Abstract
The arenavirus envelope glycoprotein (GPC) initiates infection in the host cell through pH-induced fusion of the viral and endosomal membranes. As in other class I viral fusion proteins, this process proceeds through a structural reorganization in GPC in which the ectodomain of the transmembrane fusion subunit (G2) engages the host cell membrane and subsequently refolds to form a highly stable six-helix bundle structure that brings the two membranes into apposition for fusion. Here, we describe a G2-directed monoclonal antibody, F100G5, that prevents membrane fusion by binding to an intermediate form of the protein on the fusion pathway. Inhibition of syncytium formation requires that F100G5 be present concomitant with exposure of GPC to acidic pH. We show that F100G5 recognizes neither the six-helix bundle nor the larger trimer-of-hairpins structure in the postfusion form of G2. Rather, Western blot analysis using recombinant proteins and a panel of alanine-scanning GPC mutants revealed that F100G5 binding is dependent on an invariant lysine residue (K283) near the N terminus of G2, in the so-called fusion peptide that inserts into the host cell membrane during the fusion process. The F100G5 epitope is located in the internal segment of the bipartite GPC fusion peptide, which also contains four conserved cysteine residues, raising the possibility that this fusion peptide may be highly structured. Collectively, our studies indicate that F100G5 identifies an on-path intermediate form of GPC. Binding to the transiently exposed fusion peptide may interfere with G2 insertion into the host cell membrane. Strategies to effectively target fusion peptide function in the endosome may lead to novel classes of antiviral agents.
Enveloped viruses enter their target cells through fusion of the virus and cell membranes, in a process promoted by the viral envelope glycoprotein. For some viruses, such as human immunodeficiency virus (HIV), entry is initiated by interaction of the envelope glycoprotein with cell surface receptor proteins. Other viruses, such as influenza virus, are endocytosed and membrane fusion is triggered by exposure to acidic pH in the maturing endosome. The subsequent merger of the viral and cell membranes is accomplished through a major structural reorganization of the envelope glycoprotein. Antiviral strategies that target virus entry by using neutralizing antibodies or small-molecule fusion inhibitors can, in many cases, prevent virus infection and disease.
The Arenaviridae comprise a diverse group of rodent-borne viruses, some of which are responsible for severe hemorrhagic fevers in humans. Lassa fever virus (LASV) is endemic in western Africa (59), and at least five New World species are recognized to cause fatal disease in the Americas, including the Argentine hemorrhagic fever virus Junín (JUNV) (63). New pathogenic arenavirus species continue to emerge from their distinct animal reservoirs (1, 11, 24). At present, there are no licensed vaccines or effective therapies to address the threat of arenavirus infection.
Arenaviruses are enveloped, negative-strand RNA viruses whose bipartite genome encodes ambisense expression of four viral proteins (12, 22). The arenavirus envelope glycoprotein, GPC, is a member of the class I virus fusion proteins (33, 40, 75), a group that includes HIV Env, influenza virus hemagglutinin (HA), and paramyxovirus F protein. These envelope glycoproteins share several salient features. The precursor glycoproteins assemble as trimeric complexes and are subsequently rendered competent for membrane fusion by a proteolytic cleavage that results in the formation of the mature receptor-binding and transmembrane fusion subunits. The GPC precursor glycoprotein is cleaved by the cellular SKI-1/S1P protease (6, 51, 54) to generate the respective G1 and G2 subunits, which remain noncovalently associated. The ectodomain of the class I fusion subunit is distinguished by the presence of two 4-3 heptad repeat (HR1 and HR2) sequences that, in the course of membrane fusion, refold to form the now-classical six-helix bundle structure, which defines this class of envelope glycoproteins. Unlike other class I fusion proteins, GPC also contains a cleaved and stable signal peptide (SSP) as a third and essential subunit in the mature complex (2, 32, 69, 77, 81).
Arenavirus infection is initiated by G1 binding to a cell surface receptor. The pathogenic clade B New World arenaviruses utilize transferrin receptor 1 (TfR1) for entry (1, 64, 65), whereas those in clades A and C, as well as the Old World viruses, bind α-dystroglycan and/or an unknown receptor (15, 34, 71). The virion particle is subsequently endocytosed (9), and membrane fusion is initiated by acidification in the maturing endosome (17, 28, 29). pH-dependent activation of GPC is modulated through a unique interaction between SSP and G2 (79, 80) and can be targeted by small-molecule inhibitors that block membrane fusion (76) and protect against arenavirus infection (8, 52).
A generally accepted model for membrane fusion by the class I envelope glycoproteins (reviewed in references 45 and 73) posits that the native complex exists in a metastable state that is established on proteolytic maturation of the biosynthetic precursor. Upon activation, whether by acidic pH in the endosome or receptor binding at the plasma membrane, the fusion subunit that was sequestered in the prefusion state is exposed and undergoes a series of dramatic conformational changes leading to membrane fusion. In this process, a hydrophobic region at or near the N terminus of the fusion subunit (the fusion peptide) inserts into the host cell membrane, thus allowing the protein to bridge the two membranes. This so-called prehairpin intermediate subsequently collapses upon itself to form the highly stable six-helix bundle structure, in which the three HR2 helices pack into hydrophobic grooves on the trimeric HR1 coiled-coil in an antiparallel manner, bringing the virus and cell membranes into apposition. Free energy made available in the formation of this stable structure is thought to drive fusion of the lipid bilayers. Peptides that correspond in sequence to HR2 (C-peptides) bind to the putative prehairpin intermediate and interfere with its refolding, thereby preventing membrane fusion (18, 57, 74). While the structure of the six-helix bundle core has been elucidated in atomic detail (45, 73), information regarding the molecular pathway leading to this postfusion state is largely indirect. Indeed, the prehairpin intermediate is conceptualized through the activity of C-peptide fusion inhibitors (57, 74).
In this report, we describe a G2-directed monoclonal antibody (MAb), F100G5, that recognizes a pH-induced intermediate of JUNV GPC and prevents GPC-mediated membrane fusion. This MAb binds at or near the internal fusion peptide of G2 and may act by interfering with its penetration into the host cell membrane. These studies highlight the feasibility of targeting short-lived GPC intermediates for inhibition of membrane fusion.
MATERIALS AND METHODS
Monoclonal antibodies.
For immunization, the G2 ectodomain sequences of JUNV (Romero strain, amino acids 255 to 417; GenBank accession number AY619641) and Machupo virus (MACV; Carvallo strain; amino acids 266 to 428; GenBank accession number AY619643) were molecularly cloned into the pQE30 Escherichia coli expression plasmid (Qiagen) The 6His-tagged fusion proteins were expressed and solubilized in QIAexpress buffers containing 8 M urea or 6 M guanidium hydrochloride (GdmHCl), bound to an Ni-nitrilotriacetic acid (NTA) matrix (Qiagen), and eluted using a low-pH buffer (buffer E; Qiagen) containing urea and 120 mM imidazole. Proteins were dialyzed against phosphate-buffered saline (PBS) and formulated with complete Freund's adjuvant for immunization. The antibody response in BALB/c mice was assessed in an enzyme-linked immunosorbent assay (ELISA) using the protein (60 ng per well) as antigen (43), and spleens from responding animals were used to generate hybridomas as previously described (5). Identification and purification of reactive hybridomas were guided by ELISA using the respective immunogens. Hybridoma cell culture supernatant was used as the source of MAb, and F100G5-containing supernatant was estimated to contain 150 μg/ml of immunoglobulin. The well-characterized G1-directed MAb GB03-BE08 (68) and the JUNV nucleoprotein-directed MAb NA05-AG12 were obtained from the CDC through the NIH Biodefense and Emerging Infectious Diseases Research Resources Repository.
