Abstract
Replication of positive-strand RNA viruses occurs through the assembly of membrane-associated viral RNA replication complexes that include viral replicase proteins, viral RNA templates, and host proteins. Red clover necrotic mosaic virus (RCNMV) is a positive-strand RNA plant virus with a genome consisting of RNA1 and RNA2. The two proteins encoded by RNA1, a 27-kDa protein (p27) and an 88-kDa protein containing an RNA-dependent RNA polymerase (RdRP) motif (p88), are essential for RCNMV RNA replication. To analyze RCNMV RNA replication complexes, we used blue-native polyacrylamide gel electrophoresis (BN/PAGE), which enabled us to analyze detergent-solubilized large membrane protein complexes. p27 and p88 formed a complex of 480 kDa in RCNMV-infected plants. As a result of sucrose gradient sedimentation, the 480-kDa complex cofractionated with both endogenous template-bound and exogenous template-dependent RdRP activities. The amount of the 480-kDa complex corresponded to the activity of exogenous template-dependent RdRP, which produced RNA fragments by specifically recognizing the 3′-terminal core promoter sequences of RCNMV RNAs, but did not correspond to the activity of endogenous template-bound RdRP, which produced genome-sized RNAs without the addition of RNA templates. These results suggest that the 480-kDa complex contributes to template-dependent RdRP activities. We subjected those RdRP complexes to affinity purification and analyzed their components using two-dimensional BN/sodium dodecyl sulfate-PAGE (BN/SDS-PAGE) and mass spectrometry. The 480-kDa complex contained p27, p88, and possible host proteins, and the original affinity-purified RdRP preparation contained HSP70, HSP90, and several ribosomal proteins that were not detected in the 480-kDa complex. A model for the formation of RCNMV RNA replication complexes is proposed.
Positive-strand RNA viruses replicate their RNA on intracellular membranes (5). One or more viral nonstructural proteins, including RNA-dependent RNA polymerase (RdRP) and accessory proteins, play essential roles in RNA replication as integral membrane proteins (13, 25, 28) or as peripheral membrane proteins (4, 6). These viral nonstructural proteins interact with each other and with host proteins and viral RNA templates to form the membrane-associated viral RNA replication complexes. The membrane-associated RNA replication complexes of positive-strand RNA viruses have been isolated from the membrane fraction of virally infected tissues by the use of detergents (2, 11, 29, 32, 40, 49, 50). The purified fractions contain viral and host proteins and retain two types of RdRP activities. One is an endogenous template-bound RdRP activity that synthesizes virus-related RNAs without adding RNA templates. The other is an exogenous template-dependent RdRP activity capable of de novo initiation of cRNA synthesis from selected RNA templates. Although there has been intensive work performed on viral RdRP complexes, their organizations, properties, and functions have not been fully characterized.
Red clover necrotic mosaic virus (RCNMV) is a positive-strand RNA plant virus and a member of the genus Dianthovirus in the family Tombusviridae. The genome of RCNMV consists of two RNA components, RNA1 and RNA2. RNA1 encodes N-teminally overlapping nonstructural proteins, a 27-kDa protein (p27) and an 88-kDa protein containing an RdRP motif (p88) (19, 53). The p88 region is translated via programmed −1-position ribosomal frameshifting (17, 52), and p88 protein is preferentially required in cis for the replication of RNA1 (30). Both p27 and p88 are required for RCNMV replication, which is strongly linked to RNA silencing suppression (42). RNA1 also encodes a coat protein (CP), which is translated from subgenomic RNA (sgRNA) (55). RNA2 encodes a movement protein (MP) required for viral movement in plants (51). Although RNA2 is not required for RNA1 replication in protoplasts, a stem-loop structure located in the MP-coding region of RNA2 is necessary for transcription of CP subgenomic RNA from RNA1 (31, 41, 43).
RNA1 and RNA2 have no cap structure at the 5′ end (27) and no poly(A) tail at the 3′ end (24, 53). Instead, there is a cis-acting RNA element that functions as a translational enhancer in the 3′ untranslated region (UTR) of RNA1 (27). Recently, a small RNA fragment (SR1f) was found to be generated from the 3′ UTR of RNA1 and to function as a negative regulator against both cap-independent and cap-dependent translation (15). In contrast to the translation mechanism of RNA1, RNA2 has no such translational enhancer. A strong link between cap-independent translation and replication of RNA2 suggests that only de novo-synthesized RNA2 can function as an mRNA template for translation (26). Both p27 and p88 are to be found in a partially purified RdRP fraction extracted from RCNMV-infected tissue (2), and green fluorescence protein (GFP)-fused p27 and p88 localize to the endoplasmic reticulum (ER) (44). These findings suggest that ER-localized p27 and p88 function coordinately in RCNMV RNA replication.
In this study, to obtain insights into the formation of RCNMV RNA replication complexes on the ER membrane and their organization, we used immunoprecipitation to analyze the interactions between replicase proteins and blue-native polyacrylamide gel electrophoresis (BN/PAGE) to analyze the replication complexes. BN/PAGE has been used in combination with detergents to investigate the properties and components of large membrane-associated protein complexes (8). We show that the RCNMV p27 and p88 replicase proteins form a tightly membrane-associated protein complex with an apparent molecular mass of 480 kDa and that this complex is associated with a template-dependent RdRP that synthesizes RNA fragments from RCNMV RNAs by recognizing their 3′-terminal core promoter sequences. Further, using mass spectrometry, we identified possible host proteins present in the RCNMV RdRP complexes, including the 480-kDa complex. Our results suggest that the 480-kDa complex, together with several host proteins, plays an essential role in the synthesis of RCNMV RNA.
MATERIALS AND METHODS
Plasmid construction.
Previously described plasmids given the prefix “pBIC” were used for Agrobacterium infiltration, and plasmids with the prefix “pUC,” “pRC,” or “pBYL” were used for in vitro transcription. The plasmids included pBIC18 (42), pBICGFP (42), pBICdsGFP (42), pBICRC1 (42), pBICRC2 (42), pBICp27 (42), pBICp88 (42), pSP64-RLUC (27), and pLUCpA60 (27). pUCR1 (42) and pRC2|G (54) were full-length cDNA clones of RNA1 and RNA2 of the Australian strain of RCNMV, respectively. The plasmid p3′-8, which contains a 60-nt poly(A) tract, was a gift from K. A. White. pUC118 was purchased from Takara Bio Inc. (Shiga, Japan). All plasmids constructed in this study were verified by sequencing.
(i) pBICp27-HA.
Two DNA fragments were amplified from pBICp27 by PCR using two sets of primers, primer pair p35S-HSR (5′-GGTGGTAAGCTTCCCGGGAACATGGTGGAGCACGAC-3′) and p27C-HA-L (5′-CCATACGATGTTCCAGATTACGCTTAGCGCTGAAATCACCAGTC-3′) and primer pair TCa-KpnL (5′-GGTGGTGGTACCTATAGGGACTTTAGGTGATC-3′) and p27C-HA-R (5′-CCATACGATGTTCCAGATTACGCTTAGCGCTGAAATCACCAGTC-3′). A recombinant PCR product was then amplified from the mixture of these two fragments by the use of the p35S-HSR and TCa-KpnL primers, digested with SmaI and KpnI restriction enzymes, and inserted into the corresponding region of pBIC18.
(ii) pBICp27-iHA.
A PCR fragment from pBICdsGFP was amplified using TOM1/int8R (5′-GTACTTAGATTCATATATCTG-3′) and TOM1/int8L (5′-CTGCAATGAAAAGAGACGA-3′). Two PCR fragments from pBICp27-HA were then amplified using two sets of primers, primer pair p35S-HSR and p27-HA/int-L (5′-CATACAGATATATGAATCTAAGTACATCGTATGGGTAAAAATCC-3′) and primer pair p27-HA/int-R (5′-AATATTCGTCTCTTTTCATTGCAGGTTCCAGATTACGCTTAGC-3′) and BICEco (5′-TGCGCAACTGTTGGGAA-3′). Then, a recombinant PCR product was amplified from the mixture of these three fragments by the use of p35S-HSR and BICEco, digested with EcoRI, and inserted into the corresponding region of pBICp27-HA.
(iii) pBICp27-iFTH.
A PCR fragment from pBICdsGFP was amplified using FLAG/int-R (5′-GATTACAAGGTACTTAGATTCATATATCTG-3′) and FLAG/int/TEV-L (5′-ACCAGCATCAACAGTTCCAGATTGGAAGTAAAGGTTTTCCTTGTCATCGTCGTCCTGCAATGAAAAGAGACG-3′). Two PCR fragments from pBICp27-HA were amplified using two sets of primers, primer pair p35S-HSR and p27-FLAG/int-L (5′-ATATGAATCTAAGTACCTTGTAATCAAAATCCTCAAGGGATTTGA-3′) and primer pair TEV/HA-R (5′-ATCTGGAACTGTTGATGCTGGTTACCCATACGATGTTCCA-3′) and BICEco. Then, a recombinant PCR product was amplified from the mixture of these three fragments by the use of p35S-HSR and BICEco, digested with EcoRI, and inserted into the corresponding region of pBICp27-HA.
