Abstract
While the simian immunodeficiency virus (SIV)-infected rhesus monkey is an important animal model for human immunodeficiency virus type 1 (HIV-1) infection of humans, much remains to be learned about the evolution of the humoral immune response in this model. In HIV-1 infection, autologous neutralizing antibodies emerge 2 to 3 months after infection. However, the ontogeny of the SIV-specific neutralizing antibody response in mucosally infected animals has not been defined. We characterized the kinetics of the autologous neutralizing antibody response to the transmitted/founder SIVmac251 using a pseudovirion-based TZM-bl cell assay and monitored env sequence evolution using single-genome amplification in four rhesus animals that were infected via intrarectal inoculations. We show that the SIVmac251 founder viruses induced neutralizing antibodies at 5 to 8 months after infection. Despite their slow emergence and low titers, these neutralizing antibodies selected for escape mutants that harbored substitutions and deletions in variable region 1 (V1), V2, and V4 of Env. The neutralizing antibody response was initially focused on V4 at 5 to 8 months after infection and then targeted V1/V2 and V4 by 16 months. These findings reveal a striking delay in the development of neutralizing antibodies in SIVmac-infected animals, thus raising questions concerning the suitability of SIVmac251 as a challenge strain to screen AIDS vaccines that elicit neutralizing antibodies as a means to prevent virus acquisition. They also illustrate the capacity of the SIVmac quasispecies to modify antigenic determinants in response to very modest titers of neutralizing antibodies.
While neutralizing antibodies (Nabs) mediate protection in humans against a diversity of viral pathogens (38, 53, 72), it is unclear how they impact human immunodeficiency virus type 1 (HIV-1) infection. One reason that the contribution of neutralizing antibodies to the control of HIV-1 remains uncertain is that HIV-specific neutralizing antibodies develop relatively late in natural infection. High titers of HIV-specific autologous neutralizing antibodies usually emerge as late as 2 to 3 months after infection and continue to evolve throughout the first years of infection (18, 25, 57, 66, 74). Such neutralizing antibodies have been shown to influence HIV-1 evolution within a host and to be responsible for viral escape mutations (47, 48, 58, 59). A better understanding of why there is a prolonged time associated with the maturation of the neutralizing antibody response in HIV-1 infection and whether conserved viral epitopes exist that could mediate antibody protection is important for the development of effective HIV-1 vaccine strategies.
The simian immunodeficiency virus (SIV)/rhesus macaque model of AIDS provides an important system to study AIDS immunopathogenesis and to evaluate HIV-1 vaccine strategies. SIVmac251, an uncloned, pathogenic, CCR5-tropic virus isolate comprised of a swarm of quasispecies that are closely related (33), and SIVmac239, an infectious molecular clone derived from SIVmac251, are the two most commonly used rhesus monkey SIV challenge viruses utilized in AIDS vaccine research in the nonhuman primate (NHP) model. SIVmac239 has been shown to be relatively resistant to antibody-mediated neutralization by both autologous antibodies and a wide range of monoclonal antibodies (29, 30). The env sequence evolution in SIVmac239-infected rhesus monkeys and SIVMne-CL8-infected pigtailed macaques has been well described (8, 50, 51). Some of these changes in Env have been shown to result in viral escape from neutralizing antibodies (7, 10, 34, 60). In particular, a recent study by Sato et al. characterized SIVmac239 env sequence changes that were associated with viral escape in a rhesus monkey with an unusually high titer of neutralizing antibodies after intravenous infection (67). However, the antibody-mediated neutralization of SIVmac251 has not been tested rigorously using standardized assays that are currently being used to measure neutralization of HIV-1, thereby precluding a direct comparison of the neutralization sensitivities of HIV-1 and SIV. Furthermore, it is also unclear whether more typical titers of neutralizing antibodies against SIV239/251 exert selection pressure on the viral population in animals that acquire infection mucosally.
The aims of this study were to elucidate the kinetics of the neutralizing antibody response against the transmitted viruses and the sequence evolution of env in association with humoral immunity in mucosally infected rhesus macaques. We hypothesized that a low titer of SIVmac Env-specific neutralizing antibodies exerts potent selection pressure on the viral quasispecies. To test this hypothesis, we utilized a pseudovirion-based TZM-bl reporter gene neutralization assay and single genome amplification (SGA) in order to characterize the humoral immune pressures driving viral sequence evolution in four rhesus monkeys that were infected with SIVmac251 via intrarectal inoculations.
MATERIALS AND METHODS
Study animals.
Four adult rhesus monkeys (Macaca mulatta) were used in this study. All animals were housed at Bioqual (Rockville, MD) and maintained in accordance with the Association for Assessment and Accreditation of Laboratory Animals with the approval of the Animal Care and Use Committees of the National Institutes of Health and Harvard Medical School.
SIV challenge stocks.
The SIVmac251 isolate used in this study is a cell-free uncloned pathogenic stock that was expanded on human peripheral blood mononuclear cells (PBMCs). The viral quasispecies in the inoculum have previously been characterized and shown to be 99% similar in Env sequence identity (33). To initiate infection, animals were intrarectally exposed to 6 weekly doses of either 6.3 × 107 or 6.3 × 106 RNA copies of SIVmac251 (39). Plasma viral-RNA (vRNA) levels after infection were measured by an ultrasensitive branched-DNA amplification assay with a detection limit of 125 copies per ml (Bayer Diagnostics, Berkeley, CA).
Viral-RNA extraction and cDNA synthesis.
Viral RNAs were extracted using the QIAamp Viral RNA Mini kit (Qiagen). RNA was eluted and immediately subjected to cDNA synthesis. Reverse transcription (RT) of RNA to single-stranded cDNA was performed using SuperScript III reverse transcription according to the manufacturer's recommendations (Invitrogen). In brief, each cDNA reaction mixture included 1× RT buffer, 0.5 mM each deoxynucleoside triphosphate, 5 mM dithiothreitol, 2 U/ml RNaseOut (RNase inhibitor), 10 U/ml of SuperScript III reverse transcriptase, and 0.25 mM antisense primer R1 (5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′). The mixture was incubated at 50°C for 60 min, followed by an increase in temperature to 55°C for an additional 60 min. The reaction mixture was then heat inactivated at 70°C for 15 min and treated with 2 U of RNase H at 37°C for 20 min. The newly synthesized cDNA was used immediately or frozen at −80°C.
SGA.
cDNA was serially diluted and distributed among wells of replicate 96-well plates to identify a dilution at which PCR-positive wells constituted <30% of the total number of reactions, as previously described (64). At this dilution, most wells contained amplicons derived from a single cDNA molecule. This was confirmed in every positive well by direct sequencing of the amplicon and inspection of the sequence for mixed bases, which would be indicative of priming from more than one original template or the introduction of PCR error in early cycles. Any sequence with mixed bases was excluded from further analysis. PCR amplification was performed in the presence of 1× High Fidelity polymerase in a 20-μl reaction mixture (Invitrogen). The first-round PCR primers included sense primer F6476 (5′-GTGTTGCATACCATTGCCAGTTTTG-3′) and antisense primer R9339 (5′-GTCATCATCTTCCTCATCTACATC-3′). PCR was performed in MicroAmp 96-well reaction plates (Applied Biosystems) with the following PCR parameters: 1 cycle of 94°C for 2 min and 35 cycles of a denaturing step of 94°C for 15 s, an annealing step of 54°C for 30 s, and an extension step of 68°C for 4 min, followed by a final extension of 68°C for 10 min. Next, 2 μl of the first-round PCR product was added to a second-round PCR mixture that included the sense primer F6484 (5′-ACCATTGCCAGTTTTGTTTTCTTA-3′) and antisense primer R9283 (5′-TCTCTTCAGCTGGGTTTCTCC-3′). The second-round PCR was performed under the same conditions used for the first-round PCR but with a total of 45 cycles. Amplicons (2.8 kb) containing tat, rev, and env were inspected on precast 1% agarose 96-well E gels (Invitrogen).
Env sequencing and analysis.
