (Fig. 4f). However, patches of protruding 100-nm-diameter spherical structures, reminiscent of spherical influenza virus particles, were visible on the plasma membrane (Fig. 4f). When infected, FIP3-depleted cells were examined, prominent bundles of long viral filaments were also absent although numerous short protrusions were still visible (Fig. 4h). At high magnification, these were seen to consist of short ∼1μm-length structures and an occasional longer filament (Fig. 4i). The greater numbers of such structures on infected cells (compare Fig. 4g, h, and i) make it likely that some are viral in nature, but the imaging technique does not unambiguously identify the origin of individual protrusions. Nevertheless, it was clear that FIP3 depletion blocked the formation of large numbers of long viral filaments, consistent with the immunofluorescence analysis. SEM analysis confirmed that siRNA targeting of FIP2 did not reduce filament formation (data not shown). Overall, we therefore conclude that Rab11 and Rab11-FIP3 are essential for formation of filamentous influenza virions. In contrast to results obtained with respiratory syncytial virus, however (51), Rab11-FIP2 is apparently nonessential.
FIG. 4.
SEM analysis of filament formation in Rab11- or FIP3-depleted (−) cells. 293T cells were transfected with siRNA sequences targeting Rab11a and Rab11b (d to f), FIP3 (g to i), or a nontargeting control (a to c), and infected with PR8 MUd at a multiplicity of infection of 5 at 72 h posttransfection. At 16 h p.i. cells were fixed and processed for SEM imaging.
FIG. 9.
SEM visualization of virus budding in Rab11-depleted cells. 293T cells were transfected with siRNA sequences targeting Rab11a and Rab11b (b and d) or a nontargeting control (a and c), and either mock infected or infected with PR8 at a multiplicity of infection of 5 at 72 h posttransfection. At 16 h p.i. cells were washed and fixed overnight on coverslips before processing for SEM. Scale bar, 2 μm.
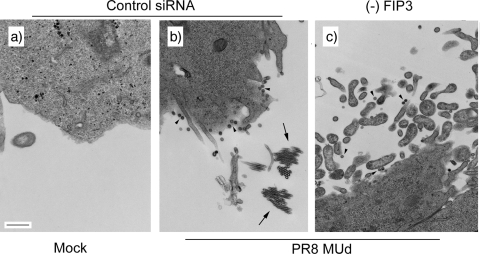
Filamentous strains of influenza A virus also produce spherical particles (43), and loss of filamentous morphology on adaptation to growth in eggs is often associated with higher titers resulting from increased production of spherical particles (31). It was therefore of interest to examine whether the loss of FIP3 blocked the production of spherical as well as filamentous particles from PR8 MUd-infected cells. To this end, we examined infected cells by TEM. Uninfected cells treated with a control siRNA exhibited relatively smooth plasma membranes, with the occasional filopodium visible in cross section (Fig. 5a). Control siRNA-treated, PR8 MUd-infected cells exhibited the expected mixture of spherical (Fig. 5b, arrowheads) and filamentous particles, which were most commonly seen in cross section through the bundles (Fig. 5b, arrows). When infected, FIP3-depleted cells were examined, released spherical particles were still visible (Fig. 5c, arrowheads), but consistent with the previous experiments, no large bundles of filaments were seen. However, the titer of infectious virus released from FIP3-depleted cells was slightly lower (on average, 80%) than that from control cells, suggesting that in this situation, loss of filaments was not associated with increased production of spherical particles. A similar outcome was seen when the experiment was repeated with the filamentous Udorn strain of virus, with FIP3 depletion blocking the formation of long filamentous bundles (data not shown) but decreasing overall titer by only around one-third. Thus, FIP3 depletion specifically blocks filamentous budding without having a major effect on spherical virus budding.
FIG. 5.
TEM analysis of viral budding in FIP3-depleted cells. 293T cells were transfected with nontargeting siRNA sequences (a and b) or siRNA against FIP3 (c) and infected or mock infected as labeled with PR8 MUd at a multiplicity of infection of 5 at 72 h posttransfection. At 16 h p.i. cells were fixed and processed for TEM imaging. Scale bar, 500 nm. Arrowheads indicate spherical virus particles; arrows indicate cross sections through bundles of filaments.
