Abstract
The discovery of a novel coronavirus (CoV) as the causative agent of severe acute respiratory syndrome (SARS) has highlighted the need for a better understanding of CoV replication. The replication of SARS-CoV is highly dependent on host cell factors. However, relatively little is known about the cellular proteome changes that occur during SARS-CoV replication. Recently, we developed a cell line expressing a SARS-CoV subgenomic replicon and used it to screen inhibitors of SARS-CoV replication. To identify host proteins important for SARS-CoV RNA replication, the protein profiles of the SARS-CoV replicon cells and parental BHK21 cells were compared using a quantitative proteomic strategy termed “stable-isotope labeling by amino acids in cell culture-mass spectrometry” (SILAC-MS). Our results revealed that, among the 1,081 host proteins quantified in both forward and reverse SILAC measurements, 74 had significantly altered levels of expression. Of these, significantly upregulated BCL2-associated athanogene 3 (BAG3) was selected for further functional studies. BAG3 is involved in a wide variety of cellular processes, including cell survival, cellular stress response, proliferation, migration, and apoptosis. Our results show that inhibition of BAG3 expression by RNA interference led to significant suppression of SARS-CoV replication, suggesting the possibility that upregulation of BAG3 may be part of the machinery that SARS-CoV relies on for replication. By correlating the proteomic data with these functional studies, the findings of this study provide important information for understanding SARS-CoV replication.
The outcome of a viral infection is regulated in part by the complex coordination of viral and host interactions that compete for the control and optimization of virus replication. Virus-host interactions are crucial determinants of virus host range, replication, and pathology. Studies of virus-host interactions have advanced understanding of viral and cellular function and can provide targets for antiviral development. One area in which the importance of host factors is increasingly emerging is the replication of positive-strand RNA [(+) RNA] viruses. (+) RNA viruses are the largest genetic class of viruses and include significant human pathogens such as severe acute respiratory syndrome coronavirus (SARS-CoV), hepatitis C virus, and West Nile virus. Defining the host factors that govern the replication of (+) RNA viruses will enhance our general understanding of their molecular biology and may have important implications for the development of novel antiviral control strategies. Whereas recent studies show that host factors are critical for (+) RNA virus genome replication and mRNA synthesis and are targeted by (+) RNA viruses to modulate host gene expression and defenses (1, 33), identifying such factors remains difficult.
In 2003, a novel coronavirus, SARS-CoV, emerged from zoonotic pools of virus in China to cause a global outbreak of SARS (10, 31). The SARS-CoV genome encompasses 29,727 nucleotides, and the genome organization is similar to that of other coronaviruses. The genome is predicted to contain 14 functional open reading frames (ORFs) (41, 60). SARS-CoV genome translation yields two large replicase polyproteins (pp1a and pp1ab) that are autoproteolytically cleaved into 16 nonstructural proteins (nsp1 to -16) by proteases residing in nsp3 and nsp5 (21, 23, 53). These 16 SARS-CoV nsps include RNA-binding protein (nsp9), RNA-dependent RNA polymerase (nsp12), helicase (nsp13), RNA synthesis proteins (nsp8 and nsp14), and several transmembrane proteins (nsp3, nsp4, and nsp6), etc. (69). They are the primary constituents of the replication/transcription complex (RTC), which is believed to be associated with characteristic double membrane vesicles (DMVs) derived from modified host cell membranes (57, 62). RNA replication is believed to occur on DMVs and uses host proteins as part of their replication strategies (57). Thus, identifying such host factors and their contributions has long been recognized as an important frontier.
Recent advances in molecular profiling technologies have allowed for advances in our understanding of the mechanisms of cellular responses to the SARS-CoV infection. Analysis of gene expression profiles during viral infection is one of the powerful approaches to probe potential cellular genes involved in viral infection and pathogenesis (37, 59), but ultimately protein expression and posttranslational modification (PTM) determine virus replication. Thus, the molecular analysis of viral infection would greatly benefit from a proteomics approach that combines the advantages of high-throughput analysis and the focus on protein levels and modifications. Proteomic techniques as a powerful research tool have recently become available for large-scale protein analysis, and stable-isotope labeling by amino acids in cell culture (SILAC) is one of the most effective methods for the simultaneous detection of diverse changes in protein expression (49). With SILAC, the entire proteome of a given cell population is metabolically labeled by heavy, nonradioactive isotopic variants of amino acids, thus making it distinguishable by mass spectrometry (MS) analysis. Thereafter, two or more distinctly SILAC-labeled cell populations can be mixed and analyzed in one MS experiment, which allows accurate quantitation of proteins from the different cellular states (49). Because of its properties of being simple, inexpensive, and accurate, SILAC is being used in the life sciences more and more extensively (4, 9, 30, 58). To date, a small but increasing number of studies have used proteomics approaches to investigate various aspects concerning the infection and pathogenesis of the SARS-CoV and the virus-host interactions (24, 67). In spite of these data, it is important to establish a comprehensive catalogue of the cellular factors interacting with the virus RNA that may regulate the SARS-CoV replication.
In our previous work, we developed the first SARS-CoV-derived replicon cell line (19). This SARS-associated replicon cell line is based on the use of SARS-CoV replicon cDNAs generated by reverse genetic techniques. The viral envelope-protein coding S, E, and M genes were replaced by a selectable marker and a green fluorescent protein (GFP) reporter gene. The nucleocapsid (N) gene was retained for efficient replication of replicon RNA. Due to the absence of some structural genes, no infectious viral particle could be produced by the cells. However, since all trans- and cis-acting components required for viral RNA synthesis are retained, these partial viral RNAs could replicate autonomously in the cells. Our data indicate that this replicon cell line can be applied to high-throughput screening for anti-SARS drugs without the need to grow infectious SARS-CoV (20).
To generate additional insights into the molecular events controlling SARS-CoV replication, in this study, the SILAC method was employed to compare the protein profiles of the SARS-CoV-derived replicon cells to those of the parental BHK21 cells. Many interesting differentially expressed proteins were identified that potentially play functional roles during virus replication. Further functional studies demonstrated that BAG3 plays an important role in SARS-CoV replication. By correlating the proteomic data with these functional studies, the current results not only provide insights into the mechanism underlying the virus-host interactions but also have direct implications for drug development for SARS-CoV.
MATERIALS AND METHODS
Cells and viruses.
The BHK21 (baby hamster kidney) and Vero E6 (African green monkey kidney) cell lines were purchased from the American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen) at 37°C in 5% CO2. The SARS-CoV replicon cell line SCR-1 stably expressing a SARS-CoV replicon encoding GFP has been described previously (19, 20), and the cells were cultured in DMEM (Gibco) containing 10% fetal calf serum (Gibco) and 10 μg/ml blasticidin (Invitrogen). A seed stock of SARS-CoV (strain SIN 2774) passaged in Vero E6 cells was used for infection. The Beaudette strain of the coronavirus infectious bronchitis virus (IBV) (ATCC VR-22) was obtained from the ATCC and was adapted to Vero E6 cells as described previously (40).
Protein analysis by SILAC labeling.