Both DNA strands on env amplicons were directly sequenced using partially overlapping fragments. Individual sequence fragments for each amplicon were assembled and edited using the Sequencher program version 4.7 (Gene Codes, Ann Arbor, MI). Inspection of individual chromatograms allowed the confirmation of amplicons derived from single versus multiple templates. The absence of mixed bases at each nucleotide position throughout env was taken as evidence of SGA from a single vRNA/cDNA template. This quality control measure allowed us to exclude from the analysis amplicons that resulted from PCR-generated in vitro recombination events or Taq polymerase errors and to obtain individual SIV env sequences that proportionately represented those circulating in vivo. All alignments were initially made with ClustalW and further codon aligned using HIVAlign (http://www.hiv.lanl.gov/content/sequence/HMM/HmmAlign.html). Each nucleotide base and amino acid residue of SIVmac251 was numbered according to the corresponding position in Env of SIVmac239 (accession no. M33262). The SIVmac251 isolate that was used to initiate infection in this cohort of animals contained three amino acid insertions in variable region 1 (V1) compared to a SIVmac239 clone at residue 134. The amino acid insertions were given the number of the residue prior to the insertion with a letter to denote each inserted residue.
A maximum-likelihood tree of the gap-stripped nucleotide sequences from all animals was constructed using PhyML (version 2.4.4) and the general time reversible plus gamma (GTR+gamma) substitution model. The locations of nonsynonymous substitutions were visualized with Highlighter plots (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter.html) without gap stripping. For clarity, identical sequences were not included. For each sequence, V1 (amino acids 114 to 153), V2 (152 to 205), V3 (312 to 343), V4 (403 to 431), and V5 (469 to 481) corresponding to the SIVmac239 envelope amino acid residues were defined based on the previously published crystal and tomographic structures of HIV-1 and SIVmac239 (11, 42). The numbers of amino acids and predicted N-linked glycosylation sites in each segment were determined through manual counting and by using the tool available at http:///www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html. Each clone was scored for the presence of the canonical PNG motif (Asn-X-Ser/Thr, where X represents any residue other than proline).
Methods for detecting positive selection.
Complementary analytic methods were used to identify nucleotide positions enriched for positive selection throughout infection in all animals. Three tests were used to identify single sites and localized regions of selection, as previously described (24, 70). The enriched-mutation statistic was computed by counting the mutations in a sliding window of 45 nucleotides (nt). The clustered-mutation statistic used a 27-nt sliding window to identify putative cytotoxic and neutralizing antibody epitopes composed of 9 amino acids. Both statistics compare observed mutations per window with null distributions by randomly permuting the locations of mutations in each sequence. For the enriched-mutation statistic, the P value is the number of randomized windows that contain at least the observed number of mutations divided by the number of permuted replicates. For the clustered-mutation statistic, P values result from Fisher's exact test. The false-discovery rate (q) was computed to correct for multiple testing, using a q value of <0.1 to detect windows where significant differences between observed and shuffled mutations occurred. The code, written in R and Perl, to perform these window-based statistics for positive selection can be found at ftp://ftp-t10.lanl.gov/pub/WindowSelectionStatistics.
Third, protein-coding regions for all Env proteins were tested for specific codons under positive selection in each animal using fixed-effects likelihood (FEL) (35, 55). A neighbor-joining tree and a nucleotide substitution model were computed for each aligned gene before FEL analysis. Sites with greater nonsynonymous than synonymous substitution rates (dN > dS) and P values of <0.2 were taken as significant for positive selection. FEL results were not corrected for multiple testing. We verified the robustness of the results in the presence of alternative substitution models, discarding sites whose significance could not be repeated using another substitution model.
Amino acid substitutions in viral variants that evolved in late infection at 16 to 22 months postinfection (p.i.) across all animals were also identified using Fisher's exact test with phylogenetic correction (3, 36). Briefly, the state of the immediate ancestor for each sequence was inferred, and a 2-by-2 contingency table of substitution counts was built for each amino acid position. Fisher's exact test was used on the contingency table to find significant associations between recurring amino acid substitutions and time in infection under the null hypothesis that the prevalence of substitutions was independent of time. To adjust for multiple tests, we computed false-discovery rates and report results for q values of <0.5% and required that substitutions be observed independently in at least 3 of 4 animals.
Construction of pseudovirus.
The representative env variants were cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA), transformed into TOP10F′ chemically competent cells, and selected with ampicillin. Inserts were confirmed by restriction digestion and direct sequencing. 293T cells were transfected with Lipofectamine (Qiagen, Valencia, CA) according to the manufacturer's instructions. Cells were plated to 80% confluence in a T75 flask, and 14 μg total DNA with a 6:1 SIVmac239Δenv backbone-to-env ratio was prepared in a mixture of Dulbecco's modified Eagle's medium (Invitrogen), as previously described (2). Viruses were harvested 24 h later, made cell free by filtration through 0.45-μm filters, and stored at −80°C until they were used. The pseudovirus was titrated on TZM-bl cells, and 200 50% tissue culture infective doses were added to each neutralization assay.
Neutralizing antibody assay.
Plasma from each animal was tested at week −1 (preinfection) and 3, 5, 8, 16, and 22 months p.i. Neutralizing antibodies were measured in a luciferase reporter gene assay that utilized TZM-bl cells as described previously (45). The 50% inhibitory dose (ID50) titer was defined as the plasma dilution that resulted in a 50% reduction in relative luminescence units (RLU) compared to virus control wells after subtraction of cell control RLU. Any sample that did not have a detectable ID50 titer at a dilution of 1:10 was reported as <10.
RESULTS
Plasma viral RNA levels following SIVmac251 intrarectal infection.
To characterize the autologous neutralizing antibody response and the env sequence evolution in animals that acquired infection mucosally, we infected a cohort of rhesus monkeys with SIVmac251 via repeated, low-dose intrarectal inoculations (39). These monkeys were members of a cohort of animals in which the transmitted/founder (T/F) viruses had been characterized (33). Each animal became infected with one T/F virus after 1 to 6 rectal exposures. The viral replication kinetics after mucosal infection was typical of SIVmac251 replication in naïve rhesus monkeys infected by the intravenous route. All animals developed uniform peak plasma viral RNA levels of 7 log units at 14 days after infection, followed by sustained viremia of 3 to 5 log units of measurable plasma viral RNA at 70 days p.i., as previously shown (33, 76). These animals were monitored for 2 years after infection and typed for major histocompatibility complex (MHC) class I alleles Mamu-A*01, Mamu-A*02, Mamu-B*08, and Mamu-B*17, and their haplotypes are indicated in Table 1.
TABLE 1.
Selection in chronic infection for SIV variants with deletions in variable loops 1 and 4
| Monkey | MHC class I allele(s)a | 16 months p.i. [no. (%)] |
22 months p.i. [no. (%)] |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔV1b | ΔV4c | ΔV1 and V4d | Totale | ΔV1 | ΔV4 | ΔV1 and V4 | Total | ||
| PBE | Mamu-A2+ | 16/57 (28) | 0/57 (0) | 38/57 (67) | 54/57 (95) | 9/18 (50) | 6/18 (33) | 2/18 (11) | 17/18 (94) |
| CPIW | Mamu-A1+/B8+ | 1/47 (2) | 34/47 (72) | 8/47 (17) | 43/47 (91) | 14/16 (88) | 0/16 (0) | 2/16 (12) | 16/16 (100) |
| CR2A | Mamu-A1+/A2+ | 3/30 (10) | 6/30 (20) | 21/30 (70) | 30/30 (100) | 0/12 (0) | 3/12 (25) | 9/12 (75) | 12/12 (100) |
| CR53 | Mamu-A1+/B17+ | 9/28 (32) | 7/28 (25) | 8/28 (29) | 24/28 (86) | 7/14 (53) | 0/14 (0) | 7/14 (47) | 14/14 (100) |
Defined alleles expressed by each monkey.
Number of viruses containing deletions in only Env V1 out of the total number of viruses sequenced.
Viral quasispecies containing deletions in only Env V4 out of the total number of quasispecies.
Number of viral clones containing deletions in both Env V1 and V4 out of the total number of clones.
Total number of viral clones containing deletions in either V1 or V4 out of the total number of quasispecies.
Autologous neutralizing antibodies to transmitted/founder SIVmac251 develop 5 to 8 months after infection.
Traditionally, studies demonstrating the relative resistance of SIV to antibody-mediated neutralization were done using primary viruses in cell-killing assays (23, 27, 46). More recently, a laboratory-adapted strain of SIVmac251 (TCLA-SIVmac251) has been utilized to measure neutralizing antibody titers to SIVmac251 (45). However, this assay likely does not accurately reflect the true neutralization capacities of antibodies, due to the extreme sensitivity of TCLA-SIVmac251 to neutralization. A more neutralization-resistant Env clone obtained from in vitro-passaged stock of SIVmac251 (SIVmac251CS.41) has also been utilized in pseudovirus-based neutralization assays (76). However, the sequence of this Env differs significantly from that of the Envs in the inoculum stock that was used in the present study. Therefore, to assess the kinetics of the autologous neutralizing antibodies to the T/F viruses that initiated infection in each animal, we generated pseudoviruses that expressed the specific T/F Env in each animal.