Reactivity of the newly developed MAbs to authentic JUNV GPC was determined by Western blot analysis using Vero cell cultures infected with the Romero strain of JUNV. All virus experiments were performed in the biosafety level 4 (BSL4) facility at the National Microbiology Laboratory of the Public Health Agency of Canada (Winnipeg, Manitoba, Canada), and cell lysates were boiled in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer prior to removal from containment. Proteins were resolved by SDS-PAGE, and Western blot reactivity was visualized by chemiluminescence. The attenuated vaccine strain of JUNV, Candid 1 (4, 58), was grown under BSL2 conditions at The University of Montana.
Dissection of the G2 six-helix bundle.
JUNV G2 ectodomain segments (residues 264 to 414 and 305 to 418; referred to as G2-151 and G2-114, respectively) were introduced into the pET3a vector (Novagen) and the recombinant proteins were expressed in E. coli BL21(DE3)/pLysS (Novagen) as previously described (57). Inclusion bodies were washed extensively with 1% Triton X-100 and solubilized in buffer containing 8 M urea. The protein was then loaded onto a DEAE-Sepharose column (Pharmacia) equilibrated with 3 M urea and eluted using a NaCl gradient (0 to 1,000 mM). Following dialysis against 5% (vol/vol) acetic acid, the protein was purified to homogeneity by reverse-phase high-performance liquid chromatography (HPLC; Waters) on a Vydac C4 preparative column (Hesperia, CA) with a water-acetonitrile gradient in the presence of 0.1% trifluoroacetic acid as previously described (70). Intramolecular disulfide bonds were formed by air oxidation in the presence of 6 M GdmHCl, and the oxidized protein was repurified by reverse-phase HPLC and lyophilized.
The G2-151 protein is largely aggregated in 50 mM Tris-HCl (pH 8.0) and was resuspended in this buffer for digestion with trypsin or proteinase K (1:200, wt/wt) for 1 h at room temperature as previously described (57). Soluble protease-resistant material was reduced with dithiothreitol, analyzed by reverse-phase HPLC on a Vydac C18 column, and identified by N-terminal sequencing and electrospray mass spectrometry (PerSeptive Biosystems Voyager Elite [Cambridge, MA]). Additional peptides were synthesized by standard fluorenylmethoxycarbonyl chemistry with an acetylated N terminus and amidated C terminus. After cleavage from the resin, the peptides were purified by reverse-phase HPLC. Protein concentrations were determined by using the method of Edelhoch (31).
Circular dichroism spectroscopy.
Circular dichroism (CD) experiments were performed on an Aviv 62A/DS spectropolarimeter (Aviv Associates, Lakewood, NJ) equipped with a thermoelectric temperature control, in phosphate-buffered saline (PBS; 50 mM sodium phosphate, 150 mM NaCl; pH 7.0). CD spectra were collected from 260 to 200 nm at 4°C, using an average time of 5 s and a bandwidth of 1 nm. A [θ]222 value of −33,000 deg cm2 dmol−1 was taken to correspond to 100% helix (21). Thermal stability was determined by monitoring the ellipticity at 222 nm of the N29/C30 complex, [θ]222, as a function of temperature in PBS (pH 7.0). Thermal melts were performed in 2° intervals with a 2-min equilibration at the desired temperature and an integration time of 30 s. Reversibility was verified by repeated scans. Superimposable folding and unfolding curves were observed, and >85% of the signal was regained upon cooling. Values of midpoint unfolding transitions (Tm) were estimated by evaluating the maximum of the first derivative of [θ]222 versus temperature (14).
Sedimentation equilibrium analysis.
Analytical ultracentrifugation measurements were carried out on a Beckman XL-A (Beckman Coulter) analytical ultracentrifuge equipped with an An-60 Ti rotor (Beckman Coulter) at 20°C. Peptide samples were dialyzed overnight against PBS, loaded at initial concentrations of 30, 100, and 300 μM, and analyzed at rotor speeds of 22 and 25 krpm. Data were acquired at two wavelengths per rotor speed setting and processed simultaneously with a nonlinear least-squares fitting routine (47). Solvent density and protein partial specific volume were calculated according to solvent and protein composition, respectively (53). The apparent molecular mass of the N29/C30 complex was within 10% of that calculated for an ideal trimer, with no systematic deviation of the residuals.
Peptide ELISA.
Ninety-six-well microtiter plates (Immulon II; ThermoLabsystems) were coated overnight with purified peptide (10 μg/ml in 50 mM Tris-HCl, pH 8.8) and subsequently washed with Dulbecco's phosphate-buffered saline (DPBS) containing 0.1% Tween 20 and blocked in the same buffer plus 1% bovine serum albumin and 5% dried milk. Control wells were coated with S-protein (Novagen). Purified MAb (200 ng/well) or hybridoma supernatant (undiluted or 1:4 dilution) in blocking buffer was incubated for 1 h at 37°C prior to washing and incubation with a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch). Reaction of the SureBlue TMB chromogenic reagent (KPL, Inc.) was stopped with the addition of 1 N HCl, and color was read at 450 nm. In our studies, specific binding was either all or none: ≥2 absorbance units or ≤0.05 absorbance units.
GPC-mediated cell-cell fusion.
The ability of MAbs to inhibit GPC-mediated membrane fusion in cell culture was determined using the molecularly cloned GPC from the MC2 strain of JUNV (GenBank accession number D10072) (79, 81). This GPC is closely related to that of the Romero strain and identical in its G2 ectodomain. Transient expression was accomplished as previously described (79, 81) using the bacteriophage T7 promoter in the pcDNA3.1-based (Invitrogen) GPC plasmid and Vero cells infected with the vTF-7 vaccinia virus expressing T7 polymerase (38). In some studies, to obviate potential concerns regarding the relative efficiency of signal peptidase cleavage, GPC expression was accomplished by cotransfection of two plasmids: one expressing SSP (in which a stop codon was introduced following the C-terminal amino acid in SSP [T58]) and the other expressing the entire G1-G2 precursor (where SSP was replaced by the conventional signal peptide of CD4) (78). The respective polypeptides have been shown to associate in trans to reconstitute the native GPC complex (32, 80).
pH-dependent membrane fusion and syncytium formation were assessed using a vaccinia virus-based β-galactosidase fusion-reporter assay (62) as previously described (80, 81). Briefly, Vero cells expressing GPC and the T7 polymerase were gently sedimented onto, and cocultured for 3 h with, target cells infected with the vCB21R-lacZ vaccinia virus bearing the β-galactosidase gene under the control of a T7 promoter (62). Syncytium formation was initiated by exposing the coculture for 20 min to low-pH medium containing 10 mM HEPES and 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (adjusted to pH 5.0) (80, 81). Cell-cell fusion was reported based on expression of β-galactosidase after a 5-h incubation at neutral pH. Membrane fusion per se is likely completed during exposure to acidic pH, as would occur in the endosome, and in this assay becomes manifest only in the subsequent incubation period. β-Galactosidase activity was quantitated using the chemiluminescent GalactoLite Plus substrate (Applied Biosystems) and a Tropix TR717 microplate luminometer. In studies examining inhibition of cell-cell fusion, MAbs were added either to GPC-expressing cells prior to plating onto the vCB21R-lacZ target cells and throughout the 3-h coculture, with a 20-min exposure to pH 5.0, or for the final 5-h incubation at neutral pH. All incubations were at 37°C, and any residual buffering or MAb carried over during medium changes are insignificant. MAb F100G5 supernatant was used at a final concentration of ∼15 μg/ml, and purified GB03-BE08 was used at 10 μg/ml.
Flow cytometry.
