Abstract
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) lef-3 is one of nine genes required for viral DNA replication in transient assays. LEF-3 is predicted to contain several domains related to its functions, including nuclear localization, single-strand DNA binding, oligomerization, interaction with P143 helicase, and interaction with a viral alkaline nuclease. To investigate the essential nature of LEF-3 and the roles it may play during baculovirus DNA replication, a lef-3 null bacmid (bKO-lef3) was constructed in Escherichia coli and characterized in Sf21 cells. The results showed that AcMNPV lef-3 is essential for DNA replication, budded virus production, and late gene expression in vivo. Cells transfected with the lef-3 knockout bacmid produced low levels of early proteins (P143, DNA polymerase, and early GP64) and no late proteins (P47, VP39, or late GP64). To investigate the functional role of domains within the LEF-3 open reading frame in the presence of the whole viral genome, plasmids expressing various LEF-3 truncations were transfected into Sf21 cells together with bKO-lef3 DNA. The results showed that expression of AcMNPV LEF-3 amino acids 1 to 125 was sufficient to stimulate viral DNA replication and to support late gene expression. Expression of Choristoneura fumiferana MNPV lef-3 did not rescue any LEF-3 functions. The construction of a LEF-3 amino acid 1 to 125 rescue bacmid revealed that this region of LEF-3, when expressed in the presence of the rest of the viral genome, stimulated viral DNA replication and late and very late protein expression, as well as budded virus production.
Members of the family Baculoviridae are large rod-shaped enveloped viruses containing a circular double-stranded DNA genome that varies in size from 80 to 180 kb (3). Baculoviruses are unique viruses that only replicate in invertebrates. In general, isolates of each baculovirus species exhibit a narrow host range. For example, Choristoneura fumiferana nucleopolyhedrovirus (CfMNPV) is known to infect only the spruce budworm (Choristoneura fumiferana), but Autographa californica multiple nucleopolyhedrovirus (AcMNPV) replicates in hosts derived from several families of Lepidoptera (14). The restriction of baculovirus replication in nonpermissive hosts has been studied, and a number of genes, expressed at different points in the virus replication cycle, have been identified as playing some role in this restriction (40). Most of these identified genes are associated with viral DNA replication and late gene expression.
Nine AcMNPV genes (ie-1, ie-2, p143, dnapol, lef-1, lef-2, lef-3, pe38, and p35) are required for directing transient replication of plasmids in transfected cells, suggesting that these genes are involved in baculovirus DNA replication (19, 27, 46). Only two of these genes, p143 and dnapol, have been shown to be essential for AcMNPV DNA replication in vivo (26, 41). Another gene, lef-11, although not essential for replication in transient assays, is also essential for DNA replication in vivo (24), indicating that questions concerning DNA replication need to be studied within the context of the whole virus genome.
LEF-3 is a single-stranded DNA-binding protein (SSB) that self-localizes to the nucleus (15, 45). LEF-3 is also responsible for transporting P143, a predicted DNA unwinding (helicase) protein, into the nucleus, where it is required for viral DNA replication (26, 29, 45). LEF-3 may also regulate the activity of a viral alkaline nuclease (AN) during viral DNA replication (32). We have previously mapped the region carrying the nuclear localization signal of LEF-3 to residues 26 to 32 within the N-terminal 56-amino-acid domain (1, 7). By fusing this domain in frame with P143 and testing the construct in transient plasmid replication assays, we showed that additional functions of LEF-3 are required during replication, in addition to interacting with P143 to transport it into the nucleus. In fact, we have demonstrated that there is a close interaction between LEF-3 and P143 (as well as the immediate-early 1 [IE-1] protein) on viral DNA in the nucleus (17), suggesting that direct interaction of LEF-3 and P143 is required during viral DNA replication. The LEF-3 domain necessary for directing P143 to the nucleus is included within the N-terminal 125 amino acids (7). Two conserved cysteine residues in this region (C82 and C106) are not essential for this function, so it is unknown which specific amino acids are involved in the LEF-3-P143 interaction (1).
In this study, a lef-3 knockout genome was constructed by exploiting a baculovirus shuttle vector (bacmid) system. Bacmids (a baculovirus genome carrying independent origins for replication in either bacteria or insect cells) were originally developed to prepare recombinant baculoviruses in Escherichia coli prior to transfection into insect cells (28). The system takes advantage of the site-specific transposition properties of the Tn7 transposon to simplify and enhance the process of generating recombinant bacmid DNA. In our case, we used the AcMNPV-derived bacmid as a template for deletion of the AcMNPV lef-3 gene and then examined the effect of this deletion on viral protein synthesis, budded virus (BV) production, and viral DNA replication. We also examined the ability of LEF-3 from another Alphabaculovirus species member, CfMNPV, to substitute for AcMNPV in a recombinant bacmid.
MATERIALS AND METHODS
Viruses and cells.
The commercially available bacmid bMON14272 (Invitrogen Life Technologies), derived from AcMNPV strain E2, was maintained in E. coli strain DH10B cells. Sf21 (Spodoptera frugiperda IPLB-Sf21) (13, 42) host insect cells were cultured at 28°C in TC100 medium supplemented with 10% fetal bovine serum (12). The HR3 strain of AcMNPV (10) and the EC1 strain of CfMNPV (25) were used as sources of the respective lef-3 genes.
Construction of lef-3 knockout and rescue bacmids.
A knockout bacmid was constructed in E. coli, replacing the AcMNPV lef-3 gene with the chloramphenicol aceytltransferase (cat) gene, using a lambda phage Red recombinase system expressed from plasmid pKD46 (9) as described for studies with other baculovirus knockouts (24, 37, 41). The cat gene was amplified from pKD3 by PCR using 70-nucleotide (nt) primers with 50 nt homologous to regions 5′ and 3′ to the lef-3 gene and 20 nt of sequence flanking the cat gene (primers C-25716 and C-25717 [Table 1]). The linear PCR product was electroporated into DH10B cells carrying the bacmid bMON14272 and plasmid pKD46 cells. Homologous recombination between the cat gene fragment and the lef-3 flanking sequences was detected by plating transformants in the presence of chloramphenicol (25 μg/ml) and kanamycin (50 μg/ml).
TABLE 1.