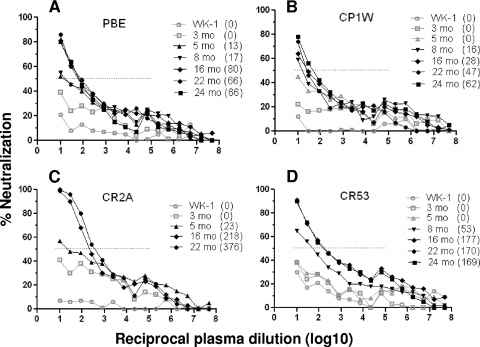
The sensitivity of the transmitted Env to neutralization was evaluated using autologous plasma samples at various intervals p.i. using TZM.bl neutralization assays. The ability of matched plasma antibody to neutralize the founder virus in each monkey is shown as percent neutralization in Fig. 1. The presence of neutralizing antibodies against the founder virus was detected as early as 3 months p.i. (open gray squares), although this neutralization did not reach the 50% inhibitory level. By 5 months p.i., plasma from monkeys PBE and CR2A demonstrated neutralization against the transmitted/founder SIVmac251 Env with ID50 titers of 13 and 23, respectively (as indicated in parentheses). Animals CP1W and CR53 developed autologous neutralizing antibodies by 8 months p.i., with ID50 titers of 16 and 53, respectively. The titer of antibodies that neutralized the founder virus increased throughout infection in all 4 animals. Monkeys CR53 and CR2A developed the highest titers of autologous neutralizing antibodies with ID50 titers of 177 and 376 at 16 and 22 months p.i., respectively. The use of the same pseudovirus-based TZM-bl neutralization as those utilized to measure the autologous neutralizing antibody titers against HIV-1 allowed us to compare the relative strengths of the neutralizing antibody responses generated to HIV-1 and SIV. Therefore, based on our data, the neutralizing antibody response to SIVmac251 was lower in titer and more delayed than that reported in HIV-1-infected humans.
FIG. 1.
Autologous neutralizing antibodies after SIVmac251 infection. Plasmas from 4 SIVmac251-infected animals, PBE (A), CP1W (B), CR2A (C), and CR53 (D), were tested at multiple time points p.i. for neutralizing antibodies against the founder env-pseudotyped virus isolated from matched animals. Percent neutralization is plotted along the y axes, and the reciprocal log10 plasma dilution (from 101 to 108) is plotted along the x axes. The dotted lines indicate 50% neutralization. The gray symbols and lines indicate time points when the reduction in neutralization was below the 50% neutralization level (ID50 titers < 10), and the black symbols and lines indicate time points when the reduction in neutralization scored above 50%. ID50 titers are indicated in parentheses.
Sequence diversity and divergence.
To investigate whether neutralizing antibodies exerted pressure on the evolution of viral quasispecies, we analyzed the env sequence evolution in all 4 animals from the time of peak viremia to 22 months p.i. Thirty to 40 env fragments were derived from plasma samples by single genome amplification at each time point p.i. Each amplicon was sequenced directly without cloning, and the sequence chromatograms were unambiguous at every position. To examine the genetic relationships among the virus sequences, a maximum-likelihood tree was constructed from env sequences present in the inoculum; at peak viremia; at 5 or 8 months p.i., when neutralizing antibodies were first detected; and at 16 and 22 months p.i., when the observed neutralizing activity reached its peak (Fig. 2). The maximum env diversity among all animals was 3.9%, while the viral sequence diversity within one animal ranged from 1.7% in animal CR53 to 2.3% in animal PBE. None of the env sequences obtained from the inoculum matched the divergent quasispecies at later times during infection, suggesting that env evolution in chronic infection did not represent the common outgrowth of minor preexisting sequences in the inoculum.
FIG. 2.
Phylogenetic analysis of env quasispecies from the 4 monkeys infected with SIVmac251. The maximum-likelihood tree is rooted on the consensus of the inoculum (blue circles). Sequences from each monkey clustered together and are represented by similarly colored symbols, as indicated. The transmitted viruses at peak viremia in each monkey are represented by open circles, viruses from 5 to 8 months (Mo) p.i. by open triangles, and quasispecies from 16 months p.i. by open squares and 22 months p.i. by closed squares. The scale bar indicates a genetic distance of 0.001 (1 nucleotide difference per 1,000 sites) in the phylogram.
Env diversification clustered in V1/2 and V4.
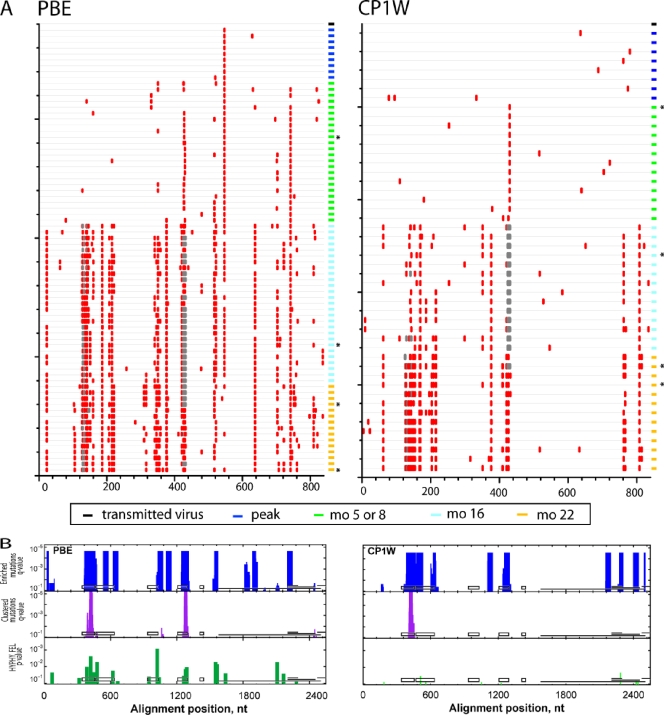
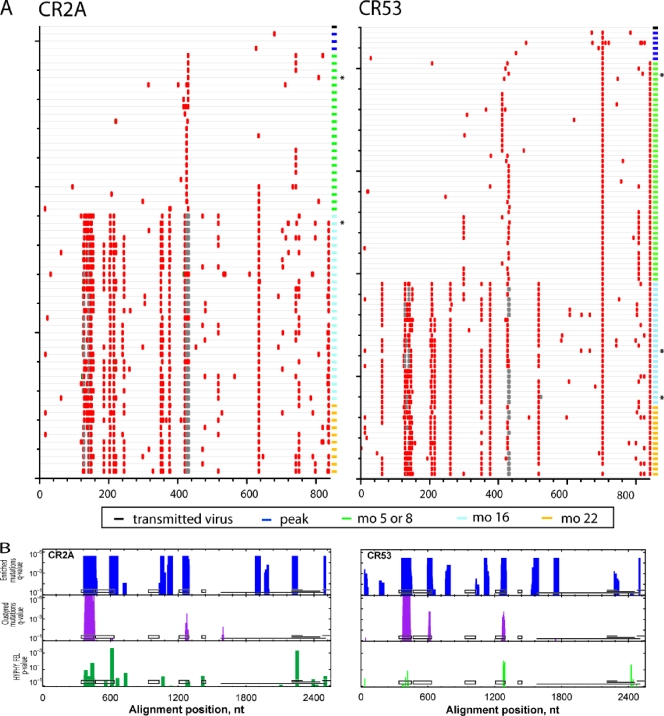
To elucidate whether viral variants that evolved in these infected monkeys shared common mutations that might be determinants for immune recognition, we used Highlighter analysis to locate nonsynonymous env mutations in animals PBE and CP1W (Fig. 3A) and CR2A and CR53 (Fig. 4A) from the time of peak viremia to 22 months p.i. For this analysis, identical sequences were not included in the Highlighter plot. Compared to viruses in the challenge stock, the transmitted viruses at ramp-up and peak viremia contained stochastic mutations without evidence for positive selection (33). However, by 5 or 8 months p.i., approximately 50% of the viral quasispecies in each animal harbored an R424Q/W substitution in V4. Although there were also other amino acid changes nearby, the R424Q/W mutation was the only substitution observed in all animals at the time points when neutralizing antibodies were first detected. Furthermore, from 5 or 8 months p.i. to 22 months p.i., there was a continued trend toward additional nonsynonymous mutations in V1/2 and V4 in all 4 animals. These data indicate sustained evolution of the variable regions in response to persistent selection pressure throughout the first 2 years of SIVmac251 infection.