To differentially label the SARS-CoV replicon cell line SCR-1 and parental BHK21 cells, the SILAC protein quantitation kit (Pierce Biotechnology, Rockford, IL) was used according to the manufacturer's instructions. In brief, cells were grown in SILAC DMEM (Pierce Biotechnology, Rockford, IL) containing 10% (vol/vol) dialyzed FBS, and either 0.1 mg/ml heavy l-[13C6]- or Light l-[12C6]lysine (Pierce Biotechnology, Rockford, IL). Cells were propagated in SILAC medium for >6 generations to ensure nearly 100% incorporation of labeled amino acids. In the forward SILAC experiment, the SCR-1 cells were cultured in light medium, whereas the BHK21 cells were cultured in heavy medium. Reverse SILAC experiments were also performed in which the SCR-1 and BHK21 cells were cultured in the heavy and light medium, respectively (Fig. 1). For protein extraction, cells were lysed in 8 M urea and 20 mM HEPES supplemented with complete protease inhibitor cocktail tablets (Roche, Nutley, NJ). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Cellular debris was removed by centrifugation for 30 min at 13,200 × g and at 4°C. Protein concentrations were measured in duplicate using the RC DC protein assay (Bio-Rad, Hercules, CA).
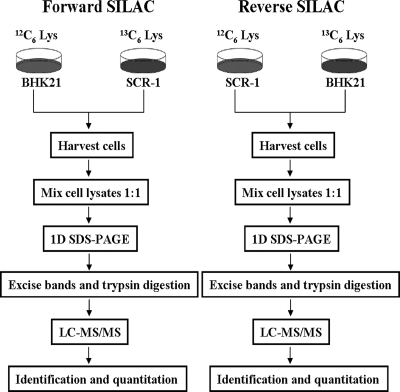
FIG. 1.
Schematic showing application of forward and reverse SILAC combined with LC-MS/MS for the comparative analysis of protein expression in SARS-CoV replicon cells (SCR-1) and their parental BHK21 cells.
Protein separation and in-gel digestion.
The light and heavy cell lysates were combined at 1:1 ratio (wt/wt) (100 μg in total), boiled in SDS-PAGE sample buffer, separated by 12% SDS-PAGE, and stained with Coomassie blue. The entire gel lane was cut into 30 sections for in-gel tryptic digestion. The excised sections were chopped into small particles and washed in water and then completely destained using 100 mM ammonium bicarbonate in 50% acetonitrile (ACN). A reduction step was performed by addition of 100 μl of 10 mM dithiothreitol (DTT) at 37°C for 1 h. The proteins were alkylated by adding 100 μl of 50 mM iodoacetamide and allowed to react in the dark at 20°C for 30 min. Gel sections were first washed in water, and then acetonitrile, and finally dried with a SpeedVac centrifuge (Thermo Fisher Scientific, Waltham, MA). Digestion was carried out using 20 μg/ml sequencing-grade modified trypsin (Promega) in 50 mM ammonium bicarbonate. Sufficient trypsin solution was added to swell the gel pieces, which were kept at 4°C for 45 min and then incubated at 37°C overnight, after which peptides were extracted from gels with 5% acetic acid in H2O and in CH3CN/H2O (1:1 [vol/vol]). The resulting peptide mixtures were dried using vacuum centrifugation and stored at −80°C for further analysis.
Protein identification and quantification.
Online liquid chromatography-tandem MS (LC-MS/MS) analysis was performed on an Agilent 6510 Q-TOF system coupled with an Agilent high-performance liquid chromatography (HPLC)-Chip Cube MS interface (Agilent Technologies, Santa Clara, CA). The sample injection, enrichment, desalting, and HPLC separation were carried out automatically on the Agilent HPLC chip with an integrated trapping column (160 nl) and a separation column (Zorbax 300SB-C18; 75 μm by 150 mm, 5-μm particle size). The peptide mixture was first loaded onto the trapping column with a solvent mixture of 0.1% formic acid in CH3CN/H2O (2:98 [vol/vol]) at a flow rate of 4 μl/min. The peptides were then separated with a 90-min linear gradient of 5 to 60% acetonitrile in 0.1% formic acid and at a flow rate of 300 nl/min. The chip spray voltage (VCap) was set as 1,950 V and varied depending on chip conditions. The temperature and flow rate of the drying gas were set at 325°C and 4 liters/min, respectively. Nitrogen was used as the collision gas, and the collision energy followed an equation with a slope of 3 V/100 Da and an offset of 2.5 V. MS/MS experiments were carried out in the data-dependent scan mode with a maximum of five MS/MS scans following each MS scan. The m/z ranges for MS and MS/MS were 300 to 2,000 and 60 to 2,000, and the acquisition rates were 6 and 3 spectra/s, respectively. Agilent MassHunter workstation software (version B.01.03) was used to extract the MS and MS/MS data. The data were converted to m/z data files with MassHunter Qualitative Analysis. Mascot Server 2.2 (Matrix Science, London, United Kingdom) was used for protein identification by searching the m/z data files against the IPI mouse protein database (version 3.21; 51,432 sequences) (28). The maximum number of miscleavages for trypsin was set as 1 per peptide. Cys (+57.0215 Da; carbamidomethylation) was set as fixed modification, whereas Met (+15.9949 Da; oxidation) and Lys (+6.0201 Da; SILAC heavy amino acid) were considered as variable modifications. The mass tolerances for MS and MS/MS were 50 ppm and 0.6 Da, respectively. Peptides identified with individual scores at or above the Mascot-assigned homology score (P < 0.01 and individual peptide score of >30) were considered as specific peptide sequences. The false discovery rates (FDR) determined by decoy database search were <0.95%. All identified peptides were subjected to relative quantification analysis using the program Census (51). Only proteins with a minimum of 2 quantifiable peptides were included in our final data set. The protein ratios were calculated from the average of all quantified peptides. The quantification was based on four independent SILAC and LC-MS/MS experiments, which included two forward and two reverse SILAC labelings, and the proteins reported here could be quantified in both forward and reverse SILAC experiments.
Protein categorization.
Differentially expressed proteins (DEPs) were classified based on the PANTHER (protein analysis through evolutionary relationships) system (http://www.pantherdb.org), which is a unique resource that classifies genes and proteins by their functions (43).
Western blot analysis.
Protein extracts (30 μg) prepared with radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 1 mM phenylmethyl sulfonyl fluoride [PMSF], 100 mM leupeptin, and 2 mg/ml aprotinin, pH 8.0) were resolved with a 10% SDS-PAGE gel and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore Corporation, Billerica, MA) by electroblotting and then blocked using Tris-buffered saline-Tween 20 (TBST) buffer containing 5% nonfat milk. The membranes were probed with rabbit anti-heat shock protein 90 (anti-HSP90) polyclonal antibody, goat anti-ENO1 polyclonal antibody, goat anti-YWHAZ polyclonal antibody, goat anti-RPS19 polyclonal antibody, rabbit anti-HSPA1A polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-BAG3 polyclonal antibody, rabbit anti-hnRNP A1 polyclonal antibody (Abcam, Inc., Cambridge, MA), mouse anti-lactate dehydrogenase B (anti-LDHB) monoclonal antibody, rabbit anti-cell division control protein 42 (anti-CDC42) polyclonal antibody (Abnova, Walnut, CA), mouse anti-tubulin monoclonal antibody (LabVision, Fremont, CA), and anti-nsp5 (3CLpro) antibody (20), respectively. After washing, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 1 h. The membranes were washed thoroughly with PBST for three times. The secondary antibody was detected by using the ChemiGlow chemiluminescence reagents (Alpha Innotech, San Leandra, CA). Finally the immunoblots were scanned, and densitometric analysis was performed using the public domain NIH Image program ImageJ (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/).
siRNA for BAG3 and transfection of siRNA.