(iv) pBICp27-iFLAG.
Two PCR fragments from pBICp27-iFTH were amplified using two sets of primers, primer pair p35S-HSR and FLAG/int-L (5′-TTCAGCGCTACTTGTCATCGTCGTCCTGCAA-3′) and primer pair FLAG/TCa-R FLAG/TCa-R (5′-CGATGACAAGTAGCGCTGAAATCACCAGTCT-3′) and BICEco. Then, a recombinant PCR product was amplified from the mixture of these two fragments by the use of p35S-HSR and BICEco. The amplified DNA was digested with EcoRI and inserted into the corresponding region of pBICp27-HA.
(v) pBICp88-T7.
Two PCR fragments from pBICp27 were amplified using two sets of primers, primer pair p35S-HSR and p88C-T7-30BL (5′-GACCTCCAGTCATAGAAGCCATTCGGGCTTTGATTAGATCTTTCTTGTACACCTCTTCAC-3′) and primer pair p88C-T7-R (5′-TGGCTTCTATGACTGGAGGTCAACAAATGGGATAACGCTGAAATCACCAGTCT-3′) and TCa-SacL (5′-GGTGGTGAGCTCTATAGGGACTTTAGGTGATC-3′). Then, a recombinant PCR product was amplified from the mixture of these two fragments by the use of p35S-HSR and TCa-SacL. The amplified DNA was digested with SmaI and SacI and subsequently cloned into pBIC18 that had been previously with the same restriction enzymes. The small SalI-BglII fragment of the resulting plasmid was replaced by the small SalI-BglII fragment of pBICp88.
(vi) pBICp88-iT7.
A PCR fragment from pBICdsGFP was amplified using TOM1/int8R and TOM1/int8L, and two PCR fragments from pBICp88-T7 were amplified using two sets of primers, primer pair p88-167R (5′-AGTGCGAGCTTCGTTGG-3′) and p88-T7/int-L (5′-CATACAGATATATGAATCTAAGTACCTGTCATAGAAGCCATTCGG-3′) and primer pair p88-T7/int-R (5′-AATATTCGTCTCTTTTCATTGCAGGTGGTCAACAAATGGGATAAC-3′) and BICEco. Then, a recombinant PCR product was amplified from the mixture of these three fragments by the use of p88-167R and BICEco, digested with XhoI and SacI, and inserted into the corresponding region of pBICp88-T7.
(vii) pBYL2.
An AscI linker (5′-AGCTGGCGCGCC-3′) was treated with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) followed by annealing. The annealed linker was inserted into p3′-8, which had been digested with HindIII, to obtain pBYL-A. A similar strategy was used to insert a NotI linker (5′-AATTGCGGCCGC-3′) into EcoRI-digested pBYL-A to obtain pBYL-AN. pBYL-AN was digested with PshBI and treated with T4 DNA polymerase (New England Biolabs). The resulting DNA fragment was inserted into SmaI-digested pUC118. The resulting plasmid, pBYL2, contained the T7 promoter sequence, an AscI site, a 60-nucleotide (60-nt) poly(A) tract, and a NotI site in the direction opposite to that of the lacZ open reading frame (ORF).
(viii) pBYLp27.
A PCR fragment from pUCR1 was amplified using AscI-p27-F (5′-AGCTGGCGCGCCATGGGTTTTATAAATCTTTC-3′) and p27/end-AscI-R (5′-AGCTGGCGCGCCGCCTAAAAATCCTCAAGG-3′), digested with AscI, and inserted into AscI-digested pBYL2.
(ix) pBYLp88.
A PCR fragment from pUCR1-p88 was amplified using AscI-p27-F and p88/end-AscI-R (5′-AGCTGGCGCGCCTTTATCGGGCTTTGA-3′), digested with AscI, and inserted into AscI-digested pBYL2.
Agrobacterium infiltration.
Nicotiana benthamiana plants and Agrobacterium tumefaciens GV3101 (pMP90) were used for infiltration experiments as previously described (42). A. tumefaciens transformed by pBICp27-iFLAG, pBICp27-iHA. and pBICp88-iT7 was used for expression of p27-FLAG, p27-HA, and p88-T7, respectively.
Coimmunoprecipitation analysis.
Agrobacterium-infiltrated leaves at 2 days post infiltration (dpi) were ground in liquid nitrogen and homogenized in three volumes of extraction buffer A (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5% glycerol, 0.5% Triton X-100, Complete Mini protease inhibitor cocktail [Roche Diagnostics, Penzberg, Germany]), followed by centrifugation at 21,000 × g for 10 min at 4°C to remove cell debris. The supernatant (1 ml) was incubated with a 25-μl bed volume of protein A Sepharose (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) together with anti-FLAG M2 monoclonal antibody (Sigma-Aldrich, St. Louis, MO) or antihemagglutinin (anti-HA) monoclonal antibody (12CA5; Roche) or anti-T7 monoclonal antibody (Merck, Darmstadt, Germany) for 4 h at 4°C. The resin was washed three times with 1 ml of washing buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5% glycerol, 0.1% Triton X-100). The bound proteins were eluted by the addition of Laemmli sample buffer (22), followed by incubation for 3 min at 95°C. The protein samples were subjected to sodium dodecyl sulfate-PAGE (SDS-PAGE) and then analyzed by Western blotting with antibodies specific to respective epitope tags.
In vitro transcription.
Plasmids containing the prefix “pUC” or “pRC” were digested with SmaI, from which RNA transcripts were synthesized. RNA transcripts from pUCR1-p27fs (15), pUCR1-p27 (42), pUCR1-p88 (42), pUCR1-d3′SLF (14), and pRC2-d3′SLF with the same mutation as pRC2-6 (45) are referred to herein as RNA1-p27fs, RNA1-p27, RNA1-p88, RNA1-dSLF, and RNA2-dSLF, respectively. p27 and p88 mRNAs were synthesized from NotI-linearized pBYLp27 and pBYLp88. Luc mRNA and RLuc mRNA were synthesized from EcoRI-linearized pLUCpA60 and pSP64-RLUC, respectively. Brome mosaic virus (BMV) RNAs were synthesized from EcoRI-linearized pB1TP3, pB2TP5, and pB3TP8 (16), and Tomato mosaic virus (ToMV) RNA was synthesized from EcoRI-linearized pToMV. Capped transcripts were prepared as previously described (14, 15).
BN/PAGE analysis.
Agrobacterium-infiltrated leaves at 2 dpi were ground in liquid nitrogen and homogenized in three volumes of extraction buffer B (50 mM Tris-HCl [pH 7.4], 10 mM KCl, 15 mM MgCl2, 0.1% 2-mercaptoethanol, 20% glycerol, Complete Mini protease inhibitor cocktail [Roche]), followed by centrifugation at 800 × g for 30 min at 4°C to remove cell debris. The supernatant was centrifuged at 21,000 × g for 30 min at 4°C to obtain the membrane fraction (P20).
Membrane fractions were also prepared from evacuolated tobacco BY-2 protoplast lysate (BYL). Capped RNA1 or its derivatives (500 ng) were added to 50 μl of BYL translation and replication mixture. Alternatively, capped p27 mRNA (500 ng) or capped p88 mRNA (500 ng) was singly added to BYL. Capped p27 and p88 mRNAs were also added to 50 μl of BYL in the absence or presence of RNA2 (500 ng) or in the presence of Luc or Rluc mRNAs with or without a cap structure. After incubation for 120 min, BYL was centrifuged at 21,000 × g for 30 min at 4°C to obtain the P20 pellet.
The P20 pellets were suspended in BN/PAGE sample buffer (50 mM Bis-Tris-HCl [pH 7.2], 50 mM NaCl, 10% glycerol, 0.001% Ponceau S, 0.5% Triton X-100) and incubated on ice for 30 min. The solubilized solution was clarified by centrifugation at 21,000 × g for 10 min at 4°C. Native PAGE (5%) G-250 sample additive (Invitrogen, Carlsbad, CA) was added at a final concentration of 0.125% to the solubilized solution before BN/PAGE. The protein samples were subjected to BN/PAGE using a NativePAGE Novex Bis-Tris gel system (Invitrogen) according to the manufacturer's instructions and then analyzed by Western blotting with appropriate antibodies.
Chemical treatment of membranes.