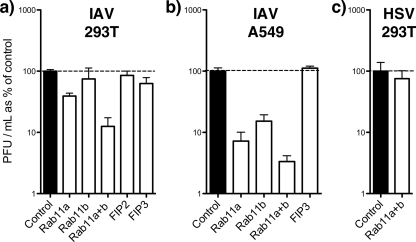
Next, we further examined the role of Rab11 and associated proteins in the production of spherical virus. To this end, 293T and A549 cells were treated with siRNAs as above and infected with a strain of influenza virus (PR8) that does not produce filamentous particles (17). The titers of released virus were then established by plaque assay. PR8 virus replicated equally well in cells treated with nontargeting siRNA pools and in cells that received no siRNA, producing titers greater than 107 PFU/ml in both cell types (Fig. 6a and b; values set as 100%). Depletion of either Rab11-FIP2 or Rab11-FIP3 had no significant effect on virus titers, regardless of cell type. Reduction of Rab11b levels in 293T cells also had little effect on virus replication but caused a more substantial 7-fold inhibition of virus production in A549 cells. Similarly, reduction of Rab11a levels caused a 2-fold decrease in titer of virus released from 293T cells but had a much more pronounced effect in A549 cells, where a greater than 10-fold decrease was seen. More striking, however, was the drop in titer of virus released from cells depleted of both Rab11a and Rab11b simultaneously, which was on average 8-fold in 293T and 30-fold in A549 cells (Fig. 6a and b). While Rab11a/b-depleted A549 cells showed consistent drops in virus titer to between 1% and 5% of the titer in control cells, the output from depleted 293T cells was more variable, ranging (in six independent experiments) from 33% to 1%. The reason for this difference in behavior between the two cell types is not clear but may reflect variations in siRNA transfection efficiency. To examine the specificity of the requirement for Rab11 in influenza A virus replication, we tested the effect of depleting cells of Rab11a and Rab11b on replication of HSV, a virus whose budding is known to involve the cellular ESCRT pathway (5, 12). In contrast to the results obtained with influenza virus, HSV replicated to similar titers in control and Rab11a/b-depleted cells (Fig. 6c).
FIG. 6.
Virus replication in siRNA-treated cells. 293T or A549 cells were transfected (or mock transfected) with siRNA sequences targeting the indicated cellular polypeptides. At 72 h posttransfection the cells were infected with PR8 virus (influenza A virus [IAV]) at a multiplicity of infection of 5 or with HSV at a multiplicity of infection of 10. Supernatants from infected cells were harvested at 24 h p.i., and titers were determined by plaque assay. The titers obtained from nontransfected cells and cells treated with nontargeting siRNAs were averaged and defined as 100%. The mean and standard error (IAV-infected 293T cells, n ≥4 replicate experiments; IAV-infected A549 cells, n ≥3; HSV-infected 293T cells, n = 2) are plotted.
As a further control, we assessed viral protein production in Rab11a/b-depleted cells to control for the possibility that reduced levels of virus replication might result from lower infection rates of the depleted cells or reduced virus gene expression. However, the levels of NP, M1, HA, and PB1 accumulation showed no significant variation between nontransfected cells or cells treated with control or Rab11 siRNAs (Fig. 2c). When NP accumulation was quantified from replicate experiments, no significant change was seen to result from Rab11/b depletion (Fig. 2b). Thus, Rab11 plays a role downstream of viral gene expression in the formation of both spherical and filamentous influenza virus particles while FIP3 is specifically required for filamentous budding.
Colocalization of Rab11 and influenza virion components.