Primer sequences
| Application | Primer | Sequence |
|---|---|---|
| Amplification of cat with AcMNPV lef-3 flanking sequences | C-25716 | 5′-ACTTTGATTAAATTATCTTCCAGCAGCATTGAGATTTGATTGAAATCCGC/GTGTAGGCTGGAGCTGCTT-3′ |
| C-25717 | 5′-CCCACATATCGACAACAGCAATATGGCGACCAAAAGATCTTTGTCTGGAG/GCCATGGTCCATATGAATA-3′ | |
| Amplification of AcMNPV lef-3 locus | C-23321 | 5′-CCTCTGACTAGTGTGAGCCGCTTATAAAAC-3′ |
| C-22915 | 5′-CGTCGAATAGAATTAAGACCCCTTTATATTGTTC-3′ | |
| cat specific | C-25459 | 5′-GCTCATGGAAAACGGTGTAACAA-3′ |
| Amplification of AcMNPV lef-3 rescue gene with pFAcT | C-24244 | 5′-AACCGCTCTAGAGC/ACGCTCAGCAAAACTATAC-3′ |
| flanking sequences | C-24249 | 5′-AGTCCGCTCGAGCGG/TTAGTACAGCAATATG-3′ |
| Amplification of CfMNPV lef-3 replacement fragment | C-24499 | 5′-AAACGTCGAATAGAA/ACCGCTTGTTTAAAGAAAAT-3′ |
| C-24246 | 5′-ATCGACAACAGCAAT/ATGATGGCCACCAAACGCGA-3′ | |
| Amplification of AcMNPV lef-3 promoter | C-24245 | 5′-TTTGGTGGCCATCAT/ATTGCTGTTGTCGATATGTG-3′ |
| Amplification of AcMNPV lef-3 poly(A) | C-24248 | 5′-CTTTAAACAAGCGGT/TTCTATTCGACGTTTGGTTG-3′ |
| CfMNPV lef-3 specific | C-6721 | 5′-GTAACACTCTTGCTCAACC-3′ |
| Fusion PCR to delete LEF-3 amino acids 126 to 385 | C-24270 | 5′-ACTAACAAACATTTTTAATTCTATTCGACGTTTG-3′ |
| C-24271 | 5′-CGTCGAATAGAATTAAAAATGTTTGTTAGTCAAAT-3′ | |
| Real-time PCR, specific for AcMNPV DNA | p95-F1 | 5′-GAAACGACTTTCACCAAGCGGCTC-3′ |
| p95-B1 | 5′-CGGCGGCACGGGAACACATTTTAG-3′ | |
| Real-time PCR, specific for Sf21 DNA | C-28093 | 5′-GAAGAAGCACTCACAGTTCATCGGC-3′ |
| C-28094 | 5′-ATGTCATCAGCATTGCGGGTCCAG-3′ |
The phenotype of the bMON14272 bacmid is polyhedrin gene negative, so all donor plasmids constructed in this study were derived from pFAcT, which carries the AcMNPV polyhedrin gene under the control of its own promoter (8). This resulted in AcMNPV bacmids that more closely represented the wild-type AcMNPV, expressing polyhedrin and allowing for visual monitoring of very late gene expression. A rescue donor plasmid carrying the AcMNPV lef-3 coding region, including its promoter and poly(A) signal region, was amplified with primers C-24244 and C-24249 and using AcMNPV genomic DNA as template. The 1.9-kb PCR product was digested with XhoI and XbaI and cloned into XhoI-XbaI-digested pFAcT to produce pFAcT-Aclef3.
LEF-3 amino acids 126 to 385 were deleted from pFAcT-Aclef3 by the PCR fusion mutagenesis protocol. Primers C-24271 and C-24244 were used to amplify products upstream of the region to be deleted, while primers C-24270 and C-24249 were used to amplify products downstream of the region to be deleted. The two PCR products were then mixed, denatured, and used as templates in a third PCR amplification with primers C-24244 and C-24249. The final PCR product was digested with XbaI and XhoI and then cloned into XbaI- and XhoI-digested pFAcT-Aclef3 to produce pFAcT-Aclef3-125.
A CfMNPV lef-3 rescue donor plasmid was constructed in which the CfMNPV lef-3 open reading frame (ORF) was flanked by the AcMNPV lef-3 promoter and poly(A) regions. This fragment was produced by fusion PCR using three overlapping PCR products. First, fragment 1, containing the AcMNPV lef-3 promoter region (346-nucleotide sequence upstream of the AcMNPV LEF-3 start codon), was amplified with primers C-24244 (which introduced a XbaI restriction site at the 5′ end of the fragment) and C-24245 (which included 15 additional nucleotides at the 5′ end homologous to the CfMNPV lef-3 ORF). Second, fragment 2, containing the CfMNPV lef-3 ORF, was amplified with primers C-24246 (which included 15 nucleotides at the 5′ end homologous to the AcMNPV lef-3 promoter region) and C-24499 [which included 15 nucleotides at the 5′ end homologous to the AcMNPV lef-3 poly(A) region and 445 nt of sequence downstream from the AcMNPV LEF-3 stop codon]. Finally, fragment 3, containing the AcMNPV lef-3 poly(A) region was amplified with primers C-24248 (which included 15 additional nucleotides at the 5′ end homologous to the CfMNPV lef-3 ORF) and C-24249 (which introduced an XhoI restriction site at the 3′ end). A mixture of fragments 1 and 2 served as template for primers C-24244 and C-24499 to produce fragment 4. A mixture of fragments 3 and 4 served as template for primers C-24244 and C-24249 to produce the final 1.9-kb PCR fragment. This product was digested with XbaI and XhoI and cloned into XbaI-XhoI-digested pFAcT to produce pFAcT-Cflef3. All plasmid constructs were confirmed by DNA sequence analysis.
DH10B E. coli cells carrying both the helper plasmid pMON7124 (Invitrogen Life Technologies) and bMON14272 or knockout bacmid DNA were transformed with specific pFAcT-based donor plasmids. The transformed cells were incubated at 37°C for 4 h in 3 ml of Super Optimal broth with catabolite repressor (SOC) medium and then plated in the presence of chloramphenicol (25 μg/ml), kanamycin (50 μg/ml), gentamicin (7 μg/ml), tetracycline (10 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 100 μg/ml), and isopropyl-β-thiogalactopyranoside (IPTG; 40 μg/ml). Plates were incubated at 37°C for a minimum of 48 h, and white colonies resistant to chloramphenicol, kanamycin, gentamicin, and tetracycline were selected and streaked onto new selection plates to confirm the phenotype. Bacmid DNA was purified from transformed bacteria using a Qiagen large-construct kit.
Bacmid growth curves and titrations.
Sf21 cells were transfected with 1 μg of purified bacmid DNA as previously described (6). At various times after transfection, supernatants were harvested and titers were determined in 50% tissue culture infective dose (TCID50) endpoint dilution assays (25) to determine the replication properties of BV produced by the various bacmids. Transfected cells were also observed microscopically for cytopathic effects, including the production of polyhedra at very late times after transfection.
Analysis of protein expression.
Sf21 cells were transfected with wild-type or mutant bacmids and various previously described LEF-3 expression plasmids (1, 7) as indicated. At 6 h posttransfection, the cells were heat shocked at 42°C for 30 min and then incubated at 28°C. Whole-cell extracts were prepared at 48 h posttransfection, and the proteins from equivalent numbers of cells were resolved by SDS-polyacrylamide gel electrophoresis followed by electroblotting to Hybond C-extra nitrocellulose membranes (GE Healthcare). Monoclonal antibodies directed against P143 (21), P47 (4), GP64 (16), and VP39 (44) (used at a dilution of 1:1,000) have been previously described. Polyclonal anti-LEF-3 (1:2,500) has been previously described (6). Monoclonal antihemagglutinin (anti-HA; Sigma) directed against amino acid residues 98 to 106 (YPYDVPDYA) of human influenza virus HA, known as the HA tag, was used at a dilution of 1:3,000. Immunoblot analysis was performed using enhanced chemiluminescence reagents (GE Healthcare).
Quantitative DNA replication assays.