SCR-1 cells were transfected with 50 nM small interfering RNA (siRNA) specific to mouse Bag3 or with nontargeting siRNA (Dharmacon, Lafayette, CO). siRNAs were introduced to cells using Lipofectamine Plus in Opti-MEM medium (Invitrogen, Carlsbad, CA). siRNA has been selected among four different siRNA sequences that have been evaluated for high specificity and lack of off-target effect at the concentration used. Forty-eight hours posttransfection, fluorescence microscopy was used to observe the changes of green fluorescence in SCR-1 cells, protein extracts were analyzed by Western blotting, and total RNA was purified and used for real-time PCR analysis.
Establishment of stable cell line expressing BAG3 shRNA.
SMARTvector 2.0 lentiviral short hairpin RNA (shRNA) particles specific to human Bag3 or nontargeting control particles (Dharmacon, Lafayette, CO) were transfected into Vero E6 cells according to the manufacturer's instructions. Transfected cells were selected by using 1 μg/ml of puromycin. One clone was selected from BAG3 shRNA-transfected Vero cells (designated as Vero-KD, for “Vero knockdown”), and one clone was selected from control shRNA-transfected Vero cells (designated as Vero-NC). BAG3 protein knockdown was assessed by Western blotting analysis.
Plaque reduction assay.
The plaque reduction assay followed the procedures previously described (20, 39). All procedures involving manipulation of live SARS-CoV were carried out in a biological safety level 3 containment laboratory.
Green fluorescence analysis.
Fluorescence microscopy was used to observe the green fluorescence of GFP expressed from the SARS-CoV replicon. The cells were observed under an Olympus IX70 inverted fluorescence microscope, and the images were recorded using Image-Pro Plus (Media Cybernetics).
Real-time PCR analysis.
Real-time reverse transcription (RT)-PCR analyses were performed to quantify the copy number of SARS-CoV RNA in SCR-1 cells or Vero cells. Primers and RT-PCR conditions were used as previously described (19). Real-time PCR signals were analyzed using the LightCycler software (version 5.32; Roche), and the sizes and uniqueness of PCR products were verified by performing both melting curves and agarose gel electrophoresis.
Statistical analysis.
All data are expressed as means ± standard deviations. Statistical significance of the in vitro data was determined by Student's t test (two tailed), while the significance of the differences between the median values of the in vivo data was determined using the two-tailed Mann-Whitney test. Statistical significance was assigned if P was <0.05.
RESULTS
Quantitative proteome analysis of SARS-CoV replicon cells.
To gain insights into the molecular pathways perturbed by SARS-CoV replication, we employed SILAC combined with LC-MS/MS to determine the differential proteomes of the SARS-CoV replicon cell SCR-1 and parental BHK21 cells. The workflow in this study is outlined in Fig. 1. To obtain reliable results, we carried out both forward and reverse SILAC experiments. After cell lysis, SDS-PAGE fractionation, in-gel digestion, LC-MS/MS analysis, and the database search, we were able to identify 1,801 proteins, among which 1,480 could be quantified. Among the quantified proteins, we include here only the quantification results for those proteins that could be quantified in both the forward and reverse SILAC. Together, this gives quantifiable results for 1,081 proteins (data not shown). Differentially expressed proteins (DEPs) were selected based on a predefined threshold of 2.0-fold change (the ratio of BHK21 to SCR-1 expression was greater than 2.0 or less than 0.5). Among the quantified proteins, 43 were upregulated and 31 were downregulated in SCR-1 cells. The quantification results for the proteins with significant changes are summarized in Table 1. (Detailed information about the quantified proteins and ratios for each experiments was obtained [data not shown].)
TABLE 1.
List of proteins with at least 2.0-fold quantitative alteration in SARS-CoV replicon cells based on SILAC analysis
| Protein type and gene accession no. | Gene product name | Description | SILAC BHK21/SCR-1 expression ratio |
|---|---|---|---|
| Nucleic acid binding | |||
| IPI00310317 | Atf3 | Activating transcription factor 3 | 0.18 ± 0.08 |
| IPI00776411 | Hcfc1 | Host cell factor C1 | 0.19 ± 0.04 |
| IPI00317794 | Ncl | Nucleolin | 0.25 ± 0.06 |
| IPI00138335 | Zfp36l1 | Zinc finger protein 36, C3H type-like 1 | 0.29 ± 0.05 |
| IPI00623284 | Sf3b1 | Splicing factor 3b, subunit 1 | 0.30 ± 0.03 |
| IPI00228457 | Myst2 | MYST histone acetyltransferase 2 | 0.41 ± 0.04 |
| IPI00113241 | Rps19 | Ribosomal protein S19 | 2.27 ± 0.27 |
| IPI00466069 | Eef2 | Eukaryotic translation elongation factor 2 | 2.29 ± 0.34 |
| IPI00268802 | Rps18 | Ribosomal protein S18 | 2.35 ± 0.41 |
| IPI00112448 | Rps10 | Ribosomal protein S10 | 2.46 ± 0.71 |
| IPI00474446 | Eif2s1 | Eukaryotic translation initiation factor 2, subunit 1 alpha | 2.83 ± 0.53 |
| IPI00404707 | Rbm14 | RNA binding motif protein 14 | 2.96 ± 0.25 |
| IPI00555113 | Rpl18 | Ribosomal protein L18 | 3.07 ± 0.19 |
| IPI00322422 | Mrpl47 | Mitochondrial ribosomal protein L47 | 3.16 ± 0.39 |
| IPI00274407 | Tufm | Tu translation elongation factor, mitochondrial | 3.18 ± 0.41 |
| IPI00133503 | Rpl27a | Ribosomal protein L27a | 3.51 ± 0.77 |
| IPI00756424 | Eif5b | Eukaryotic translation initiation factor 5B | 4.08 ± 0.60 |
| IPI00230679 | Rpl36 | Ribosomal protein L36 | 4.08 ± 0.71 |
| IPI00387566 | Ubd | Ubiquitin D | 5.40 ± 0.85 |
| IPI00400432 | Eif4a2 | Eukaryotic translation initiation factor 4A2 | 7.33 ± 1.77 |
| Select regulatory molecule | |||