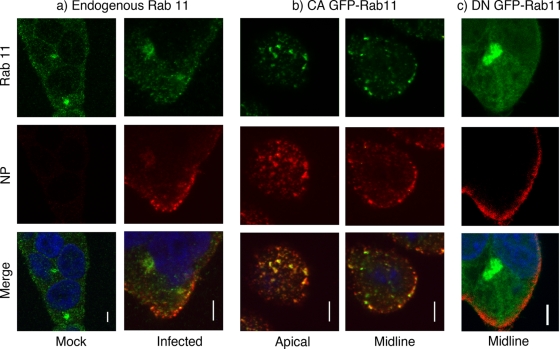
Since Rab11 is involved in directing vesicular traffic to and from the recycling endosome and the plasma membrane (49), it could potentially play roles in trafficking viral components to the site of budding, as well as (or in addition to) the final membrane scission step. We therefore explored the localization of Rab11 in relation to the viral nucleoprotein. 293T cells were either infected with PR8 or mock infected, and the localizations of endogenous Rab11 and influenza NP were visualized at 6 h p.i. by immunofluorescence. In mock-infected cells, speckles of Rab11 were found throughout the cytoplasm along with a predominant perinuclear focus (Fig. 7a), in agreement with a prior study showing its localization to the recycling endosome and TGN as well as on cytoplasmic cargo vesicles (9). In infected cells, NP localized to cytoplasmic puncta that, as expected (6), tended to concentrate at the plasma membrane although some NP accumulation was observed at a perinuclear structure (Fig. 7a). Virus infection was not associated with any obvious change in Rab11 localization at 6 h p.i. However, a striking degree of colocalization between Rab11 and NP was seen, both for the perinuclear structure and for the cytoplasmic puncta (Fig. 7a). Next, we tested for colocalization of NP and the GFP-tagged Rab11 CA and DN mutants as previous work reports distinct localization patterns for the two activity states. The CA (GTP-bound) form of Rab11 localizes to the recycling endosome and more-peripheral vesicular structures, while DN (GDP-bound) Rab11 is primarily associated with the TGN (9, 44). 293T cells were transfected with the GFP-tagged Rab11 constructs, infected with PR8, and stained at 6 h p.i. for NP before visualization by confocal microscopy. CA GFP-Rab11 localized to dispersed cytoplasmic structures (Fig. 7b), often with a perinuclear accumulation in the lower midbody of the cells (data not shown), consistent with previous reports (9, 44). Moreover, these punctate accumulations of CA Rab11 showed a high degree of colocalization with NP, especially toward the apical cell surface (Fig. 7b). In contrast, the majority of the GDP-bound DN Rab11 fusion protein localized to a perinuclear compartment likely to be the TGN (9, 44) based on its colocalization with the marker TGN46 (data not shown), and although some diffuse cytoplasmic staining was visible, this did not colocalize significantly with NP (Fig. 7c).
FIG. 7.
Colocalization of Rab11 and influenza virus NP. (a) 293T cells were mock infected or infected as labeled with PR8 at a multiplicity of infection of 5, fixed at 6 h p.i., and stained for Rab11 (green), NP (red), and DNA (blue). (b and c) Cells were transfected with plasmids encoding GFP-tagged CA or DN Rab11 proteins, infected 18 h posttransfection, and stained at 6 h p.i. for NP and DNA. Single optical slices are shown. Scale bar, 4 μm.
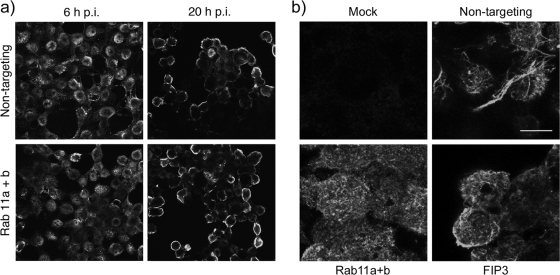
Given the extensive colocalization between Rab11 and NP, it was possible that the reduction in virus replication seen after siRNA depletion of Rab11 resulted from disruption of normal RNP trafficking. To test this, we examined NP localization in siRNA-treated 293T cells. In cells treated with control siRNAs, NP exhibited the expected distribution of a mixture of nuclear and cytoplasmic staining at 6 h p.i., followed by concentration at the plasma membrane late in infection (Fig. 8a, upper panels). No major changes to this pattern were seen in cells depleted of Rab11a and Rab11b (Fig. 8a, lower panels). Similar results were obtained when infection was carried out with the filamentous PR8 MUd virus (data not shown). Rab11 function has previously been found nonessential for surface transport of HA (9, 11), consistent with the normal levels of surface HA we saw in the presence of mutant forms of Rab 11 (Fig. 1a). Nevertheless, to test whether this was also the case after siRNA treatment, we examined surface HA staining in PR8 MUd-infected cells depleted of Rab11a/b or FIP3. Abundant staining was seen in all infected cells, whether treated with specific or control siRNAs though, as expected, large bundles of viral filaments were seen only in the control cells (Fig. 8b).
FIG. 8.