Vero cells expressing GPC were resuspended in growth medium, or in medium adjusted to pH 5.0, and incubated with intermittent swirling for 10 min at 37°C. Cells were rapidly neutralized by the addition of ice-cold DPBS containing 2% fetal bovine serum (FBS), collected by centrifugation, and resuspended in buffer containing MAb (∼30 μg/ml F100G5 or 10 μg/ml GB03-BE08) and 0.01% sodium azide. Note that GPC is irreversibly inactivated by transient exposure to acidic pH in the absence of target cells and, at this point in the assay, is no longer able to mediate membrane fusion (reference 29 and unpublished data). After 1 h on ice, the cells were washed and incubated with a fluorescein-conjugated goat anti-mouse secondary antibody and propidium diiodide prior to fixation using 4% formaldehyde. Populations were analyzed using a FACSCalibur flow cytometer and CellQuest software (BD Biosciences) as previously described (75). In some studies, GPC-expressing Vero cells were treated at neutral pH with 50 μM small-molecule fusion inhibitor ST-294 (8) prior to acidification and subsequent incubation with MAb (76). ST-294 was kindly provided by Sean Amberg (SIGA Technologies, Inc.).
Dissection of MAb epitopes.
The G2 ectodomain region of the molecularly cloned JUNV GPC (amino acids 252 to 424) was appended in frame to the C terminus of maltose-binding protein (MBP) in the pMAL-c2E vector (New England Biolabs) using standard PCR and molecular cloning techniques. Expression in E. coli TB1 cells (New England Biolabs) was induced by the addition of isopropylthiogalactoside (ITPG), and cells were disrupted using Bugbuster (Pierce) containing 250 units/ml Benzonase (Pierce) and protease inhibitors (leupeptin, pepstatin, aprotinin, and phenylmethylsulfonyl fluoride). Aliquots of the cleared lysate were incubated with a 50% slurry of amylose resin (New England BioLabs) for 2 h at 4°C to purify the MBP fusion proteins. The resin was subsequently washed, and bound fusion proteins were eluted with maltose. Proteins were resolved using 4-to-12% NuPAGE bis-Tris gels (Invitrogen) and detected by SYPRO Red protein stain (Molecular Probes) or by Western blot analysis. N- and C-terminal portions of the G2 ectodomain (amino acids 252 to 316 and 317 to 424, respectively) were similarly appended to MBP for analysis.
GPC mutants were used to further dissect MAb-binding epitopes. GPC mutants were created using QuikChange mutagenesis (Stratagene), replacing highly conserved charged residues in the N-terminal portion of G2 with alanine. These mutants (D260A, K283A, K292A, H297A, E300A, D303A, R306A, D309A, D339A, K344A, R348A, and K360A) were characterized as previously described (79) and used in Western blot experiments to assess MAb binding. All but 3 of the 12 mutants were expressed as wild-type GPC and were competent to mediate cell-cell fusion. Mutations specifically included within the N-terminal G2-ecto MBP fusion protein recognized by F100G5 are further described below.
MAb-mediated virus neutralization.
A predetermined amount of JUNV Candid 1 virus was incubated with MAb in growth medium for 10 min, and the mixture was then placed onto Vero cells for 8 h at 37°C. Final concentrations of F100G5 and GB03-BE08 were ∼30 μg/ml and 10 μg/ml, respectively. Cells were then washed, harvested with trypsin-EDTA (Invitrogen), and distributed to six-well culture plates, where infection was allowed to proceed for an additional 16 h. Cultures were then fixed with cold methanol-acetone (1:1), and the number of infected (multicellular) foci was determined by immunohistochemical staining using the nucleoprotein MAb NA05-AG12 (68), horseradish peroxidase-conjugated sheep anti-mouse secondary antibody, and diaminobenzidine substrate.
RESULTS
Monoclonal antibodies directed against the ectodomain of G2.
Recombinant forms of the G2 ectodomain of JUNV and MACV (residues 255 to 417 and 266 to 428, respectively) (Fig. 1) were expressed in E. coli and purified by affinity chromatography using an N-terminal 6His tag. JUNV and MACV are two closely related pathogenic New World arenaviruses, and the sequences included within the recombinant constructs were 88% identical (with largely conservative amino acid changes at other positions). Mice were immunized with the recombinant proteins (referred to as JUNV G2* and MACV G2*, respectively) and used to generate immunogen-specific hybridomas (5) (Table 1).
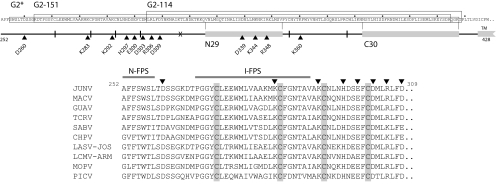
FIG. 1.
Schematic representation of the JUNV G2 ectodomain and fusion peptide region. The amino acid sequence of the JUNV G2 ectodomain is shown on top, in text, and as a line drawing (residues 252 to 428; MC2 strain, accession number D10072). Small dots above the text are spaced 10 amino acids apart, starting with position 260. The boxed sequences comprise peptides G2*, G2-151, and G2-114 (see text). The nominal start of the G2 transmembrane domain is indicated at residues 425 to 428. Cysteines are marked by vertical lines in the schematic, and gray boxes represent HR1 and HR2, as defined by the N29 and C30 peptides (see text). The X marks the division between N- and C-terminal regions in MBP fusion proteins, and arrowheads indicate alanine mutations used in this work. The positions chosen for mutation are identically conserved or invariant in charge among arenaviruses. Below, fusion peptide sequences are compared among arenaviruses (JUNV residues 252 to 309). N-terminal and internal fusion peptide regions (N-FPS and I-FPS, respectively) are indicated and are based on the work of Klewitz et al. (50). Conserved cysteines are highlighted in gray, and arrowheads represent the alanine mutations studied. Accession numbers for other arenavirus glycoproteins are as follows: Machupo virus (MACV), AAT40455; Guanarito virus (GUAV), AAS55656; Tacaribe virus (TCRV), NP_694849; Sabiá virus (SABV), YP_089665; Chapare virus (CHPV), YP_001816782; LASV-Jos, NP_694870; lymphocytic choriomeningitis virus ([LCMV] LCMV-Arm), NP_694851; Mopeia virus (MOPV), YP_170709; and Pichinde virus (PICV), AAB58484.
TABLE 1.
Properties of monoclonal antibodies directed to the G2 ectodomains of JUNV and MACVa
| Immunogen | MAb | MAb isotype | Western blot reactivityb | ELISA result |
Flow cytometry result atd: |
||
|---|---|---|---|---|---|---|---|
| Six-helix bundle peptidec | G2-114 peptide | Neutral pH | pH 5.0 | ||||
| JUNV G2* | F100G1 | IgG1/κ | − | − | +++ | − | − |
| F100G2 | IgG1/κ | − | − | − | − | − | |
| F100G3 | IgG1/κ | − | − | − | − | − | |
| F100G4 | IgG1/κ | ++ | +++ | +++ | − | − | |
| F100G5 | IgG1/κ | ++ | − | − | − | +++ | |
| F100G6 | IgG2a/κ | − | − | +++ | − | − | |
| F109G1 | IgG2b/κ | − | − | +++ | − | − | |
| MACV G2* | F106G1 | IgG1/κ | − | − | − | − | ± |
| F106G2 | IgG1/κ | − | − | − | − | − | |
| F106G3 | IgG1/κ | +++ | − | − | − | ± | |
| F106G4 | IgG2b/κ | − | − | − | − | − | |
| F106G5 | IgG2b/κ | − | − | − | − | − | |
| F106G6 | IgG2a/κ | − | − | − | − | − | |
| F111G2 | IgG1/κ | − | − | − | − | − | |
| F111G3 | IgG1/κ | − | − | − | − | − | |
| F111G4 | IgG1/κ | − | − | − | − | − | |
| JUNV virione | GB03-BE08 | IgG2a/ND | + | − | − | +++ | +++ |
Reactivity in all assays was scored relative to the maximal response in the panel of G2 MAbs, with semiquantitative quantiles reported as follows: from − (no response) to ±, +, ++, and +++ (maximal response).