FIG. 3.
Accumulation of env nucleotide sequence changes in replicating viruses after intrarectal SIVmac251 infection. (A) Highlighter alignment of env genes that were transmitted from the inoculum (black rectangle at top right of each alignment), at peak viremia (dark blue rectangles), or 5 or 8 (green rectangles), 16 (light blue rectangles), and 22 (yellow rectangles) months p.i. in animals PBE and CP1W. Each horizontal line on the vertical axis depicts individual Env variants sequenced at various time points postinfection. Identical sequences were removed for clarity. The red tic marks indicate differences that are nonsynonymous in the env coding frame. Deletions are indicated by gray tics. The horizontal axis indicates the amino acid position in the Env alignment. (B) Identification of putative sites of positive selection using three independent tests. The horizontal axis indicates the nucleotide positions in env, aligned to SIVmac239 (nt 6604 to 9108), and the vertical axis indicates the statistical significance for each of the three tests. The tests for enriched mutations and clustered mutations (turquoise and purple bars, respectively) used sliding windows of 45 and 27 nt, respectively. The FEL test from HYPHY is a tree-based statistic that identifies single codons under positive selection (green bars). For reference, the black boxes depict the locations of V1 to V5 in gp120, and the horizontal lines depict the locations of gp41, tat exon 2, rev exon 2, and nef (in reading frames 1, 3, 2, and 3, respectively). The asterisks denote Env variants from each animal that were cloned for further neutralization testing.
FIG. 4.
Accumulation of env nucleotide sequence changes in replicating viruses after infection. (A) Highlighter alignment of env genes from animals CR2A and CR53. The time points p.i. when env genes were sequenced are indicated. The red tic marks represent nonsynonymous mutations, while deletions are indicated by gray tics. (B) Identification of putative sites of positive selection using nonrandom-mutation and clustered-mutation statistics and the FEL test. The asterisks denote Env variants from each animal that were cloned for further neutralization testing.
We were interested in identifying common antibody recognition motifs shared by all animals that could have mediated viral escape from neutralizing antibodies. Therefore, four distinct statistical tests were used to identify sites of positive selection in viral variants from peak viremia to 22 months p.i. (Fig. 3B and 4B). In the enriched-mutation test (turquoise peaks), a sliding window of 45 bases was used to identify concentrated nucleotide mutations. The clustered-mutation test (purple peaks) identified clustered nucleotide mutations in a 27-base sliding window, thereby detecting positive selection in potential T-cell or B-cell epitopes. These 2 tests showed that clustered amino acid changes in V1/2, constant region 3 (C3), and V4 were positively selected in all 4 animals.
Additionally, we used a tree-based statistic, HYPHY FEL, that detects increases in the ratio of nonsynonymous to synonymous substitution rates at each codon with a likelihood ratio test to identify viral codons under positive selection. Using this test, 14 discrete codons in env (green peaks) showed evidence of strong mutational selection in at least 3 of the 4 animals by 16 to 22 months p.i. Further analysis using Fisher's exact test confirmed residues 127, 128, 134, 135, 136, 138, 345, 415, and 416 as sites of recurring mutations in Env in all animals. Of note, residues 420, 421, and 424 were identified by FEL statistics as positively selected. However, these residues were not evaluated using Fisher's test, since the majority of the studied clones had deletions at these residues. Taken together, the common mutations that evolved in all 4 animals by 22 month p.i. appeared to cluster in V1/2, C3, and V4. Very few changes accumulated in V3 or V5, suggesting that substitutions in these regions did not confer a selective advantage for the virus during in vivo replication.
Env mutations resulted in shortened V1/2 and V4 loops and shifts in predicted glycans.
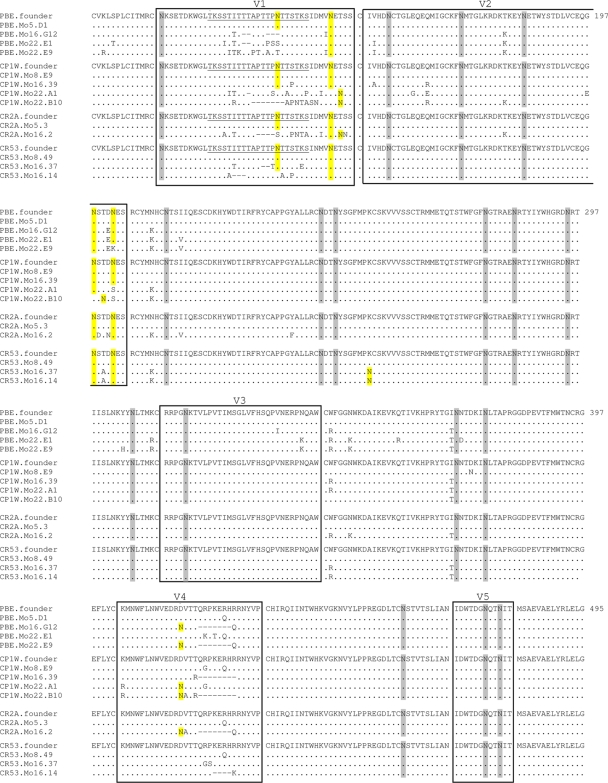
We observed a consistent shortening of V1 and V4 of SIVmac251 env over the course of infection in these monkeys (gray bars in Fig. 3A and 4A). At 16 and 22 months p.i., the majority of circulating Env variants in each animal contained 3 to 5 amino acid deletions in V1, V4, or both compared to Env from the time of peak viremia (Table 1). There was considerable variability in the deletions in V1 and relative homogeneity in the deletions in V4 of these viruses (Fig. 3 to 5). To compare the lengths of env V1 and V4 of the Env variants isolated at 16 and 22 months p.i. to those isolated at peak infection, we counted the amino acids in these regions and summarized the distributions as minimum, median, and maximum lengths of V1 and V4 per animal (Table 2). The minimum and median numbers of amino acids were lower in V1 and V4 in viruses circulating at 16 to 22 months p.i. than in viruses at the time of peak viremia. Moreover, consistent with previous reports characterizing the env sequence evolution in SIVmac species, we observed that P421 in V4 of Env was deleted in the majority of the viral clones isolated during chronic infection (67). The lengths of V2, V3, and V5 remained constant throughout infection in all 4 monkeys.
FIG. 5.
Comparison of the amino acid sequences of the variable regions of SIVmac251 gp120. The sequences of representative Env clones obtained throughout infection are aligned to the Env that was transmitted to each animal from the SIVmac251 inoculum, which is shown at the top. The first amino acid residue in the alignment represents residue number 101 in the SIVmac239 Env. The name of each Env sequence identifies the monkey and the time point when the variant was isolated. The dots represent amino acid identity and the dashes represent deletions. Variable regions V1 through V5 are boxed. The amino acid residue number is on the right of each row. Yellow highlights amino acids that represent a consensus sequence for PNGs that have been shifted, deleted, or added. PNGs in variable regions that remained unchanged throughout infection are highlighted in gray. A potential O-linked glycan attachment site in the V1 region is underlined. These Env variants were cloned for further neutralization testing.
TABLE 2.
V1 and V4 lengths of SIVmac251 at peak viremia and during chronic infection
| Variable region | Animal | Peak viremia |
Chronic infectiona |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of sequencesb | Medianc | Minimumd | Maximume | No. of sequences | Median | Minimum | Maximum | ||
| V1 | PBE | 24 | 40 | 40 | 40 | 42 | 39 | 34 | 40 |
| CP1W | 42 | 40 | 40 | 40 | 27 | 39 | 33 | 40 | |
| CR2A | 26 | 40 | 40 | 40 | 36 | 36 | 35 | 41 | |
| CR53 | 30 | 40 | 40 | 40 | 38 | 39 | 33 | 43 | |
| V4 | PBE | 24 | 29 | 29 | 29 | 42 | 22 | 22 | 29 |
| CP1W | 42 | 29 | 29 | 29 | 27 | 21 | 21 | 29 | |
| CR2A | 26 | 29 | 29 | 29 | 36 | 22 | 22 | 29 | |
| CR53 | 30 | 29 | 29 | 29 | 38 | 25 | 25 | 29 | |
env clones from 16 and 22 months postinfection.