NP and HA localization in Rab11- or FIP3-depleted cells. 293T cells were transfected with siRNA sequences targeting Rab11a and Rab11b, FIP3, or a nontargeting control as indicated and infected (or mock infected) with PR8 at a multiplicity of infection of 5 at 72 h post transfection. (a) At 6 and 20 h p.i. cells were fixed and permeabilized before staining for NP. Single optical slices are shown. Each panel represents a 150- by 135-μm window. (b) At 16 h p.i. cells were fixed and stained for HA. Maximum intensity projections of a set of optical sections through the upper halves of the cells are shown. Scale bar, 10 μm.
Next, we therefore investigated whether the drop in virus titer after Rab11 depletion was the result of a defect in budding. SEM was used to examine the surface topography of mock- or PR8-infected cells treated with nontargeting control or Rab11a/b siRNA sequences at 16 h postinfection. As before, depletion of Rab11 did not noticeably alter the appearance of mock-infected cells; both control and Rab11 siRNA-treated cells formed filopodia and microvilli but largely lacked 100-nm-diameter spherical structures that could be mistaken for virus particles (Fig. 9a and b). Such spherical structures of the correct size to be influenza virions were, however, observed on the surface of PR8-infected cells treated with a nontargeting siRNA control, as expected (Fig. 9c). The density of budding varied from cell to cell, from sparse to profuse, but a typical cell harbored a few hundred well-spaced virions (Fig. 9c). Similar size protrusions were also seen on the surface of infected, Rab11-depleted cells (Fig. 9d), but the overall density of virus particles was higher, with many Rab11-depleted cells showing excessive retention of what appeared to be fully formed virus particles on the plasma membrane, to the point that, in certain regions, there were so many virions in close proximity that they formed a pavement-like array (Fig. 9d, inset).
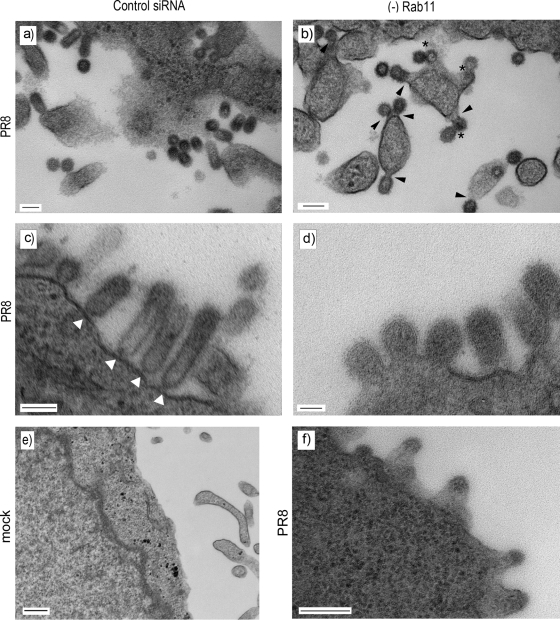
To examine the morphology of these virus-like structures in more detail, the budding of individual virions from Rab11-depleted cells was examined using thin-section TEM. The plasma membrane of mock-infected cells treated with control siRNA appeared relatively smooth though filopodia were observed in cross section (Fig. 10e). In PR8-infected cells treated with the nontargeting siRNA control, abundant spherical influenza virions were visible (Fig. 10a and c). Consistent with previous observations (10), the majority of virus particles had apparently either completely separated from the plasma membrane and presented a circular cross section (Fig. 10a) or were attached by thin “necks” and retained an elongated morphology (Fig. 10c, white arrowheads). However, in cells depleted of Rab11 we observed a large number of virions whose appearance suggested slowed or abnormal budding events (Fig. 10b, d, and f). In some cases, budding virions appeared to consist of doubled, perhaps “daisy-chained,” particles budding from a single site (Fig. 10b, asterisks). These structures resembled late-stage budding-defective HIV particles formed in cells overexpressing dominant negative components of the ESCRT pathway (35, 52). In many other cases, the budding virions remained attached to the cell by thick stalks of membrane, in clear contrast to the narrow structures normally seen tethering immature virions to the cell surface (Fig. 10b, arrowheads, and d; compare with panel c). In apparently more extreme cases, what appeared to be budding virus particles protruded from broad platforms of the plasma membrane (Fig. 10f). Taking these data in conjunction with the decrease in virus titer and the number of virions seen by SEM to be retained on the cell surface of Rab11 siRNA-treated cells, it appears that the depletion of Rab11 results in a late-stage defect in virus budding.
FIG. 10.