Reactivity to GPC in JUNV-infected cell lysates.
Equimolar N29 and C30 peptides were mixed prior to coating plates for the ELISA.
Selected histograms are shown in Fig. 3A.
MAb recognition of the authentic JUNV GPC was determined by Western blot analysis of JUNV-infected cells (Romero strain) (Table 1). Two of seven MAbs from JUNV G2*-immunized mice (F100G4 and F100G5) and one of nine from MACV G2*-immunized mice (F106G3) were able to recognize JUNV GPC in this context.
Identification of the G2 six-helix bundle core.
Ectopic expression of the class I virus fusion protein ectodomain in E. coli typically yields insoluble peptide aggregates which must be solubilized and refolded in vitro to attain a more native structure, that of the stable postfusion six-helix bundle (7, 57). MAbs directed against this structure in other envelope glycoproteins have proven invaluable for studying structure-function relationships in membrane fusion (44, 46, 48, 55), and we were interested to identify any such MAbs in our panel. Although prior studies by Eschli and colleagues had demonstrated that G2 can adopt an α-helical trimeric structure comprising heterodimeric HR1 and HR2 sequences (33), we sought to precisely define the extent of the six-stranded helical structure so as to identify specific MAbs. The recombinant peptide G2-151, comprising the JUNV G2 ectodomain without the extreme N- and C-terminal regions (residues 264 to 414) (Fig. 1), was expressed in E. coli and harvested as insoluble inclusion bodies. G2-151 was solubilized using 8 M urea and purified by reverse-phase chromatography as described in Materials and Methods. Following air oxidation, to allow formation of a disulfide-bonded loop that typically separates HR1 and HR2 regions in class I fusion proteins, the protein was subjected to refolding by stepwise dilution into PBS supplemented with 1.5 M GdmCl. In this buffer at a 10 μM protein concentration and 4°C, the CD spectrum of G2-151 exhibited the characteristic signature of an α-helical conformation, with minima at 222 and 208 nm (data not shown). Sedimentation equilibrium measurements revealed a concentration-dependent apparent molecular weight and a systematic trend in the residuals between the data and the linear fit, suggesting that G2-151 tended to aggregate.
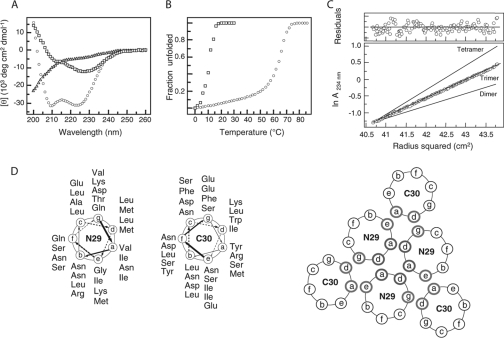
G2-151 becomes largely insoluble on complete removal of GdmCl, and this protein aggregate was subjected to limited proteolysis by trypsin or proteinase K in order to identify a soluble protease-resistant core. This method of protein dissection has frequently been used to define the well-folded helical substructure in class I virus fusion proteins (7, 20, 57). Both proteases generated two similarly sized peptide fragments from G2-151: trypsin digestion resulted in two predominant fragments corresponding to residues 325 to 360 (36 amino acids; denoted N36) and residues 382 to 417 (36 amino acids; C36), whereas proteinase K digestion yielded two slightly shorter fragments corresponding to residues 325 to 353 (N29) and residues 382 to 411 (C30) (Fig. 1). The similarity between fragments generated by each enzyme suggested a common protease-resistant core structure. Because proteinase K is a less specific protease than trypsin, the proteolytic fragments N29 and C30 may more accurately delimit the core region. This empirically determined core includes key residues previously identified by mutagenesis (75) and is somewhat offset from the HR peptides described previously (33).
In isolation, neither the synthetic N29 nor the C30 peptide is helical (Fig. 2A). C30 peptide displays little secondary structure in CD studies, and N29 shows a strong minimum at 227 nm, which is not typical of α-helical signal. Although the thermal unfolding transition of N29 was highly cooperative, the Tm was low (<12°C at 100 μM peptide concentration), and the solution became turbid upon heating, indicating that the isolated N29 peptide is not well structured and has a strong tendency to aggregate. Importantly, however, an equimolar mixture of the N29 and C30 peptides displayed a CD spectrum diagnostic of an α-helix, with characteristic minima at 222 and 208 nm (Fig. 2A). The [θ]222 indicated greater than 90% helical content at 4°C in PBS.
FIG. 2.
Solution properties of the N29/C30 complex. (A) CD spectra in PBS (pH 7.0) at 4°C and 100 μM peptide concentration. (B) Thermal melts monitored by CD at 222 nm. (C) Sedimentation equilibrium data for the N29/C30 complex (100 μM) at 20°C and 22 krpm in PBS (pH 7.0). The data fit closely to a trimeric complex. Curves expected for dimeric and tetrameric models are indicated for comparison. The deviation in the data from the linear fit for a trimer is plotted (upper). (D) Helical wheel diagrams of the six-helix bundle. The antiparallel N and C helices are drawn looking down toward the membrane. The register of the respective coils was assigned to maximize hydrophobicity at interhelical a and d positions.
The stability of the α-helical N29/C30 complex was assessed by monitoring the change in [θ]222 as a function of temperature. The thermal unfolding transition was cooperative and reversible, with a Tm of 64°C at 100 μM peptide concentration in PBS (pH 7.0) (Fig. 2B). Sedimentation equilibrium experiments indicated that the N29 and C30 peptides form a trimer of heterodimers: the observed molecular mass 21.2 (kDa) was consistent with the predicted molecular mass for a trimer of 6.95 kDa monomeric N29/C30 heterodimers (Fig. 2C). We concluded that the N29 and C30 peptides associate to form a discrete, stable α-helical trimer of heterodimers, which closely defines the extent of the six-helix bundle in the postfusion form of JUNV GPC.
MAbs directed against postfusion forms of G2.
Using the N29/C30 peptides to form a six-helix bundle, we identified only one MAb (F100G4) that recognized this structure in an ELISA (Table 1). Because this MAb was also able to bind isolated C41 peptide (data not shown), we concluded that F100G4 is directed against a linear rather than conformational epitope.
In the class I virus envelope glycoprotein, the soluble six-helix bundle is part of a postfusion trimer-of-hairpins structure that includes the disulfide-bonded hinge region between HR1 and HR2. To determine whether this loop is recognized by any of the MAbs, we engineered a single peptide (G2-114) that included HR1 and HR2 as well as the intervening amino acids (residues 305 to 418). Upon refolding and oxidation, G2-114 was similar to G2-151 in α-helical content yet, importantly, remained soluble in PBS (data not shown). Thus, G2-114 likely represents a soluble model for the JUNV trimer-of-hairpins structure.
Using G2-114 as antigen in an ELISA, we identified three additional MAbs (F100G1, F100G6, and F109G1) that recognized the intervening region in the trimer-of-hairpins structure (Table 1). As in other class I fusion proteins, this region likely forms a disulfide-bonded loop in GPC. Consistent with this notion, serine substitutions at C356 and/or C377 (Fig. 1) result in severe misfolding of the mutant GPC (J. York and J. H. Nunberg, unpublished data).
MAb F100G5 recognizes a pH-induced epitope on cells expressing GPC.