The P20 pellet extracted from Agrobacterium-infiltrated leaves was resuspended in high-salt extraction buffer (50 mM Tris-HCl [pH 7.4], 15 mM MgCl2, 1 M KCl, 0.1% 2-mercaptoethanol), alkaline extraction buffer (0.1 M Na2CO3 [pH 10.5]), or urea extraction buffer (25 mM HEPES-KOH [pH 6.8], 4 M urea). The samples were incubated on ice for 30 min with occasional mixing and centrifuged at 21,000 × g for 30 min at 4°C to obtain the supernatant and the pellet fractions. Both fractions were subjected to SDS-PAGE, followed by Western blotting with appropriate antibodies. Alternatively, the pellet fractions were resuspended in BN/PAGE sample buffer and subjected to BN/PAGE followed by Western blotting.
Sucrose gradient sedimentation analysis.
The P20 pellets extracted from 0.8-g aliquots of Agrobacterium-infiltrated leaves were resuspended in 800 μl of resuspension buffer (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 1 mM dithiothreitol [DTT], 0.5% Triton X-100, Complete Mini protease inhibitor cocktail), incubated for 30 min at 4°C, and centrifuged at 21,000 × g for 10 min at 4°C. The supernatant was loaded onto 8.8-ml lots of 5 to 40% continuous sucrose gradients and centrifuged at 170,000 × g for 16 h at 4°C. Fractions of 800 μl each were collected and analyzed by Western blotting after SDS-PAGE or BN/PAGE. The RdRP activity of each fraction was assayed as described below.
Affinity purification of RCNMV RdRP complexes.
The P20 pellet extracted from 1 g of Agrobacterium-infiltrated leaves was resuspended in 1 ml of resuspension buffer, incubated for 30 min at 4°C, and centrifuged at 21,000 × g for 10 min at 4°C. Then, the supernatant was incubated with a 50-μl bed volume of anti-FLAG M2-agarose affinity gel (Sigma-Aldrich) with gentle rotation at 4°C for 1 h. The resin was washed three times with 1 ml of resuspension buffer supplemented with 500 mM NaCl and washed once with 1 ml of resuspension buffer. The bound proteins were eluted in 250 μl of resuspension buffer containing 150 μg/ml 3× FLAG peptide (Sigma-Aldrich). The eluates were subjected to BN/PAGE and SDS-PAGE, followed by Western blotting with appropriate antibodies. If required, micrococcal nuclease (MNase) was added in the presence of 2 mM CaCl2 to remove endogenous RNAs. EGTA was added at a 5 mM final concentration to inactivate the MNase. The RdRP activity of the eluates was assayed as described below.
For two-dimensional BN-SDS-PAGE analysis, strips from the first-dimensional BN/PAGE were excised and incubated in denaturing buffer (62.5 mM Tris-HCl [pH 6.8], 1% SDS, 1% 2-mercaptoethanol) for 5 min at 50°C. The strips were layered onto SDS-PAGE gels and electrophoresed. The separated proteins were analyzed by Western blotting with appropriate antibodies.
RdRP assay.
RdRP reactions were performed by adding 5× RdRP buffer (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 5 mM [each] ATP, CTP, and GTP, 50 μM UTP, 1 Ci/liter [32P]UTP) to RdRP preparations in the absence or presence of RNA templates in a total volume of 25 μl. RNA templates were added at the final concentration of 20 ng/μl. Reaction mixtures were incubated at 25°C for 1 h. The RdRP products were isolated by phenol-chloroform extraction and ethanol precipitation. When required, the RdRP products were resuspended in 50 μl of buffer (30 mM NaOAc, 280 mM NaCl, 1 mM ZnSO4, 1 U/μl S1 nuclease). These samples were incubated for 30 min at 37°C, extracted with phenol-chloroform, and precipitated with ethanol. All RdRP products were loaded onto denaturing agarose gels containing formaldehyde. Gels were dried and exposed to an imaging plate. Radioactive signals were detected using a FLA-5100 imaging system (Fujifilm, Tokyo, Japan).
Western blot analysis.
Western blot analysis was performed essentially as previously described (42). Protein samples were analyzed on 10% SDS-PAGE gels or on 4 to 16% BN/PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, MA). Anti-FLAG M2 monoclonal antibody, anti-HA monoclonal antibody (3F10, Roche), anti-T7 monoclonal antibody, anti-p27 antisera (42), and anti-ER-localized luminal binding protein (BiP) antibody (9) were used as the primary antibodies. Alkaline phosphatase (AP)-conjugated anti-rabbit IgG antibody (Cell Signaling Technology, Beverley, MA), AP-conjugated anti-mouse IgG antibody (KPL, Gaithersburg, MD), and AP-conjugated anti-rat IgG antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) were used as secondary antibodies. The signals were visualized with CDP-star (Roche) and detected using a luminescent-image analyzer (LAS 1000 plus; Fujifilm).
Mass spectrometry.
The affinity-purified RdRP preparation and its control preparation were subjected to SDS-PAGE and BN/PAGE. After visualization of protein bands on the gels with a silver staining kit (Wako, Osaka, Japan), protein bands of interest detected using SDS-PAGE and a protein band corresponding to the 480-kDa complex on BN/PAGE were excised and subjected to in-gel digestion with trypsin. The resulting peptides were extracted from the gels and subjected to liquid chromatography-tandem mass spectrometry (LC/MS/MS) analysis. Protein bands in control lanes of the same size as those in sample lanes were also analyzed by LC/MS/MS to identify proteins specifically present in the affinity-purified RdRP preparation. Peak lists obtained from the MS/MS spectra and the Mascot search engine (Matrix Science, London, United Kingdom) were used to identify proteins.
RESULTS
p27 interacts with both p27 and p88.
The coexistence of p27 and p88 in the purified RdRP fraction from RCNMV-infected tissue (2) suggests that the formation of functional RCNMV RNA replication complexes requires interactions between p27 and p88. To address this issue, we analyzed interactions between RCNMV replicase proteins by coimmunoprecipitation using C-terminally hemagglutinin (HA)-tagged p27 (p27-HA), C-terminally FLAG-tagged p27 (p27-FLAG), and C-terminally T7-tagged p88 (p88-T7). These epitope-tagged replicase proteins were functional: Agrobacterium-mediated expression of these proteins in appropriate combinations supported the accumulation of positive- and negative-strand RNA2 in coinfiltrated N. benthamiana leaves at levels comparable to those seen with wild-type p27 and p88 (data not shown).
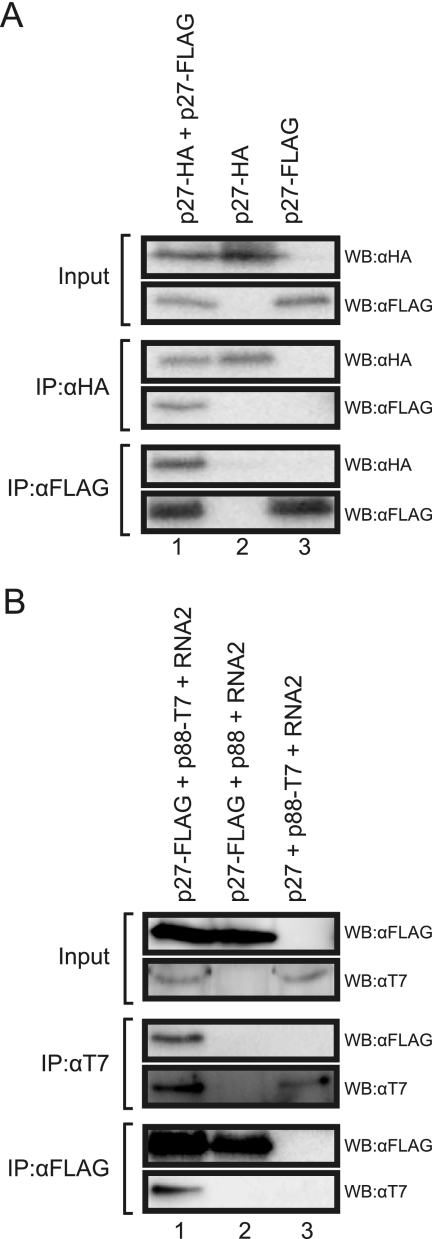
First, we tested whether p27 interacts with itself. p27-HA and p27-FLAG were coexpressed in N. benthamiana by Agrobacterium-mediated expression, and clarified leaf extracts were used for immunoprecipitation analysis. The anti-FLAG antibody copurified p27-HA together with p27-FLAG, and the anti-HA antibody copurified p27-FLAG together with p27-HA (Fig. 1A, lane 1). When p27-HA or p27-FLAG was expressed singly, these proteins were not detected after immunoprecipitation with anti-FLAG or anti-HA antibodies, respectively (Fig. 1A, lanes 2 and 3). These results indicate that p27 interacts with itself.
FIG. 1.
