Abstract
Liver-related mortality is increased in the setting of HIV-hepatitis B virus (HBV) coinfection. However, interactions between HIV and HBV to explain this observation have not been described. We hypothesized that HIV infection of hepatocytes directly affects the life cycle of HBV. We infected human hepatic cell lines expressing HBV (Hep3B and AD38 cells) or not expressing HBV (Huh7, HepG2, and AD43 cells) with laboratory strains of HIV (NL4-3 and AD8), as well as a vesicular stomatitis virus (VSV)-pseudotyped HIV expressing enhanced green fluorescent protein (EGFP). Following HIV infection with NL4-3 or AD8 in hepatic cell lines, we observed a significant increase in HIV reverse transcriptase activity which was infectious. Despite no detection of surface CD4, CCR5, and CXCR4 by flow cytometry, AD8 infection of AD38 cells was inhibited by maraviroc and NL4-3 was inhibited by AMD3100, demonstrating that HIV enters AD38 hepatic cell lines via CCR5 or CXCR4. High-level infection of AD38 cells (50%) was achieved using VSV-pseudotyped HIV. Coinfection of the AD38 cell line with HIV did not alter the HBV DNA amount or species as determined by Southern blotting or nucleic acid signal amplification. However, coinfection with HIV was associated with a significant increase in intracellular HBsAg when measured by Western blotting, quantitative HBsAg, and fluorescence microscopy. We conclude that HIV infection of HBV-infected hepatic cell lines significantly increased intracellular HBsAg but not HBV DNA synthesis and that increased intrahepatic HBsAg secondary to direct infection by HIV may contribute to accelerated liver disease in HIV-HBV-coinfected individuals.
Approximately 5 to 10% of individuals with HIV are coinfected with hepatitis B virus (HBV) but this may be as high as 20% in parts of Africa. The natural history of HBV infection is altered in HIV-HBV coinfection, and liver-related mortality is significantly higher in HIV-HBV-coinfected individuals than in those infected with either HIV or HBV alone (28). How HIV infection accelerates the progression of HBV-related liver disease is not known but is likely to be multifactorial. HIV-HBV coinfection is associated with higher levels of HBV DNA and lower alanine aminotransferase (ALT) levels (11). The lower ALT levels are indicative of less hepatocyte destruction, possibly due to a depressed HBV-specific T-cell response (9), suggesting that other factors may be involved in driving liver disease progression.
HBV is a noncytopathic virus that directly infects hepatocytes. Hepatitis B surface antigen (HBsAg), the viral envelope, comprises large (L), medium (M), and small (S) HBsAg. Each surface protein shares the same stop codon, but translation of each begins from different start codons; S is encoded by the S gene, the N-terminal extension of M is encoded by the upstream pre-S2 gene, and the additional N-terminal extension of L is encoded by pre-S1. The HBV surface proteins envelop nucleocapsids to form virions. In addition, noninfectious spherical and filamentous subviral particles, denoted HBsAg, are secreted and can exceed the HBV virion level by at least 1,000-fold.
HIV has been shown to infect multiple cells in the liver, including hepatocytes (reviewed in reference 3). HIV DNA has been detected by PCR and HIV RNA detected by in situ hybridization in hepatocytes and Kupffer cells, and HIV capsid antigen (p24) has been detected in Kupffer cells by immunohistochemistry in liver specimens from HIV-infected individuals (7). HIV RNA has also been detected in Kupffer cells, sinusoidal cells, and portal mononuclear inflammatory cells, in addition to hepatocytes (7). In vitro studies of HIV infection of liver cells support the in vivo findings using primary human Kupffer cells, primary endothelial cells, and a number of hepatic cell lines (derived from hepatoma and hepatoblastoma cells) (8).
Few studies have examined the interaction between HIV and HBV in vitro. We hypothesized that HIV infection of hepatocytes has a direct effect on the HBV viral life cycle. To examine this, we developed an in vitro model to assess the interaction of HIV and HBV in hepatic cell lines and found that HIV coinfection of an HBV-expressing hepatic cell line led to an increase in intracellular HBsAg. Given that intracellular HBsAg can facilitate hepatocyte toxicity, this interaction may potentially explain enhanced liver disease progression in HIV-HBV coinfection.
MATERIALS AND METHODS
Cell lines.
The human hepatic cell lines expressing HBV antigens (Hep3B cells, which express HBsAg, and AD38 cells, which express proteins, RNA, and DNA intermediates characteristic of HBV replication [including HBsAg, HBcAg, and HBV DNA, although HBeAg production is reduced] [18]) or not expressing HBV (Huh7, HepG2, and AD43 cells) were cultured at 37°C in minimal essential media (MEM) or Dulbecco's modified Eagle medium (DMEM) (Gibco, Invitrogen, Grand Island, NY). The medium was supplemented with 10% heat-inactivated fetal calf serum (Progen, Darra, Australia), 100 U/ml penicillin G, 100 U/ml streptomycin, and l-glutamine (Gibco, Invitrogen). AD38 and AD43 cells were grown in DMEM with nutrient mixture F-12 (DMEM/F-12) with 400 μg/ml G418 (Geneticin; Gibco-BRL) and 2 μg/ml tetracycline (Sigma, St. Louis, MO). TZM-bl cells (a HeLa-derived indicator cell line which express surface CD4, CXCR4, CCR5, and the β-galactosidase and luciferase genes under the control of the HIV-1 promoter) were obtained through the NIH AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH) (provided by John C. Kappes, Xiaoyun Wu, and Tranzyme Inc.). TZM-bl cells, a nonhepatic cell line, were used as a positive control for HIV replication. Culture of AD38 cells in the presence of 1% dimethyl sulfoxide (DMSO) was used as a positive control for HBV DNA and HBsAg production, as this has been shown previously to increase HBV replication in cell lines (16).