| IPI00131870 | Cops3 | COP9 signalosome complex subunit 3 | 0.21 ± 0.09 |
| IPI00127408 | Rac1 | RAS-related C3 botulinum substrate 1 | 0.22 ± 0.09 |
| IPI00315100 | Rhoa | Ras homolog gene family, member A | 0.31 ± 0.05 |
| IPI00754880 | Arhgef12 | Rho guanine nucleotide exchange factor (GEF) 12 | 0.40 ± 0.09 |
| IPI00114560 | Rab1 | RAB1, member RAS oncogene family | 0.46 ± 0.08 |
| IPI00762919 | Hip1 | Huntingtin interacting protein 1 | 3.50 ± 0.52 |
| IPI00113849 | Cdc42 | Cell division cycle 42 | 4.16 ± 0.15 |
| IPI00138143 | Cdkn2b | Cyclin-dependent kinase inhibitor 2B | 4.58 ± 0.70 |
| Chaperone | |||
| IPI00229080 | Hsp90ab1 | Heat shock protein 90 kDa alpha (cytosolic) | 0.11 ± 0.03 |
| IPI00323357 | Hspa8 | Heat shock protein 8 | 0.18 ± 0.05 |
| IPI00116498 | Ywhaz | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 0.23 ± 0.04 |
| IPI00669474 | Hspa1a | Heat shock protein 1A | 0.24 ± 0.07 |
| IPI00331556 | Hspa4 | Heat shock protein 4 | 0.31 ± 0.06 |
| IPI00230707 | Ywhag | 3-Monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | 0.31 ± 0.07 |
| IPI00331334 | Bag3 | Bcl2-associated athanogene 3 | 0.37 ± 0.04 |
| IPI00153740 | Ahsa1 | AHA1, activator of heat shock protein ATPase homolog 1 (yeast) | 0.41 ± 0.07 |
| Kinase | |||
| IPI00316677 | Pik3cd | Phosphatidylinositol 3-kinase catalytic delta polypeptide | 0.18 ± 0.01 |
| IPI00119772 | Stk4 | Serine/threonine kinase 4 | 0.24 ± 0.06 |
| IPI00229884 | Pak2 | P21 (CDKN1A)-activated kinase 2 | 0.25 ± 0.03 |
| IPI00283633 | Limk2 | LIM motif-containing protein kinase 2 | 0.25 ± 0.06 |
| IPI00556823 | Prkaa1 | Protein kinase, AMP-activated, alpha 1 catalytic subunit | 0.29 ± 0.05 |
| IPI00119663 | Mapk1 | Mitogen-activated protein kinase 1 | 0.29 ± 0.07 |
| Transferase | |||
| IPI00119772 | Stk4 | Serine/threonine kinase 4 | 0.24 ± 0.06 |
| IPI00269091 | Trit1 | TRNA isopentenyltransferase 1 | 0.41 ± 0.03 |
| IPI00228457 | Myst2 | MYST histone acetyltransferase 2 | 0.41 ± 0.04 |
| IPI00323353 | Upp1 | Uridine phosphorylase 1 | 2.37 ± 0.37 |
| Transcription factor | |||
| IPI00310317 | Atf3 | Activating transcription factor 3 | 0.18 ± 0.08 |
| IPI00119059 | Nr2f1 | Nuclear receptor subfamily 2, group F, member 1 | 0.31 ± 0.10 |
| IPI00135883 | Gata3 | GATA binding protein 3 | 0.32 ± 0.04 |
| Transporter | |||
| IPI00130924 | Slc27a2 | Solute carrier family 27 (fatty acid transporter), member 2 | 0.33 ± 0.05 |
| IPI00120572 | Nup50 | Nucleoporin 50 | 0.33 ± 0.15 |
| IPI00122648 | Laptm4a | Lysosomal-associated protein transmembrane 4A | 4.53 ± 1.03 |
| Cytoskeletal | |||
| IPI00515564 | Kifc1 | Kinesin family member C1 | 2.90 ± 0.34 |
| IPI00762919 | Hip1 | Huntingtin interacting protein 1 | 3.50 ± 0.53 |
| IPI00229647 | Tln2 | Talin 2 | 5.33 ± 0.55 |
| Oxidoreductase | |||
| IPI00318108 | Acox3 | Acyl-coenzyme A oxidase 3, pristanoyl | 2.42 ± 0.44 |
| IPI00229510 | Ldhb | Lactate dehydrogenase B | 2.43 ± 0.37 |
| IPI00222809 | H6pd | Hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) | 4.77 ± 0.49 |
| Signaling molecule | |||
| IPI00320634 | Elk3 | ELK3, member of ETS oncogene family | 0.20 ± 0.05 |
| Synthase and synthetase | |||
| IPI00331707 | Hmgcs1 | 3-Hydroxy-3-methylglutaryl-coenzyme A synthase 1 | 0.21 ± 0.03 |
| Select calcium binding protein | |||
| IPI00317309 | Anxa5 | Annexin A5 | 0.43 ± 0.09 |
| Receptor | |||
| IPI00119059 | Nr2f1 | Nuclear receptor subfamily 2, group F, member 1 | 0.30 ± 0.10 |
| Membrane trafficking | |||
| IPI00266752 | Cpne3 | Copine-3 | 0.22 ± 0.06 |
| Lyase | |||
| IPI00318496 | Gad1 | Glutamic acid decarboxylase 1 | 0.30 ± 0.03 |
| IPI00462072 | Eno1 | Enolase 1, alpha non-neuron | 2.06 ± 0.13 |
| Protease | |||
| IPI00128154 | Ctsl | Cathepsin L | 2.78 ± 0.46 |
| IPI00626909 | Capn1 | Calpain 1 | 4.32 ± 0.88 |
| Phosphatase | |||
| IPI00116554 | Ptpn11 | Protein tyrosine phosphatase, non-receptor type 11 | 0.28 ± 0.04 |
| IPI00130507 | Dusp7 | Dual-specificity phosphatase 7 | 2.58 ± 0.35 |
| IPI00330483 | Ppp1r10 | Protein phosphatase 1, regulatory subunit 10 | 3.06 ± 0.39 |
| IPI00177072 | Pfkfb3 | 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 3.09 ± 0.33 |
| Miscellaneous function | |||
| IPI00222515 | Psmd11 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | 0.26 ± 0.05 |
| IPI00669522 | Clasp1 | CLIP associating protein 1 | 0.34 ± 0.06 |
| Molecular function unclassified | |||
| IPI00169634 | Snapc1 | Small nuclear RNA activating complex, polypeptide 1 | 0.21 ± 0.08 |
| IPI00315187 | 2400001E08Rik | RIKEN cDNA 2400001E08 gene | 0.22 ± 0.03 |
| IPI00126917 | Phb | Prohibitin | 0.37 ± 0.06 |
| IPI00654076 | F630110N24Rik | RIKEN cDNA F630110N24 gene | 0.40 ± 0.07 |
| IPI00187289 | 2310028N02Rik | RIKEN cDNA 2310028N02 gene | 3.00 ± 0.23 |
Functional categories of DEPs.
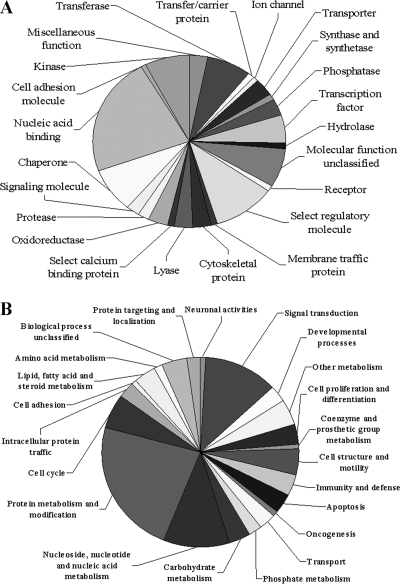
In order to understand the biological relevance of the changes in protein expression in response to SARS-CoV replication, PANTHER classification system was used to classify the 74 DEPs according to their functions. The PANTHER classification system revealed that the DEPs can be classified into 23 groups according to their functional properties (Fig. 2). These proteins are implicated in a broad range of cellular activities (Fig. 2). Proteins involved in nucleic acid binding account for the largest portion (27%). There are also a significant number of proteins involved in regulatory molecule (13%), chaperone (10%), and kinase (9%) activities. Among these DEPs, of particular interest are upregulated chaperones and downregulated host translational machinery. We expect that these host factors play important roles in SARS-CoV replication.