Interaction of p27 with both p27 and p88. (A) Coimmunoprecipitation analysis of the interaction between p27-HA and p27-FLAG. Protein extracts from Agrobacterium-infiltrated leaves expressing viral proteins were subjected to immunoprecipitation with anti-HA antibody (IP:αHA) or anti-FLAG antibody (IP:αFLAG), followed by Western blotting using anti-HA antibody (WB:αHA) and anti-FLAG antibody (WB:αFLAG). (B) Coimmunoprecipitation analysis of the interaction between p27-FLAG and p88-T7. Protein extracts from Agrobacterium-infiltrated leaves expressing viral proteins and RNA2 were subjected to immunoprecipitation with anti-FLAG antibody (IP:αFLAG) or anti-T7 antibody (IP:αT7), followed by Western blotting using anti-FLAG antibody (WB:αFLAG) and anti-T7 antibody (WB:αT7).
Next, to test whether p27 interacts with p88, we expressed p27-FLAG and p88-T7 in the presence and absence of RNA2. In the presence of RNA2, p27-FLAG copurified p88-T7 and vice versa (Fig. 1B, lane 1). p27-FLAG and p88-T7 were not immunoprecipitated with the anti-T7 and anti-FLAG antibodies, respectively (Fig. 1B, lanes 2 and 3), ruling out nonspecific copurification. These results indicate that p27 interacts with p88 in vivo. In the absence of RNA2, we failed to detect p88-T7 in the total fraction and even in the affinity-purified fraction (data not shown). These results suggest that replication of RNA2 is required for p88 to accumulate to a level sufficient for analysis of the p27-p88 interaction in vivo.
RCNMV replicase proteins form two types of complexes of different sizes.
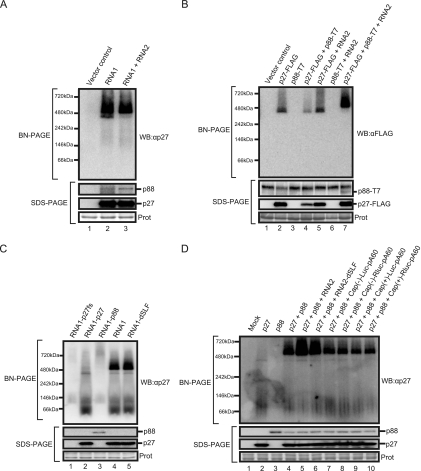
Interactions between RCNMV replicase proteins (Fig. 1) and their localization to the ER (44) suggested that p27 and p88 might form replication complexes on the ER membrane. To characterize these formations, we performed BN/PAGE analysis. RNA1 alone and RNA1 plus RNA2 were expressed in N. benthamiana by Agrobacterium-mediated expression. The membrane fraction prepared from Agrobacterium-infiltrated leaves was solubilized with Triton X-100 and subjected to BN/PAGE, followed by Western blotting with anti-p27 antisera. Two specific bands of about 380 kDa and about 480 kDa were detected (Fig. 2A, lanes 2 and 3). Western blot analysis of the same samples after SDS-PAGE confirmed the accumulation of both p27 and p88 in these leaves (Fig. 2A). These results suggest that RCNMV replicase proteins form two differently sized complexes in infected plants.
FIG. 2.
RCNMV replicase proteins form two different patterns of high-molecular-mass complexes. Membrane fractions were prepared from Agrobacterium-infiltrated leaves expressing RCNMV RNAs (A), from Agrobacterium-infiltrated leaves expressing various combinations of p27-FLAG, p88-T7, and RNA2 (B), from BYL incubated with RNA1 derivatives (C), or from BYL incubated with various combinations of p27 mRNA, p88 mRNA, RNA2, RAN2-dSLF, or uncapped or capped Luc and RLuc mRNAs (D). All membrane fractions were solubilized with Triton X-100 and subjected to BN/PAGE (upper panel) and SDS-PAGE (lower panel). Western blots for BN/PAGE and SDS-PAGE were performed using appropriate antibodies. Coomassie brilliant blue (CBB)-stained cellular proteins on SDS-PAGE gels are shown below the Western blots as a loading control (Prot).
To investigate these complexes in more detail, we expressed p27-FLAG, p88-T7, and RNA2 in various combinations in N. benthamiana. Protein complexes were separated by BN/PAGE and probed with an anti-FLAG antibody. Western blot analysis showed that the 380-kDa complex accumulated in leaves expressing p27-FLAG alone (Fig. 2B, lane 2). RNA2 did not affect the accumulated level of the 380-kDa complex (Fig. 2B, lane 5). These results indicate that p27 forms the 380-kDa complex in the absence of p88 and RCNMV RNAs. Expression of p27-FLAG plus p88-T7 resulted in the accumulation of the 380-kDa but not the 480-kDa complex (Fig. 2B, lane 4). Neither the 380-kDa nor the 480-kDa complex was detected in leaves expressing p88-T7 or p88-T7 plus RNA2 (Fig. 2B, lanes 3 and 6). Interestingly, the 480-kDa complex accumulated predominantly in leaves expressing p27-FLAG together with p88-T7 and RNA2 (Fig. 2B, lane 7). Neither the 380-kDa nor the 480-kDa complex was detected with an anti-T7 antibody (data not shown). Western blot analysis with an anti-T7 antibody after SDS-PAGE demonstrated that p88-T7 was detected only in the presence of p27-FLAG and RNA2 (Fig. 2B, lane 7), suggesting that p88 is unstable in the absence of p27 and viral RNAs in vivo. Together, these results indicate that both the 380-kDa and 480-kDa complexes contain p27 and suggest that the formation of the 480-kDa complex requires three viral components: p27, p88, and viral RNAs.
The formation of the 480-kDa complex does not require negative-strand RNA synthesis in vitro.
It remained to be determined whether the accumulation of p88 or viral RNA replication is responsible for the formation of the 480-kDa complex. To address this issue, we analyzed protein complexes formed in a BYL in vitro translation-replication system. This has previously been used successfully to analyze the translation and replication mechanisms of plant RNA viruses, including RCNMV (14, 18, 27). RNA1 mutants expressing p27 or p88 alone, or no functional protein, and an RNA1 mutant expressing both p27 and p88 but lacking a 3′-terminal stem-loop structure were incubated in BYL. Protein samples prepared from the membrane fractions of BYL were subjected to BN/PAGE and analyzed by Western blotting with anti-p27 antisera. Accumulation of the 380-kDa but not the 480-kDa complex was detected after incubation with RNA1-p27, which expresses only p27, confirming the results of in vivo BN/PAGE (Fig. 2C, lane 2). After incubation with RNA1-p88, which expresses only p88, a huge (∼720 kDa) complex was detected. SDS-PAGE and Western blot analysis showed a stable accumulation of p88 (Fig. 2C, lane 3), suggesting that overexpression of p88 might have been responsible for its aggregation. As expected, both the 380-kDa and 480-kDa complexes were detected in incubation with wild-type RNA1 (Fig. 2C, lane 4). Interestingly, these complexes were also detected in incubation with RNA1-dSLF (Fig. 2C, lane 5), which lacks the 3′-terminal stem-loop structure required for negative-strand RNA synthesis of RNA1 (14). These results suggest that formation of the 480-kDa complex requires both p27 and p88 in the presence of RNA1 but does not require the initiation of negative-strand RNA synthesis.
To further investigate the viral RNA elements required for the accumulation of the 480-kDa complex, we expressed p27 and p88 from capped mRNAs with poly(A) in BYL. The 480-kDa complex accumulated in BYL when p27 and p88 were expressed from these capped mRNAs, even in the absence of RNA2 (Fig. 2D, lane 4). This confirmed the lack of a requirement for negative-strand RNA synthesis for accumulation of the 480-kDa complex. It is possible that viral RNA elements in the ORFs of p27 and p88 play roles in the formation of the 480-kDa complex. Interestingly, the accumulated level of the 480-kDa complex was enhanced by adding either RNA2 or RNA2-dSLF in BYL but not by adding nonviral Luc or RLuc mRNAs with or without a cap structure (Fig. 2D). These results suggest that viral nucleotide sequences other than the 3′-terminal conserved sequences of RNA2 are involved in forming the 480-kDa complex.
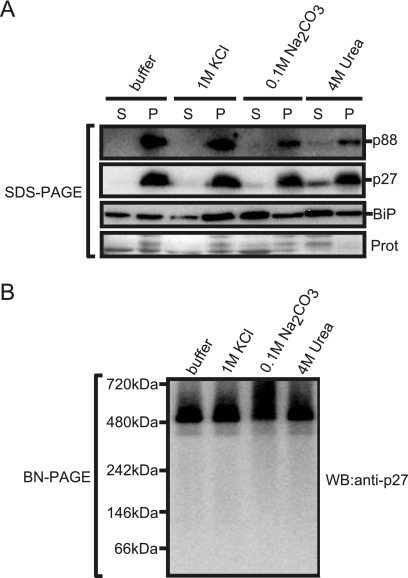
Both the 380-kDa and 480-kDa complexes tightly associate with membranes.