Number of env sequences per animal amplified by single-genome amplification.
Median number of amino acids in V1 and V4 per animal.
Mininum number of amino acids in V1 and V4 per animal.
Maximum number of amino acids in V1 and V4 per animal.
To assess the statistical significance of the lengths of V1 and V4 over the course of infection, we used a 2-sided binomial test on the direction of changes in the lengths of the variable regions. This test does not assume independence among sequences from the same animal. We counted the distinct changes (insertions or deletions) and compared the observed proportion of deletion events to the null expectation of 50%. In V1, we observed 3 distinct insertion events and 22 deletion events. Therefore, the observed V1 changes are biased toward deletions with statistical significance (P = 0.00016; 95% confidence interval [CI] = 69 to 97%). Similarly, we observed 3 different deletion events and no insertions in V4, with a trend toward statistical significance (P = 0.25).
In addition to playing an important role in the structure and function of Env, carbohydrates have been shown to modulate the antigenicity and sensitivity of HIV-1/SIV to neutralizing antibodies (6, 10, 15, 26, 67, 74). Therefore, we determined whether the amino acid substitutions/deletions observed in Env variants isolated during chronic infection resulted in changes in potential N- and O-linked glycosylation sites. We compared the number of predicted potential N-linked glycosylation sites (PNGs) in env V1 to V5 of viruses isolated during peak viremia and chronic infection at 16 and 22 months p.i. (Fig. 5 and Table 3). Seventeen of the 21 glycans in Env were conserved throughout the course of infection in all animals, consistent with the hypothesis that lentiviral envelopes maintain a core set of glycans that are important in producing a functional protein (4). Fourteen of the 17 conserved glycans were in the constant regions of Env. As Env accrued mutations during the course of infection, substitutions eliminating N-glycan attachment sites were associated with the coincidental creation of new attachment sites. Interestingly, with the exception of the PNG introduced by the K254N substitution in C2 of animal CR53, nearly all of the changes in PNGs occurred within limited windows in V1/2 and V4 of SIV Env. Several PNG sites were either shifted or deleted in V1 and V2 (residues 135, 138, 146, 148, 198, 200, and 202), while PNGs were added in V4 due to a change of aspartic acid to asparagine at amino acid position 415. Env sequences at peak viremia contained a median of 8 PNGs in V1/2 and no PNGs in V4 (Table 3). In contrast, Env sequences isolated from chronic infection had 1 or 2 fewer PNGs in V1/2 and 1 more PNG in V4 in Env. There were no changes in PNGs in V3 and V5 between peak viremia and chronic infection. Additionally, changes in residues 127, 128, 129, 130, 131, 132, and 134 in many Env variants sampled during chronic infection may have resulted in potential losses and additions of O-linked glycans in the serine/threnonine-rich stretch within V1 (124TKSSTITTTAPTTPNTTSTKS141).
TABLE 3.
Potential N-linked glycosylation sites in SIVmac251 at peak viremia and during chronic infection
| Variable region | Animal | Peak viremia |
Chronic infectiona |
||||
|---|---|---|---|---|---|---|---|
| Medianc | Minimumd | Maximume | Median | Minimum | Maximum | ||
| V1b | PBE | 3 | 3 | 3 | 2 | 2 | 3 |
| CP1W | 3 | 3 | 3 | 2 | 1 | 3 | |
| CR2A | 3 | 3 | 3 | 3 | 2 | 3 | |
| CR53 | 3 | 3 | 3 | 3 | 2 | 3 | |
| V2 | PBE | 5 | 5 | 5 | 5 | 4 | 5 |
| CP1W | 5 | 5 | 5 | 5 | 4 | 5 | |
| CR2A | 5 | 5 | 5 | 5 | 4 | 5 | |
| CR53 | 5 | 5 | 5 | 4 | 4 | 5 | |
| V3 | PBE | 1 | 1 | 1 | 1 | 1 | 1 |
| CP1W | 1 | 1 | 1 | 1 | 1 | 1 | |
| CR2A | 1 | 1 | 1 | 1 | 1 | 1 | |
| CR53 | 1 | 1 | 1 | 1 | 1 | 1 | |
| V4 | PBE | 0 | 0 | 0 | 1 | 0 | 1 |
| CP1W | 0 | 0 | 0 | 0 | 0 | 1 | |
| CR2A | 0 | 0 | 0 | 1 | 1 | 1 | |
| CR53 | 0 | 0 | 0 | 0 | 0 | 1 | |
| V5 | PBE | 2 | 2 | 2 | 2 | 2 | 2 |
| CP1W | 2 | 2 | 2 | 2 | 2 | 2 | |
| CR2A | 2 | 2 | 2 | 2 | 2 | 2 | |
| CR53 | 2 | 2 | 2 | 2 | 2 | 2 | |
env sequences from 16 and 22 months postinfection.
Potential N-linked glycosylation motifs (NXS/T) in V1 to V5 were deduced and counted.
Median number of PNG motifs per animal.
Mininum number of PNG motifs per animal.
Maximum number of PNG motifs per animal.
We also used a binomial test on the changes in the number of PNG sites in each animal over the course of infection. In V1, all 4 animals contained env genes with fewer PNGs in chronic infection than in peak viremia (P = 0.125). Similarly, each animal exhibited at least one env with one additional PNG site in V4 during chronic infection (P = 0.125). Although the observations regarding the evolution of the lengths of the variable loops and PNGs over the course of infection were not all statistically significant due to the small number of animals, these data suggest that mutations in V1/2 and V4 likely conferred a fitness advantage on SIV variants that resulted in their accumulation and eventual predominance as major variants in chronic infection.
Viral variants evade recognition by autologous neutralizing antibodies.
The emergence of Env variants with nonsynonymous nucleotide substitutions and deletions in V1/2 and V4 suggested that substantial selection pressure was being exerted on the replicating virus population in vivo. Since these animals had different MHC class I haplotypes and therefore likely restricted divergent cytotoxic-T-lymphocyte (CTL) epitopes, we hypothesized that the common Env mutations in V1/2 and V4 observed in all 4 animals likely emerged as a result of viral escape from neutralizing antibodies. To determine whether the diversification of env was a consequence of selective pressure from the host humoral immune response, we tested the abilities of autologous plasma samples to neutralize representative Env variants isolated during the intermediate stage (5 or 8 months p.i.) and late stage (16 and 22 months p.i.) of infection from each monkey. Representative Env variants were selected because they contained the most prevalent mutations in each animal at the chosen time points. The Env variants from each animal that were tested in neutralizing assays are denoted by asterisks in Fig. 3 and 4.
At 5 months p.i., when the neutralizing antibody titer to the founder Env in animal PBE was 13, 50% of Env variants circulating in the animal harbored an R424Q substitution in V4 (Fig. 3A). This mutation was also observed in the other 3 animals at the time when neutralizing antibodies against their respective founder viruses were detected. An Env variant isolated from PBE (PBEΔV4.Mo5.D1) containing this mutation was not neutralized by contemporaneous plasma or by later plasma at 8 months p.i., when the ID50 titer to the founder Env increased to 17 (Fig. 6A). Although the difference in the ID50 titers to the wild-type and variant R424Q Env proteins is small due to the low titers of neutralizing antibodies, their kinetics and the env sequence evolution suggest that an R424Q mutation in Env might have mediated early viral escape from the humoral immune response. Interestingly, this Env escape variant became sensitive to neutralization by plasma obtained later at 16 months p.i., indicating that neutralizing antibodies evolved to contain the R424Q Env variant that emerged earlier at 5 months p.i. Neutralizing antibodies that targeted early viral escape mutants appeared to exert further immune pressure on the viral population, as demonstrated by the continued env sequence evolution between 5 and 16 months p.i. To determine whether Env variants with additional substitutions and deletions in V1/2 and V4 were neutralization escape mutations, we generated pseudoviruses with representative env variants at 16 and 22 months p.i. from animal PBE. Env variant PBEΔV1/V4.Mo16.G12 represented 60% of the clones at 16 months p.i., while PBEΔV1/V4.Mo22.E1 and PBEΔV1/V4.Mo22.E9 were codominant clones at 22 months p.i. These Env variants isolated from PBE during chronic infection were not recognized by contemporaneous plasma samples, indicating that they were also neutralization escape mutants (Fig. 6A).