Replication of bacmid DNA in Sf21 cells was monitored by quantitative real-time PCR (qPCR). Sf21 cells (2 × 106) were transfected with 1 μg of bacmid DNA. At various times posttransfection, the cells were harvested into 1 ml of phosphate-buffered saline (BAC-PBS; 4 mM KCl, 14 mM NaCl, 0.1 mM Na2HPO4·7H2O, 1.05 mM KH2PO4, pH 6.2). Total DNA was extracted with an Easy-DNA kit (Invitrogen) and resuspended in 100 μl of elution buffer (10 mM Tris-HCl, pH 8.5). Prior to PCR, 4 μg of total DNA was digested with 20 units of DpnI (New England Biolabs) overnight to digest input DNA. DNA samples were diluted, and 3 μl was used for qPCR. Primers C-28093 and C-28094, designed to amplify a unique segment of genomic DNA (264 bp), were derived from the Sf21 hsp90 gene (20). Primers p95-F1 and p95-B1, designed to amplify a segment of AcMNPV (369 bp), were derived from sequences upstream of the AcMNPV hr3 region. We previously reported that the genome complement of Sf21 cells is 1.6 pg/cell (4 genome copies per cell) (5). This value was used in determining the cell numbers and then the ratio of viral to cell copy numbers. PCR was performed in a Rotogene 3000 cycler as previously described (5).
RESULTS
Construction of AcMNPV lef-3 knockout and rescue bacmids.
To investigate the in vivo function of LEF-3, we constructed a knockout AcMNPV bacmid in which a cat gene cassette replaced the lef-3 gene. To preserve the adjacent orf66, 99 bp of the 3′ end of the lef-3 coding region were retained in the bacmid. The 5′ end of lef-3 overlaps the other adjacent orf68 and its potential promoter region (Fig. 1A). Since the N-terminal 56 amino acids of LEF-3 contain a known nuclear localization signal domain and may be involved in P143 interaction (1, 7), it was considered important to completely remove this region from the lef-3 knockout. This meant that the potential promoter and 5′ end of orf68 were deleted. No differences in production of infectious budded virus, occlusion body formation, or the production of nucleocapsids were detected between an orf68 knockout and a wild-type bacmid (23). The lef-3 knockout bacmid was named bKO-lef3-cat (Fig. 1A). A polyhedrin-positive lef-3 knockout bacmid was then constructed by transposition between pFAcT and bKO-lef3-cat to produce bKO-lef3 (Fig. 1B). As a control for all subsequent experiments, a complete wild-type AcMNPV bacmid (bAcP), carrying the polyhedrin gene, was also constructed by transposing the polyhedrin gene in pFAcT into the polyhedrin locus of bMON14272. The presence of the AcMNPV lef-3 gene in bAcP was confirmed by PCR with primers C-22915 and C-23321 (Fig. 1C, Ac-lef3 [lane 1]). The deletion of lef-3 in bKO-lef3 was confirmed by PCR using two different primer pairs. Primers C-22915 and C-23321 confirmed the loss of the lef-3 coding region from bKO-lef3 (Fig. 1, Aclef3, compare lane 1 with lane 2), while primers C-25459 and C-24244 confirmed the recombination junction site between the cat gene and the viral sequences within orf68 (an expected 94- bp fragment when cat was present) (Fig. 1C, cat-lef3, compare lane 1 with lane 2). An AcMNPV lef-3 rescue bacmid was constructed by transposing the entire AcMNPV lef-3 ORF and its endogenous promoter from the donor plasmid pFAcT-Aclef3 into the polyhedrin locus of bKO-lef-3-cat. This produced a polyhedrin-positive bacmid expressing the AcMNPV lef-3 gene (bKO-lef3-Ac). The presence of AcMNPV lef-3 and the original cat gene in bKO-lef3-Ac were confirmed by PCR using primers C-22915 and C-23321 (Fig. 1C, Ac-lef3 [lane 3] and cat-lef3 [lane 3]).
FIG. 1.
Construction of lef-3 knockout and rescue AcMNPV bacmids. (A) Schematic drawing of construction of a lef-3 knockout bacmid (bKO-lef3-cat) from bMON14272. The relative location and orientations of orf66, orf68, and orf69 in the lef-3 locus of bMON14272 (AcMNPV bacmid) are indicated. The cat gene flanked by the FLP recognition target was amplified by PCR using primers C-25716 and C-25717, and the product was electroporated into DH10Bac-pKD46 cells to replace the lef-3 by homologous recombination (bKO-lef3-cat). The location and orientation of primers used to confirm the constructs are also shown. (B) Schematic diagram of construction of polyhedrin-positive bacmid bKO-lef3 and polyhedrin (polh)-positive rescue bacmids bKO-lef3-Ac and bKO-lef3-Cf from bKO-lef3-cat. The names and locations of primers used in the donor plasmid construction and the location of the selection antibiotic (Gen) are shown for each construct. (C) Results of PCR confirmation of bacmid sequence modifications. Purified bacmid DNA from bAcP (lane 1), bKO-lef3 (lane 2), bKO-lef3-Ac (lane 3), and bKO-lef3-Cf (lane 4) were used as templates with primer pairs specific for AcMNPV lef-3 (Ac-lef3; C-23321and C-22915) or the cat gene in the appropriate locations (cat-lef3; C-25459 and C-24244) or CfMNPV lef-3 (Cf-lef3; C-6721 and C-24244). Primers C-23321 and C-22915 were used to confirm the loss of the lef-3 coding region. Primers C-25459 and C-24244 were used to confirm the recombination junction of flanking regions.
To investigate viral species differences in LEF-3 function, an AcMNPV bacmid expressing the CfMNPV lef-3 gene was constructed. The CfMNPV lef-3 gene under the control of the AcMNPV lef-3 promoter from pFAcT-Cflef3 was transposed into the polyhedrin locus of bKO-lef3-cat to produce bKO-lef3-Cf (Fig. 1B). The presence of CfMNPV lef-3 (using primers C-24244 and C-6721) and the absence of the AcMNPV lef-3 gene (using primers C-22915 and C-23321) in bKO-lef3-Cf were confirmed by PCR (Fig. 1C, Aclef3 [lane 4] and Cf-lef3 [lane 4]).
DNA replication in the lef-3 knockout bacmid.
Because lef-3 has been shown to be essential for DNA replication in transient assays, the ability of the lef-3 knockout and rescue bacmids to replicate viral DNA in insect cells was determined. Various purified bacmid DNAs were transfected into Sf21 cells. At 24 and 48 h posttransfection, total intracellular DNA was harvested, and the amount of virus-specific DNA was quantified by qPCR. To ensure that only replicated DNA was amplified in the reactions, input bacmid DNA was eliminated by DpnI digestion of the total DNA samples prior to the PCR. The viral region targeted for PCR carries two DpnI sites. The amount of viral and cellular DNA for each bacmid transfection was determined using template-specific primers (Table 1) (5), and the relative viral genome copy numbers (viral genome copies per cell) of each bacmid at 24 and 48 h posttransfection were calculated (Fig. 2). The lef-3 knockout bKO-lef3 did not produce any detectable viral DNA at either 24 or 48 h posttransfection. Replication function was restored to the lef-3 knockout by adding back AcMNPV lef-3 in bKO-lef3-Ac, demonstrating for the first time that AcMNPV DNA replication in vivo is absolutely dependent upon lef-3 expression. As expected, adding the CfMNPV lef-3 gene to the lef-3 knockout did not restore any viral DNA replication. Clearly, CfMNPV LEF-3 even in the presence of the rest of the ACMNPV genome cannot complement AcMNPV LEF-3 in supporting efficient AcMNPV DNA replication in Sf21 cells, as previously suggested by transient plasmid replication assays (6).