To investigate the nature of the association of p27 and p88 with ER, the membrane fraction prepared from Agrobacterium-infiltrated leaves expressing RNA1 plus RNA2 was treated with agents that were expected to remove proteins weakly or peripherally associated with the membrane. The ER-localized luminal binding protein (BiP) was used as a control. After treatments with the agents, the membrane fractions were collected by centrifugation and analyzed by Western blotting with anti-p27 antisera or anti-BiP antibody. As shown in Fig. 3A, the association of BiP with ER was not affected by treatment with 1 M KCl but was affected by treatment with 0.1 M Na2CO3 (pH 10.5) or 4 M urea. In contrast, neither treatment significantly affected the association of p27 and p88 with ER. Furthermore, the 380-kDa and the 480-kDa complexes were detected in Triton X-100-solubilized membranes after these treatments (Fig. 3B). Together, these results suggest that p27 and p88 tightly associate with the ER as the 380-kDa and 480-kDa complexes.
FIG. 3.
Effects of membrane washes on association of p27 and p88 and their complexes with the ER. (A) Membrane fractions prepared from Agrobacterium-infiltrated leaves expressing RNA1 plus RNA2 were incubated on ice for 30 min with 1 M KCl, 0.1 M Na2CO3 (pH 10.5), or 4 M urea. After incubation, samples were centrifuged to obtain the supernatant (lanes S) and the pellet (lanes P) fractions. Both fractions were subjected to SDS-PAGE, followed by Western blotting with anti-p27 antisera and anti-BiP antibody. CBB-stained cellular proteins are shown below the Western blots as a loading control. (B) The pellet fractions were solubilized with 0.5% Triton X-100 and subjected to BN/PAGE, followed by Western blotting with anti-p27 antisera.
The 480-kDa complex cofractionates with both endogenous template-bound and exogenous template-dependent RdRP activities.
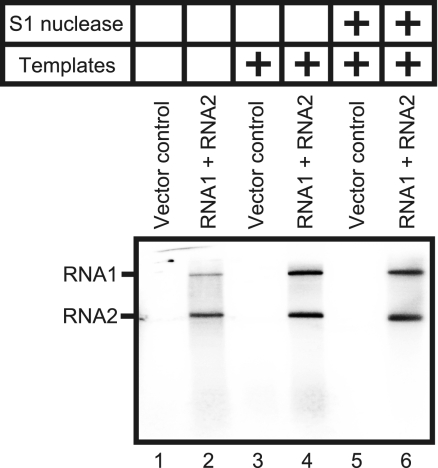
The membrane fraction extracted from Agrobacterium-infiltrated leaves expressing RNA1 plus RNA2 were treated with Triton X-100 and centrifuged to obtain a solubilized fraction. The RdRP activity of the fraction was assayed in the presence or absence of added RNA templates. Two labeled products corresponding to RNA1 and RNA2 in size were detected in the absence of added RNA templates (Fig. 4, lane 2). Addition of RNA templates increased incorporation of 32P into these products, which were resistant to S1 nuclease (Fig. 4, lanes 4 and 6). These products were not detected in the Triton X-100-solubilized fraction prepared from leaves infiltrated with an Agrobacterium strain carrying an empty vector (Fig. 4, lanes 1, 3, and 5). These results suggest that this solubilized fraction prepared from Agrobacterium-infiltrated leaves expressing RNA1 plus RNA2 contains both an endogenous template-bound RdRP and an exogenous template-dependent RdRP.
FIG. 4.
RdRP activity of the solubilized membrane fractions prepared from Agrobacterium-infiltrated leaves expressing RNA1 plus RNA2. The RdRP assays were performed without RNA templates (lanes 1 and 2) and with RCNMV RNA1 and -2 (lanes 3 to 6). The RdRP products represented in lanes 5 and 6 were treated with S1 nuclease.
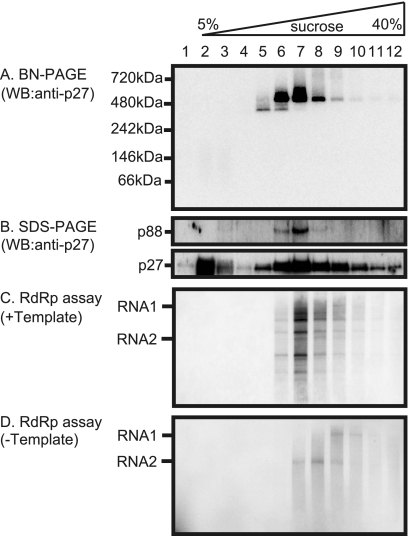
To further characterize the RCNMV RNA replication complexes, a Triton X-100-solubilized fraction prepared from Agrobacterium-infiltrated leaves expressing RNA1 plus RNA2 was subjected to 5 to 40% continuous sucrose gradient centrifugation. Aliquots of 12 fractions were analyzed by Western blotting with anti-p27 antisera after SDS-PAGE or BN/PAGE, and the RdRP activity of each fraction was assayed in the presence or absence of RCNMV RNAs. After BN/PAGE, the 380-kDa complex and the 480-kDa complex were detected mainly in fractions 5 and 6 and in fractions 6 to 8, respectively, whereas the faint band of the 480-kDa complex was detected in fractions 5 and 9 to 12 (Fig. 5A). SDS-PAGE analysis showed that the p88 component was mainly detected in fractions 6 to 8 and that its accumulating levels paralleled those of the 480-kDa complex. Both exogenous template-dependent RdRP activities (Fig. 5C, lanes 6 to 12) and endogenous template-bound RdRP activities (Fig. 5D, lanes 7 to 10) cofractionated with the 480-kDa complexes (Fig. 5). The accumulating levels of the 480-kDa complex were found to be more nearly parallel with template-dependent RdRP activities than with endogenous template-bound RdRP activities. Fraction 6 exibited only template-dependent RdRP activity but no detectable endogenous template-bound RdRP activity (Fig. 5C and D, lanes 6). The labeled RNA products of endogenous template-bound RdRP were full-length RNAs, whereas the products of template-dependent RdRP were RNA fragments. Interestingly, the products of endogenous template-bound RdRP in the lower-density fractions corresponded to RNA2 in size (Fig. 5D, lanes 7 to 9) and those in higher density fractions corresponded to RNA1 (Fig. 5D, lanes 9 and 10). This could reflect the differences in size between RNA1 (3890 nt) and RNA2 (1448 nt). No RdRP activity was detected in fraction 5 (Fig. 5C and D, lanes 5), suggesting that the 380-kDa complex retained no RdRP activity. Together, these results suggest that the 480-kDa complexes contain both p27 and p88 and contribute to exogenous template-dependent RdRP activities.
FIG. 5.
Sucrose gradient sedimentation analysis of RCNMV replicase proteins, the 380-kDa and the 480-kDa complexes, and the assay for their RdRP activity. (A and B) The solubilized membrane fractions prepared from Agrobacterium-infiltrated leaves expressing RNA1 plus RNA2 were loaded onto 5 to 40% continuous sucrose gradients and centrifuged at 170,000 × g for 16 h at 4°C. Twelve fractions (numbered 1 to 12 from the top to the bottom of the mixture) were collected and subjected to BN/PAGE (A) and SDS-PAGE (B), followed by Western blotting with anti-p27 antisera. (C and D) The RdRP activity of each fraction was assayed with or without RCNMV RNA templates.
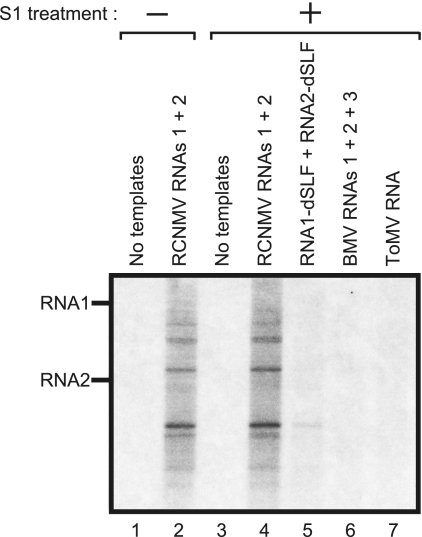
To characterize the template-dependent RdRP activity of the 480-kDa complexes, fraction 6 of the sucrose gradients was used to assess the template specificity of the complex. The templates used were RCNMV RNAs (RNA1, RNA2, RNA1 plus RNA2, and RNA1-dSLF plus RNA2-dSLF) and the genomic RNAs of BMV and ToMV. Both RCNMV RNA1 and RNA2 served as efficient templates in producing S1 nuclease-resistant RNA fragments (Fig. 6, lanes 2 and 4, and data not shown). RNA1-dSLF and RNA2-dSLF, which lack the 3′-terminal stem-loop structure required for negative-strand synthesis (14, 45), did not serve as efficient templates (Fig. 6, lane 5). No detectable RNA synthesis was observed with the BMV or ToMV RNAs (Fig. 6, lanes 6 and 7). These results suggest that the 480-kDa complex specifically recognizes the 3′-terminal core promoter sequences of RCNMV genomic RNAs to synthesize viral RNA fragments.