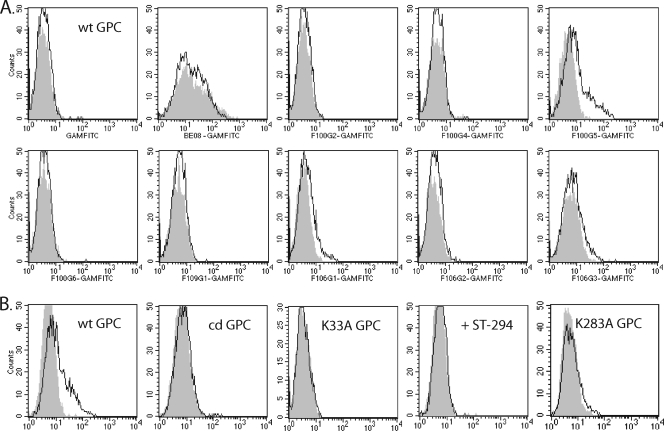
Despite reactivity of some of the MAbs in Western blotting and ELISA experiments, flow cytometry revealed that none of the 16 MAbs was able to recognize JUNV GPC expressed on the surface of transfected Vero cells (Table 1; Fig. 3A). GPC expression in these experiments was confirmed by using the well-characterized G1-directed MAb GB03-BE08 (68, 81) (Fig. 3A) and by the ability of the cell monolayers to form syncytia in response to acidic pH (81). The failure of the G2 MAbs to detect GPC on the cell surface may reflect a number of factors, including the likelihood that the recombinant peptide immunogen contained predominantly postfusion structures that would not be found in the prefusion complex.
FIG. 3.
Flow cytometric analysis of MAb binding to cell surface GPC. (A) Selected MAbs were incubated with Vero cells expressing wild-type (wt) JUNV GPC that had been exposed to neutral pH (gray histogram) or to pH 5.0 (black line histogram). The MAb (and secondary antibody) used is shown below each histogram. Abbreviations: GAMFITC, fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody; BE08, MAb GB03-BE08. (B) MAb F100G5 was incubated with cells expressing wt or mutant GPC that had been exposed to neutral pH (gray histogram) or to pH 5.0 (black line histogram). Mutants are cleavage-defective (cd GPC [81]), K33A GPC (80), and K283A GPC (see text). + ST-294 indicates that cells expressing wt GPC were first incubated with a 50 μM concentration of the small-molecule fusion inhibitor ST-294 (8, 76) prior to exposure to neutral or acidic pH.
We therefore asked whether any of the MAbs might be able to recognize the postfusion form of GPC on the cell surface. In order to avoid syncytium formation in these experiments, GPC-expressing cells were harvested and resuspended prior to exposure to low-pH medium. In the absence of a monolayer of abutting cells, membrane fusion in suspension is induced but proceeds nonproductively. This process is irreversible, and pH treatment of virions in the absence of target cells is known to destroy infectivity (29). For flow cytometric analysis, after 10 min at low pH, the cell suspension was returned to neutral pH by the addition of an excess of cold DPBS and then incubated with MAb.
In contrast to its lack of reactivity with prefusion GPC, MAb F100G5 was able to recognize a form of the protein arising upon acidification (Fig. 3A). This new epitope might develop through pH-induced structural changes in G2 or through exposure of a previously sequestered region of the protein. Upon neutralization, however, the epitope is stably expressed on the inactivated GPC product. All other G2 MAbs again failed to bind, including those that recognize postfusion determinants in the peptide ELISA (F100G1, F100G4, F100G6, and F109G1). The G1-directed MAb BE08 bound well to pH-treated cells, with some reduction in maximal signal due to pH-induced shedding of G1 from mature GPC (compared to coexisting GPC precursor) on the cell surface (76).
Fusion competence is required for F100G5 binding.
In order to assess whether the newly presented F100G5 epitope arises along the conformational pathway leading to membrane fusion or simply as an incidental consequence of exposure to acidic pH, we examined formation of the epitope in several forms of GPC that are specifically unable to promote membrane fusion. A cleavage-defective GPC mutant in which the SKI-1/S1P cleavage site has been altered is unable to mediate cell-cell fusion (75) and, as shown in Fig. 3B, was unable to induce F100G5 binding upon exposure to low pH. Similarly, the fusion-defective K33A mutant, containing a mutation in SSP that stabilizes the prefusion GPC complex against acidic pH (80), was unable to generate the F100G5 epitope. Finally, incubation of wild-type GPC with the small-molecule fusion inhibitor ST-294 (8), which binds to stabilize the prefusion GPC complex against pH-induced activation (76), likewise prevented exposure of the F100G5 epitope. Taken together, these results indicate that formation of the F100G5 epitope is specific and dependent on the ability of GPC to initiate membrane fusion activity.
F100G5 prevents cell-cell fusion when present during activation.
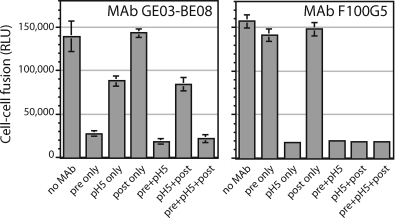
To further explore the relationship of the F100G5 epitope to the pH-induced structural changes leading to membrane fusion, we asked whether F100G5 binding could inhibit GPC-mediated syncytium formation. Specifically, we examined the effect of F100G5 addition during each of three phases in the cell-cell fusion assay (81): during the initial coculture of GPC-expressing and target cells, during acidification at pH 5.0, and upon subsequent incubation at neutral pH when cell-cell fusion becomes evident. As a control, we also examined the behavior of the virus-neutralizing G1-directed MAb GB03-BE08 (68). GB03-BE08 binds to GPC-expressing cells at neutral pH (81), and we predicted that inhibition of syncytium formation by this MAb would require incubation prior to acidification. Because F100G5 binding requires prior exposure to low pH, we reasoned that if the MAb were to inhibit syncytium formation it would do so only during the low-pH pulse.
The virus-neutralizing MAb GB03-BE08 was able to completely inhibit cell-cell fusion when added to the coculture prior to acidification (Fig. 4). Only a small residuum of inhibition remained when the MAb was added with the pH pulse, possibly reflecting the relative rates of pH-induced activation versus MAb binding. Incubation after the coculture was returned to neutral pH had no effect on syncytium formation. These findings suggest that the period of vulnerability to inhibition by MAb GB03-BE08 had largely closed upon acidification. This behavior is consistent with MAb binding to the native, prefusion GPC complex in such a way as to prevent pH-induced membrane fusion.
FIG. 4.
MAb F100G5 inhibits cell-cell fusion when added on exposure to acidic pH. Cells expressing wild-type GPC were incubated with MAb GB03-BE08 (left; 10 μg/ml) or F100G5 (right; ∼30 μg/ml) either during the initial coculture with target cells at neutral medium (pre), during acidification at pH 5.0 (pH 5), or upon the return of the culture to neutral medium, in which cell-cell fusion becomes manifest (post) (81). Syncytium formation was reported by expression of β-galactosidase and quantitated by chemiluminescence (in relative light units [RLU]). Error bars represent 1 standard deviation among quadruplicate wells. Missing bars were not rendered at the scale of the graph. The experiment shown is representative of five independent repetitions.
Interestingly, F100G5 was also able to inhibit syncytium formation, but only when present during the low-pH pulse (Fig. 4). Incubation with the MAb either before acidification or after neutralization had no effect on cell-cell fusion. This result is in keeping with the requirement for pH-induced activation for F100G5 binding (Fig. 3A). Importantly, inhibition by F100G5 indicates that the MAb targets an intermediate form of GPC that is on-path to membrane fusion. Binding to an off-path intermediate, or to a terminal postfusion structure, would not be expected to interfere with syncytium formation.