FIG. 2.

Viral DNA replication in bacmid-transfected Sf21 cells. Total intracellular DNA from Sf21 cells transfected with bAcP, bKO-lef3, bKO-lef3-Ac, or bKO-lef3-Cf was isolated at 24 and 48 h posttransfection. The DNA was digested with DpnI for 16 h and analyzed by real-time PCR using host genome- and virus-specific primer pairs. The relative viral genome copy number per cell was calculated for each bacmid transfection. The results are expressed as the number of viral genome equivalents per cell and are displayed as averages of samples analyzed in triplicate from transfections performed in duplicate. Error bars indicate standard deviations.
Budded virus production in bacmid-transfected cells.
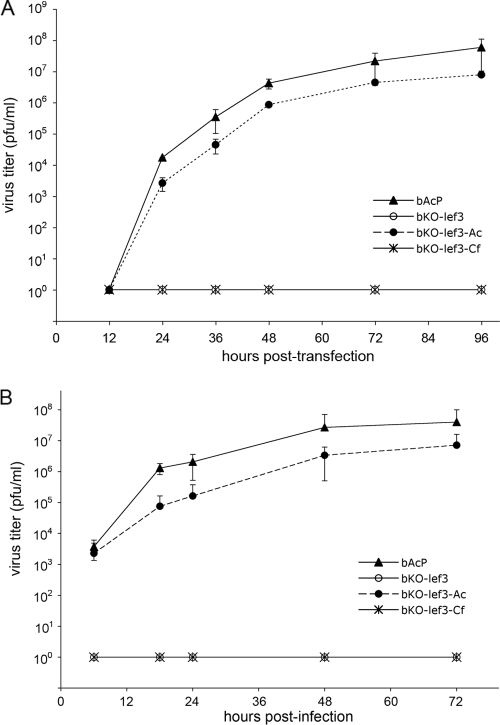
The process of viral DNA replication is considered essential for late gene expression leading to BV formation. To examine the ability of the lef-3 knockout and rescue bacmids to produce BV, Sf21 cells were transfected with 1 μg of each bacmid construct. At various times posttransfection, the supernatants were harvested, and titers were determined in TCID50 assays (Fig. 3A). The lef-3 knockout bacmid bKO-lef3 produced no detectable infectious virus, confirming that the expression of AcMNPV lef-3 and DNA replication are also essential for BV production. Transfection with bKO-lef3-Ac resulted in a normal virus infection cycle, with BV detectable by 24 h posttransfection with increasing titers to 96 h. Transfection with bKO-lef3-Cf resulted in no detectable BV production, even at 96 h, indicating that on its own, CfMNPV lef-3 was sufficiently different that it could not complement the AcMNPV homologue in supporting DNA replication or BV production.
FIG. 3.
Budded virus production in bacmid-transfected and infected cells. (A) Sf21 cells were transfected with bAcP, bKO-lef3, bKO-lef3-Ac, or bKO-lef3-Cf bacmid DNA. Cell culture supernatants were harvested at 12, 24, 36, 48, 72, and 96 h posttransfection and titrated by TCID50 endpoint dilution assays for the presence of infectious budded virus. The results represent the average titers derived from three independent assays. Error bars represent standard deviations. (B) Sf21 cells were infected (multiplicity of infection of 1) with supernatants harvested at 96 h posttransfection from cells transfected with bAcP, bKO-lef3, bKO-lef3-Ac, or bKO-lef3-Cf bacmid DNA. Cell culture supernatants from these infections were harvested and titrated by TCID50 assays for the presence of infectious budded virus. The results represent the average titers derived from three independent assays. Error bars represent standard deviations.
Differences in DNA preparations could affect the transfection efficiency and productivity in these assays. To demonstrate that the growth curves were an accurate representation of BV production from the rescue bacmids, supernatants from the original transfection mixtures were collected at 96 h posttransfection. Supernatants from cells transfected with bAcP, bKO-lef3, bKO-lef3-Ac,or bKO-lef3-Cf were used to conduct a second series of virus growth curve experiments. The results using BV were very similar to those from the transfection experiments (Fig. 3B). The wild-type bacmid bAcP and rescue bacmid bKO-lef3-Ac produced similar BV growth curves with titer increases between 12 and 24 h postinfection and continued to increase BV production until 72 h. The CfMNPV LEF-3 rescue bacmid bKO-lef3-Cf, as expected, did not produce any detectable BV over the same time period.
Deletion mutants of LEF-3 can support late gene expression.
The LEF-3 knockout bacmid provided us with a tool to investigate the functional role of LEF-3 during the virus replication cycle. We previously showed that amino acids 26 to 32 are essential for nuclear localization of LEF-3 and are also required for interaction with P143 (1). We have also demonstrated that the LEF-3 N-terminal 125 amino acids carry a domain required for nuclear transport of P143 (7). However, both of these studies were based on transient assays with transfected plasmids expressing different regions of AcMNPV LEF-3 and could not provide any information on the possible role that LEF-3 may play in expression of other viral genes. We therefore investigated the expression levels of a number of viral early and late genes in various bacmid-transfected cells.
Sf21 cells were transfected with bAcP, bKO-lef3, or bKO-lef3-Ac. At 48 h posttransfection, cell extracts were harvested and analyzed by immunoblotting with antibodies specific for early (P143 and DNA polymerase [DNApol]), early/late (GP64), and late (P47 and VP39) proteins. All of these proteins were easily detected in both bAcP and bKO-lef3-Ac, and the levels of protein expression were similar with both bacmids (Fig. 4A, lanes 1 and 2). However, in bKO-lef3-transfected cells, only the early proteins P143 and DNApol and early expression of GP64 were detected. Their levels were reduced compared to the amounts produced during infection with either bAcP or bKO-lef3-Ac (Fig. 4A, lane 3). This level of early gene products apparently represented the basal level of expression of P143 and DNApol in the absence of viral DNA replication. Although the gp64 gene codes for a structural envelop protein, it is transcribed at both early and late times after infection (2, 43). High levels of late expressed GP64 were detected in both bAcP- and bKO-lef3-Ac-transfected cells. Only a very weak early GP64 signal and no P47 or VP39 were observed in bKO-lef3-transfected cells, indicating that their late expression was dependent upon the presence of LEF-3.
FIG. 4.
Viral protein expression of bKO-lef3-transfected cells in the absence or presence of various truncations of LEF-3. (A) Sf21 cells were transfected with bacmid bAcP (WT; lane 1), AcMNPV lef-3 rescue bacmid (Ac-R; lane 2), or AcMNPV lef-3 knockout bacmid (KO; lane 3). Transfected cells were harvested at 48 h posttransfection and analyzed by immunoblotting for the expression of early and late viral proteins. Antibodies used as probes on the immunoblots were directed against the specific proteins indicated on the right. (B) The AcMNPV lef-3 knockout bacmid bAcKO-lef3 was transfected alone (lane 1) or cotransfected with plasmids expressing the full-length LEF-3 (lane 2), LEF-3 amino acids (aa) 2 to 189 (lane 3), LEF-3 aa 2 to 125 (lane 4), LEF-3 aa 2 to 83 (lane 5), or LEF-3 aa 190 to 385 (lane 5). At 48 h posttransfection, cell extracts were prepared and analyzed by immunoblotting for the expression of early and late viral proteins. Antibodies used as probes were directed against the specific proteins indicated on the right. The lower panel shows the results of probing the immunoblot with antibody directed against the HA tag fused to the various truncated LEF-3 proteins. The results of probing mock-transfected cells are also shown (lane 7).
