Abstract
Experiments in cell-free systems have demonstrated that the VP5* cleavage fragment of the rotavirus spike protein, VP4, undergoes a foldback rearrangement that translocates three clustered hydrophobic loops from one end of the molecule to the other. This conformational change resembles the foldback rearrangements of enveloped virus fusion proteins. By recoating rotavirus subviral particles with recombinant VP4 and VP7, we tested the effects on cell entry of substituting hydrophilic for hydrophobic residues in the clustered VP5* loops. Several of these mutations decreased the infectivity of recoated particles without preventing either recoating or folding back. In particular, the V391D mutant had a diminished capacity to interact with liposomes when triggered to fold back by serial protease digestion in solution, and particles recoated with this mutant VP4 were 10,000-fold less infectious than particles recoated with wild-type VP4. Particles with V391D mutant VP4 attached normally to cells and internalized efficiently, but they failed in the permeabilization step that allows coentry of the toxin α-sarcin. These findings indicate that the hydrophobicity of the VP5* apex is required for membrane disruption during rotavirus cell entry.
Cell entry by nonenveloped viruses requires disruption or perforation of a membrane and translocation of a modified virion or an infectious genome into the cytosol (30). A variety of mechanisms have evolved to carry out these steps. Viruses with double-stranded RNA (dsRNA) genomes, such as rotaviruses and orthoreoviruses, deliver an inner capsid particle to the cytosol of the target cell. The rotavirus inner capsid particle, known as a “double-layered particle” (DLP) because of its two-shell structure (Fig. 1), contains the 11 viral genome segments and the enzymes required for RNA synthesis and capping (13). The DLP remains intact throughout the infection, and new plus-sense RNA strands are made, capped, and extruded from the particle (17, 23). The outer layer of the virion (“triple-layered particle” [TLP]) contains two protein species, VP4 and VP7, which provide the molecular apparatus for cell attachment and membrane penetration.
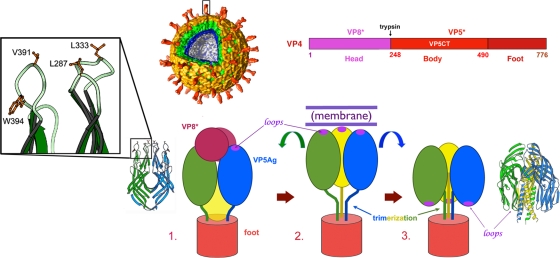
FIG. 1.
Structures and model for conformational rearrangements of VP4. (Top center) Surface rendering from electron cryomicroscopy of a three-dimensional reconstruction of the rotavirus particle. A trypsin-cleaved VP4 spike (red) is boxed. The cutaway shows the multiple layers of the TLP. The VP7 layer is in yellow. The layers of the DLP are in green (VP6) and blue (VP2). (Top right) The VP4 primary structure indicating the boundaries of proteolytic products. (Bottom) Model for VP4 conformational rearrangements accompanying membrane penetration. (Step 1) Trypsin-activated VP4, in a schematic representation of a spike in roughly the orientation of the boxed spike in the rendering of a virion. The VP4 trimer has a 3-fold-symmetric “foot” but an asymmetrically organized projection. The ribbon diagram shows a dimeric form of the VP5 β-barrel domain (or antigen domain), which fits the dimer-clustered “body” of the projection, and the inset shows details of the three conserved hydrophobic loops that cap the β-barrel domain of VP5*. The hydrophobic residues mutated in this study are labeled. (Step 2) Dissociation of VP8* exposes the hydrophobic loops (shown as purple ovals) of VP5*. VP5* extends and engages a target membrane with the hydrophobic loops, probably from all three subunits. (Step 3) VP5* folds back to a stable trimeric structure, represented by the VP5CT crystal structure. This foldback is proposed to drive membrane penetration.
VP4 makes up the “spikes,” which are evident on mature rotavirus particles only after tryptic cleavage of VP4 into fragments VP8* and VP5* (Fig. 1). This cleavage activates virions for efficient infectivity (12). Prior to cleavage, the outer parts of VP4 are probably flexibly linked to the “foot” (10), which is clamped by VP7 onto the underlying DLP. Each spike contains three copies of VP4. The virion-distal part of the spike appears to be dimer clustered and displaced from the local axis; electron cryomicroscopy has shown the foot to be a 3-fold-symmetric trimer (18). This unusual mismatch of symmetries suggests that the spike structure may be metastable and that a suitable trigger may induce it to rearrange further.
Structural analyses of various VP4 domains, of VP7, and of DLPs and TLPs (2, 6, 10, 11, 18, 31, 33), together with biochemical studies of VP7, VP4, and VP4 fragments (8, 9, 28, 29, 32), suggest the model illustrated in Fig. 1. Trypsin-cleaved VP4 forms the spike, in which the “body” regions of two of the three VP5* fragments cluster together; the two associated VP8* fragments cover the hydrophobic tips of these clustered VP5* β-barrel domains (designated in previous papers “antigen domains” [VP5Ag]). All three subunits contribute to the C-terminal foot. VP7, a calcium-stabilized trimer, locks the VP4 foot in place. Dissociation of VP7 (“uncoating”), induced by a lowered calcium concentration, allows VP5* to rearrange further into a symmetrical trimer, with the VP5* antigen domains rotated by roughly 180°, so that their hydrophobic tips point toward the foot. This step probably requires loss of VP8* and formation of a transient extended intermediate.
The properties of recombinant VP4 in solution correlate with the steps of the model described above. Full-length recombinant VP4 is predominantly monomeric in solution (8). Successive cleavages with chymotrypsin and trypsin produce “VP5CT,” a fragment that coincides with VP5* at its N terminus but has lost residues corresponding roughly to the foot at its C terminus (Fig. 1) (8). It is an SDS-resistant trimer that remains associated unless it is heated to 95°C in SDS-PAGE sample buffer (8, 32). Authentic VP5* released from uncoated virions also forms an SDS-resistant trimer (32).
Studies of the properties of VP4 fragments, prepared by proteolysis of monomeric VP4 or by release from virions, provide evidence for a transient, extended intermediate of VP5* (Fig. 1) and for its interaction with synthetic membranes (29). Digestion of recombinant VP4 with chymotrypsin and trypsin in the presence of liposomes leads to membrane association of the resulting VP5CT, but preformed VP5CT does not associate with liposomes added after cleavage and trimerization are complete. Authentic VP5* has similar properties: if released from virions (by chelating Ca2+) in the presence of liposomes, it associates with them, but it does not do so if the liposomes are added after uncoating. In both cases, the lipid bilayer appears to have captured a transient intermediate in the rearrangement to the folded back, trimeric species revealed by the VP5CT crystal structure.