HIV viral stocks.
HIV viral stocks were produced by transfecting 293T cells with CXCR4-using NL4-3 and a recombinant virus expressing enhanced green fluorescent protein (EGFP) in an NL4-3 backbone (NLEGFP) (31) or the CCR5-using AD8 DNA, using FuGENE 6 (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions. In some experiments we also used a virus that expressed EGFP in place of the nef gene (NL4-3ΔnefEGFP, a kind gift from Damien Purcell, University of Melbourne, Melbourne, Australia). Pseudotyped virus containing vesicular stomatitis virus (VSV) envelope and HIV and with EGFP in place of nef (VSV-NLNE) was produced in the same manner (2). Mock infection was carried out with supernatant from 293T cells cotransfected with VSV and pNLA1, a plasmid which expresses HIV Env and accessory proteins (but not Gag and GagPol) under the control of the HIV long terminal repeat (LTR) but does not produce replication-competent virus (17) (a kind gift from Johnson Mak, Burnet Institute, Melbourne, Australia). In some experiments, an inhibitor of HIV replication was added at 2 to 4 h prior to infection. These inhibitors included lamivudine (LMV) (Sigma, St. Louis, MO), AMD3100 (12), and maraviroc (Pfizer Ltd., Sandwich, United Kingdom).
Micro-RT assay.
HIV replication was determined by measuring HIV reverse transcriptase (RT) activity via a micro-RT assay as previously described (6).
Fluorescence microscopy.
AD38 cells were grown in Ibidi eight-well μ-slides (Integrated BioDiagnostics, Munich, Germany). Cells were infected with VSV-NLNE or mock infected with VSV-NLA1 and maintained in culture for at least 48 to 72 h. Staining for nuclear DNA was performed with Hoechst 33258 (Invitrogen, Mount Waverley, Australia). Cells were then fixed in 4% formaldehyde for no longer than 10 min, rinsed with phosphate-buffered saline (PBS) deficient in calcium and magnesium, and permeabilized in 0.1% Triton X-100 for 10 min. Prevention of nonspecific binding was achieved by incubation in blocking solution containing 3% (wt/vol) casein in PBS with 0.05% Tween 20 (PBS-T) for 30 min at 37°C prior to incubation with primary antibody diluted 1:100 in 0.05% PBS-T with 1% (wt/vol) casein for 1 h at 37°C. Cells were gently rinsed with PBS-T before incubation with secondary antibody diluted 1:1,000 in 0.05% PBS-T with 1% (wt/vol) casein for 1 h at 37°C. Cells were then rinsed three times in PBS-T before a final rinse in PBS. Primary monoclonal mouse antisera raised to HBsAg (Abbott Diagnostics, Abbott Park, IL), polyclonal rabbit antisera to HBcAg (Dako), and Texas Red-conjugated secondary antibodies raised to mouse and rabbit immunoglobulins (Invitrogen) were used. Images were captured on a charge-coupled device camera (CoolSnap HQ; Photometric) through 40× 0.65- to 1.35-numerical-aperture or 100× 1.4-numerical-aperture oil immersion lenses on a DeltaVision microscope (Applied Precision, Issaquah, WA) and deconvolved using softWoRx deconvolution software. The intensity of the HBsAg staining was quantified using the data inspector function of softWoRx software. The data were derived from a volume compression of a z-stack of 12 images taken at a step size of 0.5-micrometer.
HBsAg quantitation.
HBsAg was quantified using the Architect chemiluminescence assay (Abbott) according to the manufacturer's instructions. Samples were diluted 1:2 in Architect HBsAg diluent for measurement of HBsAg.
Western blotting.
Western blot analysis was performed on lysed cell monolayers as described previously (30). Primary monoclonal mouse antisera raised to HBsAg (Abbott) were diluted 1:1,000 in PBS-T with 1% (wt/vol) casein (30). The HBsAg antibody was specific for all three surface proteins, as shown previously (30). Horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:2,000 were used for detection via enhanced chemiluminescence (Perkin-Elmer, Waltham, MA). Image intensity was compared using Quantity One software package (Bio-Rad, Richmond, CA).
HBV DNA quantitation by Southern blotting.
Intracellular HBV DNA replicative intermediates were isolated from cytoplasmic core particles by incubating cell monolayers in lysis buffer containing 0.5% NP-40. After removal of the nuclear pellet by centrifugation, cell lysate supernatants were incubated with 10 units of DNase I (Roche Diagnostics) at 37°C before addition of 0.5% SDS and 25 mM EDTA in Tris (pH 7.5) and incubation with 0.5 mg/ml proteinase K (Roche Diagnostics) at 37°C overnight. Extractions with Tris-saturated phenol and chloroform were performed, and nucleic acids were then recovered by precipitation with 1 volume of isopropanol. Pellets containing viral DNA were dried and then redissolved in 10 mM Tris-10 mM EDTA before analysis by electrophoresis on 1% Tris-acetate-EDTA (TAE) agarose gels and transfer to a nylon membrane (Amersham Life Sciences, Braunschweig, Germany). A radiolabeled DNA probe was prepared using the random primer extension labeling system (NEN Life Science Products, Boston, MA) and [32P]dCTP as described previously (4).
HBV DNA quantitation by bDNA assay.