TEM visualization of budding virions in Rab11-depleted cells. 293T cells were transfected with siRNA sequences targeting Rab11a (b), Rab11a and Rab11b (d and f), or a nontargeting control (a, c, and e), and mock infected (e) or infected with PR8 at a multiplicity of infection of 5 at 72 h posttransfection. At 16 h p.i. cells were fixed and processed for TEM. Scale bar, 100 nm.
DISCUSSION
All enveloped viruses face the problem of separating viral and cellular membranes to release a fully formed virion. In some cases, this energetically unfavorable process is thought to be primarily overcome by “pulling” forces generated by interactions between the viral capsid and membrane proteins (54). Another (not mutually exclusive) paradigm holds that membrane scission is driven by an external “push” from coopted cellular proteins, most notably the ESCRT machinery (7). In the case of influenza virus, the lack of a rigid internal capsid and the extreme variation in particle lengths produced by filamentous strains perhaps argue against sole use of a pulling mechanism. Equally, however, evidence argues strongly against use of the best-characterized cellular system for providing a push (5, 8). We provide evidence here that influenza virus utilizes the Rab11 pathway for budding, a cellular system whose normal role, like that of the ESCRT machinery, is in directing vesicular traffic.
We base our conclusions on the findings that GTP-cycling mutants of Rab11 (but not other Rab proteins) interfere with filamentous budding (Fig. 1), that siRNA depletion of Rab11 reduces the titer of released virus (Fig. 6), and, most importantly, that EM imaging of virus budding in Rab11-depleted cells shows structures consistent with defective budding (Fig. 4, 9, and 10). Similar occurrences of what are clearly virus particles that have yet to start the pinching-off process have been observed previously in normal infected cells, where they were interpreted as representing intermediate steps in the budding process (10). We also saw these types of budding events in normal (as well as control siRNA-treated) cells, but in agreement with previous work (10), they were rare occurrences, with the majority of particles being either completely detached or attached by a narrow stalk (Fig. 10 and data not shown). Accordingly, it is the much larger numbers of such apparent “intermediate” structures in Rab11-depleted cells that lead us to define the outcome as defective budding. We did not observe complete cessation of viral release in Rab11-depleted cells as assessed by viral titer, and while this may reflect only a partial reduction in Rab11 expression, it would be interesting to know whether the incomplete budding events we observed represent truly stalled episodes of particle formation or just slower assembly kinetics.
Our data can be interpreted in light of a pulling model, where virus budding is driven largely by forces generated by self-assembly of viral proteins within the nascent particle, or of a pushing model, where cellular functions are redirected to the viral bud. The pulling model would predict that virus budding is reduced because of insufficient delivery of crucial viral structural proteins to the bud zone. The pushing model predicts that the absence of Rab11 leads to a failure to recruit the cellular machinery responsible for membrane scission. Both models are compatible with the normal cellular functions of Rab11 and its interaction partners in regulating vesicular traffic to and from the plasma membrane, TGN, and recycling endosome as well as in cytokinesis (22, 58).
The pulling model is consistent with the extensive colocalization we observe between Rab11 and NP (Fig. 7) as the coincidence of the two polypeptides suggests a mechanism for transporting RNPs across the cytoplasm to the apical plasma membrane, which accordingly could be perturbed by disruption of Rab11 function. However, we saw no major change in NP localization in the presence of Rab11 mutants or in Rab11-depleted cells (Fig. 7 and 8). This apparently normal localization of NP after Rab11 depletion is perhaps surprising, but one possible explanation for this is that Rab11 levels were not reduced sufficiently to cause a trafficking defect. We also saw normal levels of surface HA in the presence of Rab11 mutants (Fig. 1) or after siRNA treatment (Fig. 8), consistent with previous studies that found Rab11 function nonessential for surface transport of HA (9, 11). Thus, while we would not rule out a role for the Rab11 pathway in trafficking of virion components to the plasma membrane, we do not think that this is the primary cause of the budding defect.