Three loops (designated BC, DE, and FG) form the hydrophobic patch that caps one end of the VP5* β-barrel domain (Fig. 1) (10, 31). The position of this patch makes it a likely candidate to mediate the membrane interactions described above; indeed, the FG loop amino acid sequence resembles that of an alphavirus fusion loop, suggesting that it could insert directly into a lipid bilayer. We report here experiments that test whether the hydrophobicity of the VP5* loops is important for rotavirus infectivity and for membrane association of the trimeric VP5* intermediate. Because sequential addition of recombinant VP4 and VP7 to rotavirus DLPs yields recoated particles (RPs) that are fully infectious after trypsin priming (28), we can incorporate VP4 with mutated hydrophobic residues to examine the progress of such particles along the cell entry pathway. We show that a modification in a VP4 hydrophobic loop reduces infectivity by blocking a membrane permeabilization step that follows cell attachment and endocytic internalization. The results support the proposal that these loops couple a conformational change in VP5* to disruption or perforation of an endosomal membrane.
MATERIALS AND METHODS
Cells.
MA104 cells were grown in Medium 199 (M199) (Invitrogen) supplemented with 7.5% fetal bovine serum (HyClone), 10 mM HEPES, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Sf9 insect cells were grown in spinner flasks in Sf900 II serum-free medium (Invitrogen).
Virus.
Rhesus rotavirus (RRV) (serotype G3, P5B[3]) was propagated in MA104 cells and purified as previously described (28). DLPs were obtained by adding 5 mM EDTA to infected cell lysates prior to particle purification. 35S-labeled rotavirus was prepared by infecting cells in medium containing 85% Dulbecco's modified Eagle's medium (DMEM) Cys− Met− (Invitrogen), 15% M199, 1 μg/ml porcine pancreatic trypsin (Sigma-Aldrich), and 10 μCi/ml 35S-labeled cysteine and methionine (EasyTag EXPRE35S35S Protein Labeling Mix; Perkin Elmer). Unlabeled TLPs from cleared, infected cell lysates were concentrated by ultrafiltration through a polyethersulfone membrane with a 50-kDa molecular mass cutoff in a stirred cell (Amicon). DLPs and 35S-labeled TLPs were concentrated by pelleting them from cleared infected cell lysates at 158,000 × g for 1 h at 4°C in a 45Ti rotor (Beckman-Coulter). The DLPs were stored in TN (20 mM Tris, pH 8.0, 100 mM NaCl) with 0.02% sodium azide. TLPs were stored in TNC (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM CaCl2). DLP concentrations were determined by absorbance at 260 nm with 1 A260 unit equal to 0.213 mg/ml DLPs (28). The TLP concentration was determined by densitometry of VP2 and VP6 bands on Coomassie-stained SDS-PAGE gels and comparison with DLP standards (28). Virus infectivity was determined by infectious-focus assay (28).
Recombinant VP4 and VP7.
Baculoviruses expressing RRV VP4 or VP7 have been described previously (14, 20). We purified recombinant VP4 from infected Sf9 cells by anion-exchange chromatography and size exclusion chromatography, according to a published protocol (8), and stored the protein in TN0.1E (20 mM Tris, pH 8.0, 100 mM NaCl, 0.1 mM EDTA). We purified recombinant VP7 from the medium of infected Sf9 cells by lectin affinity, immunoaffinity, and size exclusion chromatography, according to a published protocol (9), and stored the protein in 0.1× TNC (2 mM Tris, pH 8.0, 10 mM NaCl, 0.1 mM CaCl2).
Mutagenesis.
Point mutants of VP4 were generated from the pFastBac-1 vector (Invitrogen) by PCR with mutagenic primers. The PCR products were flanked with BamHI and NotI restriction sites and ligated into a suitably digested pFastBac-1 vector. After isolation of clones from Escherichia coli DH5α, we verified the DNA sequences of all constructs across the entire VP4 coding region.
Production of VP5CT.
VP5CT was produced from purified recombinant VP4 as described previously (8). In brief, 10 μg of VP4 at a concentration of 1 mg/ml in TN0.1E was digested with TLCK (1-chloro-3-tosylamido-7-amino-2-heptanone)-treated chymotrypsin (Worthington Biochemical) at 4.2 μg/ml for 30 min at 37°C. Following a 10-min incubation on ice, the reaction mixture was further digested with TPCK [l-(tosylamido-2-phenyl) chloromethyl ketone]-treated trypsin (Worthington Biochemical) at 3.6 μg/ml for 1 h at room temperature (RT). Proteolysis was quenched by the addition of phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich) to 1 mM and incubation for at least 10 min on ice. For SDS-PAGE, half of the sample was heated to 95°C in 1× reducing SDS sample buffer and the other half was incubated in 1× reducing SDS sample buffer at RT.
Liposome association of VP5CT and VP5*.
Liposome association reactions were performed as described previously (29). In brief, phosphatidylcholine (PC) (from chicken eggs; Avanti Polar Lipids) liposomes were formed in HN (20 mM HEPES, pH 7.3, 140 mM NaCl) at a concentration of 10 mg/ml. To assay for liposome association of VP4 cleaved with chymotrypsin and trypsin, 10 μg of purified recombinant VP4 was mixed with 12.5 μl of liposomes in a total volume of 150 μl HN and digested with 1 μg/ml chymotrypsin for 30 min at 37°C, followed by digestion with 1 μg/ml trypsin for an additional 30 min at 37°C. PMSF was then added to 1 mM to quench proteolysis. For the sucrose gradient, the reaction mixture was mixed with 200 μl of 70% (wt/vol) sucrose to yield 350 μl of a 40% solution. The 40% sucrose solution was overlaid with 1 ml of 25% (wt/vol) sucrose and 150 μl of 5% (wt/vol) sucrose and centrifuged at 180,000 × g in an SW-55 rotor (Beckman-Coulter) for 2.5 h at 4°C. Fractions were collected from the top with a wide-bore pipette tip and stored at −80°C. To assay for liposome association of VP5*, 10 μg of trypsin-activated recoated particles was mixed with 12.5 μl of liposomes and brought to a total volume of 150 μl with HN buffer. EDTA was added to 1 mM, and the reaction mixture was incubated for 1 h at 37°C to induce uncoating. After uncoating, 1 mM PMSF was added to block proteolysis of VP5*. Separation on the sucrose gradient and fractionation were performed as described above. All sucrose solutions were made in HN buffer containing 1 mM EDTA.