F100G5 binds at or near the N terminus of G2.
To better understand the nature of the F100G5 epitope, we sought to define amino acid sequences required for binding. For these studies, we took advantage of the reactivity of F100G5 in Western blot analysis. As the MAb recognizes authentic G2 in lysates of JUNV-infected cells (Table 1) and in GPC-expressing Vero cells (see Fig. 6, below), we found that F100G5 also binds an E. coli-expressed fusion protein containing the G2 ectodomain (residues 252 to 424) appended to the C terminus of MBP. Despite the susceptibility of the MBP fusion protein to proteolytic degradation (Fig. 5), full-length molecules were detectible by protein staining of the affinity-purified protein in SDS-PAGE (left panel) and were clearly recognized by MAb F100G5 (right panel). Binding does not require that GPC be glycosylated.
FIG. 6.
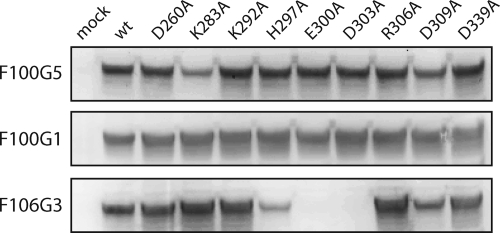
Mapping the F100G5 epitope using GPC mutants. Vero cells expressing the indicated wild-type (wt) or alanine mutant GPC were solubilized using 1% Triton X-100, and proteins were resolved by SDS-PAGE and detected by Western blot analysis using the indicated MAbs. MAb F100G1 recognizes the C-terminal trimer-of-hairpins structure and thus serves to control for potential differences in expression among the mutants. Only the uncleaved G1-G2 precursor is shown, to simplify the analysis of MAb binding; the mature G2 subunit is less abundant, and staining intensity varies depending on the extent of proteolytic maturation in each mutant.
FIG. 5.
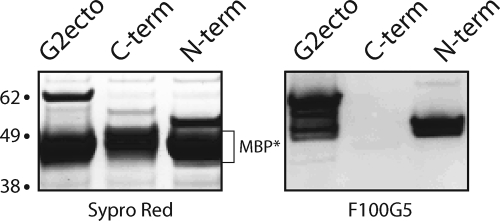
Western blot analysis of F100G5 binding to MBP fusion proteins. Sequences encoding the entire ectodomain of JUNV G2 (G2ecto; residues 252 to 424), the N-terminal region (N-term; residues 252 to 316), or the C-terminal region (C-term; residues 317 to 424) were appended to the C terminus of MBP, and the recombinant fusion proteins were expressed in E. coli. Affinity-purified proteins were resolved by SDS-PAGE and detected by using Sypro Red protein stain (left) or by Western blot analysis using MAb F100G5 (right). The fusion proteins were proteolytically unstable, and the major band detected by protein staining is the stable MBP core (MBP*); the full-length fusion proteins are visible in decreasing order of molecular mass (62, 55, and 50 kDa). F100G5 binds only to the full-length proteins and to proteolytic fragments of G2-ecto. Molecular mass markers are indicated in kilodaltons.
Based on this reactivity with recombinant protein, we further partitioned the G2 ectodomain to generate fusion proteins containing either the N- or C-terminal regions (residues 252 to 316 or 317 to 424, respectively). The larger C-terminal construct included both HR1 and HR2. Western blot analysis of the purified fusion proteins showed that MAb F100G5 recognized only the N-terminal portion of G2 (Fig. 5, right). Thus, binding requires sequences that lie between the N terminus of G2 and the region upstream of HR1. The failure of F100G5 to bind the C-terminal portion of the G2 ectodomain is consistent with its inability to recognize the six-helix bundle and G2-114 peptides in ELISA (Table 1).
Further mapping of the F100G5 epitope made use of a panel of GPC mutants. Eight highly conserved positions in the N-terminal portion of GPC (D260, K283, K292, H297, E300, D303, R306, and D309) were replaced with alanine, and the mutant proteins were expressed in Vero cells. Of the eight mutants, all but two were efficiently assembled, processed, and transported to the cell surface; E300A and D303A were misfolded and remained in the endoplasmic reticulum (York and Nunberg, unpublished). The properly folded mutants were also able to promote cell-cell fusion at levels comparable to wild-type GPC (80 to 160% of wild-type activity) (data not shown). Only D309A was somewhat debilitated (30% of wild-type activity). Nonetheless, all eight GPC mutants were expressed well (Fig. 6) and could be used to evaluate recognition by F100G5 in Western blot analysis. Of the mutants, only K283A GPC was significantly reduced in its reactivity with F100G5 (Fig. 6). As anticipated, MAb F100G1, which recognized the trimer-of-hairpins structure in the G2-114 peptide (Table 1), bound equally to all the N-terminal GPC mutants. This finding also confirms that the reduction in F100G5 binding to K283A was not due to differences in the level of protein expression. Recognition by another MAb, F106G3, was highly dependent on positions H297, E300, and D303 (Fig. 6). Although this MAb was originally derived from MACV G2*, this span of amino acids is identically conserved in JUNV GPC. Indeed, MAb F106G3 recognized all other JUNV mutants, including K283A, as strongly as the wild type. Taken together, these findings demonstrate that MAb F100G5 binding is dependent on the K283 side chain located near the N terminus of G2. Consistent with this conclusion, binding of F100G5 to the pH-induced form of the K283A GPC on the cell surface was also abolished (Fig. 3B). Interestingly, GPC of the related Tacaribe virus (TCRV), which differs from JUNV at three positions immediately N-terminal to K283 (Fig. 1), was not bound by F100G5, whereas MACV GPC, which is identical to JUNV at these positions, is recognized (data not shown). These observations suggest that the F100G5 epitope may include the three adjacent residues (A280, K281, and M282). The MAb F106G3 appears to bind an epitope 15 to 20 residues C-terminal to F100G5.
DISCUSSION
In this report, we describe a MAb that recognizes a pH-induced intermediate form of GPC so as to disrupt the conformational cascade leading to membrane fusion. The MAb F100G5 epitope is not present in the prefusion GPC complex, but rather forms upon pH-induced activation and prior to the completion of cell-cell fusion. Bracketing formation of the F100G5 binding site along the fusion reaction pathway focuses attention on the complex series of on-path intermediates that enable the productive completion of membrane fusion.
Exposure of the fusion peptide provides a target for fusion inhibition.
One clue to the placement of the F100G5-defined intermediate along the pathway of GPC-mediated membrane fusion, and the mechanism of fusion inhibition, derives from our finding that F100G5 binds at or near the N terminus of G2, a region that typically contains the envelope glycoprotein fusion peptide. In the well-accepted model for membrane fusion by class I virus proteins (30, 45, 73), the N-terminal fusion peptide is sequestered in the prefusion state (19) and, upon activation, is inserted into the target-cell membrane via a “spring-loaded” mechanism (16). Collapse of the prehairpin intermediate relocates the fusion peptide to the membrane-proximal base of the postfusion six-helix bundle structure, bringing the virus and target cell membranes into apposition (13, 16, 56). While fusion of the lipid bilayers is energetically coupled to formation of the stable six-helix bundle, the mechanism whereby the respective membranes are destabilized for fusion is unclear. In this context, binding of F100G5 near the G2 fusion peptide (50) might be envisioned to prevent insertion of the fusion peptide into the target cell membrane, or to interfere with the subsequent collapse of the prehairpin intermediate.