FIG. 2.
Pie chart representations of the distribution of differentially expressed proteins according to their molecular functions (A) and biological processes (B). Categorizations were based on information provided by the online resource the PANTHER classification system.
Validation of differential protein expression.
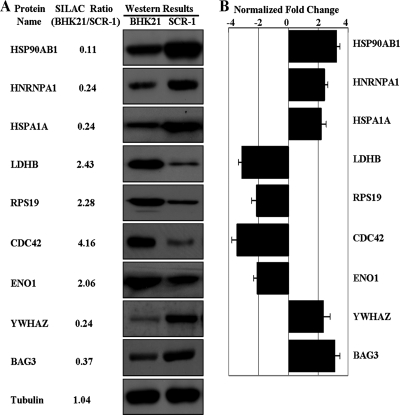
To further confirm the SILAC ratios we observed with MS, we used Western blot analysis to examine the expression of a selected panel of proteins, including HSPA1A, HSP90AB1, hnRNPA1, lactate dehydrogenase B (LDHB), RPS19, YWHAZ, ENO1, cell division control protein 42 (CDC42), and BAG3. As shown in Fig. 3, for all of the selected proteins, the Western blotting and densitometric analysis results showed the same pattern of expression as that obtained from SILAC experiments.
FIG. 3.
Western blot and densitometric analysis of nine DEPs and internal control protein tubulin. (A) Western blot images for nine DEPs and internal control protein tubulin. (B) Relative expression of nine DEPs (normalized to tubulin band) was determined using image densitometry and expressed in a bar chart format. The error bars represent the standard deviation of the mean for three representative analyses.
BAG3 is essential for efficient SARS-CoV replication.
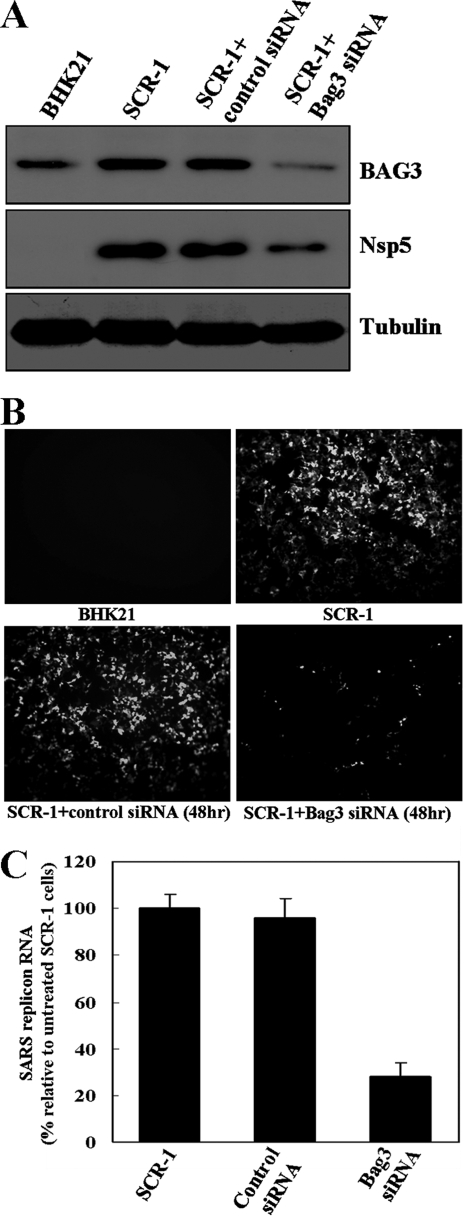
To evaluate the functional requirement of BAG3 for the replication of SARS-CoV, we used RNA interference (RNAi) to reduce cellular BAG3 levels and examine the effect of BAG3 knockdown. The effect of the BAG3 siRNA in silencing the BAG3 gene in SCR-1 cells was examined by directly measuring changes in BAG3 protein levels. As shown in Fig. 4B, transfection of replicon cells with the BAG3 siRNA resulted in a significant decrease in the level of BAG3 protein, whereas mock transfection with a random siRNA had no such effect. This effect was specific because the BAG3 siRNA did not change the levels of tubulin protein (Fig. 4A).
FIG. 4.
Effects of BAG3 knockdown on SARS-CoV replicon cells. (A) The knockdown of BAG3 in SCR-1 cells was confirmed by Western blotting analysis. Western blotting results indicated that BAG3 is upregulated in SCR-1 cells, and knockdown of BAG3 led to reduction of SARS-CoV proteins in SCR-1 cells. (B) Inhibition of BAG3 expression led to suppressed GFP fluorescence in SCR-1 cells compared to untreated SCR-1 cells or cells transfected with control siRNA. (C) Quantification of SARS-CoV mRNA revealed markedly reduced mRNA in BAG3 knockdown SCR-1 cells compared to untreated cells or cells transfected with control siRNA.
To analyze whether BAG3 knockdown has an effect on SARS-CoV replication in SCR-1, the green fluorescence analysis, quantitative real-time PCR, and Western blotting were performed. As shown in Fig. 4, knockdown of BAG3 resulted in a marked decrease in GFP fluorescence signal, replicon RNA levels, and SARS-CoV nsp5 protein level in SCR-1 cells. All of these data indicate that BAG3 knockdown can inhibit SARS-CoV replication and protein synthesis.
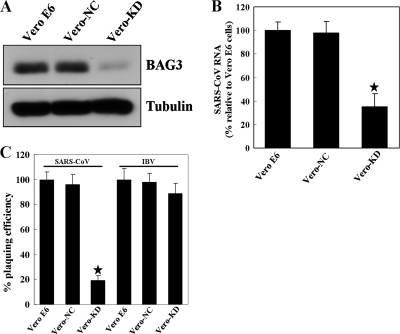
To evaluate the functional requirement of BAG3 for the replication of SARS-CoV in Vero E6 cells, we constructed cell lines stably expressing siRNAs targeting the BAG3 mRNA. The knockdown of BAG3 in Vero-KD cells was confirmed by Western blotting analysis (Fig. 5A). The apoptosis and cell growth assay revealed that reduction of BAG3 had no significant influence on Vero cell growth or apoptosis (data not shown). The resulting cell lines were tested for the ability of SARS-CoV to replicate using real-time PCR and plaque reduction assay. Reduction of BAG3 levels resulted in a significant decrease in SARS-CoV RNA and virus titer compared to the parental Vero E6 and Vero-NC cells (Fig. 5B and C). To evaluate BAG3's specificity for SARS-CoV replication, we asked if growth of another coronavirus, IBV, was altered in the Vero-KD cells. In contrast to SARS-CoV, plaquing efficiency of IBV was unaffected by decreased BAG3 levels (Fig. 5C). Thus, our data indicate that BAG3 is a host protein that is specifically required for efficient replication of SARS-CoV and not IBV.
FIG. 5.
BAG3 is essential for efficient SARS-CoV replication. Columns are the means of three independent experiments; error bars represent ± standard deviation. The asterisks indicate significant difference (P < 0.05). (A) Confirmation of BAG3 knockdown in Vero-KD cells by Western blotting analysis. (B) Quantification of SARS-CoV RNA revealed markedly reduced SARS-CoV RNA in Vero-KD cells compared to parental Vero E6 or Vero-NC cells. Quantitative PCR was performed as described in Materials and Methods. (C) The reduction in SARS-CoV or IBV titers was determined by plaque assays. Vero E6, Vero-NC, and Vero-KD cells were infected with SARS-CoV or IBV, and the plaque numbers in the different cell lines were then normalized to the number of plaques that formed in the Vero E6 cells.