Instead, we currently favor the hypothesis that Rab11 plays a role in influenza virus budding by recruiting the cellular machinery necessary for assisting in the separation of viral and cellular membranes. This is consistent with the high densities of virus particles visible on the surface of Rab11-depleted cells by SEM (Fig. 9) and the common occurrence of finding budding particles in cross section that often appeared reasonably normal except for the width of the neck between virion content and cytoplasm (Fig. 10b and d). The identity of the actual protein(s) responsible for pinching off the membrane in this model is unknown; although Rab proteins are accepted to have functions in both membrane fusion and membrane scission, the former process is much better characterized (22, 58). Nevertheless, the yeast homologues of Rab11, Ypt31 and Ypt32, have been shown to be involved in exocytic traffic from the Golgi stack, probably at the point of membrane scission (2). Although not topologically equivalent to virus budding, the function is nevertheless suggestive. Further work is required to determine whether Rab11 plays a direct role in separating viral and cellular membranes or an indirect one via the recruitment of other cellular factors.
The identity of the Rab11 FIP(s) used during influenza virus budding is also uncertain. Unlike the case with RSV (51), we saw no role for FIP2, either in supporting spherical particle release or in filamentous virion formation. While we saw a strong dependence on FIP3 for production of influenza virus filaments (Fig. 3 to 5), this was not mirrored by any requirement for spherical virus budding, as assessed by the titer of replicated virus (Fig. 6). This does, however, indicate that filament and spherical virus budding are biochemically separable events, consistent with their differential dependence on the actin cytoskeleton (42, 48). Indeed, FIP3 also interacts with the actin-regulating protein Arf6 at the plasma membrane (16, 25), so it is possible that the differential requirements for FIP3 and the actin cytoskeleton during viral morphogenesis are part of the same process.
We have previously considered the hypothesis that filamentous influenza virions are produced when a budding particle fails to engage the cellular machinery necessary for membrane scission (5). Evidence in support of such a mechanism exists for RSV, where expression of a DN FIP2 polypeptide inhibited the release of infectious virus but actually made filaments longer (51). However, for influenza virus, loss of FIP3 or Rab11 abolished the filamentous phenotype (Fig. 3 to 5 and 9) rather than making viral budding hyperfilamentous. Similarly, loss of FIP3 or Rab11 did not convert PR8 budding into a filamentous phenotype, but, instead, Rab11 depletion led to slow or stalled production of spherical particles (Fig. 9 and 10). As discussed above, this could reflect profound differences in the cellular pathways used for filamentous and spherical budding by influenza virus rather than that the two forms of virion represent different ends of the same spectrum. An alternative possibility is that filament formation and spherical particle formation are mechanistically linked but that forming a filament requires more than one Rab11-dependent step. Work is ongoing to test between these possibilities.
Acknowledgments
We thank Richard Compans, Stephen Ferguson, Folma Buss, and Rytis Prekeris for the gift of reagents. We also acknowledge Jeremy Skepper, Janet Powell, and Evi Miklejewska from the Multi-Imaging Centre, University of Cambridge, for assistance with TEM and SEM sample preparation and imaging.
This work was supported by a grant from the MRC (grant G0700815). E.A.B. is supported by a studentship from the Gates Cambridge Trust.
Footnotes
Published ahead of print on 31 March 2010.
REFERENCES
- 1.Amorim, M. J., E. K. Read, R. M. Dalton, L. Medcalf, and P. Digard. 2007. Nuclear export of influenza A virus mRNAs requires ongoing RNA polymerase II activity. Traffic 8:1-11. [DOI] [PubMed] [Google Scholar]
- 2.Benli, M., F. Doring, D. G. Robinson, X. Yang, and D. Gallwitz. 1996. Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. EMBO J. 15:6460-6475. [PMC free article] [PubMed] [Google Scholar]
- 3.Bourmakina, S. V., and A. Garcia-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. [DOI] [PubMed] [Google Scholar]
- 4.Brock, S. C., J. R. Goldenring, and J. E. Crowe, Jr. 2003. Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 100:15143-15148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce, E. A., L. Medcalf, C. M. Crump, S. L. Noton, A. D. Stuart, H. M. Wise, D. Elton, K. Bowers, and P. Digard. 2009. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology 390:268-278. [DOI] [PubMed] [Google Scholar]