For fusion inhibition to occur, MAb F100G5 must bind within the time frame of the pH-induced structural changes. If the fusion peptide is only transiently exposed in productive membrane fusion, then the time window for F100G5 binding is even shorter. Single-particle measurements of influenza virus fusion with defined lipid bilayers have shown that the initial merger of the outer membrane leaflets (hemifusion) occurs within 15 to 20 s of exposure to low pH at 23°C and can be kinetically modeled to comprise three intermediate steps (35). Transition to a stable fusion pore, in which both layers of the membranes have fully coalesced, follows with a half-life of roughly 2 s. In contrast, HIV Env-mediated membrane fusion in cells takes place over the course of 15 to 60 min at 37°C (37, 41, 66), and the prehairpin intermediate can be operationally trapped at temperatures below 23°C (60, 61). The extended lifetime of the HIV prehairpin intermediate may be critical for C-peptide binding and its inhibition of HIV entry (18, 36, 57, 60, 66, 74). These microscopic measurements of overt fusion subsume multiple events and thus provide only a lower limit on the rates of conformational changes in the envelope glycoprotein. While the time frame for membrane fusion by GPC is not known, syncytium formation is readily induced by short exposure to acidic pH (5 min) and proceeds well at 23°C (York and Nunberg, unpublished). These observations suggest that GPC-mediated membrane fusion may share elements with the pH-induced process in HA.
The internal fusion peptide of GPC.
In the prototypic class I virus fusion proteins (influenza HA, HIV Env), the fusion peptide is located at the N terminus of the fusion subunit and comprises a hydrophobic region of variable length and sequence (15 to 25 amino acids) (recently reviewed in reference 23). In contrast, the N-terminal sequence of JUNV G2 contains only short hydrophobic regions that are interspersed with charged residues and is thus not readily identified as a prototypic fusion peptide (Fig. 1). Nonetheless, scanning mutagenesis by Klewitz and colleagues (50) has identified positions in the N-terminal region of the LASV G2 that define a putative fusion peptide. Key amino acids were shown to cluster in a short N-terminal sequence (residues 252 to 258 in JUNV) as well as in a neighboring internal region (residues 268 to 290). Internal fusion peptide regions have also been described in other class I proteins, such as the Ebola virus glycoprotein (72) and the avian sarcoma/leukosis virus (ASLV) Env (25, 27, 39). The K283 position recognized by MAb F100G5 lies within the internal fusion peptide domain of JUNV and is conserved among all arenaviruses.
Also invariant in the arenavirus fusion peptide is an array of four cysteine residues (C271, C284, C293, and C302) that together point to the potential for disulfide bonding. In LASV GPC (50), a deficiency engendered by the C292A mutation (homologous to JUNV C284) is consistent with global misfolding of the protein that arises as a consequence of an unpaired cysteine. While the pattern of connectivity among the six invariant cysteines in the G2 ectodomain is unknown, it is likely that the two cysteines between HR1 and HR2 (C356 and C377) form the disulfide-bonded loop region commonly found in class I fusion proteins. This implies that the four cysteine side chains at the N terminus of G2 are available to form a tight disulfide-bonded loop structure in the internal fusion peptide. This putative architecture is distinct from the linear N-terminal fusion peptides of HA and Env, but perhaps similar to the disulfide-bonded internal fusion peptides found in Ebola virus (72) and ASLV (25, 27, 39) glycoproteins. Structural constraints imposed through disulfide bonding may facilitate membrane insertion of the fusion peptide or its role in destabilizing the apposed membranes for fusion.
Interestingly, the fusion peptides of so-called class II and class III fusion proteins (e.g., of flaviviruses [10] and rhabdoviruses [67], respectively) are also presented as disulfide-bonded loops, albeit they are formed at the tips of anti-parallel β-sheet structures internal to the protein. Much as the recently described class III fusion proteins appear to contain postfusion structural elements of both class I and class II proteins (recently reviewed in references 3 and 73), the internal fusion peptides of certain class I proteins may share a constrained loop structure with those of the other classes. Many of these internal fusion peptides, including that of GPC (Fig. 1), contain a central proline residue that has been suggested to introduce a reverse turn into the disulfide-bonded loop (26). Because fusion peptides have not been directly observed participating in membrane fusion, the structural basis for fusion peptide function is not well understood.
Virus neutralization in the endosome.
As expected, F100G5 was unable to inhibit virus infection in cell culture. Incubation of the attenuated Candid 1 isolate of JUNV (4, 58) with ∼30 μg/ml of F100G5 had no effect on the number of infected cell foci (data not shown). In contrast, virus neutralization using MAb GB03-BE08, which binds G1 in the prefusion GPC complex, was essentially complete. Based on the size of the endocytic vesicle (diameter, ∼100 nm [49]), one can calculate that only a small fraction of virus-containing vesicles will capture F100G5 by passive fluid-phase endocytosis. For instance, in 100 μg/ml of F100G5, fewer than one in five endocytic vesicles will contain even one immunoglobulin molecule. If the volume of the endocytosed virion is considered (diameter of also 100 nm), this fraction is greatly reduced. Therefore, antiviral activity in the endosome is essentially precluded for MAbs that do not recognize the native virion particle. This observation suggests that one might be able to increase the antiviral potency of F100G5 by engineering bifunctional reagents that also bind to the virion, or to nascent endosomal patches on the cell surface (42). While F100G5 is not an immediate candidate for use in antiviral intervention, structures identified by the MAb at or near the G2 fusion peptide may serve as viable targets for small-molecule compounds that effectively enter the endosome.
Acknowledgments
We are grateful to Lisa Schmidt (Public Health Agency of Canada) for expert technical assistance. The following reagents were generated by Tony Sanchez and colleagues at the Centers for Disease Control and Prevention and obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: monoclonal anti-Junín virus, clone GB03-BE08 (IgG, mouse), NR-2564; monoclonal anti-Junín virus, clone NA05-AG12 (IgG, mouse), NR-2582. We are grateful to Sean Amberg (SIGA Technologies, Inc., Corvallis, OR) for providing ST-294.
Financial support for the work was provided by the Special Pathogens Program of the Public Health Agency of Canada (to H.F.) and by NIH grants R01 AI042382 (to M.L.) and R01 AI059355 (to J.H.N.) and by U54 AI065357 (Rocky Mountain Regional Center of Excellence in Biodefense and Emerging Infectious Disease Research; subaward to J.H.N.).
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Abraham, J., J. A. Kwong, C. G. Albariño, J. G. Lu, S. R. Radoshitzky, J. Salazar-Bravo, M. Farzan, C. F. Spiropoulou, and H. Choe. 2009. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic new world clade B arenaviruses. PLoS Pathog. 5:e1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnihothram, S. S., J. York, M. Trahey, and J. H. Nunberg. 2007. Bitopic membrane topology of the stable signal peptide in the tripartite Junín virus GP-C envelope glycoprotein complex. J. Virol. 81:4331-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backovic, M., and T. S. Jardetzky. 2009. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 19:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrera Oro, J. G., and K. T. McKee, Jr. 1991. Toward a vaccine against Argentine hemorrhagic fever. Bull. Pan Am. Health Org. 25:118-126. [PubMed] [Google Scholar]
- 5.Berry, J. D., S. Jones, M. A. Drebot, A. Andonov, M. Sabara, X. Y. Yuan, H. Weingartl, L. Fernando, P. Marszal, J. Gren, B. Nicolas, M. Andonova, F. Ranada, M. J. Gubbins, T. B. Ball, P. Kitching, Y. Li, A. Kabani, and F. Plummer. 2004. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods 120:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyer, W. R., D. Popplau, W. Garten, D. von Laer, and O. Lenz. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77:2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blacklow, S. C., M. Lu, and P. S. Kim. 1995. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry 34:14955-14962. [DOI] [PubMed] [Google Scholar]
- 8.Bolken, T. C., S. Laquerre, Y. Zhang, T. R. Bailey, D. C. Pevear, S. S. Kickner, L. E. Sperzel, K. F. Jones, T. K. Warren, S. Amanda Lund, D. L. Kirkwood-Watts, D. S. King, A. C. Shurtleff, M. C. Guttieri, Y. Deng, M. Bleam, and D. E. Hruby. 2006. Identification and characterization of potent small molecule inhibitor of hemorrhagic fever New World arenaviruses. Antiviral Res. 69:86-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borrow, P., and M. B. A. Oldstone. 1994. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology 198:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briese, T., J. T. Paweska, L. K. McMullan, S. K. Hutchison, C. Street, G. Palacios, M. L. Khristova, J. Weyer, R. Swanepoel, M. Egholm, S. T. Nichol, and W. I. Lipkin. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 5:e1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchmeier, M. J., M. D. Bowen, and C. J. Peters. 2001. Arenaviruses and their replication, p. 1635-1668. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 14.Cantor, C., and P. Schimmel. 1980. Biophysical chemistry, Part III. W. H. Freeman and Co., New York, NY.