DISCUSSION
The emergence of SARS-CoV underscores the importance of advancing understanding of virus-host interactions. Increasing evidence emphasizes comparative proteomics to screen the differentially expressed proteins associated with host cellular pathophysiological processes of virus infection (42, 54, 63). From the literature, very few studies have been performed to analyze the interaction between CoV and host cells using proteomics analysis. This study is the first to employ the SILAC technique to globally search for the dysregulated host proteins in SARS-CoV replicon cells. In the present work, a total of 74 differentially expressed host proteins were identified and differential expression levels of nine DEPs were confirmed by Western blotting and densitometric analysis. The PANTHER classification system revealed that the proteins can be classified into 23 groups according to their biological process or molecular functions (Fig. 2). Based on the identified proteins in the present work, we obtained an overview of the altered protein expression of host cells responding to SARS-CoV replication.
By comparison with the previous genomic study that analyzed the SARS-CoV-infected cells using mRNA microarray approaches (37), among 74 proteins identified in the current study, 14 of them were also found to be altered at the mRNA level, namely, Rps10, Rps19, Laptm4a, Rac1, Ctsl, Rpl27a, Myst2, Rpl36, Rps18, Trit1, Ppp1r10, Rpl18, Hspa1a, and Clasp1. Most of the remaining identified proteins are newly discovered. The limited overlap between two studies is expected to reflect, at least in part, the different effects between viral infection and replicon RNA on the host cell. In addition, the poor correlation between proteomic and genomic results was not unexpected, as previous studies showed that there exists an approximately 60 to 80% discordance between mRNA and protein abundances (47, 64). This lack of correlations could be the result of mRNA degradation, alternative splicing, and posttranscriptional regulation of gene expression. Therefore, the current proteomic study represents a complementary strategy for studies of mechanisms that underlie the replication of SARS-CoV.
Among all DEPs, significantly upregulated BAG3, a member of the BAG cochaperone protein family, was selected for further functional studies. Our results demonstrate that BAG3 knockdown results in greatly diminished virus replication, suggesting the possibility that upregulation of BAG3 may be part of the machinery that SARS-CoV relies on for replication. However, the mechanisms by which BAG3 regulates SARS-CoV replication remain unknown. BAG proteins participate in a wide variety of cellular processes, including cell survival, cellular stress response, proliferation, migration, and apoptosis (14, 25). BAG3 is implicated in the pathogenesis of neoplasia via its ability to regulate stress-induced apoptosis in a prosurvival fashion. This regulation occurs at a number of levels, including cytochrome c release, apoptosome assembly, and others (5, 55). Other cellular signaling molecules that have been reported to be regulated by BAG3 include Raf-1, cyclin-dependent kinase 4 (CDK-4), and epidermal growth factor receptor (EGFR) (13), as well as focal adhesion kinase (27). Notably, there is increasing evidence that BAG3 is required for efficient growth of different viruses, including varicella-zoster virus (VZV) (35), polyomavirus JC (3), Epstein-Barr virus (EBV) (66), herpes simplex virus (HSV) (36), and HIV (56) and depletion of BAG3 by small interfering RNA results in inhibition of virus replication (35).
Recent evidence implicates an additional function of BAG3 in the regulation of the autophagy pathway. These findings indicate that autophagosome formation and turnover may depend on BAG3 and BAG3 can stimulate autophagy processes (7, 8, 18). Autophagy is a cellular stress response that functions to recycle proteins and organelles and an intracellular catabolic transport route conserved among all eukaryotic cells (22, 65). One of its functions is to act as an immune mechanism against intracellular pathogens (38, 50). Bacteria and viruses targeted for destruction are sequestered into large double-membrane vesicles (DMV) called autophagosomes and subsequently delivered to the lysosomes, where they are degraded by hydrolases. However, it has become clear that the autophagy pathway is often exploited by viruses to facilitate their entry or replication (29, 32). Importantly, the laboratory of Mark Denison has shown that the DMVs induced by infection with SARS-CoV and the coronavirus mouse hepatitis virus (MHV) could be autophagosomes, while MHV infection appears to induce autophagy and inhibition of autophagy leads to suppression of MHV growth (52, 53, 68). Their results demonstrate that the autophagy pathway is required for formation of a viral replication complex and for efficient viral growth. The significant conservation of replicase gene organization and probable functions of SARS-CoV and MHV replicase proteins further suggest that SARS-CoV may share with MHV the strategy of induction and use of the autophagy pathway for efficient replication (53).
Based on the critical role of BAG3 in the stimulation of the autophagy pathway, it is tempting to suggest that BAG3 might be recruited to and activate the autophagy machinery to facilitate the efficient replication of SARS-CoV. Therefore, the increase in BAG3 expression is likely part of the cell's response to SARS-CoV infection and appears to represent a novel mechanism for maintaining SARS-CoV replication in host cells. This speculative idea, however, is not yet supported by experimental data, and further experiments will be required to determine the functional implication of BAG3 and the autophagy pathway in the life cycle of SARS-CoV.
In the present work, the most distinguishable proteins to be downregulated in SARS-CoV replicon cells were those involved in the host translational machinery, including 40S ribosomal proteins (RPS10, RPS18, and RPS19), 60S ribosomal proteins (RPL18, RPL27A, and RPL36), and eukaryotic translation initiation factors (EIF2S1, EIF5B, and EIF4A2). Such host translational downregulation is frequently observed, as evident in infections with herpes simplex virus (15), poliovirus (34), and SARS-CoV (37). Recently, it was reported that SARS-CoV nsp1 protein uses a two-pronged strategy to inhibit host translation and gene expression by binding to the 40S ribosomal subunit and inactivating the translational activity of the 40S subunits (26). Thus, downregulation of the host translational machinery may play an important role in SARS-CoV replication in host cells.
Two regulatory molecules, Cdc42 and RhoA, were found to be downregulated in replicon cells. Cdc42 and RhoA belong to the Rho family of small GTPases. These proteins play a role in cell migration, membrane traffic, and actin cytoskeleton reorganization (6). It has also been shown that Cdc42 regulates the phosphatidylinositol 3-kinase (PI3K)-mTOR pathway (6, 16) and PI3K/Akt signaling pathways are important for establishing persistent SARS-CoV infection in Vero E6 cells (44, 45). Therefore, modulation of SARS-CoV replication by Cdc42 and RhoA may be mediated by the PI3K-mTOR pathway and the PI3K-mTOR pathway may be one of the key factors for understanding persistence of SARS-CoV replicon RNA in host cells.
Other proteins of interest uncovered in this study include 14-3-3 proteins. The 14-3-3 proteins have emerged as critical regulators of diverse cellular responses in eukaryotic organisms (2, 17, 46). In mammalian cells, seven different isoforms have been identified (ζ, β, γ, ɛ, σ, η, and θ), with each isoform having distinct tissue localization and function. Through interactions with more than 400 target proteins identified so far, these 14-3-3 proteins are known to be involved in widespread biological processes such as signal transduction, cell cycle control, apoptosis, cellular metabolism, proliferation, cytoskeletal regulation, transcription, redox regulation, stress response, etc. (11, 61). Interesting evidence has been reported indicating that 14-3-3 might be important for host translational shutoff, the shutoff of minus strand synthesis, or other processes that are time dependent in virus-infected cells (12). Recently, it was also reported that 14-3-3 proteins are involved in the activation of signaling cascades during viral infection (48). Therefore, the increase in 14-3-3 protein expression is likely part of the cell's response to SARS-CoV replication and appears to represent a homeostatic mechanism for cell defense.