FIG. 6.
The RdRP activity of the 480-kDa complex exhibits high template specificity. The RdRP activity of fraction 6 of the sucrose gradients was assayed without templates (lanes 1 and 3), with RCNMV RNA1 and -2 (lanes 2 and 4), with RCNMV RNA1-dSLF and RNA2-dSLF (lane 5), with BMV RNA1, -2, and -3 (lane 6), and with ToMV RNA (lane 7). The RdRP products represented in lanes 3 to 7 were treated with S1 nuclease before electrophoresis.
Identification of the components of the 480-kDa complex.
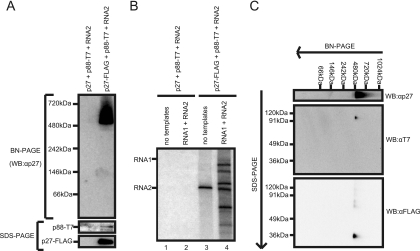
To further investigate the characteristics and protein components of the 480-kDa complexes, the Triton X-100-solubilized fraction prepared from Agrobacterium-infiltrated leaves expressing p27-FLAG together with p88-T7 and RNA2 was subjected to FLAG tag-based affinity purification as described in Materials and Methods. The affinity-purified fraction contained p27-FLAG, p88-T7, and both the 380-kDa and the 480-kDa complexes (Fig. 7A). This affinity-purified fraction also showed an endogenous template-bound RdRP activity for synthesizing an RNA corresponding to RNA2 in size (Fig. 7B, lane 3) and template-dependent RdRP activity on RCNMV RNAs, as observed in fractions 6 to 12 of the sucrose gradients (Fig. 5). These results suggest that the 480-kDa complex purified with FLAG tag contributes to both endogenous template-bound RdRP activity and exogenous template-dependent RdRP activity to synthesize RNA fragments from RCNMV RNA templates.
FIG. 7.
Identification of the components of the 480-kDa complex. The solubilized membrane fractions prepared from Agrobacterium-infiltrated leaves expressing p27 plus p88-T7 plus RNA2 or p27-FLAG plus p88-T7 plus RNA2 were subjected to affinity purification with anti-FLAG antibody-conjugated agarose beads. (A) The affinity-purified fractions were subjected to BN/PAGE and SDS-PAGE, followed by Western blotting with anti-p27 antisera. (B) The RdRP activity of the affinity-purified fractions was assayed in the absence or presence of RCNMV RNA templates. (C) The affinity-purified fraction prepared from Agrobacterium-infiltrated leaves expressing p27-FLAG plus p88-T7 plus RNA2 was subjected to two-dimensional BN/SDS-PAGE, followed by Western blotting with anti-T7 and anti-FLAG antibodies.
To confirm the presence of both p27 and p88 in the 480-kDa complexes, we carried out two-dimensional BN/SDS-PAGE analyses using the affinity-purified fraction. After electrophoresis, the separated proteins were blotted onto membranes and probed with anti-T7 or anti-FLAG antibodies. Spots corresponding to p88-T7 and p27-FLAG in size were detected by the anti-T7 and anti-FLAG antibodies, respectively, along the vertical line below the position of the 480-kDa complexes in BN/PAGE (Fig. 7C). This result indicates that both p27 and p88 are present in the 480-kDa complex.
Next, to identify host proteins included in the 480-kDa complexes, we prepared the affinity-purified 480-kDa complex from solubilized membrane fractions extracted from Agrobacterium-infiltrated N. benthamiana leaves expressing p27-FLAG together with RNA1-p88 and RNA2. We chose this combination because replication of both RNA1 and RNA2 reflects RCNMV replication in nature and because only RNA1—from which p88 is translated—can be an efficient template for viral RNA replication in the presence of p27. As a control, we prepared similar preparations from solubilized membrane fractions extracted from Agrobacterium-infiltrated leaves expressing p27 together with RNA1-p88 and RNA2. The affinity-purified RCNMV RdRP preparation retained the activity to produce RNA1- and RNA2-sized products without adding RNA templates and the ability to produce RNA fragments when we added RCNMV RNAs (data not shown).
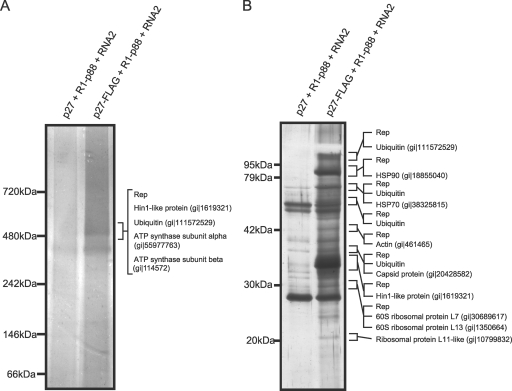
LC/MS/MS analysis of a band corresponding to the affinity-purified 480-kDa complex on BN/PAGE identified RCNMV p27 and p88 replicase proteins, ATP synthase alpha and beta subunits, harpin-induced protein 1 (Hin1)-like protein, and ubiquitin (Fig. 8A). This result suggests that several host proteins, including ATP synthase subunits, Hin1-like protein, and ubiquitin, are candidates for the components of the 480-kDa complex, although further experiments to confirm the involvement of these proteins in the complex are needed.
FIG. 8.
Identification of proteins copurified with RCNMV RdRP complexes. The solubilized membrane fractions prepared from Agrobacterium-infiltrated leaves expressing p27 plus RNA1-p88 plus RNA2 or p27-FLAG plus RNA1-p88 plus RNA2 were subjected to affinity purification with anti-FLAG antibody-conjugated agarose beads. (A) The affinity-purified fractions were subjected to BN/PAGE and stained with MS-compatible silver staining. A protein band corresponding to the 480-kDa complex was excised, subjected to in-gel digestion, and analyzed by tandem mass spectrometry. Proteins that showed Mascot search scores above 30 and that were absent in control protein bands are indicated on the right side of the panel. (B) The affinity-purified fractions were subjected to SDS-PAGE and stained using MS-compatible silver staining. Protein bands of interests were excised, subjected to in-gel digestion, and analyzed by tandem mass spectrometry. Proteins that showed Mascot search scores above 50 and that were absent in control protein bands, and proteins that showed significantly higher scores than control proteins, are indicated on the right sides of the panels. The NCBI accession numbers of the identified proteins are also indicated. Rep, RCNMV replicase proteins.
We also performed LC/MS/MS analysis of protein bands from SDS-PAGE separations of the affinity-purified RdRP preparations and identified at least 12 proteins. These included RCNMV p27 and p88 replicase proteins, RCNMV CP, heat shock protein 70, heat shock protein 90, HinI-like protein, actin, ubiquitin, and several ribosomal proteins (Fig. 8B). It should be noted that this preparation contained several proteins that were not detected in a band corresponding to the 480-kDa complex separated on BN/PAGE.
DISCUSSION
This study has shown that RCNMV p27 and p88 replicase proteins, together with possible host proteins, form a tightly membrane-associated protein complex with an approximate size of 480 kDa. This complex contributes to the template-dependent RdRP activity that synthesizes RNA fragments by specifically recognizing core promoter sequences located at the 3′ end of RCNMV genomic RNAs. These data suggest that the 480-kDa complex plays an essential role in RCNMV RNA synthesis.