Recoating.
DLPs were recoated with VP4 and VP7 essentially as described previously (28), with the following modification. To achieve a concentration of 1 mg/ml of VP4 during recoating, 1.5 μg of DLPs in 3 μl of TN with 0.02% sodium azide was mixed with 20 μg of VP4 (corresponding to 48.6 molecules for each binding site) in 10 μl of TN0.1E. The solution was adjusted to pH 5.2 with 1.44 μl (0.1 volume) of 0.5 M sodium acetate and incubated for 1 h at RT (approximately 20°C). VP7 was then added in approximately 5-fold molar excess (3 μg in 3 μl 0.1× TNC, 2 μl calcium buffer [20 mM Tris, pH 8.0, 10 mM CaCl2], and 0.56 μl 0.5 M sodium acetate), and the reaction mixture was incubated for an additional 1 h at RT. To match wild-type VP4 (VP4WT) and VP4L333D occupancy, less VP4WT was used. VP4WT concentrations in these reaction mixtures ranged from 0.08 to 1.0 mg/ml, but the total reaction volume remained the same. The products of all recoating reactions were trypsin activated, as described previously (28).
Immunoblotting.
Samples were reduced in 5% β-mercaptoethanol at RT or 95°C, separated on 10% SDS-PAGE gels, and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories). VP4 and VP5* were detected by monoclonal antibody (MAb) HS2 (22), VP4, serially digested with chymotrypsin and trypsin, was detected by MAb 4D8 (32), followed by peroxidase-conjugated goat anti-mouse IgG (Kirkegaard and Perry Laboratories, Inc.). All immunoblots were developed using a chemiluminescent peroxidase substrate (ECL Plus; GE Healthcare) and exposure to film.
Virus binding to cells.
35S-labeled TLPs or RPs (∼5 × 104 particles/cell) in cold binding buffer (M199, 1% bovine serum albumin) were added to prechilled confluent monolayers of MA104 cells in 48-well plates with a final well volume of 200 μl and incubated for 1 h at 4°C with continuous rocking. The cells were then washed with cold TNC and lysed in 1% SDS for 30 min at 4°C. The lysates were mixed with Opti-Fluor scintillation fluid (Perkin Elmer), and the amount of virus that remained bound was determined by liquid scintillation counting. Binding was expressed as the percentage of input virus that remained associated with the cells after they were washed. In some samples, cells were pretreated with 200 mU/ml of Vibrio cholerae neuraminidase (Sigma-Aldrich) for 1 h at 37°C and washed in cold TNC before labeled virus was added. In other samples, virus was preincubated with MAb 7A12 (1 mg/ml) (25) for 1 h at 37°C before being added to cells.
Virus internalization.
35S-labeled RPs (∼5 × 104 particles/cell) were added in cold binding buffer (see above) to prechilled confluent monolayers of MA104 cells in 24-well plates with a final well volume of 0.5 ml for 1 h at 4°C with continuous rocking. The cells were then washed with cold M199 to remove unbound virus, overlaid with M199 prewarmed to 37°C, and incubated at 37°C for 45 min to allow internalization. The cells were then washed with cold digestion buffer (M199, 7 mM EDTA) to remove unattached virus and digested with proteinase K (0.5 mg/ml in digestion buffer) for 45 min at 4°C. Proteolysis was quenched by adding 1 mM PMSF (diluted from a 100 mM stock in ethanol) for 10 min at 4°C. Following digestion and quenching, the cells were pelleted in a microcentrifuge at 1,500 rpm for 5 min at 4°C. The pelleted cells were washed once with cold M199 and lysed in 1% Triton X-100 for 30 min at 4°C. The lysates were mixed with Opti-Fluor scintillation fluid, and the amount of cell-associated virus remaining was determined by liquid scintillation counting. For the 0-min time point, cells were not warmed to 37°C but were otherwise processed as described above. Parallel samples at each time point that were not treated with proteinase K were assayed to determine total amounts of cell-associated virus. Internalization is expressed as the percentage of bound virus at each time point that remained cell associated after proteinase K treatment.
α-Sarcin coentry.
The α-sarcin coentry assay was performed as described previously (19). Confluent layers of MA104 cells in 96-well plates were incubated in DMEM Cys− Met− for 1.5 h at 37°C. TLPs, RPs, DLPs (∼5 × 104 particles/cell), or no particles were added with or without α-sarcin from Aspergillus giganteus (100 μg/ml; Sigma-Aldrich) in DMEM Cys− Met− for 1 h at 37°C. The cells were washed and transferred to DMEM containing 35S-labeled cysteine and methionine (1 μCi/ml; EasyTag EXPRE35S35S Protein Labeling Mix;, Perkin Elmer) for 1 h at 37°C. The cells were washed again, and proteins were precipitated by the addition of 10% trichloroacetic acid (TCA) to the wells for 1 h at 4°C. Following precipitation, the wells were washed twice with ethanol, air dried, and dissolved overnight with 0.5% SDS in 0.1 M NaOH at room temperature. The lysates were mixed with Opti-Fluor scintillation fluid to measure radiolabel incorporation by liquid scintillation counting. The label incorporation in uninfected cells that were not treated with α-sarcin was set as 100%.
RESULTS
Design, expression, and purification of mutant VP4s.
We designed four mutant RRV VP4s. In each, a conserved hydrophobic residue in one of the three hydrophobic loops at the apex of the VP5* body domain was replaced by a residue with a charged side chain (Fig. 1, box). Aspartic acid replaced L287 at the tip of the BC loop, L333 at the tip of the DE loop, and V391 at the tip of the FG loop. These leucine and valine side chains are solvent exposed and do not contribute to hydrophobic cores. We therefore anticipated that replacement with a charged residue would not cause misfolding. W394 on the side of the FG loop makes a hydrogen bond to the carbonyl of P390 on the antiparallel strand of the loop, stabilizing the local conformation. As a replacement for this tryptophan, we chose glutamine, which might also donate a hydrogen bond to the P390 carbonyl, substituting for the stabilizing effect of the hydrogen bond donated by tryptophan while eliminating a prominently exposed aromatic group.