HBV DNA quantitation in cell lysates was determined using Bayer Versant bDNA 3.0 assay (Bayer Diagnostics, Tarrytown, NY) according to the manufacturer's instructions. Samples for bDNA assay were removed prior to incubation with proteinase K as described above and diluted 1:10 in PBS.
Flow cytometry.
Infected hepatic cells were washed thoroughly in ice-cold PBS that contained 1% fetal bovine serum and 1 mM EDTA. Cells were then washed in PBS and treated with directly conjugated antibody to CCR5 (BD Biosciences, San Jose, CA), CXCR4 (BD Biosciences), or CD4 (BD Biosciences), primary antibody to major histocompatibility complex (MHC) class I (W6/32; ATCC, Manassas, VA), or the relevant isotype controls (BD Biosciences) diluted 1: 30. Cells underwent further washing in PBS containing 1% fetal bovine serum and 1 mM EDTA before being suspended in PBS containing 1% formaldehyde. All data were acquired on a FACSCalibur and were analyzed using CellQuest (BD Biosciences).
MTS Assay.
Cell toxicity and decreased cellular metabolic activity were determined using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) colorimetric assay (Promega, Madison, WI).
Statistical analysis.
Results were compared using the Mann-Whitney test, Kruskal-Wallis analysis of variance, and two-way analysis of variance where appropriate, using SPSS version 16.0 (Chicago, IL) and GraphPad Prism 5 (GraphPad, La Jolla, CA). A P value of <0.05 was considered significant.
RESULTS
Low-level HIV infection of hepatic cell lines.
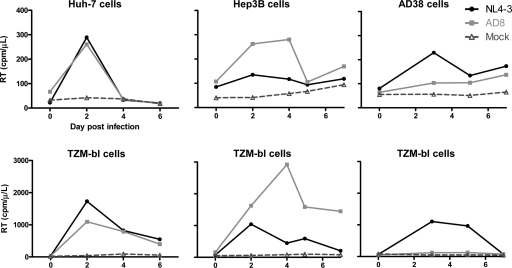
We infected HBV-expressing hepatic cell lines (AD38 and Hep3B cells) and a non-HBV-expressing hepatic cell line (Huh7) with either NL4-3 or AD8 and demonstrated a significant but short-lived and low-level increase in HIV RT activity in cell culture supernatant, indicative of infection (Fig. 1). The peak HIV RT level was approximately 10-fold lower than that following infection of the TZM-bl cell line. The HIV RT level in the TZM-bl cell line did vary with increasing passage number (Fig. 1, lower panels), as did the levels of CD4, CCR5, and CXCR4 expression (data not shown). There was no difference in the kinetics of the increase in HIV RT between HBV-expressing and non-HBV-expressing hepatic cell lines. We also performed the same experiments using NLEGFP, and although the frequency of infection was low, EGFP-expressing hepatic cells were clearly detected (data not shown). In addition, the supernatant from these HIV-infected HBV-expressing hepatic cell lines was infectious, as indicated by detection of β-galactosidase activity following incubation of supernatants with TZM-bl cells (data not shown).
FIG. 1.
HIV infection of hepatic cell lines. Reverse transcriptase (RT) in supernatants was quantified following infection with either NL4-3 or AD8 or mock infection of noninfected (Huh7) and HBV-infected (Hep3B and AD38) hepatic cell lines (upper panels) and TZM-bl cell lines (lower panels). Results from a representative example of three separate experiments are shown.
Given that peak HIV RT levels were quite low and to demonstrate that the increase in RT represented active rounds of replication and not just residual input virus, we also performed HIV infection of hepatic cell lines (HepG2 and AD38 cells) in the presence of the nucleoside reverse transcriptase inhibitor lamivudine (LMV). Infection was inhibited in the presence of LMV at concentrations of 5 μM and 50 μM (Fig. 2), confirming that hepatic cell lines infected with HBV were permissive to HIV infection, although at low levels.
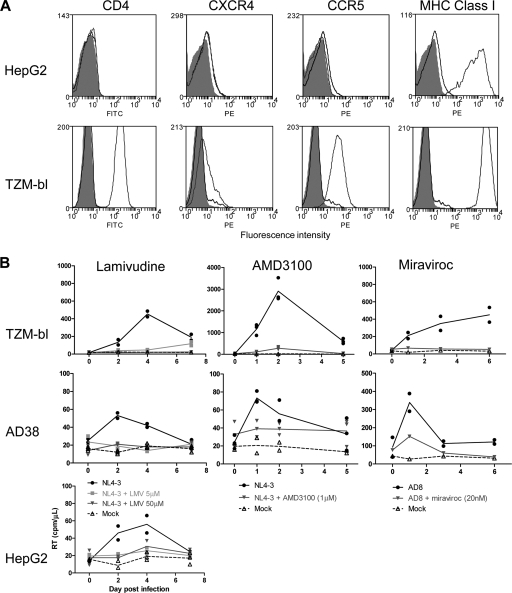
FIG. 2.
Expression and function of HIV coreceptors on hepatic cell lines. (A) Expression of CD4, CXCR4, and CCR5 was quantified using flow cytometry. Histograms of fluorescence are shown for cells alone (solid gray), isotype control (black line), and cells stained with fluorescently labeled antibodies to CD4, CXCR4, CCR5, and MHC class I (dark gray line) in HepG2 (upper row) and TZM-bl (lower row) cells. (B) Infection of AD38, HepG2, and TZM-bl cells was performed with either NL4-3 or AD8 in the absence presence of drug, or mock infection was performed. HIV reverse transcriptase (RT) in cell culture supernatant was measured. Lamivudine (at either 5 μM or 50 μM (left panels) or AMD3100 (1 μM) (middle panels) was incubated with cell lines for 24 h prior to addition of NL4-3. Maraviroc (20 nM) (right panels) was incubated with cell lines for 24 h prior to addition of AD8. Duplicates of the same experiment are shown as symbols. The line represents the mean of the duplicates.