RCNMV RdRP has previously been isolated from RCNMV-infected Nicotiana cleveladii leaves and characterized (2). Solubilization of the membrane-bound RdRP with dodecyl-β-d-maltoside (DoM) followed by chromatography purifications produced an endogenous template-bound RdRP fraction that contained p27, p88, and possible host proteins. Treatment of the template-bound RdRP with MNase converted the RdRP to a template-dependent RdRP that was capable of full-length RNA synthesis when RNA templates were added (2). We used Triton X-100 to solubilize the membrane fractions prepared from RCNMV-infected N. benthamiana tissues. This solubilized membrane fraction contained both an endogenous template-bound RdRP and an exogenous template-dependent RdRP (Fig. 4). RNA products of both RdRP fractions were full-length and S1 nuclease-resistant RNAs (Fig. 4). However, we failed to obtain RdRP fractions synthesizing full-length RNAs in a template-dependent manner by sucrose gradient centrifugation, although the endogenous template-bound RdRP still had the ability to synthesize full-length RNAs (Fig. 5C and D). Similar results were obtained in the sucrose gradient sedimentation analysis of the DoM-solubilized membrane fractions (data not shown). Furthermore, the FLAG tag-based affinity-purified RdRP fraction also showed endogenous template-bound and template-dependent RdRP activities, which synthesized full-length RNA and RNA fragments, respectively (Fig. 7B). Together, these results suggest that the failure of these RdRP preparations to synthesize full-length viral RNAs in a template-dependent manner might have been caused by the lack of some factors present in endogenous template-bound RdRP complexes. These factors might have been removed from the replication complexes during the purification processes, including sucrose gradient centrifugation. It should be noted that MNase treatment of our affinity-purified RdRPs did not make the RdRPs capable of synthesizing full-length RNAs. This result is inconsistent with that reported by Bates et al. (2), possibly due to differences in experimental conditions such as purification procedures and/or differences in the nature of the RdRP preparations. Their preparation contained only endogenous template-bound RdRP, whereas ours contained both template-bound and template-dependent RdRPs. The coexistence of template-bound and template-dependent RdRPs may mask the effect of MNase treatment.
In the sucrose gradient sedimentation analysis, both endogenous template-bound and exogenous template-dependent RdRP activities cofractionated with the 480-kDa complexes containing p27 and p88 (Fig. 5 and 7C). These results suggest that both the endogenous template-bound RdRP and the template-dependent RdRP complexes are associated with the 480-kDa complex. This leaves one question to be addressed: how can endogenous template-bound RdRP complexes synthesize full-length RNAs? If the template-bound RdRP complexes contain full-length RCNMV genomic RNAs, they should be much larger than 480 kDa, because the predicted molecular masses of RNA1 and RNA2 are 1,340 kDa and 500 kDa, respectively. We failed to detect such complexes by BN/PAGE. Hence, we suspect that (i) such complexes were too large to enter the BN/PAGE gel, (ii) the accumulation levels of such complexes were under the detection limits, or (iii) the 480-kDa complexes detected by BN/PAGE analysis were the endogenous template-bound RdRP complexes from which genomic RNAs were released during electrophoresis (12). The third hypothesis might best explain why the 480-kDa complexes were detected even in relatively high-density fractions such as fractions 10, 11, and 12, as shown in Fig. 5A.
Interestingly, fraction 6 of the sucrose gradients containing the 480-kDa complex exhibited only template-dependent but not template-bound RdRP activity (Fig. 5, lane 6), suggesting that the 480-kDa complexes retained no endogenous RNA templates. This template-dependent RdRP activity was partial but exhibited marked template specificity (Fig. 6). These results suggest that the 480-kDa complex is an RCNMV-specific template-dependent RdRP and that it requires some additional factor(s) to synthesize full-length products. The additional factor(s) required for full-length RNA synthesis could be present in endogenous template-bound RdRP complexes as a result of binding to the endogenous RCNMV RNA templates. In fact, it has been reported that some host-encoded proteins bind to viral RNAs and thereby affect viral RNA replication (10, 23, 56). For instance, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) preferentially binds to an AU pentamer sequence in the negative-strand RNA of Tomato bushy stunt virus (TBSV) to control asymmetric RNA synthesis (47). GAPDH is known to bind to AU-rich sequences at the 3′ terminus of mRNAs to enhance their stability in addition to its central role in cytosolic energy production (3). Thus, it is possible that the 480-kDa complex is engaged in RCNMV RNA synthesis together with some host proteins that bind to RCNMV RNA templates.
HSP70, HSP90, actin, and ribosomal proteins could be candidates that promote full-length RCNMV RNA synthesis, because they were not detected in the 480-kDa complex but were instead associated with the affinity-purified RdRP complexes with FLAG-tagged p27 that could synthesize full-length RNA. For instance, some ribosomal proteins bind to viral genomic RNAs and promote viral RNA replication (21, 23, 33). HSP70s also play important roles in viral RNA replication (35, 40, 48). Recently, Wang et al. (48) reported that yeast HSP70 (Ssa1p) is required during the TBSV replicase assembly process but not during RNA synthesis.
Copurification of RCNMV CP with viral RdRP complexes is an interesting finding (Fig. 8B), although CP is dispensable for RCNMV RNA replication (30, 42). It is possible that CP was copurified with RCNMV RdRPs via its binding to viral RNAs. Basnayake et al. (1) demonstrated that the trans-activator (TA) element, a multifunctional 34-nt SL structure within the RNA2 MP ORF, is an essential minimal element for RCNMV virion assembly, suggesting binding of CP to the TA element. Also, the TA element serves as a cis-acting replication element (43) and functions in CP sgRNA transcription from RNA1 by base pairing with the TA binding site found within the RNA1 CP subgenomic promoter (41). These findings, together with that of the copurification of CP with RCNMV RdRP complexes, may imply that CP sgRNA transcription, CP translation, and RCNMV encapsidation occur close to the site for RCNMV RNA replication.
Using coimmunoprecipitation and BN/PAGE analyses, we have shown that p27 interacts with both p27 and p88 (Fig. 1) and that these replicase proteins form the 380-kDa and the 480-kDa complexes (Fig. 2), suggesting that these complexes are assembled through p27-p27 and p27-p88 interactions. p27 could form the 380-kDa complex in the absence of p88 and viral RNAs (Fig. 2B). This suggests that the 380-kDa complex is an oligomeric form of p27 or a multiprotein complex that includes p27 and host protein(s). On the other hand, the 480-kDa complex contained both p27 and p88 and was associated with possible host proteins, including ATP synthase subunits, Hin1-like protein, and ubiquitin (Fig. 7C and 9A). However, the 480-kDa complex formed by p27-FLAG and p88-T7 in the crude membrane fraction could not be detected by Western blotting with anti-T7 antibody (Fig. 2B and data not shown). It is possible that it is difficult for the antibody to access the C-terminal epitope in p88-T7 under native conditions. Alternatively, it is likely that the amount of the 480-kDa complex in the crude membrane fraction is not sufficient for detection of the p88-T7 component by Western blotting, probably because only limited amounts of protein samples can be applied to BN/PAGE gels. Consistent with this possibility, we failed to detect p88-T7 when the 480-kDa complex in the crude membrane fraction was resolved by two-dimensional BN/SDS-PAGE (data not shown), although we could detect the protein when the affinity-purified samples were used for BN/SDS-PAGE analysis (Fig. 7C).
The 480-kDa complex accumulated in BYL when p27 and p88 were produced from capped mRNA with poly(A) in the absence of RNA2 or from a replication-deficient RNA1 mutant (Fig. 2C). These results indicate that negative-strand RNA synthesis is not essential for the formation of the 480-kDa complex in BYL. It is possible that viral RNA elements in protein-coding regions and noncoding regions other than the 3′-terminal SL sequences are involved in forming the 480-kDa complex. This possibility is supported by the findings that the addition of RNA2 or replication-deficient RNA2-dSLF, but not of nonviral RNAs, enhanced the accumulation of the 480-kDa complex in BYL (Fig. 2D). The 480-kDa complex might be formed at sites other than a core promoter prior to negative-strand RNA synthesis. The similar accumulation levels of the 480-kDa product produced by RNA1 and RNA1-dSLF in BYL also support this idea. The involvement of RNA elements other than 3′-proximal SL sequences in the RdRP complex formation has previously been proposed for BMV, Cucumber necrosis virus, and Alfalfa mosaic virus (34, 36, 37, 46). An internal RNA element in a protein-coding region plays an important role in the assembly of the tombusvirus replicase (36).
Tight associations with intracellular membranes seem to be common features of the replicase proteins of positive-strand RNA viruses. Hepatitis C virus NS4B induces the formation of distinct membrane structures, such as membranous webs derived from ER membranes (7, 13), and Flock house virus protein A induces membranous spherules derived from outer mitochondrial membranes (20, 25). In the case of BMV, replicase protein 1a induces spherical invaginations or appressed double membranes of ER (38, 39). Furthermore, 1a recruits 2a polymerase and RNA templates into the 1a-induced membrane structures to establish functional RNA replication complexes (38, 39). RCNMV p27 and p88 replicase proteins were also tightly associated with membranes as part of the 380-kDa and the 480-kDa complexes (Fig. 3). Previous studies have shown that RCNMV replicase proteins localize to the ER and that RCNMV infection causes morphological changes in ER membranes, including the formation of large aggregates near the nuclei (44). Together, the 380-kDa and the 480-kDa complexes are thought to be components of such aggregates, which could be the site for RCNMV RNA replication.