We expressed the four mutant VP4s in recombinant baculovirus-infected Sf9 insect cells and purified them by the protocol used to prepare VP4WT (see Materials and Methods). The properties of the mutant proteins on anion-exchange and size exclusion chromatography were similar to those of VP4WT, indicating that the folding and stability of the mutant and wild-type VP4s are similar (data not shown).
Rearrangement of mutant VP4s after protease digestion in solution.
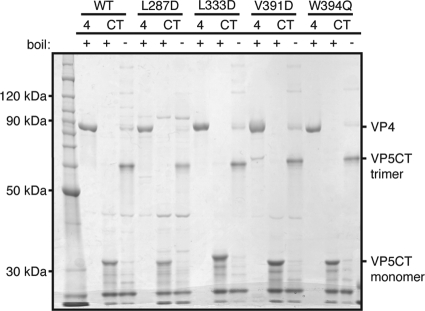
The formation of SDS-resistant VP5CT trimers by serial digestion of recombinant, soluble VP4 with chymotrypsin and trypsin provides an assay for protease-triggered rearrangement of soluble VP4 (8, 28). After this serial protease digestion, VP5CTL287D, VP5CTL333D, VP5CTV391D, and VP5CTW394Q all migrated more slowly in SDS-PAGE of unheated samples than in SDS-PAGE of heated samples, and all comigrated with VP5CTWT prepared similarly (Fig. 2). This electrophoretic behavior suggests that the cleavage fragments of the mutant VP4s had rearranged into the umbrella-shaped trimer seen in the X-ray crystal structure of VP5CTWT (8, 10). The data indicate that the mutations at the apex of the VP5* antigen domain do not affect the oligomerization or foldback rearrangement triggered by serial protease digestion of VP4 in solution.
FIG. 2.
VP5CT formation by VP4 mutants. VP4 mutants serially digested with chymotrypsin and trypsin (CT) were mixed with reducing SDS-PAGE sample buffer and either heated to 95°C (+) or incubated at RT (−) before separation by SDS-PAGE and Coomassie staining. Purified VP4 (1 μg), heated and reduced, was loaded in lanes 4. A protein molecular mass ladder was loaded in the left lane.
Liposome association of mutant VP4s after protease digestion in solution.
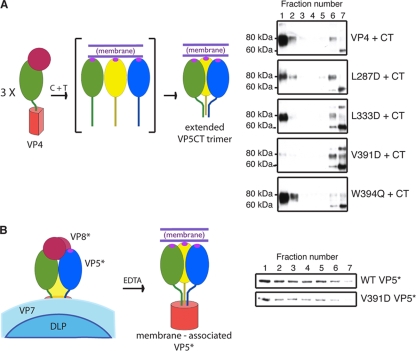
Serial chymotrypsin and trypsin digestion of VP4WT in the presence of liposomes yields liposome-associated VP5CTWT, as assayed by cosedimentation on a sucrose gradient (29). When digested in the presence of liposomes, each of the mutant VP4s produced a VP5CT-like molecule that was recognized by the trimer-specific MAb 4D8 (32) in immunoblots of unheated samples from a sucrose gradient separation of the digestion products (Fig. 3 A, right). VP5CTW394Q associated with the liposomes to the same degree as VPCTWT (as indicated by colocalization in the least dense fraction of the gradient [Fig. 3A, right, top and bottom]), but very little VP5CTV391D (Fig. 3A, right, fourth from top) and intermediate amounts of VP5CTL287D and VP5CTL333D (Fig. 3A, right, second and third from top) associated with the liposomes. The data indicate that the V391D mutation, although it does not block trimerization and the foldback rearrangement, does inhibit the interaction of VP5CTV391D with membranes and that the L287D and L333D mutations probably also reduce this interaction.
FIG. 3.
Liposome interaction of VP4 loop mutants. (A) Liposome interaction of VP4 mutants serially digested with chymotrypsin and trypsin. (Left) Schematic of the digestion reaction showing the starting reactants and the putative liposome-interacting product. (Right) VP4 was serially digested in the presence of liposomes, and the mixtures were separated on discontinuous sucrose gradients (29). Samples of gradient fractions from top (lane 1) to bottom (lane 7) were not heated before separation by SDS-PAGE and immunoblotting with MAb 4D8 to detect VP5CT. (B) Liposome interaction of V391D VP5*. (Left) Schematic of the uncoating reaction showing the starting reactants and the putative liposome-interacting product. (Right) Purified RPs were uncoated in the presence of liposomes by adding 1 mM EDTA, and the mixtures were separated over discontinuous sucrose gradients. Samples of gradient fractions from top (lane 1) to bottom (lane 7) were heated prior to separation by SDS-PAGE and immunoblotting with MAb HS2 to detect VP5*.
In the 4D8 immunoblots, the liposome-associated VP5CT from each VP4 (wild type or mutant) had an apparent mass of 80 kDa; much of the remainder, like VP5CT, produced by serial digestion in the absence of liposomes (not shown), had an apparent mass of 60 kDa. Insufficient quantities of the pure 80-kDa species were obtained to understand the basis for its slower migration.
Efficiency of recoating DLPs with mutant VP4s.
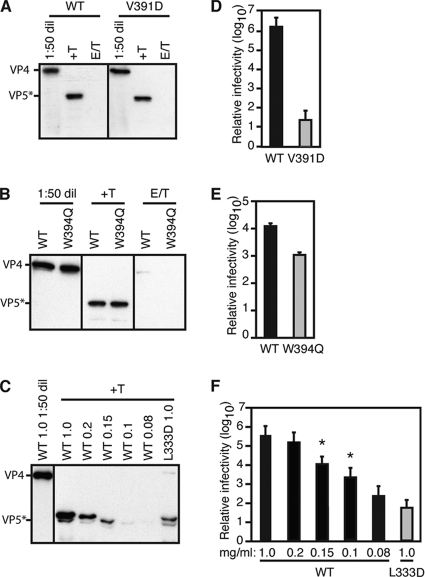
We recoated rotavirus DLPs with recombinant wild-type VP7 and wild-type or mutant VP4 (Fig. 4 A to C) using a previously described technique (28). The efficiency of proper VP4 incorporation can be determined by digesting RPs with trypsin, which cleaves assembled VP4 selectively to VP8* and VP5* but degrades unassembled VP4 more extensively (8, 28). Thus, immunoblots of trypsin-treated RPs using antibodies specific for VP5* were used to assess the amount of VP4 correctly assembled into particles. Because the efficiency of in vitro VP4 assembly depends strongly on its concentration during recoating (28), we chose VP4 concentrations at which wild-type and mutant VP4s had comparable occupancies on RPs. Recoating with 1 mg/ml VP4V391D or VP4W394Q yielded particles with VP4 occupancy equivalent to that obtained by recoating with 1 mg/ml VP4WT (Fig. 4A and B). Incorporation of VP4L333D was less efficient. Recoating with 1 mg/ml VP4L333D yielded RPs with a VP4 occupancy equivalent to those from recoating reactions with 0.1 to 0.15 mg/ml VP4WT (Fig. 4C). Therefore, both of these lower-occupancy wild-type RPs were used as controls for L333D RPs in subsequent experiments. Recoating with VP4L287D was too inefficient for further characterization of L287D RPs.