We then investigated if specific regions of LEF-3 were capable of rescuing the ability of bKO-lef3 to stimulate late gene expression. Cells were cotransfected with bKO-lef3 and plasmids expressing various AcMNPV LEF-3 deletions (1), and the resulting cell extracts were examined by immunoblotting. As expected, the expression of all early and late proteins examined was stimulated to the same level as that observed with the bKO-lef3-Ac by cotransfecting a plasmid expressing the whole LEF-3 ORF (Fig. 4B, lane 2). These results also demonstrated that this approach could be used to examine the role that other regions might have in stimulating gene expression. Cotransfection of bKO-lef3 and plasmids expressing LEF-3 amino acids 1 to 83, or amino acids 190 to 385, resulted in levels of P143, DNApol, and GP64 similar to or less than that observed with bKO-lef3 alone, while no P47 or VP39 was detected (Fig. 4B, lanes 5 and 6). Expression of the LEF-3 N-terminal 189 amino acids stimulated low but detectable amounts of P47, GP64, and VP39 (Fig. 4B, lane 3). Surprisingly, expression of the LEF-3 N-terminal 125 amino acids enhanced the expression of P143 and DNApol and also stimulated detectable levels of P47 and high levels of GP64 and VP39 (Fig. 4B, lane 4). These data suggest that a region including the N-terminal 125 amino acids is required and sufficient to stimulate late gene expression. To confirm that the differences in the abilities of the different plasmids to support DNA replication were not an artifact related to inefficient protein expression, cell extracts of similar cotransfections were examined by immunoblotting using antibodies directed against the HA tag present on each construct (Fig. 4B, HA). The results demonstrated high levels of the full-length and various deletion mutant LEF-3 proteins, indicating that the lack of late protein synthesis was functionally related to the different expressed regions of LEF-3.
We also investigated the abilities of these same expression vectors to support very late gene expression by examining similarly cotransfected cells by using phase-contrast microscopy for the presence or absence of polyhedra. Polyhedra were seen in cells cotransfected with bKO-lef3 and plasmids expressing full-length LEF-3, LEF-3 amino acids 1 to 125, or LEF-3 amino acids 1 to 189 (data not shown). No polyhedra were observed in cells cotransfected with bKO-lef3 and plasmids expressing LEF-3 amino acids 1 to 83 or 190 to 385 (data not shown). Together, these data demonstrate that the LEF-3 region that includes amino acids 1 to 125 was able to rescue LEF-3 function responsible for late and very late gene expression, while amino acids 1 to 83 could not. Interestingly, there was a consistently lower level of early and late gene expression in the presence of amino acids 1 to 189 versus 1 to 125 (Fig. 4B, lane 3 versus lane 4).
Expression of LEF-3 amino acids 1 to 125 region can stimulate DNA replication.
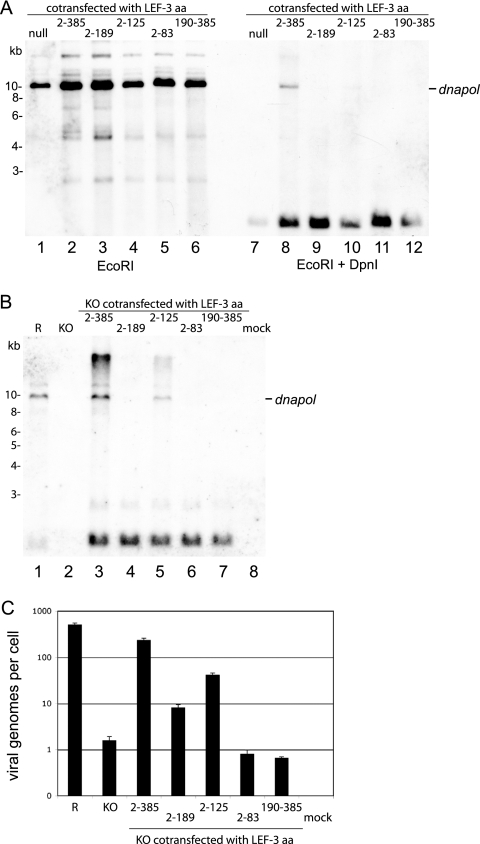
Since late baculovirus gene expression is somehow coupled to viral DNA replication, and based on the results presented above, we investigated the ability of the N-terminal 125 amino acids of LEF-3 to also support viral DNA replication using transient-replication assays. Two approaches were taken. First, Sf21 cells were cotransfected with the library of plasmids expressing a set of the minimum genes required for viral DNA replication (ie-1, ie-2, pe38, lef-1, lef-2, p143, dnapol, and p35) plus plasmids expressing various lef-3 constructs. At 48 h posttransfection, total intracellular DNA was purified and analyzed for plasmid DNA replication by DpnI digestion and Southern blotting, using a portion of the viral DNA polymerase gene as a probe. Full-length LEF-3 was able to stimulate DNA replication (Fig. 5A, lane 8), while very small amounts of replicated plasmid DNA could be detected in the presence of LEF-3 amino acids 1 to 125 (Fig. 5A, lane 10) or, to a lesser extent, with amino acids 1 to 189 (Fig. 5A, lane 9). No plasmid replication was detected when cells expressed LEF-3 regions 1 to 83 or 190 to 385 (Fig. 5A, lanes 11 and 12). Immunoblotting of similarly transfected cells, probed with an anti-HA antibody, confirmed the appropriate expression of the various LEF-3 construct proteins (data not shown).
FIG. 5.
Transient viral DNA replication assays. (A) Sf21 cells were transfected with a collection of plasmids, which together expressed the AcMNPV genes necessary for plasmid DNA replication (ie-1, dnapol, lef-1, lef-2, p35, pe38, and ie-2) in the absence of lef-3 (lane 1) or in the presence of plasmids expressing the full-length LEF-3 (lanes 2 and 8), LEF-3 amino acids (aa) 2 to 189 (lanes 3 and 9), LEF-3 aa 2 to 125 (lanes 4 and 10), LEF-3 aa 2 to 83 (lanes 5 and 11), or LEF-3 aa 190 to 385 (lanes 6 and 12). Following incubation for 48 h, total intracellular DNA was prepared and digested with EcoRI to linearize the plasmids or with EcoRI and DpnI to detect replicated plasmid DNA. Restricted DNA was prepared by Southern blotting, and replicated DNA was detected with a probe prepared from the viral DNA polymerase gene (dnapol). (B) Sf21 cells were transfected with rescue bacmid bKO-lef3-Ac (lane 1), bKO-lef3 (lane 2), or bKO-lef3 cotransfected with plasmids expressing the full-length LEF-3 (lane 3), LEF-3 aa 2 to 189 (lane 4), LEF-3 aa 2 to 125 (lane 5), LEF-3 aa 2 to 83 (lane 6), or LEF-3 aa 190 to 385 (lane 7). A control transfected with only herring sperm DNA was included (lane 8). At 48 h, total intracellular DNA was prepared and probed as described above. DpnI-resistant DNA was detected with a probed prepared from the viral DNA polymerase gene (dnapol). (C) The same DNA samples shown in panel B were digested with DpnI for 16 h and analyzed by real-time PCR using host genome- and virus-specific primer pairs. The relative viral copy number per cell was calculated for each bacmid transfection. The results are expressed on a log10 scale as the number of viral genome equivalents per cell and are displayed as averages of samples analyzed in triplicate. Error bars indicate standard deviations.