Expression of HIV coreceptors.
To determine the mechanism of HIV entry, we first evaluated expression of CD4, CXCR4, and CCR5 on the hepatic cell lines Huh7, HepG2, Hep3B, and AD38 using flow cytometry. We found no expression of CD4, CXCR4, or CCR5 on any of the cell lines (Fig. 2A [data are from HepG2 cells only]). Expression of CD4, CXCR4, and CCR5 was clearly demonstrated in TZM-bl cells (Fig. 2A, lower panels). To determine if these coreceptors mediated infection despite lack of detectable surface expression, we infected AD38 cells with either NL4-3 or AD8 in the presence or absence of a CXCR4 antagonist, AMD3100, or a CCR5 antagonist, maraviroc (Fig. 2B). Following infection with NL4-3, infection was inhibited with AMD3100 but not maraviroc, demonstrating entry via CXCR4. Similarly, following infection with AD8, infection was inhibited with maraviroc but not AMD3100. These data demonstrate that HIV enters AD38 hepatic cell lines via CXCR4 or CCR5, despite the inability to detect surface expression of each coreceptor.
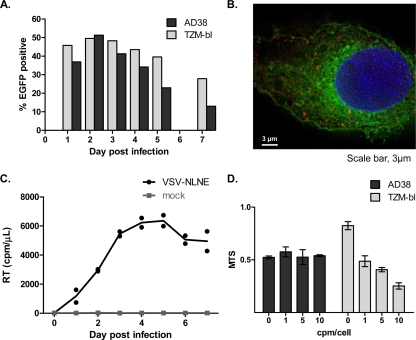
High-level HIV infection with VSV-NLNE-pseudotyped virus.
We then wanted to determine what effects HIV replication within a hepatocyte would have on the HBV life cycle. In order to increase the level of HIV infection and to directly visualize HIV-infected hepatic cells, we infected hepatic cell lines with VSV-NLNE-pseudotyped virus. High-level infection was determined by measurement of HIV RT in addition to detection of EGFP by flow cytometry and fluorescence microscopy (peak = 51%) (Fig. 3A) and was achieved at levels similar to that seen in the TZM-bl cell line (data not shown). EGFP expression in VSV-NLNE-infected AD38 cells also expressing HBcAg was demonstrated using fluorescence microscopy (Fig. 3B). Following infection of the AD38 cell line (n = 3) with VSV-NLNE, the peak RT level (mean ± standard error [SE]) was 6,366 ± 534 cpm (Fig. 3C). Despite this high-level infection, cell proliferation determined using the MTS assay was similar to that in uninfected AD38 cells (Fig. 3D), although we did not measure apoptosis specifically. In contrast, we observed toxicity in TZM-bl cells at higher concentrations of VSV-NLNE (Fig. 3D). Therefore, the use of VSV-NLNE allowed for high-level infection of HBV-expressing hepatic cell lines without significant cell death. This model was then used to evaluate any changes in the HBV life cycle following HIV coinfection.
FIG. 3.
Infection of hepatic cell lines with VSV-NLNE. (A and B) AD38 and TZM-bl cells were infected with VSV-NLNE, and infection was quantified by detection of EGFP using flow cytometry (A) or fluorescence microscopy (B). An AD38 cell is shown. Hoechst staining of nucleus is blue, HBcAg staining using primary polyclonal rabbit antibody (Dako) and Texas Red-conjugated secondary goat anti-rabbit antibody (Invitrogen) is red, and EGFP expression in the HIV-infected cell is green. Magnification, ×100. (C) RT in cell culture supernatant was measured following infection of AD38 cells with VSV-NLNE or VSV-NLA1. Duplicate results from the same experiment are shown as symbols. The line represents the mean of the duplicates. Data are representative of three separate experiments. (D) MTS in AD38 cells and TZM-bl cells was measured at different multiplicities of infection (from 0 to 10 cpm/cell). The mean ± standard error (SE) for separate experiments performed in triplicate is shown.
Effect of high-level HIV infection on HBV expression.
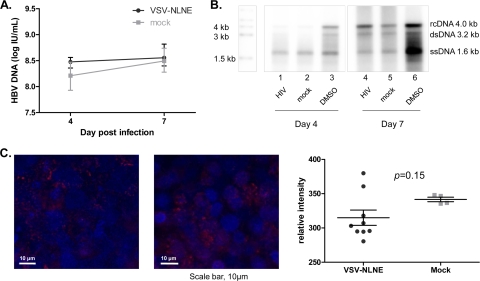
To determine whether HIV coinfection of hepatic cell lines altered HBV replication, we first assessed HBV DNA by Southern blotting as well as signal amplification (using the bDNA assay) in AD38 cells infected with VSV-NLNE and compared the results to those with mock-infected controls (Fig. 4A and B). We found no significant differences in total HBV DNA in the cytosolic fraction of cell lysates in the presence or absence of HIV coinfection. In addition, the relative appearances of relaxed circular DNA (rcDNA), double-stranded DNA (dsDNA), and single-stranded DNA (ssDNA) were unchanged in the presence or absence of coinfection with HIV. Incubation with DMSO was used as a positive control and showed a clear and significant increase in all HBV DNA intermediates, as previously described (16). We also performed fluorescence microscopy for HBcAg in AD38 cells coinfected with HIV and found no difference in the intensity of HBcAg staining (Fig. 4C). Overall, these data suggest that high-level HIV coinfection of hepatic cell lines did not alter HBV DNA production, although any affect on HBV RNA intermediates or HBV DNA secretion has not been assessed.