FIG. 4.
VP4 incorporation into and infectivity of particles recoated with loop mutants of VP4. (A to C) Trypsin cleavage of assembled VP4 on RPs containing VP4V391D (A), VP4W394Q (B), and VP4L333D (C). Before separation of equivalent volumes by SDS-PAGE, the recoating products were digested with 5 μg/ml trypsin in 1 mM calcium (+T) or 5 mM EDTA (to uncoat the particles) (E/T) or were not digested but were diluted 1:50 (1:50 dil). VP4 and VP5* bands were detected by immunoblotting with MAb HS2. No VP4 or VP5* bands were detected after the VP4L333D recoating reaction products were incubated with EDTA and digested with trypsin (not shown). In panels C and F, VP4L333D concentrations (mg/ml) during VP4 recoating are indicated. (D to F) Infectivities of particles recoated with VP4V391D (D), VP4W394Q (E), and VP4L333D (F) and then primed with trypsin. Infectivity is reported relative to that of DLPs. In panel F, the recoating VP4WT concentrations at which VP4WT occupancy was equivalent to VP4L333D occupancy are indicated by an asterisk above each bar. The error bars represent standard deviations.
Infectivity of RPs with mutant VP4s.
Analysis of recoated particles allowed correlation of the biochemical behavior of the VP4 mutants in cell-free systems with the behavior of rotavirus particles bearing these altered VP4s during infection of cells. We tested the infectivity of the VP4 mutant RPs by infectious-focus assays in MA104 cells. RPs with mutant VP4s were substantially less infectious than wild-type RPs (Fig. 4D to F). The infectivity of VP4V391D RPs was reduced by a factor of 104 to 105 relative to the wild type; the infectivities of VP4L333D and VP4W394D RPs were reduced at least 10-fold each. The effect of the VP4 mutations on infectivity suggests that the hydrophobicity of the VP5* loops is important for viral replication, most likely for a step in the cell entry pathway. The V391D mutation caused the greatest reduction in liposome binding upon serial digestion of VP4 in solution and also caused the greatest reduction of infectivity, suggesting a common mechanism for the phenomena. Because VP4V391D assembled efficiently into RPs and produced the greatest reduction in infectivity, we chose the corresponding mutant RP for further characterization.
Liposome association of mutant VP5*s released from uncoating RPs.
VP5*WT, released from virions in the presence of liposomes by EDTA-triggered uncoating, associates with the liposomes (29), as does VP5*V391D released from RPs in the presence of liposomes (Fig. 3B). Thus, the V391D mutation, which prevents liposome association of VP5CTV391D during serial proteolysis of soluble VP4V391D (Fig. 3A), does not prevent membrane binding when intact VP5*V391D is released from uncoating particles.
The different oligomeric states of released VP5* and VP5CT during proteolytic processing may account for their different membrane association properties. VP5* on virions is a trimer, stabilized by contacts in the foot domain, which is absent from VP5CT. If the foot domain does not unfold, VP5* may retain its oligomeric state when released from virions by EDTA. VP5CT is a trimer stabilized by a coiled coil, which forms only after the serial proteolysis that generates VP5CT from monomeric VP4. Thus, VP5CT must undergo a monomer-to-trimer conversion during its formation (8, 10). Liposome association of intermediate (probably monomeric) states of VP5CT may be more sensitive to mutation of the hydrophobic loops than association of released (probably trimeric) VP5*.
Attachment of VP4V391D RPs to cells.
We measured particle attachment to cells by incubating confluent MA104 cell monolayers for 1 h at 4°C with 35S-labeled purified authentic virions (TLPs) or with RPs prepared by recoating 35S-labeled DLPs with VP4WT or VP4V391D. The cells were washed and lysed, and the amount of bound label was determined by scintillation counting. VP4WT RPs and VP4V391D RPs bound MA104 cells with similar efficiencies (11 to 12% of input counts bound), TLPs bound cells less efficiently (5 to 6% of input counts bound), and DLPs did not bind cells appreciably (Fig. 5 A). The diminished attachment of purified TLPs relative to RPs was proportional to the lower specific infectivity of purified TLPs. Comparison of particle infectivity to the estimated particle count (see Materials and Methods) yielded a specific infectivity of 2,000 TLPs per focus-forming unit (FFU) and 1,000 VP4WT RPs per FFU. The presence of 35S in the outer capsid of TLPs, but not of RPs, may also contribute to the difference in the measured binding of TLPs and RPs by this assay.
FIG. 5.