In the second approach, Sf21 cells were cotransfected with bKO-lef3 bacmid DNA and the same series of LEF-3 expression plasmids to determine the effect of expressing the various LEF-3 domains in the presence of the rest of the viral genome. At 48 h posttransfection, total intracellular DNA was harvested and analyzed for viral DNA replication by DpnI digestion and Southern blotting. As expected, viral DNA replication was detected with the rescue bacmid (Fig. 5B, bKO-lef3-Ac, lane 1). Viral DNA replication was also detected with the knockout bacmid bKO-lef3 in the presence of full-length LEF-3 (Fig. 5B, lane 3), as well as LEF-3 amino acids 1 to 125 (Fig. 5B, lane 5). A very faint band representing replicated DNA was observed with long exposures for LEF-3 amino acids 1 to 189 (Fig. 5B, lane 4), but no replication was detected in the presence of LEF-3 amino acids 1 to 83 or 190 to 385 (Fig. 5B, lanes 7 and 8).
The levels of detection of replicated DNA based on Southern blotting were low. Therefore, the total intracellular DNA from cells cotransfected with bKO-lef3 and the various LEF-3-expressing plasmids was also examined by qPCR to monitor the level of viral DNA replication. As expected, the highest level of replicated DNA was detected with full-length LEF-3 (about 240 copies per cell) (Fig. 5C, column 3). This was about 5-fold higher than the level seen with LEF-3 amino acids 1 to 125 (about 42 copies per cell) (Fig. 5C, column 5) and 25-fold higher than the level seen with LEF-3 amino acids 1 to 189 (about 8 copies per cell) (Fig. 5C, column 4). However, these levels were higher than background levels (about one copy or less per cell) of viral DNA detected in cells transfected with bKO-lef3 alone or with bKO-lef3 plus LEF-3 amino acids 1 to 83 (Fig. 5C, column 6) or 190 to 385 (Fig. 5C, column 7). Together these data demonstrated that the amino acids 1 to 125 region of LEF-3 was capable of supporting viral DNA replication in the presence of the minimum library of genes or in the presence of the viral genome (as a cotransfected lef-3 KO bacmid). The results also demonstrated that expressing LEF-3 amino acids 1 to 189 resulted in less efficient DNA replication than expressing the region including a smaller region, amino acids 1 to 125. These results were also consistent with the protein expression levels detected in experiments described above (Fig. 4).
Optimal viral DNA replication requires full-length LEF-3.
To investigate the role that the 1-125 LEF-3 domain had in supporting DNA replication in vivo, we constructed a rescue bacmid expressing this particular domain. The construct pFAcT-Aclef3-125 was introduced into the polyhedrin locus of bKO-lef3-cat to produce a polyhedrin-positive bacmid expressing only the N-terminal 125 amino acids of the AcMNPV lef-3 gene (bKO-lef3-125) (Fig. 6A). To evaluate the ability of this N-terminal LEF-3 region to support viral DNA replication in the context of the rest of the genome, quantitative DNA replication assays were performed. Replicated bAcP DNA was detectable by 12 h, and the level increased steadily to 24 h and then dramatically increased at 36 and 48 h. The rescue bacmid bKO-lef3-Ac produced a similar pattern of DNA replication, but the amount of DNA at each time point was less than in bAcP-transfected cells. By 48 h, the amount of viral DNA produced was about 60% of the bAcP level, consistent with data shown in Fig. 2. The bacmid expressing only the N-terminal 125 amino acids of LEF-3 (bKO-lef3-125) produced a profile almost identical to that of the rescue bacmid at 36 h, but by 48 h the amount of replicated viral DNA in bKO-lef3-125-transfected cells was about 30% of the amount seen in bAcP and about 50% of that seen with the rescue bacmid. These results clearly demonstrated that expression of only the N-terminal 125 amino acids of LEF-3 resulted in substantial levels of replicated viral DNA.
FIG. 6.
Construction and characterization of bKO-lef3-125. (A) Schematic diagram describing the construction of the LEF-3 amino acid 1 to 125 rescue bacmid, bKO-lef3-125. The location and orientation of primers used in donor plasmid constructions and the diagnostic PCR are shown as arrows with names. (B) Viral DNA replication in bacmid-transfected Sf21 cells. Total intracellular DNA was isolated from Sf21 cells transfected with bAcP, bKO-lef3-Ac, bKO-lef3-125, or bKO-lef3-Cf at various times posttransfection. The DNA was digested with DpnI for 16 h and analyzed by real-time PCR. The relative viral copy number per cell was calculated for each bacmid transfection. The results are expressed as the number of viral genome equivalents per cell and are displayed as averages of samples analyzed in triplicate from transfections performed in duplicate. Error bars indicate standard deviations. (C) Budded virus growth curves in Sf21 cells transfected with bAcP or bKO-lef3-125 DNA. Supernatants were harvested at various times posttransfection and titrated for the presence of infectious virus in TCID50 endpoint dilution assays. (D) The virus supernatants harvested at 96 h from bacmid transfection mixtures were used to infect Sf21 cells (multiplicity of infection, 0.1). At various times after infection, cell culture supernatants were harvested and titer were determined in TCID50 assays. Virus titers represent the averages derived from three independent assays. Error bars represent standard deviations.
We then examined the ability of bKO-lef3-125 to produce BV by transfecting Sf21 cells with bKO-lef3-125 or wild-type bacmid bAcP DNA and determining the BV titers at various times posttransfection (Fig. 6C). Transfection with bKO-lef3-125 resulted in a normal virus infection cycle, with BV production levels very similar to that of bAcP to 36 h. However, there was little or no increase in infectious BV to 96 h, and the final titer reached was about 10-fold lower than with bAcP. To confirm the infectivity of bKO-lef3-125 budded virus, Sf21 cells were infected with virus supernatants harvested from the original transfection mixtures at 96 h. Both bAcP and bKO-lef3-125 produced similar BV growth curves, with an initial increase seen by 24 h and a steady increase in titer to about 72 h postinfection. The final titer of BV produced by bKO-lef3-125 was about 40-fold lower than with the wild type (Fig. 6D). Clearly, deleting amino acids 126 to 385 of LEF-3 did not completely inhibit either viral DNA replication or BV production.
DISCUSSION
In this study, we have shown for the first time that LEF-3 is essential for AcMNPV DNA replication in vivo. By taking advantage of the AcMNPV bacmid system, we demonstrated that lef-3 is essential for all aspects of virus replication investigated, including DNA replication, BV production, and polyhedra formation, because all of these functions were absent in cells transfected with the lef-3 knockout bacmid. Repairing the lef-3 gene function by adding back the AcMNPV gene into the polyhedrin gene region of the rescue bacmid was successful in regenerating infectious virus, demonstrating that this single gene was responsible for the observed phenotype. Repairing the LEF-3 KO with the CfMNPV lef-3 gene did not rescue viral DNA replication or late gene expression, consistent with our previous results (6).