FIG. 4.
Changes in HBV DNA expression following HIV coinfection. (A) HBV DNA in cytoplasmic extracts from AD38 cells collected 4 and 7 days after infection with VSV-NLNE or mock infection was quantified. The median and range from four different experiments are shown. (B) Southern blot analysis of cell lysates of AD38 cells 4 days after infection with VSV-NLNE (lane 1), mock infection (lane 2), or treatment with 1% DMSO (lane 3) and 7 days after infection with VSV-NLNE (lane 4), mock infection (lane 5), or treatment with 1% DMSO (lane 6). Data are representative of six independent experiments. Markers show the sizes of rcDNA, dsDNA, and ssDNA. (C) HBcAg expression in AD38 cell monolayers 3 days after infection with VSV-NLNE (left panel) or mock infection (right panel) using fluorescence microscopy. Hoechst staining of nuclei is blue, and HBcAg staining is red. Magnification, ×40. Right panel, quantification of the relative intensity of HBcAg staining using softWoRx software (Applied Precision, Issaquah, WA) following infection with VSV-NLNE or mock infection. Each symbol represents a separate field from the same experiment. The black lines show the mean ± SE.
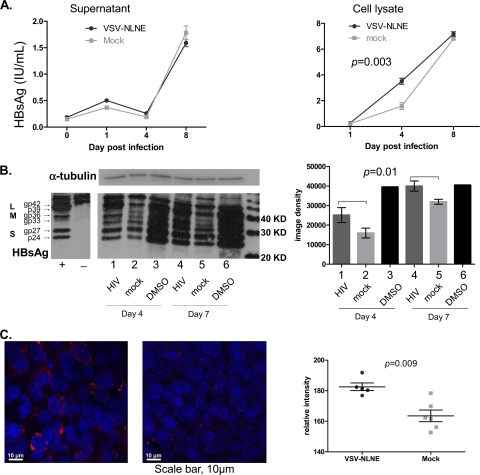
We then quantified HBsAg in cell lysates and supernatants by either quantitative HBsAg (Architect assay; Abbott), Western blotting, or fluorescence microscopy (Fig. 5). When we compared quantitative HBsAg in supernatants and cell lysates from HIV-infected AD38 cells and mock-infected cells, we found no significant difference in quantitative HBsAg in supernatants (P = 0.83) but significantly elevated quantitative HBsAg in cell lysates in HIV-infected cells compared with mock-infected cells (P = 0.003) (Fig. 5A). The difference between the levels in HIV-infected and mock-infected AD38 cells was greatest at day 4 postinfection but was not observed by day 8. This may be a result of declining HIV replication by day 8 (Fig. 3A), given that the VSV-pseudotyped virus would only have a single round and not multiple rounds of infection. These findings were consistent with findings from Western blotting. When we quantified glycosylated and nonglycosylated L, M, or S proteins in cell lysates (Fig. 5B), we found that there was a significant increase in all HBsAg proteins, i.e., L, M, and S proteins, in HIV-coinfected cells. Western blotting of HepG2 cell lysates was performed as a negative control, and no bands were observed (Fig. 5B). Our findings from fluorescence microscopy also confirmed a significant increase in intracellular HBsAg in HIV-infected AD38 cells compared to mock-infected cells (P = 0.009) (Fig. 5C). Both HBsAg and HBcAg were present predominantly in the cytoplasm in monoinfected and coinfected AD38 cells (Fig. 4C and 5C). These findings therefore demonstrated that HIV coinfection of AD38 cells lead to increased intracellular L, M, and S HBsAg.
FIG. 5.
Expression of HBsAg following HIV coinfection. (A) HBsAg in cell culture supernatant (left panel) and cell lysates (right panel) from AD38 cells infected with VSV-NLNE or mock infected was quantified using the Architect assay (Abbott). The median and range for duplicate samples are shown. Statistical significance was determined by two-way analysis of variance (ANOVA) of duplicate samples from one experiment. A similar trend was seen in two further separate experiments. (B) HBsAg was quantified by Western blotting (left panel) of AD38 cell lysates 4 days (lanes 1 to 3) and 7 days (lanes 4 to 6) after infection with VSV-NLNE (lanes 1 and 4) or mock infection (lanes 2 and 5) or in the presence of 1% DMSO (lanes 3 and 6). Specificity of HBsAg staining is demonstrated in the positive (AD38 cells) and negative (HepG2 cells) controls from a separate gel. Input protein was normalized and confirmed by expression of α-tubulin. Quantification of HBsAg using image density (right panel) is also shown for the median and SE from five experiments. Statistical significance was determined by two-way ANOVA of results from three separate experiments. (C) HBsAg in AD38 cell monolayers was quantified 3 days after infection with VSV-NLNE (left panel) or mock (right panel). Hoechst staining of nuclei is shown in blue. HBsAg is shown in red. Magnification, ×40. Right panel, quantification of the relative intensity of HBsAg staining using softWoRx software (Applied Precision, Issaquah, WA) following infection with VSV-NLNE or mock infection. symbols). Each symbol represents a separate high-power field from the same experiment. The black lines show the mean ± SE.
DISCUSSION
The mechanism for how HIV coinfection leads to accelerated HBV-related liver disease remains unknown. We developed a novel model to evaluate interactions between HIV infection and HBV replication in vitro. We show here that HIV can infect multiple hepatic cell lines and that following high-level in vitro infection, the major effect on the HBV life cycle was an increase in the intracellular concentration of HBsAg.