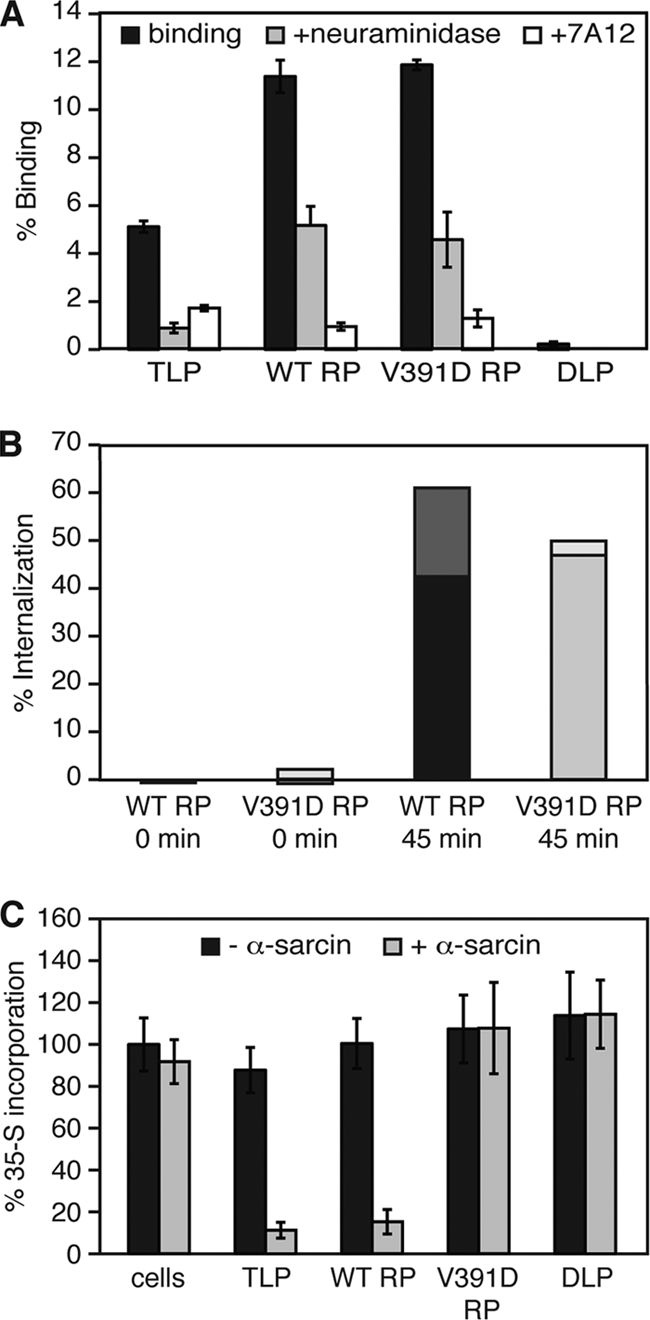
VP4V391D RP binding, internalization, and membrane permeabilization during cell entry. (A) VP4V391D RP binding to cells. 35S-labeled TLPs or RPs (∼5 × 104 particles/cell) were bound to confluent MA104 cell monolayers for 1 h at 4°C. After washing and cell lysis in 1% SDS, the amount of bound virus was quantitated by liquid scintillation. Cells were pretreated with 200 mU/ml of V. cholerae neuraminidase for 1 h at 37°C before the addition of labeled virus (+neuraminidase), or particles were preincubated with MAb 7A12 (1 mg/ml) for 1 h at 37°C before being added to the cells (+7A12). The radioactivity detected from input particles was set as 100%. The error bars represent standard deviations. (B) Internalization of VP4V391D RPs. 35S-labeled VP4WT RPs or VP4V391D RPs (∼5 × 104 particles/cell) were incubated with MA104 cell monolayers for 1 h at 4°C and washed. Some monolayers were then digested with 0.5 mg/ml proteinase K (0 min); others were incubated at 37°C for an additional 45 min before proteinase K digestion (45 min). The cells were lysed in 1% Triton X-100, and the associated label was quantitated by liquid scintillation. At each time point, the signal from parallel samples not digested with proteinase K was set as 100%. Values for duplicate trials are indicated by the dual shading of each bar. (C) α-Sarcin coentry. TLPs, VP4WT RPs, VP4V391D RPs, or DLPs (∼5 × 104 particles/cell) or no particles, with or without α-sarcin (100 μg/ml), were incubated with confluent MA104 cell monolayers for 1 h at 37°C. Inocula were removed, and the cells were washed and then incubated in medium containing 35S-labeled Cys/Met (1 μCi/ml) for 1 h at 37°C. Following washing and TCA precipitation, radiolabel incorporation was quantitated by liquid scintillation counting. The amount of label incorporation in cells that were not exposed to α-sarcin or particles was set as 100%. The error bars represent standard deviations.
We tested whether the measured attachment was mediated by VP8* binding to cell surface sialosides, as expected for RRV TLPs and RPs (25). Before binding, cells were incubated with neuraminidase (removing sialic acid) or particles were incubated with the VP8*-specific MAb 7A12, which neutralizes by blocking attachment (25). Neuraminidase treatment of cells reduced binding of TLPs, VP4WT RPs, and VP4V391D RPs to 20%, 45%, and 40% of control levels, respectively; preincubation of these particles with 7A12 reduced particle binding to 35%, 10%, and 10% of control levels, respectively (Fig. 5A). We conclude that the V391D mutation in VP4 inhibits RP infectivity by blocking a postattachment step in cell entry and that most of the particles attach to MA104 cells through VP8* interactions with cell surface sialosides.
Internalization of VP4V391D RPs.
We assayed the internalization of bound 35S-labeled particles by measuring the label that remained cell associated after proteinase K digestion of particles exposed on the surfaces of intact MA104 cells (Fig. 5B). Less than 1% of the label in VP4WT RPs or VP4V391D RPs remained cell associated following incubation with cells for 1 h at 4°C to allow attachment, washing to remove unbound particles, and addition of proteinase K. If the cells were incubated for an additional 45 min at 37°C to allow internalization before proteinase K was added, approximately 50% of the bound label in either VP4WT RPs or VP4V391D RPs remained cell associated. These results indicate that wild-type and mutant RPs are both internalized efficiently. We conclude that the VP4 V391D mutation inhibits infectivity by blocking a postinternalization step, assuming that most rotavirus particles are internalized by a potentially productive entry pathway.
Coentry of α-sarcin with VP4V391D RPs.
The 17-kDa fungal protein toxin α-sarcin irreversibly inhibits protein synthesis but cannot cross intact cellular membranes (4). When added to cells together with rotavirus, the toxin gains access to the cytosol, presumably because the virus perforates the membrane of a compartment that also contains the toxin (19). MA104 cells were incubated with α-sarcin alone or together with TLPs, DLPs, VP4WT RPs, or VP4V391D RPs for 1 h at 37°C. The cells were washed and then incubated for 1 h in medium containing [35S]methionine and [35S]cysteine, but neither toxin nor particles. Cellular protein synthesis was assayed by the amount of radiolabel incorporated into TCA-precipitable material. When α-sarcin was added to cells together with TLPs or VP4WT RPs, incorporation of label was reduced to less than 20% of the levels we measured when α-sarcin was absent, when α-sarcin was added alone to cells, or when α-sarcin was added to cells together with DLPs or VP4V391D RPs (Fig. 5C). These results show that TLPs and VP4WT RPs mediate coentry of α-sarcin but DLPs and VP4V391D RPs do not. We conclude that the V391D mutation reduces infectivity by blocking permeabilization of an internal membrane or a step before membrane permeabilization but after attachment and internalization.