- 6.Carrasco, M., M. J. Amorim, and P. Digard. 2004. Lipid raft-dependent targeting of the influenza A virus nucleoprotein to the apical plasma membrane. Traffic 5:979-992. [DOI] [PubMed] [Google Scholar]
- 7.Chen, B. J., and R. A. Lamb. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, B. J., G. P. Leser, E. Morita, and R. A. Lamb. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81:7111-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, W., Y. Feng, D. Chen, and A. Wandinger-Ness. 1998. Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol. Biol. Cell 9:3241-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compans, R. W., and N. J. Dimmock. 1969. An electron microscopic study of single-cycle infection of chick embryo fibroblasts by influenza virus. Virology 39:499-515. [DOI] [PubMed] [Google Scholar]
- 11.Cresawn, K. O., B. A. Potter, A. Oztan, C. J. Guerriero, G. Ihrke, J. R. Goldenring, G. Apodaca, and O. A. Weisz. 2007. Differential involvement of endocytic compartments in the biosynthetic traffic of apical proteins. EMBO J. 26:3737-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crump, C. M., C. Yates, and T. Minson. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dale, L. B., J. L. Seachrist, A. V. Babwah, and S. S. Ferguson. 2004. Regulation of angiotensin II type 1A receptor intracellular retention, degradation, and recycling by Rab5, Rab7, and Rab11 GTPases. J. Biol. Chem. 279:13110-13118. [DOI] [PubMed] [Google Scholar]
- 14.de Wit, E., M. I. Spronken, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2004. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 103:155-161. [DOI] [PubMed] [Google Scholar]
- 15.Digard, P., V. C. Blok, and S. C. Inglis. 1989. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology 171:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson, J. G. 2003. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 278:41573-41576. [DOI] [PubMed] [Google Scholar]
- 17.Elleman, C. J., and W. S. Barclay. 2004. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology 321:144-153. [DOI] [PubMed] [Google Scholar]
- 18.Elton, D., M. Simpson-Holley, K. Archer, L. Medcalf, R. Hallam, J. McCauley, and P. Digard. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75:408-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 20.Gentet, L. J., G. J. Stuart, and J. D. Clements. 2000. Direct measurement of specific membrane capacitance in neurons. Biophys. J. 79:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grosshans, B. L., D. Ortiz, and P. Novick. 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U. S. A. 103:11821-11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hales, C. M., R. Griner, K. C. Hobdy-Henderson, M. C. Dorn, D. Hardy, R. Kumar, J. Navarre, E. K. Chan, L. A. Lapierre, and J. R. Goldenring. 2001. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276:39067-39075. [DOI] [PubMed] [Google Scholar]
- 24.Hales, C. M., J. P. Vaerman, and J. R. Goldenring. 2002. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J. Biol. Chem. 277:50415-50421. [DOI] [PubMed] [Google Scholar]
- 25.Horgan, C. P., and M. W. McCaffrey. 2009. The dynamic Rab11-FIPs. Biochem. Soc. Trans. 37:1032-1036. [DOI] [PubMed] [Google Scholar]
- 26.Horgan, C. P., A. Oleksy, A. V. Zhdanov, P. Y. Lall, I. J. White, A. R. Khan, C. E. Futter, J. G. McCaffrey, and M. W. McCaffrey. 2007. Rab11-FIP3 is critical for the structural integrity of the endosomal recycling compartment. Traffic 8:414-430. [DOI] [PubMed] [Google Scholar]
- 27.Horgan, C. P., T. H. Zurawski, and M. W. McCaffrey. 2005. Purification and functional properties of Rab11-FIP3. Methods Enzymol. 403:499-512. [DOI] [PubMed] [Google Scholar]
- 28.Hutchinson, E. C., M. D. Curran, E. K. Read, J. R. Gog, and P. Digard. 2008. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 82:11869-11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jing, J., E. Tarbutton, G. Wilson, and R. Prekeris. 2009. Rab11-FIP3 is a Rab11-binding protein that regulates breast cancer cell motility by modulating the actin cytoskeleton. Eur. J. Cell Biol. 88:325-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Junutula, J. R., E. Schonteich, G. M. Wilson, A. A. Peden, R. H. Scheller, and R. Prekeris. 2004. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 279:33430-33437. [DOI] [PubMed] [Google Scholar]