Increased intrahepatic HBsAg without a corresponding increase in HBsAg release in the cell culture supernatant could indicate accumulation of HBsAg due to enhanced production or impaired release. Intracellular accumulation of HBsAg has been shown to result from mutations which cause truncation of HBsAg proteins (30). as well as pre-S2 deletion mutants, which appear frequently during chronic HBV infection. The consequent intracellular accumulation of HBV proteins correlates with more rapid disease progression (33). This may relate to pre-S mutations causing increased endoplasmic reticulum stress and hepatocyte apoptosis (29). Pre-S mutations may also lead to the appearance of “ground-glass hepatocytes” which are important histological markers of chronic HBV infection and may be associated with the development of hepatocellular carcinoma (26). Ground-glass hepatocytes are characterized by increased endoplasmic reticulum surrounded by HBsAg particles, giving the cytoplasm a clouded or “glassy” appearance. In addition, intracellular retention of the HBV L protein has also been shown to induce cellular apoptosis and vacuolization in vitro (14) and causes severe liver disease, eventually leading to neoplasia, in transgenic mice (10). Despite the elevated intracellular HBsAg, we did not see morphological changes or evidence of cell toxicity in our model using the MTS assay. It is possible that the morphological changes of ground-glass hepatocytes and cytotoxic effects may not be due to HBsAg alone or that these effects may not be seen in immortalized cell lines.
HBsAg production is dependent on the translation of L HBsAg from the 2.4-kb mRNA transcript and of M and S proteins from the 2.1-kb mRNA (reviewed in reference 21). Expression of these transcripts is dependent on the pre-S and S promoters, respectively. L and S, but not M, are necessary for viral assembly and release. The activity of the S promoter usually greatly exceeds that of the upstream pre-S promoter, so there is usually an excess of S relative to L (32). An increase in production of L relative to S results in retention of L, infective virions, and subviral particles (5). Regulation of HBsAg production can occur via pre-S and S promoters (27), although regulation at the translational level has also been described (24). Although we found an overall increase in intracellular HBsAg, we found no difference in the relative amounts of L, S, and M proteins, suggesting that HIV altered transcription from the pre-S and S promoters, or translation of the corresponding transcripts, equally.
The AD38 cell line was created by stably transfecting HepG2 cells with a single copy of the cDNA of the pregenomic RNA (pgRNA) of HBV under the control of the tetracycline-responsive cytomegalovirus promoter (18). In this system, pre-S and S mRNAs are expressed from two separate promoters on the viral genome, unlike pgRNA, and are not expected to be regulated by tetracycline. It is still possible that the increase in HBsAg in this model may have been secondary to direct activity of HIV on the tetracycline promoter. However, this is unlikely because a corresponding increase in HBV DNA via increased pgRNA transcription would be expected, and this was not evident. In addition, in other control experiments, VSV-NLNE had no effect on β-galactosidase in AD43 cells, which is also under the control of the same tetracycline-responsive cytomegalovirus promoter (results not shown).
The mechanism(s) by which HIV infection may alter transcription or translation of the HBsAg is unknown. While it has been previously shown that the HBV X protein acts synergistically with the HIV Tat protein to induce HIV replication (15), the effect of this synergy on the HBV life cycle is unknown. It is possible that HIV directly influences transcription of the HBsAg mRNA, similar to the effect of the HBV X protein on HIV LTR transcription (25). Alternatively, there may be competition between HIV and HBV subviral particle secretion. HBV surface proteins and HIV gp160 envelope glycoprotein are membrane-associated proteins that are translated at the endoplasmic reticulum membrane. There may be competition for host cellular machinery associated with secretion via multivesicular bodies such as ESCRT, Nedd4, Vps4B, or ALIX/AIP1, which are all thought to have a role in both HIV and HBV secretion (19, 23). This interaction may specifically affect HBV subviral particle secretion, which is thought to differ from virion secretion (19). Experiments to investigate this interaction will form the basis of future studies. The absence of a direct effect on HBV replication in our studies was interesting, as HIV-HBV coinfection is associated with higher levels of HBV DNA in patients (11). This suggests that factors other than a direct HIV-HBV interaction are contributing to the increased HBV DNA levels in coinfected individuals.
We demonstrated low-level HIV infection of HBV-expressing and nonexpressing hepatic cell lines. This was in contrast to previous studies where high levels of p24 were detected in culture supernatant following infection of hepatic cell lines with HIV (8). Cao and colleagues infected HepG2, Huh7, and Hep3B cells with CXCR4- and CCR5-using strains of HIV and demonstrated highly productive infection independent of CD4 (8). We did not measure p24 activity in our study, but we detected low levels of HIV RT activity in the hepatic cell lines tested using a similar multiplicity of infection. This low level of infection may be related to inefficient viral entry due to low-level surface expression of receptors for HIV.
We were unable to detect expression of CD4, CCR5, or CXCR4 on the surfaces of multiple hepatic cell lines, although we clearly showed using flow cytometry that HIV used both CCR5 and CXCR4 to enter these cell lines. Previous studies using slot blot hybridization have shown that CD4 was not expressed on any of the hepatic cell lines tested (8). Other studies using more sensitive methods such as confocal microscopy and semiquantitative reverse transcriptase PCR have reported the presence of HIV coreceptors CXCR4 and CCR5 on hepatic cell lines where flow cytometry was negative (22). Increased apoptosis in Huh7 cells in the presence of soluble gp120 was blocked by pretreatment with AMD3100, consistent with expression of CXCR4 on Huh7 cells (1). Taking these results together with our findings that HIV infection of AD38 cells could be blocked with either the CXCR4 antagonist AMD3100 or the CCR5 antagonist maraviroc, it is likely that hepatic cell lines express both HIV coreceptors but at levels below or at the limit of detection using flow cytometry. These in vitro data, together with previous in vivo studies demonstrating HIV in liver tissue, imply that HIV and HBV could replicate together in hepatocytes.