In conclusion, the quantitative proteomics analysis described here identified many host factors that potentially affect SARS-CoV replication and implicated previously unconsidered pathways in the virus replication. By using siRNA, we also obtained evidence that depletion of BAG3 results in inhibition of virus replication. Our study provides important information for understanding SARS-CoV replication. Further studies will determine more directly how implicated host factors affect the virus and how such effects illuminate cellular functions and pathways.
Acknowledgments
We thank Ming Xue for technical support in operating the LC-MS/MS system.
This work was supported by funding from the Biomedical Research Council, Singapore, a Singapore Millennium Foundation Postdoctoral Fellowship (to L.Z.), and the Open Research Fund Program of the State Key Laboratory of Virology of China (grant 2009003).
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitken, A. 2006. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 16:162-172. [DOI] [PubMed] [Google Scholar]
- 3.Basile, A., N. Darbinian, R. Kaminski, M. K. White, A. Gentilella, M. C. Turco, and K. Khalili. 2009. Evidence for modulation of BAG3 by polyomavirus JC early protein. J. Gen. Virol. 90:1629-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blagoev, B., S. E. Ong, I. Kratchmarova, and M. Mann. 2004. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22:1139-1145. [DOI] [PubMed] [Google Scholar]
- 5.Bonelli, P., A. Petrella, A. Rosati, M. F. Romano, R. Lerose, M. G. Pagliuca, T. Amelio, M. Festa, G. Martire, S. Venuta, M. C. Turco, and A. Leone. 2004. BAG3 protein regulates stress-induced apoptosis in normal and neoplastic leukocytes. Leukemia 18:358-360. [DOI] [PubMed] [Google Scholar]
- 6.Buchsbaum, R. J. 2007. Rho activation at a glance. J. Cell Sci. 120:1149-1152. [DOI] [PubMed] [Google Scholar]
- 7.Carra, S., J. F. Brunsting, H. Lambert, J. Landry, and H. H. Kampinga. 2009. HspB8 participates in protein quality control by a non-chaperone-like mechanism that requires eIF2{alpha} phosphorylation. J. Biol. Chem. 284:5523-5532. [DOI] [PubMed] [Google Scholar]
- 8.Carra, S., S. J. Seguin, H. Lambert, and J. Landry. 2008. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J. Biol. Chem. 283:1437-1444. [DOI] [PubMed] [Google Scholar]
- 9.Chen, N., W. Sun, X. Deng, Y. Hao, X. Chen, B. Xing, W. Jia, J. Ma, H. Wei, Y. Zhu, X. Qian, Y. Jiang, and F. He. 2008. Quantitative proteome analysis of HCC cell lines with different metastatic potentials by SILAC. Proteomics 8:5108-5118. [DOI] [PubMed] [Google Scholar]
- 10.Christian, M. D., S. M. Poutanen, M. R. Loutfy, M. P. Muller, and D. E. Low. 2004. Severe acute respiratory syndrome. Clin. Infect. Dis. 38:1420-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coblitz, B., M. Wu, S. Shikano, and M. Li. 2006. C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 580:1531-1535. [DOI] [PubMed] [Google Scholar]
- 12.Cristea, I. M., J. W. Carroll, M. P. Rout, C. M. Rice, B. T. Chait, and M. R. MacDonald. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 281:30269-30278. [DOI] [PubMed] [Google Scholar]
- 13.Doong, H., K. Rizzo, S. Fang, V. Kulpa, A. M. Weissman, and E. C. Kohn. 2003. CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation: accumulation of poly-ubiquitinated Hsp90 client proteins. J. Biol. Chem. 278:28490-28500. [DOI] [PubMed] [Google Scholar]
- 14.Doong, H., A. Vrailas, and E. C. Kohn. 2002. What's in the ‘BAG’? A functional domain analysis of the BAG-family proteins. Cancer Lett. 188:25-32. [DOI] [PubMed] [Google Scholar]
- 15.Everly, D. N., Jr., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 76:8560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang, Y., I. H. Park, A. L. Wu, G. Du, P. Huang, M. A. Frohman, S. J. Walker, H. A. Brown, and J. Chen. 2003. PLD1 regulates mTOR signaling and mediates Cdc42 activation of S6K1. Curr. Biol. 13:2037-2044. [DOI] [PubMed] [Google Scholar]
- 17.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617-647. [DOI] [PubMed] [Google Scholar]
- 18.Gamerdinger, M., P. Hajieva, A. M. Kaya, U. Wolfrum, F. U. Hartl, and C. Behl. 2009. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 28:889-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge, F., Y. Luo, P. X. Liew, and E. Hung. 2007. Derivation of a novel SARS-coronavirus replicon cell line and its application for anti-SARS drug screening. Virology 360:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge, F., S. Xiong, F. S. Lin, Z. P. Zhang, and X. E. Zhang. 2008. High-throughput assay using a GFP-expressing replicon for SARS-CoV drug discovery. Antiviral Res. 80:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath, R. J., and R. J. Xavier. 2009. Autophagy, immunity and human disease. Curr. Opin. Gastroenterol. 25:512-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain, S., J. Pan, J. Xu, Y. Yang, Y. Chen, Y. Peng, Y. Wu, Z. Li, Y. Zhu, P. Tien, and D. Guo. 2006. Identification and characterization of severe acute respiratory syndrome coronavirus subgenomic RNAs. Adv. Exp. Med. Biol. 581:85-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, X. S., L. Y. Tang, J. Dai, H. Zhou, S. J. Li, Q. C. Xia, J. R. Wu, and R. Zeng. 2005. Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches: implications for cellular responses to virus infection. Mol. Cell. Proteomics 4:902-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabbage, M., and M. B. Dickman. 2008. The BAG proteins: a ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 65:1390-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamitani, W., C. Huang, K. Narayanan, K. G. Lokugamage, and S. Makino. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kassis, J. N., E. A. Guancial, H. Doong, V. Virador, and E. C. Kohn. 2006. CAIR-1/BAG-3 modulates cell adhesion and migration by downregulating activity of focal adhesion proteins. Exp. Cell Res. 312:2962-2971. [DOI] [PubMed] [Google Scholar]
- 28.Kersey, P. J., J. Duarte, A. Williams, Y. Karavidopoulou, E. Birney, and R. Apweiler. 2004. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4:1985-1988. [DOI] [PubMed] [Google Scholar]
- 29.Kirkegaard, K. 2009. Subversion of the cellular autophagy pathway by viruses. Curr. Top. Microbiol. Immunol. 335:323-333. [DOI] [PubMed] [Google Scholar]
- 30.Kratchmarova, I., B. Blagoev, M. Haack-Sorensen, M. Kassem, and M. Mann. 2005. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308:1472-1477. [DOI] [PubMed] [Google Scholar]