As far as we are aware, this is the first study in which BN/PAGE was applied to investigate the replication complexes of positive-strand RNA viruses associated with intracellular membranes. Our approach revealed that (i) the 480-kDa complex contains RCNMV p27 and p88 replicase proteins and was associated with possible host proteins, (ii) the 480-kDa complex contributes to exogenous template-dependent RdRP activities, and (iii) the 480-kDa complex specifically recognizes RCNMV RNA templates to produce viral RNA fragments. We conclude that the 480-kDa complex is an RCNMV RdRP complex, although it requires an additional host factor(s) for complete functionality in RCNMV RNA synthesis. Furthermore, our study confirms that BN/PAGE, in combination with mass spectrometric analysis, can be a powerful tool to identify and characterize replication complexes of positive-strand RNA viruses.
Acknowledgments
We thank S. A. Lommel for the original RNA1 and RNA2 cDNA clones of RCNMV Australian strain, R. S. Boston for the anti-BiP antibody, K. A. White for the plasmid p3′-8, and the Radioisotope Research Center of Kyoto University for assistance with radioisotope experiments. We are also grateful to H. Iwakawa for helpful discussions.
This work was supported in part by a Grant-in-Aid for Scientific Research (A) (18208004) and by a Grant-in-Aid for Scientific Research (A) (13306005) from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 7 April 2010.
REFERENCES
- 1.Basnayake, V. R., T. L. Sit, and S. A. Lommel. 2009. The Red clover necrotic mosaic virus origin of assembly is delimited to the RNA-2 trans-activator. Virology 384:169-178. [DOI] [PubMed] [Google Scholar]
- 2.Bates, H. J., M. Farjah, T. A. M. Osman, and K. W. Buck. 1995. Isolation and characterization of an RNA-dependent RNA polymerase from Nicotiana clevelandii plants infected with red clover necrotic mosaic dianthovirus. J. Gen. Virol. 76:1483-1491. [DOI] [PubMed] [Google Scholar]
- 3.Bonafé, N., M. Gilmore-Hebert, N. L. Folk, M. Azodi, Y. Zhou, and S. K. Chambers. 2005. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 65:3762-3771. [DOI] [PubMed] [Google Scholar]
- 4.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. Hellings, H. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 5.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Boon, J. A., J. B. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eubel, H., H. Braun, and A. Millar. 2005. Blue-native PAGE in plants: a tool in analysis of protein-protein interactions. Plant Methods 1:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontes, E. B. P., B. B. Shank, R. L. Wrobel, S. P. Moose, G. R. Obrian, E. T. Wurtzel, and R. S. Boston. 1991. Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell 3:483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujisaki, K., and M. Ishikawa. 2008. Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication. Virology 380:402-411. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, R. J., and K. W. Buck. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63:363-368. [DOI] [PubMed] [Google Scholar]
- 12.Hellman, L. M., and M. G. Fried. 2007. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2:1849-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hügle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 14.Iwakawa, H. O., M. Kaido, K. Mise, and T. Okuno. 2007. cis-Acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3′-untranslated region of Red clover necrotic mosaic virus RNA1. Virology 369:168-181. [DOI] [PubMed] [Google Scholar]
- 15.Iwakawa, H. O., H. Mizumoto, H. Nagano, Y. Imoto, K. Takigawa, S. Sarawaneeyaruk, M. Kaido, K. Mise, and T. Okuno. 2008. A viral noncoding RNA generated by cis-element-mediated protection against 5′→3′ RNA decay represses both cap-independent and cap-dependent translation. J. Virol. 82:10162-10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 17.Kim, K. H., and S. A. Lommel. 1994. Identification and analysis of the site of -1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology 200:574-582. [DOI] [PubMed] [Google Scholar]
- 18.Komoda, K., S. Naito, and M. Ishikawa. 2004. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. U. S. A. 101:1863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 20.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:2022-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kushner, D. B., B. D. Lindenbach, V. Z. Grdzelishvili, A. O. Noueiry, S. M. Paul, and P. Ahlquist. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 100:15764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Li, Z., J. Pogany, T. Panavas, K. Xu, A. Esposito, T. Kinzy, and P. D. Nagy. 2009. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385:245-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lommel, S. A., M. Westonfina, Z. Xiong, and G. P. Lomonossoff. 1988. The nucleotide sequence and gene organization of red clover necrotic mosaic virus RNA-2. Nucleic Acids Res. 16:8587-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, D. J., and P. Ahlquist. 2002. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizumoto, H., H. O. Iwakawa, M. Kaido, K. Mise, and T. Okuno. 2006. Cap-independent translation mechanism of Red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication. J. Virol. 80:3781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizumoto, H., M. Tatsuta, M. Kaido, K. Mise, and T. Okuno. 2003. Cap-independent translational enhancement by the 3′ untranslated region of Red Clover Necrotic Mosaic Virus RNA1. J. Virol. 77:12113-12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro, B., L. Rubino, and M. Russo. 2004. Expression of the Cymbidium ringspot virus 33-kilodalton protein in Saccharomyces cerevisiae and molecular dissection of the peroxisomal targeting signal. J. Virol. 78:4744-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishikiori, M., K. Dohi, M. Mori, T. Meshi, S. Naito, and M. Ishikawa. 2006. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 80:8459-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto, K., H. Nagano, H. Iwakawa, H. Mizumoto, A. Takeda, M. Kaido, K. Mise, and T. Okuno. 2008. cis-Preferential requirement of a-1 frameshift product p88 for the replication of Red clover necrotic mosaic virus RNA1. Virology 375:205-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osman, T. A. M., and K. W. Buck. 1987. Replication of red clover necrotic mosaic virus RNA in cowpea protoplasts: RNA1 replicates independently of RNA 2. J. Gen. Virol. 68:289-296. [Google Scholar]
- 32.Osman, T. A. M., and K. W. Buck. 1997. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J. Virol. 71:6075-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panavas, T., E. Serviene, J. Brasher, and P. D. Nagy. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 102:7326-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panaviene, Z., T. Panavas, S. Serva, and P. D. Nagy. 2004. Purification of the Cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 78:8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogany, J., J. Stork, Z. H. Li, and P. D. Nagy. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. U. S. A. 105:19956-19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pogany, J., K. A. White, and P. D. Nagy. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 79:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. U. S. A. 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz, M., J. B. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz, M., J. B. Chen, W. M. Lee, M. Janda, and P. Ahlquist. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. U. S. A. 101:11263-11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serva, S., and P. D. Nagy. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 80:2162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 42.Takeda, A., M. Tsukuda, H. Mizumoto, K. Okamoto, M. Kaido, K. Mise, and T. Okuno. 2005. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 24:3147-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatsuta, M., H. Mizumoto, M. Kaido, K. Mise, and T. Okuno. 2005. The Red clover necrotic mosaic virus RNA2 trans-activator is also a cis-acting RNA2 replication element. J. Virol. 79:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner, K. A., T. L. Sit, A. S. Callaway, N. S. Allen, and S. A. Lommel. 2004. Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320:276-290. [DOI] [PubMed] [Google Scholar]
- 45.Turner, R. L., and K. W. Buck. 1999. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of red clover necrotic mosaic virus. Virology 253:115-124. [DOI] [PubMed] [Google Scholar]
- 46.Vlot, A. C., L. Neeleman, H. J. M. Linthorst, and J. F. Bol. 2001. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 75:6440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, R. Y. L., and P. D. Nagy. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3:178-187. [DOI] [PubMed] [Google Scholar]
- 48.Wang, R. Y. L., J. Stork, and P. D. Nagy. 2009. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 83:3276-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waris, G., S. Sarker, and A. Siddiqui. 2004. Two-step affinity purification of the hepatitis C virus ribonucleoprotein complex. RNA 10:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe, T., A. Honda, A. Iwata, S. Ueda, T. Hibi, and A. Ishihama. 1999. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J. Virol. 73:2633-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong, Z., K. H. Kim, D. Giesmancookmeyer, and S. A. Lommel. 1993. The roles of the red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology 192:27-32. [DOI] [PubMed] [Google Scholar]
- 52.Xiong, Z., K. H. Kim, T. L. Kendall, and S. A. Lommel. 1993. Synthesis of of the putative red clover necrotic mosaic virus RNA-dependent RNA polymerase by ribosomal frameshifting in vitro. Virology 193:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong, Z., and S. A. Lommel. 1989. The complete nucleotide sequence and genome organization of red clover necrotic mosaic virus RNA-1. Virology 171:543-554. [DOI] [PubMed] [Google Scholar]
- 54.Xiong, Z. G., and S. A. Lommel. 1991. Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology 182:388-392. [DOI] [PubMed] [Google Scholar]
- 55.Zavriev, S. K., C. M. Hickey, and S. A. Lommel. 1996. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology 216:407-410. [DOI] [PubMed] [Google Scholar]
- 56.Zhu, J., K. Gopinath, A. Murali, G. H. Yi, S. D. Hayward, H. Zhu, and C. C. Kao. 2007. RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. U. S. A. 104:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]