Potential limitations of this study include the use of hepatic cell lines which are widely used as a convenient alternative to primary hepatocytes for in vitro studies. It is highly likely that after multiple passages, expression of both surface receptors and other cellular genes is altered. We favored AD38 cells over HepG2.2.15 cells because HepG2.2.15 cells contain a retroviral LTR promoter instead of a cytomegalovirus promoter and therefore would not be an appropriate model for understanding intracellular interactions between HIV and HBV. Both AD38 and HepG2.2.15 cells mirror the HBV life cycle more closely than Hep3B cells, which do not produce infectious virions.
Inclusion of EGFP in the VSV-NLNE was helpful in identifying efficiency of infection, although it is possible that the EGFP itself may have interfered with HBV replication. However, to our knowledge there have been no reports of the effect of EGFP on HBV replication, although there have been reports of cell toxicity related to EGFP in some circumstances (20). In addition, pseudotyped HIV may not represent what happens in vivo, given the likely low level of HIV infection of hepatocytes. However, VSV-pseudotyped viruses have been used previously to understand the pathological effects in other cell types subject to low-level HIV infection, such as astrocytes (13).
In summary, we demonstrated that HIV clearly infects hepatic cell lines but at low levels and that infection is noncytopathic and short lived. In addition, we established a robust in vitro model to examine HIV-HBV interactions within hepatic cell lines using a pseudotyped virus and showed that HIV had a significant effect on increasing intracellular HBsAg but no effect on HBV DNA synthesis. An increase in intracellular HBsAg secondary to direct infection of hepatocytes by HIV may potentially contribute to the adverse effects of HIV on the natural history of HBV infection.
Acknowledgments
This work was supported by a Postgraduate Scholarship from the National Health and Medical Research Council (NHMRC) of Australia (D.M.I.). S.R.L. is an NHMRC Practitioner Fellow.
No conflicts of interest exist.
Footnotes
Published ahead of print on 31 March 2010.
REFERENCES
- 1.Babu, C., K. Suwansrinon, G. Bren, A. Badley, and S. Rizza. 2009. HIV induces TRAIL sensitivity in hepatocytes. PLoS One 4:e4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacsi, A., P. Ebbeson, J. Szabo, Z. Beck, I. Andirko, E. Csoma, and F. Toth. 2001. Pseudotypes of vesicular stomatitis virus-bearing envelope antigens of certain HIV-1 strains permissively infect human syncytiotrophoblasts cultured in vitro: implications for in vivo infection of syncytiotrophoblasts by cell-free HIV-1. J. Med. Virol. 64:387-397. [DOI] [PubMed] [Google Scholar]
- 3.Blackard, J., and K. Sherman. 2008. HCV/HIV co-infection: time to re-evaluate the role of HIV in the liver? J. Viral Hepat. 15:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock, C. T., H. L. Tillmann, J. Torresi, J. Klempnauer, S. Locarnini, M. P. Manns, and C. Trautwein. 2002. Selection of hepatitis B virus polymerase mutants with enhanced replication by lamivudine treatment after liver transplantation. Gastroenterology 122:264-273. [DOI] [PubMed] [Google Scholar]
- 5.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. U. S. A. 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron, P., P. Freudenthal, J. Barker, S. Gezelter, K. Inaba, and R. Steinman. 1992. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 257:383-387. [DOI] [PubMed] [Google Scholar]
- 7.Cao, Y., D. Dieterich, P. Thomas, Y. Huang, M. Mirabile, and D. Ho. 1992. Identification and quantitation of HIV-1 in the liver of patients with AIDS. AIDS 6:65-70. [DOI] [PubMed] [Google Scholar]
- 8.Cao, Y., A. Friedman-Kien, Y. Huang, X. Li, M. Mirabile, T. Moudgil, D. Zucker-Franklin, and D. Ho. 1990. CD4-independent, productive human immunodeficiency virus type 1 infection of hepatoma cell lines in vitro. J. Virol. 64:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, J. J., F. Wightman, A. Bartholomeusz, A. Ayres, S. J. Kent, J. Sasadeusz, and S. R. Lewin. 2005. Reduced hepatitis B virus (HBV)-specific CD4+ T-cell responses in human immunodeficiency virus type 1-HBV-coinfected individuals receiving HBV-active antiretroviral therapy. J. Virol. 79:3038-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chisari, F. V., K. Klopchin, T. Moriyama, C. Pasquinelli, H. A. Dunsford, S. Sell, C. A. Pinkert, R. L. Brinster, and R. D. Palmiter. 1989. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell 59:1145-1156. [DOI] [PubMed] [Google Scholar]
- 11.Colin, J. F., D. Cazals-Hatem, M. A. Loriot, M. Martinot-Peignoux, B. N. Pham, A. Auperin, C. Degott, J. P. Benhamou, S. Erlinger, D. Valla, and P. Marcellin. 1999. Influence of human immunodeficiency virus infection on chronic hepatitis B in homosexual men. Hepatology 29:1306-1310. [DOI] [PubMed] [Google Scholar]