SSBs from other better-characterized systems are important multifunctional proteins which physically associate with a large number of different cellular genome maintenance proteins, likely defining their role as nucleoprotein substrates for DNA replication, recombination, DNA repair, and replication restart processes. SSBs can modulate the activity of other proteins by their ability to bind to single-stranded DNA (ssDNA), melting out double-stranded regions by contiguous cooperative binding but also by direct protein-protein interactions with proteins of the same replication system. These functions of SSBs appear to be conserved throughout all phyla and are essential for survival (39). The SSB family is characterized structurally by their oligonucleotide oligosaccharide-binding fold (OB fold), which binds to single-stranded DNA substrates (35). The primary sequence homology and peptide-binding homology of SSB OB folds are negligible, but structural analysis reveals common domains that can occur once (e.g., E. coli) or as multiple OB folds (e.g., mammalian RPA) (38). Another important aspect of SSB structures is their oligomeric form, which appears to bring together different numbers of OB folds to the protein's active form (18). Viruses with DNA genomes also require SSBs for replication of their genomes, and several have been identified in such diverse eukaryotic virus families as Adenoviridae, Herpesviridae, and Poxviridae. LEF-3 shares properties with these other well-characterized SSBs: it binds ssDNA (15, 45); it oligomerizes in solution (33, 34); it interacts with the helicase P143 (7, 45); it closely associates with the viral DNA polymerase and the potential initiator IE-1 on viral DNA (17); it localizes to the nucleus (1); it can both unwind DNA duplexes and assist in annealing cDNA strands (31).
Several functional domains in AcMNPV LEF-3 have been identified, including a nuclear localization signal (amino acids 26 to 32) (1), ssDNA-binding domains (amino acids 28 to 326) (33), an oligomerization region (amino acids 1 to 385) (11), a P143-interacting domain (amino acids 1 to 125) (7), and a region that interacts with the viral alkaline nuclease (amino acids 1 to 311) (30). If all of these functional domains are essential for LEF-3 function during the AcMNPV replication cycle, the deletion of any domain might cause a significant disruption to its structure, thus affecting the ability of LEF-3 to function normally, as suggested by an earlier mutagenesis analysis (11). Hence, we expected that any lef-3 deletion construct would not be able to support a complete replication cycle when cotransfected with the LEF-3 knockout bacmid. Thus, the results of both the cotransfection experiments and analysis of a bacmid expressing only the N-terminal 125 amino acids showing that this region could support viral DNA replication, late gene expression, and BV production were surprising.
Mutants of LEF-3 have been previously prepared and examined for their ability to support viral DNA replication (11). The results indicated that deleting 77 amino acids from the N terminus of LEF-3 was enough to inhibit its ability to support DNA replication in a transient assay, in agreement with our data which showed that up to 125 amino acids are required. However, in that study, deleting 71 amino acids from the C terminus also rendered LEF-3 unable to support DNA replication in transient assays (11). We did not construct a similar expression vector, but our data with the expression of the 189-amino-acid region suggest that there may be inhibitory effects seen if the complete LEF-3 ORF is not included in addition to the first 125 amino acids.
Little is known about the mechanism of baculovirus DNA replication, but most experimental data suggest that the process includes a rolling circle mechanism combined with recombination (22, 36, 47). Analysis of bacmids where the viral an gene has been knocked out reveals that the level of DNA replicated is normal but the by-products of replication are subgenomic branched molecules that may not be properly packaged into BV (36). In addition, no BV are produced in the absence of AN (37). LEF-3 interacts with AN, and the activity of AN is regulated in part by this interaction (30). LEF-3 has also been shown to bind viral DNA in a complex that includes at least P143 and IE-1 (17). Thus, LEF-3 appears to play a significant role in several aspects of baculovirus replication by interacting with itself as well as different proteins, including IE-1, P143, AN, and presumably the host cell karyopherins that are likely necessary for recognizing the LEF-3 nuclear localization signal and actively transporting it through nuclear pores. We have now shown that the region of LEF-3 involved in nuclear localization and interaction with P143 is sufficient to support viral replication and BV production. Since P143 is likely involved in unwinding double-stranded DNA and LEF-3 is then required to stabilize the resulting single-stranded DNA, the close interaction between P143 and LEF-3 may be essential for the correct coordination of these processes during replication. The reduced efficiency of replication observed with the bKO-lef3-125 construct may be related to its inefficient binding to ssDNA. One or more LEF-3 domains may be required for maximum DNA-binding efficiency, or other regions may be required for cooperative binding of additional LEF-3 molecules. It is also possible that other factors interact with LEF-3 during replication and that these interactions are also critical for maximizing the efficiency of the replication process, as suggested in other more-well-defined replication systems (18). The failure of CfMNPV LEF-3 to complement its AcMNPV homologue may be an indication that this ability to closely interact with other viral factors is critical to their function during replication. Further work is necessary to determine the role that other regions of LEF-3 play in making this process more efficient.
Acknowledgments
We gratefully acknowledge the technical assistance of Maike Bossert. We also thank Loy Volkman for providing VP39 antibody, Gary Blissard for providing GP64 antibody, and David Theilmann for providing the transfer plasmid pFAcT.
This research was supported by a grant from the Canadian Institutes of Health Research (FRN7624).
Footnotes
Published ahead of print on 31 March 2010.