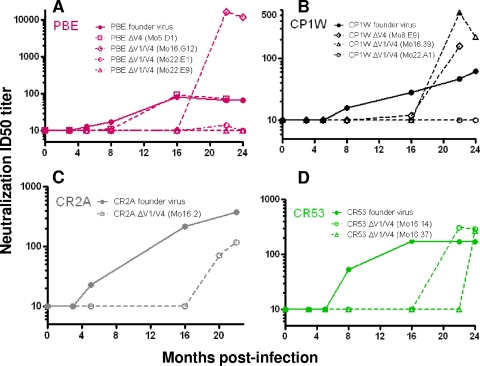
FIG. 6.
Neutralizing antibodies against autologous viral variants. The panels show the ability of autologous plasma sampled over time following infection to neutralize the founder virus and mutant viruses in animals PBE (A), CP1W (B), CR2A (C), and CR53 (D). The x axes indicate the times when plasma samples were collected, and ID50 titers are shown on the y axes. The ID50 titers against the autologous founder Envs over the course of infection are represented by solid lines and filled circles, while the neutralization titers against the mutant Env are depicted by dotted lines and open symbols. The time points p.i. when viral variants were isolated and the mutations/deletions these mutant viruses contain are indicated in parentheses.
In animal CP1W, a variant Env (CP1WΔV4.Mo8.E9) harboring R420G and R424Q mutations in V4 was isolated at 8 months p.i., when the autologous neutralizing antibodies were first detected in the animal. This mutant Env was not neutralized by contemporaneous plasma from monkey CP1W but was sensitive to neutralizing antibodies at later time points (Fig. 6B). An Env variant containing a deletion in V4 isolated later at 16 months p.i. (CP1WΔV1/V4.Mo16.39) also represented an escape variant. This virus was not neutralized by plasma sampled at 16 months p.i. but was sensitive to de novo neutralizing antibody responses at later time points. Finally, we did not detect neutralizing antibodies against an Env variant isolated at 22 months p.i. that harbored substitutions and deletions in V1 and V4 (CP1WΔV1/V4.Mo22.A1), indicating that this Env was an escape mutant that arose late in infection. We were not able to test the neutralization sensitivity of ΔV1 Env pseudoviruses from CP1W at 22 months p.i. because we could not generate these pseudoviruses at a high titer.
A similar pattern of sequential viral escapes from the autologous neutralizing antibodies was also observed in animals CR2A and CR53. We were not able to recover infectious pseudoviruses containing variant Envs from 5 to 8 months p.i. from animals CR2A and CR53, at the time when autologous neutralizing antibodies against the founder Envs were first observed. An Env clone from animal CR2A (CR2AΔV1/V4.Mo16.2) that contained deletions in both V1 and V4 was not neutralized by contemporaneous plasma (Fig. 6C). In animal CR53, Env variants CR53ΔV1/V4.Mo16.14 and CR53ΔV1/V4.Mo16.37, from 16 months p.i., were also not neutralized by contemporaneous plasma from the matched animals (Fig. 6D). These data demonstrate that Env clones containing deletions/mutations in V1, V4, or both regions were neutralization escape variants and that escape mutants were neutralized only by plasmas obtained a number of months after their emergence.
Interestingly, the escape Env variants in 3 of 4 animals (CP1W, PBE, and CR53) induced titers of neutralizing antibodies higher than those generated against the founder Env (Fig. 6A, B, and D), suggesting that mutations in V1/2 and V4 of SIVmac251 are associated with an increase in the neutralization sensitivity of the Env. These data raise the possibility that the amino acid substitutions that allowed the transmitted SIVmac251 to escape immune recognition by autologous antibodies paradoxically rendered it more susceptible to subsequent antibody neutralization. The emergence of novel escape viruses beyond 16 months p.i. in these animals and the increased susceptibility of the escape variants to subsequent neutralization indicate that neutralizing antibodies specific for escape variants of SIVmac251 exerted continuous selective pressure that shaped the evolution of the viral quasispecies even during chronic infection.
DISCUSSION
Rhesus monkeys infected with a genetically defined quasispecies of SIVmac251 provide an important model system for studying the relationship between an infecting primate immunodeficiency virus, viral evolution, and immune responses. Using a standardized pseudovirion-based TZM-bl reporter gene assay that is widely used to measure neutralizing antibodies to HIV-1, we demonstrated that autologous neutralizing antibodies directed against the transmitted/founder SIVmac251 strains were not detected until 5 to 8 months p.i. in mucosally infected monkeys. Combining single-genome amplification sequencing with statistical analyses and phenotypic testing, we showed that autologous neutralizing antibodies exerted selection pressure on SIVmac251 despite their low titers and late emergence. The Env sequence evolution was uniquely mapped in relation to the transmitted/founder viruses from a quasispecies inoculum. Finally, we demonstrated that secondary de novo antibody responses to escape variants spurred continuous Env evolution throughout infection.
The results presented in this study show that antibody-mediated neutralization of SIV is a complex and dynamic process. This evolution is similar to what has been shown recently in a cohort of drug-naïve, chronically HIV-1-infected individuals who control viral replication (44). Both HIV-1- and SIV-specific neutralizing antibodies appear to influence Env sequence evolution in vivo, even at low titers. Viral evolution and the dynamic production of autologous Nabs continue in both chronic HIV-1 and SIVmac infections (22, 40, 44, 48, 59). Similar to the autologous neutralizing antibody response and viral evolution in HIV controllers, we also observed increases in autologous neutralizing antibody titers against SIVmac251 escape variants isolated at 5 to 8 months p.i., suggesting that the development of autologous neutralizing antibodies limits certain Env variants over the course of infection. These intermediate escape variants were neutralized by emerging neutralizing antibodies that drove the continuous evolution of novel escape Env variants with common sequence mutations in V1/2 and V4 at 16 and 22 months p.i. in all 4 animals. Some of these late escape Env variants remained resistant to autologous neutralization, while others remained sensitive to Nabs at the later time points. The present data are also consistent with previous studies demonstrating that sequence evolution in V1/2 and V4 resulted in the loss of Env recognition by at least one class of conformation-dependent antibodies that neutralized HIV-1 and SIV infectivity (1, 4, 7, 9, 10, 13, 17, 23, 28, 29, 31, 32, 34, 37, 47, 48, 52, 58-61, 65, 67-69, 71, 75).
A constellation of changes was observed throughout Env at 16 months p.i. It is possible that some of these mutations reflected selection by Fcγ receptor-mediated antibody activity, CTL, and changes in cell tropism (16, 19, 21, 43, 49, 54). The Env mutations in the present study most likely do not represent genetic imprints from antibody-dependent cell-mediated cytotoxicity, because they emerged late in infection. It is also unlikely that the common mutations in V1/2 and V4 emerged as a result of viral escape from cytotoxic T lymphocytes, since these animals had disparate MHC class I haplotypes. However, since there are known CD8+ T-cell epitopes in the C2, C3, and gp41 regions of Env, some of the substitutions we observed in these regions may reflect CTL escape mutations (56). Although mutations in gp41 have also been shown to affect the sensitivity of Env to antibody neutralization (20), we did not directly evaluate the contribution of gp41 polymorphisms in mediating escape from neutralizing antibodies, since there were no common amino acid residues in the gp41 region that were temporally selected in all animals. Similarly, we also chose not to evaluate substitutions that occurred in a region where env and nef reading frames overlapped, since it is unclear how these polymorphisms relate to neutralizing antibodies.
In the transmission of clade A, C, and D HIV-1 from donors to recipients, a shortening of the Env variable loops has been associated with increased virus sensitivity to antibody-mediated neutralization (12, 17, 62). This phenomenon is followed by a lengthening of the env variable regions in HIV, simian-human immunodeficiency virus (SHIV), and certain strains of SIV during infection that is associated with a greater resistance to antibody neutralization (4, 41, 51, 58, 60, 63). In contrast, we observed that V1/2 and V4 shortened to escape from antibody recognition as the infection progressed from the acute to the chronic phase in SIVmac251-infected animals. In this cohort of animals, the shortening of the Env variable loops throughout infection was associated with decreased virus sensitivity to antibody neutralization rather than increased sensitivity, as observed in HIV-1. Nevertheless, despite the strain-specific sequence changes in the variable region, the Env changes in the genetically diverse primate immunodeficiency viruses all resulted in alterations in the Env glycosylation pattern.