- 12.Donzella, G., D. Schols, S. Lin, J. Este, K. Nagashima, P. Maddon, G. Allaway, T. Sakmar, G. Henson, E. De Clercq, and J. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 13.Dou, H., J. Morehead, J. Bradley, S. Gorantla, B. Ellison, J. Kingsley, L. Smith, W. Chao, G. Bentsman, D. Volsky, and H. Gendelman. 2006. Neuropathologic and neuroinflammatory activities of HIV-1-infected human astrocytes in murine brain. Glia 54:81-93. [DOI] [PubMed] [Google Scholar]
- 14.Foo, N. C., B. Y. Ahn, X. Ma, W. Hyun, and T. S. Yen. 2002. Cellular vacuolization and apoptosis induced by hepatitis B virus large surface protein. Hepatology 36:1400-1407. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Gonzalo, M., M. Carretero, J. Rullas, E. Lara-Pezzi, J. Aramburu, B. Berkhout, J. Alcami, and M. Lopez-Cabrera. 2001. The hepatitis B virus X protein induces HIV-1 replication and transcription in synergy with T-cell activation signals: functional roles of NF-kappaB/NF-AT and SP1-binding sites in the HIV-1 long terminal repeat promoter. J. Biol. Chem. 276:35435-35443. [DOI] [PubMed] [Google Scholar]
- 16.Gripon, P., C. Diot, A. Corlu, and C. Guguen-Guillouzo. 1989. Regulation by dimethylsulfoxide, insulin, and corticosteroids of hepatitis B virus replication. J. Med. Virol. 28:193-199. [DOI] [PubMed] [Google Scholar]
- 17.Jones, K. L., S. Sonza, and J. Mak. 2008. Primary T-lymphocytes rescue the replication of HIV-1 DIS RNA mutants in part by facilitating reverse transcription. Nucleic Acids Res. 36:1578-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladner, S., M. Otto, S. Barker, K. Zaifert, G.-H. Wang, J. Guo, C. Seeger, and R. King. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 41:1715-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert, C., T. Doring, and R. Prange. 2007. Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma2-adaptin. J. Virol. 81:9050-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, H.-S., M.-S. Jan, C.-K. Chou, P.-H. Chen, and N.-J. Ke. 1999. Is green fluorescent protein toxic to the living cells? Biochem. Biophys. Res. Commun. 260:712-717. [DOI] [PubMed] [Google Scholar]
- 21.Locarnini, S. 2004. Molecular virology of hepatitis B virus. Semin. Liver Dis. 24:3-10. [DOI] [PubMed] [Google Scholar]
- 22.Munshi, N., A. Balasubramanian, M. Koziel, R. K. Ganju, and J. E. Groopman. 2003. Hepatitis C and human immunodeficiency virus envelope proteins cooperatively induce hepatocytic apoptosis via an innocent bystander mechanism. J. Infect. Dis. 188:1192-1204. [DOI] [PubMed] [Google Scholar]
- 23.Popov, S., E. Popova, M. Inoue, and G. Gottlinger. 2008. Human immunodeficiency virus type 1 gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 82:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price, P., A. Mohamed, A. Zelent, A. Neurath, and G. Acs. 1988. Translational selection in the expression of the hepatitis B virus envelope proteins. DNA 7:417-422. [DOI] [PubMed] [Google Scholar]
- 25.Seto, E., T. B. Yen, B. Peterlin, and J.-H. Ou. 1988. Trans-activation of the human immunodeficiency virus long terminal repeat by the hepatitis B virus X protein. Proc. Natl. Acad. Sci. U. S. A. 85:8286-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su, I.-J., H.-C. Wang, H.-C. Wu, and W.-Y. Huang. 2008. Ground glass hepatocytes contain pre-S mutants and represent preneoplastic lesions in chronic hepatitis B virus infection. J. Gastroenterol. Hepatol. 23:1169-1174. [DOI] [PubMed] [Google Scholar]
- 27.Tacke, F., C. Liedtke, S. Bocklage, M. Manns, and C. Trautwein. 2005. CREB/PKA sensitive signalling pathways activate and maintain expression levels of the hepatitis B virus pre-S2/S promoter. Gut 54:1309-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thio, C., E. Seaberg, R. J. Skolasky, J. Phair, B. Visscher, A. Munoz, D. Thomas, and the Multicenter AIDS Cohort Study. 2002. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS). Lancet 360:1921-1926. [DOI] [PubMed] [Google Scholar]
- 29.Wang, H.-C., H.-C. Wu, C.-F. Chen, N. Fausto, H.-Y. Lei, and I.-J. Su. 2003. Different types of ground glass hepatocytes in chronic Hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am. J. Pathol. 163:2441-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warner, N., and S. Locarnini. 2008. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology 48:88-98. [DOI] [PubMed] [Google Scholar]
- 31.Weber, J., J. Weberova, M. Carobene, M. Mirza, J. Martinez-Picado, P. Kazanjian, and M. Quinones-Mateu. 2006. Use of a novel assay based on intact recombinant viruses expressing green (EGFP) or red (DsRed2) fluorescent proteins to examine the contribution of pol and env genes to overall HIV-1 replicative fitness. J. Virol. Methods 136:102-117. [DOI] [PubMed] [Google Scholar]
- 32.Xu, A., G. Jensen, and T. B. Yen. 1997. Activation of hepatitis B virus S promoter by the large surface protein via induction of stress in the endoplasmic reticulum. J. Virol. 71:7387-7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, Z., and T. S. Yen. 1996. Intracellular retention of surface protein by a hepatitis B virus mutant that releases virion particles. J. Virol. 70:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]