DISCUSSION
Our previous work in cell-free systems provided strong evidence that VP4 and its fragments undergo a series of conformational changes that resemble the fusogenic rearrangements of enveloped virus fusion proteins. If liposomes are present during the rearrangement of VP5* from the primed spike on virions to folded back, tightly associated trimers, VP5* binds lipid bilayers (29). The structural elements most likely to engage a target membrane are three hydrophobic loops at one end of the VP5* β-barrel domain. By mutating specific residues in these loops and incorporating the mutant VP4 into recoated particles, we have shown that their hydrophobicity is important, both for lipid binding and for viral infectivity. Thus, we have been able to link biochemical and structural findings in cell-free systems with events during entry into cells.
None of the loop mutations prevent the VP5CT fragment from undergoing the foldback rearrangement when soluble VP4 is serially digested with chymotrypsin and trypsin (Fig. 2). A BC loop mutation (L287D) prevents VP4 from recoating DLPs, a DE loop mutation (L333D) greatly decreases recoating efficiency, and two FG loop mutations (V391D and W394Q) decrease recoating efficiency slightly (Fig. 4A to C). In the cleaved spike conformation modeled by the dimeric form of VP5Ag (31), the BC and DE loops appear to be buried by the VP8* heads, and the FG loop is exposed, so that mutations in the FG loop are less likely than mutations in the other loops to interfere with intramolecular interactions. Structural data on the conformation of uncleaved VP4 would be needed to explain more reliably the differential effects of the mutations on recoating efficiency.
RPs bearing VP4V391D have greatly diminished infectivity; RPs bearing the other VP4 mutants that are competent to recoat have less pronounced infectivity defects (Fig. 4D to F). Analysis of entry by VP4V391D RPs revealed no defect in cell attachment or internalization (Fig. 5A and B). The mutant particles were defective for coentry of α-sarcin, however, consistent with the interpretation that a membrane penetration defect causes the infectivity defect (Fig. 4D and 5C).
Mutations at the tips of each of the three loops interfere with liposome binding of VP5CT, the fragment generated by cleavage of soluble VP4; the V391D mutation shows the greatest reduction in lipid binding relative to the WT (Fig. 3A). In contrast, full-length VP5* with the V391D mutation, when released from recoated particles, bound liposomes at levels similar to released WT VP5*. Thus, the membrane interaction defect associated with the V391D mutation is partial (Fig. 3B). In the case of released VP5*, avidity effects may allow more stable binding of the V391D mutant than in the case of VP5CT. VP5* is probably released as a trimer, with the foot domain determining trimer stability. VP5CT lacks the foot domain. It must undergo a monomer-to-trimer transition during its formation by cleavage of VP4. Only the transient, extended intermediate is likely to associate readily with a bilayer (29). We suggest that the extended intermediate produced during rearrangement of cleaved VP4 in solution is a monomer and that the intermediate produced during release of VP5* from virions is a trimer, accounting for the observed difference in liposome association. The correspondence between the degree of lipid binding and infectivity is not precise. VP5CT from VP4W394Q interacted with liposomes as well as did VP5CT from VP4WT, but the mutation led to a >10-fold decrease in viral infectivity (Fig. 3A and 4E). Nevertheless, the observation that the V391D mutation causes the greatest liposome binding defect and the greatest infectivity defect suggests that a strong interaction with lipids is critical for downstream events leading to eventual perforation or penetration of the cell membrane.
The structure of the hydrophobic end of VP5Ag suggests that the loops insert only partly into the outer leaflet of the bilayer. Just proximal to the hydrophobic side chains at the tips of the BC, DE, and FG loops are charged resides, such as Q393 (adjacent to V391 and G392). Structural studies of flavivirus E glycoproteins suggest that residues such as a tryptophan at the tips of their fusion loops insert at the interface between the polar head groups and the aliphatic fatty acyl chains of a lipid bilayer (21). The hydrophobic loops of VP5* probably penetrate to a similar extent, and in any case no more than one leaflet deep into the target membrane. The three loops cluster at one end of the VP5* β-barrel domain, and they may all insert into the membrane together. Thus, for one VP5* trimer, a total of nine loops would engage a target membrane. Insertion of all nine loops might perturb the outer membrane leaflet of the membrane in such a way as to facilitate its eventual disruption. The current results support the model of VP5*-mediated membrane penetration illustrated in Fig. 1. Uncoating of VP7 allows VP5* to extend its hydrophobic loops, embed them in the target membrane, and then fold back into a VP5CT-like structure. Membrane penetration may occur as a result of this foldback process.
Where does penetration occur? Coentry of α-sarcin indicates that the virus creates openings in a cell membrane large enough to admit the 16-kDa toxin (diameter, ∼30 Å) (24). Rotavirus probably makes these openings in the membrane of an enclosed endosomal space (30), to which α-sarcin also has access. The V391D mutation, which interferes with bilayer interactions in vitro and greatly diminishes viral infectivity, also blocks α-sarcin coentry, but not viral uptake. VP7 uncoating, brought about by removal of Ca2+, presumably accompanies penetration and might help trigger it. Treatments that prevent decreases in intraendosomal calcium concentrations have been reported to block rotavirus infectivity and α-sarcin coentry (5).
An initial step in membrane penetration by a number of other nonenveloped viruses, including the picornaviruses, nodaviruses, and orthoreoviruses, is release of a pore-forming peptide (e.g., picornavirus VP4 and reovirus μ1N, both of which are myristoylated) (1, 3, 7, 16, 26, 27). In the case of picornaviruses, exposure of additional hydrophobic segments appears to be critical (15). The proposed conformational changes in VP5* and its mode of engaging a target membrane appear instead to resemble the corresponding activities of viral fusion proteins. Our results suggest that a direct analysis of the extended VP5* intermediate and of its interaction with lipid bilayers can reveal how VP5* conformational changes and membrane insertion lead to membrane permeabilization and to translocation of the rotavirus DLP into the cytosol.