The mechanisms underlying the escape of SIV from neutralizing antibodies are complex. Mutations can facilitate virus escape from neutralizing antibodies by altering the antibody binding domains through changes in glycosylation or direct alteration of neutralization epitopes (59). Alternatively, mutations can also alter regions of the virus that are distant from the antibody binding site and, in so doing, abrogate antibody binding by altering the envelope protein conformation (14, 73). We were not able to spatially model the amino acid residues that were identified as being involved in neutralizing antibody recognition in SIVmac251 Env, because the sites we identified as significant for selection in V1/2 and V4 were deleted in the SIVmac239 gp120 crystal structure of Chen et al. (11). Furthermore, our attempts to functionally delineate the specific residues in V1/2 and V4 that were directly responsible for resistance to antibody-mediated neutralization have been complicated by the context-dependent nature of the variable loop domains (data not shown). These findings emphasize the importance of the interactions between noncontiguous regions of the virus in forming quaternary epitopes (1, 14).
Paradoxically, variant SIVs that successfully evaded antibody neutralization became more sensitive to recognition by neutralizing antibodies that developed later in infection. This observation is consistent with previous reports of emerging HIV-1/SIV variants in infected individuals (5, 37). These data indicate that mutations in V1/2 and V4 of SIV Env may have induced a conformational change in the trimeric Env spike structure that increased the accessibility of the variable loops to antibody-mediated neutralization or led to the exposure of conserved domains for antibody recognition.
The present findings have important implications for HIV-1 vaccine development. SIVmac251 appears to be more difficult to neutralize than HIV-1, given that autologous neutralizing antibodies emerge later and at lower titers than have been described for HIV-1. This raises the possibility that the use of SIVmac251 as a challenge virus for AIDS vaccine studies in NHPs may be too stringent, because it may not accurately predict the ability of vaccine candidates to elicit neutralizing antibodies that protect against virus acquisition. Therefore, the results of preclinical vaccine trials evaluating the acquisition of infection utilizing SIVmac251 as a challenge virus should be interpreted accordingly. These data also suggest that HIV-1/SIV evolve in relatively similar patterns, and common antibody recognition motifs that may serve as common vulnerabilities can be identified. In addition, there may be constraints on the ability of primate immunodeficiency viruses to diverge to evade neutralizing antibodies. The need to retain envelope function and mask important epitopes of the protein likely constrains the plasticity of Env. Therefore, the stereotypic pattern of viral escape mutations may provide avenues for designing vaccination strategies that force the immune evasion of viral quasispecies into pathways that either exact a fitness cost or lead to the exposure of critical epitopes for immune recognition. Ultimately, the elucidation of the genetic determinants for viral escape relative to the transmitted/founder viruses in infected individuals will yield a broader understanding of antigenic variation and immune selection. Our results demonstrate the value of combining virological, immunological, and bioinformatics analyses in identifying important properties of the neutralizing antibody response during HIV-1/SIV infection.
Acknowledgments
TZM.bl cells were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We are grateful to Xinzhen Yang for helpful discussions regarding the evolution of HIV-1 Env. We thank the Roadrunner Open Science Advanced Simulation and Computing Program for use of its computer for the analysis of recurrent mutations.
This work was supported by NIH NIAID PHS grants K08-AI069995 (W.W.Y.), AI087383 (G.M.S.), and AI067854 (I.R., B.H.H., B.T.K., and N.L.L.); the Center for HIV/AIDS Vaccine Immunology; and Bill and Melinda Gates Collaboration for AIDS Vaccine Discovery Vaccine Immune Monitoring Consortium Grant 38619 (M.S.S.).
Footnotes
Published ahead of print on 31 March 2010.
REFERENCES
- 1.Babas, T., B. Belhadj-Jrad, R. Le Grand, D. Dormont, L. Montagnier, and E. Bahraoui. 1995. Specificity and neutralizing capacity of three monoclonal antibodies produced against the envelope glycoprotein of simian immunodeficiency virus isolate 251. Virology 211:339-344. [DOI] [PubMed] [Google Scholar]
- 2.Basavapathruni, A., W. W. Yeh, R. T. Coffey, J. B. Whitney, P. T. Hraber, A. Giri, B. T. Korber, S. S. Rao, G. J. Nabel, J. R. Mascola, M. S. Seaman, and N. L. Letvin. 2010. Envelope vaccination shapes viral envelope evolution following simian immunodeficiency virus infection in rhesus monkeys. J. Virol. 84:953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583-1586. [DOI] [PubMed] [Google Scholar]
- 4.Blay, W. M., S. Gnanakaran, B. Foley, N. A. Doria-Rose, B. T. Korber, and N. L. Haigwood. 2006. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J. Virol. 80:999-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bontjer, I., A. Land, D. Eggink, E. Verkade, K. Tuin, C. Baldwin, G. Pollakis, W. A. Paxton, I. Braakman, B. Berkhout, and R. W. Sanders. 2009. Optimization of human immunodeficiency virus type 1 envelope glycoproteins with V1/V2 deleted, using virus evolution. J. Virol. 83:368-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braibant, M., H. Agut, C. Rouzioux, D. Costagliola, B. Autran, and F. Barin. 2008. Characteristics of the env genes of HIV type 1 quasispecies in long-term nonprogressors with broadly neutralizing antibodies. J. Acquir. Immune Defic. Syndr. 47:274-284. [DOI] [PubMed] [Google Scholar]
- 7.Burns, D. P., C. Collignon, and R. C. Desrosiers. 1993. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns, D. P., and R. C. Desrosiers. 1991. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J. Virol. 65:1843-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 12.Chohan, B., D. Lang, M. Sagar, B. Korber, L. Lavreys, B. Richardson, and J. Overbaugh. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 79:6528-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi, W. S., C. Collignon, C. Thiriart, D. P. Burns, E. J. Stott, K. A. Kent, and R. C. Desrosiers. 1994. Effects of natural sequence variation on recognition by monoclonal antibodies neutralize simian immunodeficiency virus infectivity. J. Virol. 68:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole, K. S., M. Alvarez, D. H. Elliott, H. Lam, E. Martin, T. Chau, K. Micken, J. L. Rowles, J. E. Clements, M. Murphey-Corb, R. C. Montelaro, and J. E. Robinson. 2001. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology 290:59-73. [DOI] [PubMed] [Google Scholar]
- 15.Cole, K. S., J. D. Steckbeck, J. L. Rowles, R. C. Desrosiers, and R. C. Montelaro. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J. Virol. 78:1525-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 18.Doria-Rose, N. A., R. M. Klein, M. M. Manion, S. O'Dell, A. Phogat, B. Chakrabarti, C. W. Hallahan, S. A. Migueles, J. Wrammert, R. Ahmed, M. Nason, R. T. Wyatt, J. R. Mascola, and M. Connors. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 83:188-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eastman, D., A. Piantadosi, X. Wu, D. N. Forthal, G. Landucci, J. T. Kimata, and J. Overbaugh. 2008. Heavily glycosylated, highly fit SIVMne variants continue to diversify and undergo selection after transmission to a new host and they elicit early antibody dependent cellular responses but delayed neutralizing antibody responses. Virol. J. 5:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glamann, J., D. R. Burton, P. W. Parren, H. J. Ditzel, K. A. Kent, C. Arnold, D. Montefiori, and V. M. Hirsch. 1998. Simian immunodeficiency virus (SIV) envelope-specific Fabs with high-level homologous neutralizing activity: recovery from a long-term-nonprogressor SIV-infected macaque. J. Virol. 72:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goonetilleke, N., M. K. Liu, J. F. Salazar-Gonzalez, G. Ferrari, E. Giorgi, V. V. Ganusov, B. F. Keele, G. H. Learn, E. L. Turnbull, M. G. Salazar, K. J. Weinhold, S. Moore, N. Letvin, B. F. Haynes, M. S. Cohen, P. Hraber, T. Bhattacharya, P. Borrow, A. S. Perelson, B. H. Hahn, G. M. Shaw, B. T. Korber, and A. J. McMichael. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray, E. S., P. L. Moore, R. A. Pantophlet, and L. Morris. 2007. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J. Virol. 81:10769-10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch, V., D. Adger-Johnson, B. Campbell, S. Goldstein, C. Brown, W. R. Elkins, and D. C. Montefiori. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javaherian, K., A. J. Langlois, S. Schmidt, M. Kaufmann, N. Cates, J. P. Langedijk, R. H. Meloen, R. C. Desrosiers, D. P. Burns, D. P. Bolognesi, et al. 1992. The principal neutralization determinant of simian immunodeficiency virus differs from that of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 89:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, W. E., H. Sanford, L. Schwall, D. R. Burton, P. W. Parren, J. E. Robinson, and R. C. Desrosiers. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurkiewicz, E., G. Hunsmann, J. Schaffner, T. Nisslein, W. Luke, and H. Petry. 1997. Identification of the V1 region as a linear neutralizing epitope of the simian immunodeficiency virus SIVmac envelope glycoprotein. J. Virol. 71:9475-9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang, S. M., F. S. Quan, C. Huang, L. Guo, L. Ye, C. Yang, and R. W. Compans. 2005. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology 331:20-32. [DOI] [PubMed] [Google Scholar]