- 31.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 32.Kudchodkar, S. B., and B. Levine. 2009. Viruses and autophagy. Rev. Med. Virol. 19:359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kushner, D. B., B. D. Lindenbach, V. Z. Grdzelishvili, A. O. Noueiry, S. M. Paul, and P. Ahlquist. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 100:15764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuyumcu-Martinez, N. M., M. E. Van Eden, P. Younan, and R. E. Lloyd. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyratsous, C. A., and S. J. Silverstein. 2007. BAG3, a host cochaperone, facilitates varicella-zoster virus replication. J. Virol. 81:7491-7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyratsous, C. A., and S. J. Silverstein. 2008. The co-chaperone BAG3 regulates herpes simplex virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:20912-20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leong, W. F., H. C. Tan, E. E. Ooi, D. R. Koh, and V. T. Chow. 2005. Microarray and real-time RT-PCR analyses of differential human gene expression patterns induced by severe acute respiratory syndrome (SARS) coronavirus infection of Vero cells. Microbes Infect. 7:248-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lerena, M. C., C. L. Vazquez, and M. I. Colombo. 2010. Bacterial pathogens and the autophagic response. Cell. Microbiol. 12:10-18. [DOI] [PubMed] [Google Scholar]
- 39.Liu, C., H. Y. Xu, and D. X. Liu. 2001. Induction of caspase-dependent apoptosis in cultured cells by the avian coronavirus infectious bronchitis virus. J. Virol. 75:6402-6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, D. X., S. Shen, H. Y. Xu, and S. F. Wang. 1998. Proteolytic mapping of the coronavirus infectious bronchitis virus 1b polyprotein: evidence for the presence of four cleavage sites of the 3C-like proteinase and identification of two novel cleavage products. Virology 246:288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 42.Maxwell, K. L., and L. Frappier. 2007. Viral proteomics. Microbiol. Mol. Biol. Rev. 71:398-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi, H., N. Guo, A. Kejariwal, and P. D. Thomas. 2007. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 35:D247-D252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizutani, T., S. Fukushi, K. Ishii, Y. Sasaki, T. Kenri, M. Saijo, Y. Kanaji, K. Shirota, I. Kurane, and S. Morikawa. 2006. Mechanisms of establishment of persistent SARS-CoV-infected cells. Biochem. Biophys. Res. Commun. 347:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizutani, T., S. Fukushi, M. Saijo, I. Kurane, and S. Morikawa. 2005. JNK and PI3k/Akt signaling pathways are required for establishing persistent SARS-CoV infection in Vero E6 cells. Biochim. Biophys. Acta 1741:4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muslin, A. J., and J. M. Lau. 2005. Differential functions of 14-3-3 isoforms in vertebrate development. Curr. Top. Dev. Biol. 65:211-228. [DOI] [PubMed] [Google Scholar]
- 47.Nie, L., G. Wu, D. E. Culley, J. C. Scholten, and W. Zhang. 2007. Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit. Rev. Biotechnol. 27:63-75. [DOI] [PubMed] [Google Scholar]
- 48.Ohman, T., N. Lietzen, E. Valimaki, J. Melchjorsen, S. Matikainen, and T. A. Nyman. 2010. Cytosolic RNA recognition pathway activates 14-3-3 protein mediated signaling and caspase-dependent disruption of cytokeratin network in human keratinocytes. J. Proteome Res. 5:1549-1564. [DOI] [PubMed] [Google Scholar]
- 49.Ong, S. E., B. Blagoev, I. Kratchmarova, D. B. Kristensen, H. Steen, A. Pandey, and M. Mann. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1:376-386. [DOI] [PubMed] [Google Scholar]
- 50.Orvedahl, A., and B. Levine. 2009. Autophagy in mammalian antiviral immunity. Curr. Top. Microbiol. Immunol. 335:267-285. [DOI] [PubMed] [Google Scholar]
- 51.Park, S. K., J. D. Venable, T. Xu, and J. R. Yates III. 2008. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ravichandran, V., and E. O. Major. 2006. Viral proteomics: a promising approach for understanding JC virus tropism. Proteomics 6:5628-5636. [DOI] [PubMed] [Google Scholar]
- 55.Rosati, A., M. Ammirante, A. Gentilella, A. Basile, M. Festa, M. Pascale, L. Marzullo, M. A. Belisario, A. Tosco, S. Franceschelli, O. Moltedo, G. Pagliuca, R. Lerose, and M. C. Turco. 2007. Apoptosis inhibition in cancer cells: a novel molecular pathway that involves BAG3 protein. Int. J. Biochem. Cell Biol. 39:1337-1342. [DOI] [PubMed] [Google Scholar]
- 56.Rosati, A., K. Khalili, S. L. Deshmane, S. Radhakrishnan, M. Pascale, M. C. Turco, and L. Marzullo. 2009. BAG3 protein regulates caspase-3 activation in HIV-1-infected human primary microglial cells. J. Cell. Physiol. 218:264-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun, Y., W. Mi, J. Cai, W. Ying, F. Liu, H. Lu, Y. Qiao, W. Jia, X. Bi, N. Lu, S. Liu, X. Qian, and X. Zhao. 2008. Quantitative proteomic signature of liver cancer cells: tissue transglutaminase 2 could be a novel protein candidate of human hepatocellular carcinoma. J. Proteome Res. 7:3847-3859. [DOI] [PubMed] [Google Scholar]
- 59.Tang, B. S., K. H. Chan, V. C. Cheng, P. C. Woo, S. K. Lau, C. C. Lam, T. L. Chan, A. K. Wu, I. F. Hung, S. Y. Leung, and K. Y. Yuen. 2005. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 79:6180-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 61.Thomas, D., M. Guthridge, J. Woodcock, and A. Lopez. 2005. 14-3-3 protein signaling in development and growth factor responses. Curr. Top. Dev. Biol. 67:285-303. [DOI] [PubMed] [Google Scholar]
- 62.van Hemert, M. J., S. H. van den Worm, K. Knoops, A. M. Mommaas, A. E. Gorbalenya, and E. J. Snijder. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viswanathan, K., and K. Fruh. 2007. Viral proteomics: global evaluation of viruses and their interaction with the host. Expert Rev. Proteomics 4:815-829. [DOI] [PubMed] [Google Scholar]
- 64.Waters, K. M., J. G. Pounds, and B. D. Thrall. 2006. Data merging for integrated microarray and proteomic analysis. Brief. Funct. Genomic Proteomic 5:261-272. [DOI] [PubMed] [Google Scholar]
- 65.Yang, Z., and D. J. Klionsky. 2009. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young, P., E. Anderton, K. Paschos, R. White, and M. J. Allday. 2008. Epstein-Barr virus nuclear antigen (EBNA) 3A induces the expression of and interacts with a subset of chaperones and co-chaperones. J. Gen. Virol. 89:866-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng, R., H. Q. Ruan, X. S. Jiang, H. Zhou, L. Shi, L. Zhang, Q. H. Sheng, Q. Tu, Q. C. Xia, and J. R. Wu. 2004. Proteomic analysis of SARS associated coronavirus using two-dimensional liquid chromatography mass spectrometry and one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by mass spectrometric analysis. J. Proteome Res. 3:549-555. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, Z., L. B. Thackray, B. C. Miller, T. M. Lynn, M. M. Becker, E. Ward, N. N. Mizushima, M. R. Denison, and H. W. t. Virgin. 2007. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3:581-585. [DOI] [PubMed] [Google Scholar]
- 69.Ziebuhr, J. 2004. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 7:412-419. [DOI] [PMC free article] [PubMed] [Google Scholar]