- 31.Kilbourne, E. D., and J. S. Murphy. 1960. Genetic studies of influenza viruses. I. Viral morphology and growth capacity as exchangeable genetic traits. Rapid in ovo adaptation of early passage Asian strain isolates by combination with PR8. J. Exp. Med. 111:387-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krzyzaniak, M. A., M. Mach, and W. J. Britt. 2009. HCMV-encoded glycoprotein M (UL100) interacts with Rab11 effector protein FIP4. Traffic 10:1439-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurejova, M., B. Uhrik, Z. Sulova, B. Sedlakova, O. Krizanova, and L. Lacinova. 2007. Changes in ultrastructure and endogenous ionic channels activity during culture of HEK 293 cell line. Eur. J. Pharmacol. 567:10-18. [DOI] [PubMed] [Google Scholar]
- 34.Latham, T., and J. M. Galarza. 2001. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 37.Matrosovich, M., T. Matrosovich, W. Garten, and H. D. Klenk. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosley, V. M., and W. G. R. Wyckoff. 1946. Electron micrography of the virus of influenza. Nature 157:263-266. [DOI] [PubMed] [Google Scholar]
- 39.Nayak, D. P., R. A. Balogun, H. Yamada, Z. H. Zhou, and S. Barman. 2009. Influenza virus morphogenesis and budding. Virus Res. 143:147-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noton, S. L., E. Medcalf, D. Fisher, A. E. Mullin, D. Elton, and P. Digard. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 88:2280-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riggs, B., W. Rothwell, S. Mische, G. R. Hickson, J. Matheson, T. S. Hays, G. W. Gould, and W. Sullivan. 2003. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J. Cell Biol. 163:143-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts, P. C., and R. W. Compans. 1998. Host cell dependence of viral morphology. Proc. Natl. Acad. Sci. U. S. A. 95:5746-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, P. C., R. A. Lamb, and R. W. Compans. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127-137. [DOI] [PubMed] [Google Scholar]
- 44.Rowe, R. K., J. W. Suszko, and A. Pekosz. 2008. Roles for the recycling endosome, Rab8, and Rab11 in hantavirus release from epithelial cells. Virology 382:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahlender, D. A., R. C. Roberts, S. D. Arden, G. Spudich, M. J. Taylor, J. P. Luzio, J. Kendrick-Jones, and F. Buss. 2005. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 169:285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seachrist, J. L., P. H. Anborgh, and S. S. Ferguson. 2000. β2-Adrenergic receptor internalization, endosomal sorting, and plasma membrane recycling are regulated by Rab GTPases. J. Biol. Chem. 275:27221-27228. [DOI] [PubMed] [Google Scholar]
- 47.Simon, G. C., and R. Prekeris. 2008. Mechanisms regulating targeting of recycling endosomes to the cleavage furrow during cytokinesis. Biochem. Soc. Trans. 36:391-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson-Holley, M., D. Ellis, D. Fisher, D. Elton, J. McCauley, and P. Digard. 2002. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 301:212-225. [DOI] [PubMed] [Google Scholar]
- 49.Ullrich, O., S. Reinsch, S. Urbe, M. Zerial, and R. G. Parton. 1996. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135:913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urbe, S., L. A. Huber, M. Zerial, S. A. Tooze, and R. G. Parton. 1993. Rab11, a small GTPase associated with both constitutive and regulated secretory pathways in PC12 cells. FEBS Lett. 334:175-182. [DOI] [PubMed] [Google Scholar]
- 51.Utley, T. J., N. A. Ducharme, V. Varthakavi, B. E. Shepherd, P. J. Santangelo, M. E. Lindquist, J. R. Goldenring, and J. E. Crowe, Jr. 2008. Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc. Natl. Acad. Sci. U. S. A. 105:10209-10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 53.Weisz, O. A., and E. Rodriguez-Boulan. 2009. Apical trafficking in epithelial cells: signals, clusters and motors. J. Cell Sci. 122:4253-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Welsch, S., B. Muller, and H. G. Krausslich. 2007. More than one door—budding of enveloped viruses through cellular membranes. FEBS Lett. 581:2089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson, G. M., A. B. Fielding, G. C. Simon, X. Yu, P. D. Andrews, R. S. Hames, A. M. Frey, A. A. Peden, G. W. Gould, and R. Prekeris. 2005. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol. Biol. Cell 16:849-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wise, H. M., A. Foeglein, J. Sun, R. M. Dalton, S. Patel, W. Howard, E. C. Anderson, W. S. Barclay, and P. Digard. 2009. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 83:8021-8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organization. 5 February 2010, posting date. Pandemic (H1N1) 2009—update 86. World Health Organization, Geneva, Switzerland. http://www.who.int/scr/disease/swineflu/laboratory05_02_2010/en/index.html.
- 58.Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107-117. [DOI] [PubMed] [Google Scholar]