- 33.Keele, B. F., H. Li, G. H. Learn, P. Hraber, E. E. Giorgi, T. Grayson, C. Sun, Y. Chen, W. W. Yeh, N. L. Letvin, J. R. Mascola, G. J. Nabel, B. F. Haynes, T. Bhattacharya, A. S. Perelson, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinsey, N. E., M. G. Anderson, T. J. Unangst, S. V. Joag, O. Narayan, M. C. Zink, and J. E. Clements. 1996. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology 221:14-21. [DOI] [PubMed] [Google Scholar]
- 35.Kosakovsky Pond, S. L., and S. D. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 36.Kulkarni, S. S., A. Lapedes, H. Tang, S. Gnanakaran, M. G. Daniels, M. Zhang, T. Bhattacharya, M. Li, V. R. Polonis, F. E. McCutchan, L. Morris, D. Ellenberger, S. T. Butera, R. C. Bollinger, B. T. Korber, R. S. Paranjape, and D. C. Montefiori. 2009. Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 385:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laird, M. E., and R. C. Desrosiers. 2007. Infectivity and neutralization of simian immunodeficiency virus with FLAG epitope insertion in gp120 variable loops. J. Virol. 81:10838-10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letvin, N. L. 2009. Virology moving forward in HIV vaccine development. Science 326:1196-1198. [DOI] [PubMed] [Google Scholar]
- 39.Letvin, N. L., S. S. Rao, V. Dang, A. P. Buzby, B. Korioth-Schmitz, D. Dombagoda, J. G. Parvani, R. H. Clarke, L. Bar, K. R. Carlson, P. A. Kozlowski, V. M. Hirsch, J. R. Mascola, and G. J. Nabel. 2007. No evidence for consistent virus-specific immunity in simian immunodeficiency virus-exposed, uninfected rhesus monkeys. J. Virol. 81:12368-12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, B., J. M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, S. Allen, E. Hunter, B. H. Hahn, G. M. Shaw, J. L. Blackwell, and C. A. Derdeyn. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahalanabis, M., P. Jayaraman, T. Miura, F. Pereyra, E. M. Chester, B. Richardson, B. Walker, and N. L. Haigwood. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montefiori, D. C. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12:Unit 12.11. [DOI] [PubMed]
- 46.Montefiori, D. C., W. E. Robinson, Jr., S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore, P. L., E. S. Gray, I. A. Choge, N. Ranchobe, K. Mlisana, S. S. Abdool Karim, C. Williamson, and L. Morris. 2008. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82:1860-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore, P. L., N. Ranchobe, B. E. Lambson, E. S. Gray, E. Cave, M. R. Abrahams, G. Bandawe, K. Mlisana, S. S. Abdool Karim, C. Williamson, and L. Morris. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogert, R. A., M. K. Lee, W. Ross, A. Buckler-White, M. A. Martin, and M. W. Cho. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J. Virol. 75:5998-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Overbaugh, J., and L. M. Rudensey. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Overbaugh, J., L. M. Rudensey, M. D. Papenhausen, R. E. Benveniste, and W. R. Morton. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petry, H., K. Pekrun, G. Hunsmann, E. Jurkiewicz, and W. Luke. 2000. Naturally occurring V1-env region variants mediate simian immunodeficiency virus SIVmac escape from high-titer neutralizing antibodies induced by a protective subunit vaccine. J. Virol. 74:11145-11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plotkin, S. A. 2008. Vaccines: correlates of vaccine-induced immunity. Clin. Infect. Dis. 47:401-409. [DOI] [PubMed] [Google Scholar]
- 54.Pollakis, G., S. Kang, A. Kliphuis, M. I. Chalaby, J. Goudsmit, and W. A. Paxton. 2001. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 276:13433-13441. [DOI] [PubMed] [Google Scholar]
- 55.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 56.Reynolds, M. R., A. M. Weiler, K. L. Weisgrau, S. M. Piaskowski, J. R. Furlott, J. T. Weinfurter, M. Kaizu, T. Soma, E. J. Leon, C. MacNair, D. P. Leaman, M. B. Zwick, E. Gostick, S. K. Musani, D. A. Price, T. C. Friedrich, E. G. Rakasz, N. A. Wilson, A. B. McDermott, R. Boyle, D. B. Allison, D. R. Burton, W. C. Koff, and D. I. Watkins. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 205:2537-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rong, R., S. Gnanakaran, J. M. Decker, F. Bibollet-Ruche, J. Taylor, J. N. Sfakianos, J. L. Mokili, M. Muldoon, J. Mulenga, S. Allen, B. H. Hahn, G. M. Shaw, J. L. Blackwell, B. T. Korber, E. Hunter, and C. A. Derdeyn. 2007. Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J. Virol. 81:5658-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rong, R., B. Li, R. M. Lynch, R. E. Haaland, M. K. Murphy, J. Mulenga, S. A. Allen, A. Pinter, G. M. Shaw, E. Hunter, J. E. Robinson, S. Gnanakaran, and C. A. Derdeyn. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rudensey, L. M., J. T. Kimata, E. M. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rybarczyk, B. J., D. Montefiori, P. R. Johnson, A. West, R. E. Johnston, and R. Swanstrom. 2004. Correlation between env V1/V2 region diversification and neutralizing antibodies during primary infection by simian immunodeficiency virus sm in rhesus macaques. J. Virol. 78:3561-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sagar, M., O. Laeyendecker, S. Lee, J. Gamiel, M. J. Wawer, R. H. Gray, D. Serwadda, N. K. Sewankambo, J. C. Shepherd, J. Toma, W. Huang, and T. C. Quinn. 2009. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J. Infect. Dis. 199:580-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sagar, M., X. Wu, S. Lee, and J. Overbaugh. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80:9586-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salazar-Gonzalez, J. F., E. Bailes, K. T. Pham, M. G. Salazar, M. B. Guffey, B. F. Keele, C. A. Derdeyn, P. Farmer, E. Hunter, S. Allen, O. Manigart, J. Mulenga, J. A. Anderson, R. Swanstrom, B. F. Haynes, G. S. Athreya, B. T. Korber, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sather, D. N., J. Armann, L. K. Ching, A. Mavrantoni, G. Sellhorn, Z. Caldwell, X. Yu, B. Wood, S. Self, S. Kalams, and L. Stamatatos. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sato, S., E. Yuste, W. A. Lauer, E. H. Chang, J. S. Morgan, J. G. Bixby, J. D. Lifson, R. C. Desrosiers, and W. E. Johnson. 2008. Potent antibody-mediated neutralization and evolution of antigenic escape variants of simian immunodeficiency virus strain SIVmac239 in vivo. J. Virol. 82:9739-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saunders, C. J., R. A. McCaffrey, I. Zharkikh, Z. Kraft, S. E. Malenbaum, B. Burke, C. Cheng-Mayer, and L. Stamatatos. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79:9069-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torres, J. V., A. Malley, B. Banapour, D. E. Anderson, M. K. Axthelm, M. B. Gardner, and E. Benjamini. 1993. An epitope on the surface envelope glycoprotein (gp130) of simian immunodeficiency virus (SIVmac) involved in viral neutralization and T cell activation. AIDS Res. Hum. Retroviruses 9:423-430. [DOI] [PubMed] [Google Scholar]
- 72.Valentine, L. E., and D. I. Watkins. 2008. Relevance of studying T cell responses in SIV-infected rhesus macaques. Trends Microbiol. 16:605-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watkins, B. A., S. Buge, K. Aldrich, A. E. Davis, J. Robinson, M. S. Reitz, Jr., and M. Robert-Guroff. 1996. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J. Virol. 70:8431-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 75.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeh, W. W., P. Jaru-Ampornpan, D. Nevidomskyte, M. Asmal, S. S. Rao, A. P. Buzby, D. C. Montefiori, B. T. Korber, and N. L. Letvin. 2009. Partial protection of simian immunodeficiency virus (SIV)-infected rhesus monkeys against superinfection with a heterologous SIV isolate. J. Virol. 83:2686-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]