REFERENCES
- 1.Au, V., M. Yu, and E. B. Carstens. 2009. Characterization of a baculovirus nuclear localization signal domain in the late expression factor 3 protein. Virology 385:209-217. [DOI] [PubMed] [Google Scholar]
- 2.Blissard, G. W., and G. F. Rohrmann. 1989. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 170:537-555. [DOI] [PubMed] [Google Scholar]
- 3.Carstens, E. B. 2009. AcMNPV as a model for baculovirus DNA replication. Virol. Sin. 24:243-267. [Google Scholar]
- 4.Carstens, E. B., A. L. Lu, and H. B. Chan. 1993. Sequence, transcriptional mapping, and overexpression of p47, a baculovirus gene regulating late gene expression. J. Virol. 67:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carstens, E. B., and Y. Wu. 2007. No single homologous repeat region is essential for DNA replication of the baculovirus Autographa californica multiple nucleopolyhedrovirus. J. Gen. Virol. 88:114-122. [DOI] [PubMed] [Google Scholar]
- 6.Chen, T., D. Sahri, and E. B. Carstens. 2004. Characterization of the interaction between P143 and LEF-3 from two different baculovirus species: Choristoneura fumiferana nucleopolyhedrovirus LEF-3 can complement Autographa californica nucleopolyhedrovirus LEF-3 in supporting DNA replication. J. Virol. 78:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z. L., and E. B. Carstens. 2005. Identification of domains in Autographa californica multiple nucleopolyhedrovirus late expression factor 3 required for nuclear transport of P143. J. Virol. 79:10915-10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, X. J., T. M. Stewart, J. A. Pathakamuri, Q. J. Li, and D. A. Theilmann. 2004. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J. Virol. 78:9633-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlandson, M. A., J. Gordon, and E. B. Carstens. 1985. Size and map locations of early transcription products on the Autographa californica nuclear polyhedrosis virus genome. Virology 142:12-23. [DOI] [PubMed] [Google Scholar]
- 11.Evans, J. T., and G. F. Rohrmann. 1997. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J. Virol. 71:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardiner, G. R., and H. Stockdale. 1975. Two tissue culture media for production of lepidopteran cells and nuclear polyhedrosis viruses. J. Invertebr. Pathol. 25:363-370. [Google Scholar]
- 13.Goodwin, R. H., J. L. Vaughn, J. R. Adams, and S. J. Louloudes. 1970. Replication of a nuclear polyhedrosis virus in an established insect cell line. J. Invertebr. Pathol. 16:284-288. [DOI] [PubMed] [Google Scholar]
- 14.Gröner, A. 1986. Specificity and safety of baculovirus, p. 177-202. In R. R. Granados and B. A. Federici (ed.), The biology of baculoviruses, vol. 1. CRC Press Inc., Boca Raton, FL. [Google Scholar]
- 15.Hang, X., W. Dong, and L. A. Guarino. 1995. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J. Virol. 69:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125:432-444. [DOI] [PubMed] [Google Scholar]
- 17.Ito, E., D. Sahri, R. Knippers, and E. B. Carstens. 2004. Baculovirus proteins IE-1, LEF-3, and P143 interact with DNA in vivo: a formaldehyde cross-linking study. Virology 329:337-347. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, X., V. Klimovich, A. I. Arunkumar, E. B. Hysinger, Y. Wang, R. D. Ott, G. D. Guler, B. Weiner, W. J. Chazin, and E. Fanning. 2006. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 25:5516-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kool, M., C. H. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl. Acad. Sci.U. S. A. 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landais, I., J. M. Pommet, K. Mita, J. Nohata, S. Gimenez, G. Devauchelle, M. Duonor-Cerutti, and M. Ogliastro. 2001. Characterization of the cDNA encoding the 90 kDa heat-schock protein in the Lepidoptera Bombyx mori and Spodoptera frugiperda. Gene 271:223-231. [DOI] [PubMed] [Google Scholar]
- 21.Laufs, S., A. Lu, L. K. Arrell, and E. B. Carstens. 1997. Autographa californica nuclear polyhedrosis virus p143 gene product is a DNA-binding protein. Virology 228:98-106. [DOI] [PubMed] [Google Scholar]
- 22.Leisy, D. J., and G. F. Rohrmann. 1993. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology 196:722-730. [DOI] [PubMed] [Google Scholar]
- 23.Li, G., J. Wang, R. Deng, and X. Wang. 2008. Characterization of AcMNPV with a deletion of ac68 gene. Virus Genes 37:119-127. [DOI] [PubMed] [Google Scholar]
- 24.Lin, G. Y., and G. W. Blissard. 2002. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J. Virol. 76:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J. J., and E. B. Carstens. 1993. Infection of Spodoptera frugiperda and Choristoneura fumiferana cell lines with the baculovirus Choristoneura fumiferana nuclear polyhedrosis virus. Can. J. Microbiol. 39:932-940. [Google Scholar]
- 26.Lu, A., and E. B. Carstens. 1991. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology 181:336-347. [DOI] [PubMed] [Google Scholar]
- 27.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDougal, V. V., and L. A. Guarino. 2000. The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. J. Virol. 74:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikhailov, V. S., K. Okano, and G. F. Rohrmann. 2003. Baculovirus alkaline nuclease possesses a 5′→3′ exonuclease activity and associates with the DNA-binding protein LEF-3. J. Virol. 77:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikhailov, V. S., K. Okano, and G. F. Rohrmann. 2005. The redox state of the baculovirus single-stranded DNA-binding protein LEF-3 regulates its DNA binding, unwinding, and annealing activities. J. Biol. Chem. 280:29444-29453. [DOI] [PubMed] [Google Scholar]
- 32.Mikhailov, V. S., K. Okano, and G. F. Rohrmann. 2004. Specificity of the endonuclease activity of the baculovirus alkaline nuclease for single-stranded DNA. J. Biol. Chem. 279:14734-14745. [DOI] [PubMed] [Google Scholar]
- 33.Mikhailov, V. S., K. Okano, and G. F. Rohrmann. 2006. Structural and functional analysis of the baculovirus single-stranded DNA-binding protein LEF-3. Virology 346:469-478. [DOI] [PubMed] [Google Scholar]
- 34.Mikhailov, V. S., A. L. Vanarsdall, and G. F. Rohrmann. 2008. Isolation and characterization of the DNA-binding protein (DBP) of the Autographa californica multiple nucleopolyhedrovirus. Virology 370:415-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murzin, A. G. 1993. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okano, K., A. L. Vanarsdall, and G. F. Rohrmann. 2007. A baculovirus alkaline nuclease knockout construct produces fragmented DNA and aberrant capsids. Virology 359:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okano, K., A. L. Vanarsdall, and G. F. Rohrmann. 2004. Characterization of a baculovirus lacking the alkaline nuclease gene. J. Virol. 78:10650-10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richard, D. J., E. Bolderson, and K. K. Khanna. 2009. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit. Rev. Biochem. Mol. Biol. 44:98-116. [DOI] [PubMed] [Google Scholar]
- 39.Shereda, R. D., A. G. Kozlov, T. M. Lohman, M. M. Cox, and J. L. Keck. 2008. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43:289-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiem, S. M. 1997. Prospects for altering host range for baculovirus bioinsecticides. Curr. Opin. Biotechnol. 8:317-322. [DOI] [PubMed] [Google Scholar]
- 41.Vanarsdall, A. L., K. Okano, and G. F. Rohrmann. 2005. Characterization of the replication of a baculovirus mutant lacking the DNA polymerase gene. Virology 331:175-180. [DOI] [PubMed] [Google Scholar]
- 42.Vaughn, J. L., R. H. Goodwin, G. L. Tompkin, and P. McCawley. 1977. The establishment of two insect cell lines from the insect Spodoptera frugiperda (Lepidoptera:Noctuidae). In Vitro 13:213-217. [DOI] [PubMed] [Google Scholar]
- 43.Whitford, M., S. Stewart, J. Kuzio, and P. Faulkner. 1989. Identification and sequence analysis of a gene encoding gp67, an abundant envelope glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 63:1393-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitt, M., and J. S. Manning. 1988. A phosphorylated 34 kDa protein and a subpopulation of polyhedrin are thiol linked to the carbohydrate layer surrounding a baculovirus occlusion body. Virology 163:33-42. [DOI] [PubMed] [Google Scholar]
- 45.Wu, Y., and E. B. Carstens. 1998. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology 247:32-40. [DOI] [PubMed] [Google Scholar]
- 46.Wu, Y., and E. B. Carstens. 1996. Initiation of baculovirus DNA replication: early promoter regions can function as infection-dependent replicating sequences in a plasmid-based replication assay. J. Virol. 70:6967-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, Y., G. Liu, and E. B. Carstens. 1999. Replication, integration, and packaging of plasmid DNA following cotransfection with baculovirus viral DNA. J. Virol. 73:5473-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]