Acknowledgments
We thank Harry B. Greenberg for antibodies against VP4.
The work was supported by NIH grant CA-13202 (to S.C.H.), by NIH grant AI-053174 and an Ellison Medical Foundation New Scholars in Global Infectious Diseases Award (to P.R.D.), and by an NSF Graduate Research Fellowship (to I.S.K.). S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 7 April 2010.
REFERENCES
- 1.Agosto, M. A., T. Ivanovic, and M. L. Nibert. 2006. Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc. Natl. Acad. Sci. U. S. A. 103:16496-16501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, S. T., E. C. Settembre, S. D. Trask, H. B. Greenberg, S. C. Harrison, and P. R. Dormitzer. 2009. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 324:1444-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bong, D. T., C. Steinem, A. Janshoff, J. E. Johnson, and M. Reza Ghadiri. 1999. A highly membrane-active peptide in Flock House virus: implications for the mechanism of nodavirus infection. Chem. Biol. 6:473-481. [DOI] [PubMed] [Google Scholar]
- 4.Carrasco, L. 1995. Modification of membrane permeability by animal viruses. Adv. Virus Res. 45:61-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemello, M. E., O. C. Aristimuno, F. Michelangeli, and M. C. Ruiz. 2002. Requirement for vacuolar H+-ATPase activity and Ca2+ gradient during entry of rotavirus into MA104 cells. J. Virol. 76:13083-13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J. Z., E. C. Settembre, S. T. Aoki, X. Zhang, A. R. Bellamy, P. R. Dormitzer, S. C. Harrison, and N. Grigorieff. 2009. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc. Natl. Acad. Sci. U. S. A. 106:10644-10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danthi, P., M. Tosteson, Q. H. Li, and M. Chow. 2003. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J. Virol. 77:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2001. Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J. Virol. 75:7339-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2000. Purified recombinant rotavirus VP7 forms soluble, calcium-dependent trimers. Virology 277:420-428. [DOI] [PubMed] [Google Scholar]
- 10.Dormitzer, P. R., E. B. Nason, B. V. Prasad, and S. C. Harrison. 2004. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dormitzer, P. R., Z. Y. Sun, G. Wagner, and S. C. Harrison. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estes, M. K., and A. Z. Kapikian. 2007. Retroviruses, p. 1918-1974. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott, Williams & Wilkins, Philadephia, PA.
- 14.Fiore, L., S. J. Dunn, B. Ridolfi, F. M. Ruggeri, E. R. Mackow, and H. B. Greenberg. 1995. Antigenicity, immunogenicity and passive protection induced by immunization of mice with baculovirus-expressed VP7 protein from rhesus rotavirus. J. Gen. Virol. 76:1981-1988. [DOI] [PubMed] [Google Scholar]
- 15.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janshoff, A., D. T. Bong, C. Steinem, J. E. Johnson, and M. R. Ghadiri. 1999. An animal virus-derived peptide switches membrane morphology: possible relevance to nodaviral transfection processes. Biochemistry 38:5328-5336. [DOI] [PubMed] [Google Scholar]
- 17.Lawton, J. A., M. K. Estes, and B. V. Prasad. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4:118-121. [DOI] [PubMed] [Google Scholar]
- 18.Li, Z., M. L. Baker, W. Jiang, M. K. Estes, and B. V. Prasad. 2009. Rotavirus architecture at subnanometer resolution. J. Virol. 83:1754-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liprandi, F., Z. Moros, M. Gerder, J. E. Ludert, F. H. Pujol, M. C. Ruiz, F. Michelangeli, A. Charpilienne, and J. Cohen. 1997. Productive penetration of rotavirus in cultured cells induces coentry of the translation inhibitor alpha-sarcin. Virology 237:430-438. [DOI] [PubMed] [Google Scholar]
- 20.Mackow, E. R., J. W. Barnett, H. Chan, and H. B. Greenberg. 1989. The rhesus rotavirus outer capsid protein VP4 functions as a hemagglutinin and is antigenically conserved when expressed by a baculovirus recombinant. J. Virol. 63:1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 22.Padilla-Noriega, L., R. Werner-Eckert, E. R. Mackow, M. Gorziglia, G. Larralde, K. Taniguchi, and H. B. Greenberg. 1993. Serologic analysis of human rotavirus serotypes P1A and P2 by using monoclonal antibodies. J. Clin. Microbiol. 31:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patton, J. T., R. Vasquez-Del Carpio, M. A. Tortorici, and Z. F. Taraporewala. 2007. Coupling of rotavirus genome replication and capsid assembly. Adv. Virus Res. 69:167-201. [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Canadillas, J. M., J. Santoro, R. Campos-Olivas, J. Lacadena, A. Martinez del Pozo, J. G. Gavilanes, M. Rico, and M. Bruix. 2000. The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity. J. Mol. Biol. 299:1061-1073. [DOI] [PubMed] [Google Scholar]
- 25.Ruggeri, F. M., and H. B. Greenberg. 1991. Antibodies to the trypsin cleavage peptide VP8 neutralize rotavirus by inhibiting binding of virions to target cells in culture. J. Virol. 65:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tosteson, M. T., M. L. Nibert, and B. N. Fields. 1993. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc. Natl. Acad. Sci. U. S. A. 90:10549-10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trask, S. D., and P. R. Dormitzer. 2006. Assembly of highly infectious rotavirus particles recoated with recombinant outer capsid proteins. J. Virol. 80:11293-11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trask, S. D., I. S. Kim, S. C. Harrison, and P. R. Dormitzer. 2010. A rotavirus spike protein conformational intermediate binds lipid bilayers. J. Virol. 84:1764-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai, B. 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 23:23-43. [DOI] [PubMed] [Google Scholar]
- 31.Yoder, J. D., and P. R. Dormitzer. 2006. Alternative intermolecular contacts underlie the rotavirus VP5* two- to three-fold rearrangement. EMBO J. 25:1559-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoder, J. D., S. D. Trask, T. P. Vo, M. Binka, N. Feng, S. C. Harrison, H. B. Greenberg, and P. R. Dormitzer. 2009. VP5* rearranges when rotavirus uncoats. J. Virol. 83:11372-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, X., E. Settembre, C. Xu, P. R. Dormitzer, R. Bellamy, S. C. Harrison, and N. Grigorieff. 2008. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. U. S. A. 105:1867-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]