- 15.Cao, W., M. D. Henry, P. Borrow, H. Yamada, J. H. Elder, E. V. Ravkov, S. T. Nichol, R. W. Compans, K. P. Campbell, and M. B. A. Oldstone. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079-2081. [DOI] [PubMed] [Google Scholar]
- 16.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 17.Castilla, V., and S. E. Mersich. 1996. Low-pH-induced fusion of Vero cells infected with Junin virus. Arch. Virol. 141:1307-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, C. H., T. J. Matthews, C. B. McDanal, D. P. Bolognesi, and M. L. Greenberg. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 69:3771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 20.Chen, J., J. J. Skehel, and D. C. Wiley. 1999. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. U. S. A. 96:8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, Y.-H., J. T. Yang, and K. H. Chau. 1974. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 13:3350-3359. [DOI] [PubMed] [Google Scholar]
- 22.Clegg, J. C. S., M. D. Bowen, M. J. Buchmeier, J.-P. Gonzalez, I. S. Lukashevich, C. J. Peters, R. Rico-Hesse, and V. Romanowski. 2000. Arenaviridae, p. 633-640. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 23.Cross, K. J., W. A. Langley, R. J. Russell, J. J. Skehel, and D. A. Steinhauer. 2009. Composition and functions of the influenza fusion peptide. Protein Pept. Lett. 16:766-778. [DOI] [PubMed] [Google Scholar]
- 24.Delgado, S., B. R. Erickson, R. Agudo, P. J. Blair, E. Vallejo, C. G. Albariño, J. Vargas, J. A. Comer, P. E. Rollin, T. G. Ksiazek, J. G. Olson, and S. T. Nichol. 2008. Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. 4:e1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delos, S. E., M. B. Brecher, Z. Chen, D. C. Melder, M. J. Federspiel, and J. M. White. 2008. Cysteines flanking the internal fusion peptide are required for the avian sarcoma/leukosis virus glycoprotein to mediate the lipid mixing stage of fusion with high efficiency. J. Virol. 82:3131-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delos, S. E., J. M. Gilbert, and J. M. White. 2000. The central proline of an internal viral fusion peptide serves two important roles. J. Virol. 74:1686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delos, S. E., and J. M. White. 2000. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J. Virol. 74:9738-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Simone, C., and M. J. Buchmeier. 1995. Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology 209:3-9. [DOI] [PubMed] [Google Scholar]
- 29.Di Simone, C., M. A. Zandonatti, and M. J. Buchmeier. 1994. Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology 198:455-465. [DOI] [PubMed] [Google Scholar]
- 30.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 31.Edelhoch, H. 1967. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6:1948-1954. [DOI] [PubMed] [Google Scholar]
- 32.Eichler, R., O. Lenz, T. Strecker, M. Eickmann, H. D. Klenk, and W. Garten. 2003. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. 4:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eschli, B., K. Quirin, A. Wepf, J. Weber, R. Zinkernagel, and H. Hengartner. 2006. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J. Virol. 80:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flanagan, M. L., J. Oldenburg, T. Reignier, N. Holt, G. A. Hamilton, V. K. Martin, and P. M. Cannon. 2008. New world clade B arenaviruses can use transferrin receptor 1 (TfR1)-dependent and -independent entry pathways, and glycoproteins from human pathogenic strains are associated with the use of TfR1. J. Virol. 82:938-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Floyd, D. L., J. R. Ragains, J. J. Skehel, S. C. Harrison, and A. M. van Oijen. 2008. Single-particle kinetics of influenza virus membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 105:15382-15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition to the fusion-active state of the HIV-1 envelope glycoprotein. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frey, S., M. Marsh, S. Günther, A. Pelchen-Matthews, P. Stephens, S. Ortlepp, and T. Stegmann. 1995. Temperature dependence of cell-cell fusion induced by the envelope glycoprotein of human immunodeficiency virus type 1. J. Virol. 69:1462-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallaher, W. R. 1996. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 85:477-478. [DOI] [PubMed] [Google Scholar]
- 40.Gallaher, W. R., C. DiSimone, and M. J. Buchmeier. 2001. The viral transmembrane superfamily: possible divergence of arenavirus and filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallo, S. A., J. D. Reeves, H. Garg, B. Foley, R. W. Doms, and R. Blumenthal. 2006. Kinetic studies of HIV-1 and HIV-2 envelope glycoprotein-mediated fusion. Retrovirology 3:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goebl, N. A., C. M. Babbey, A. Datta-Mannan, D. R. Witcher, V. J. Wroblewski, and K. W. Dunn. 2008. Neonatal Fc receptor mediates internalization of Fc in transfected human endothelial cells. Mol. Biol. Cell 19:5490-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gubbins, M. J., J. D. Berry, C. R. Corbett, J. Mogridge, X. Y. Yuan, L. Schmidt, B. Nicolas, A. Kabani, and R. S. Tsang. 2006. Production and characterization of neutralizing monoclonal antibodies that recognize an epitope in domain 2 of Bacillus anthracis protective antigen. FEMS Immunol. Med. Microbiol. 47:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gustchina, E., J. M. Louis, S. N. Lam, C. A. Bewley, and G. M. Clore. 2007. A monoclonal Fab derived from a human nonimmune phage library reveals a new epitope on gp41 and neutralizes diverse human immunodeficiency virus type 1 strains. J. Virol. 81:12946-12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrison, S. C. 2005. Mechanism of membrane fusion by viral envelope proteins. Adv. Virus Res. 64:231-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the HIV-1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson, M. L., J. J. Correia, D. A. Yphantis, and H. R. Halvorson. 1981. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 36:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kilgore, N. R., K. Salzwedel, M. Reddick, G. P. Allaway, and C. T. Wild. 2003. Direct evidence that C-peptide inhibitors of human immunodeficiency virus type 1 entry bind to the gp41 N-helical domain in receptor-activated viral envelope. J. Virol. 77:7669-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Killisch, I., P. Steinlein, K. Römisch, R. Hollinshead, H. Beug, and G. Griffiths. 1992. Characterization of early and late endocytic compartments of the transferrin cycle. J. Cell Sci. 103:211-232. [DOI] [PubMed] [Google Scholar]
- 50.Klewitz, C., H. D. Klenk, and J. ter Meulen. 2007. Amino acids from both N-terminal hydrophobic regions of the Lassa virus envelope glycoprotein GP-2 are critical for pH-dependent membrane fusion and infectivity. J. Gen. Virol. 88:2320-2328. [DOI] [PubMed] [Google Scholar]
- 51.Kunz, S., K. H. Edelmann, J.-C. de la Torre, R. Gorney, and M. B. A. Oldstone. 2003. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314:168-178. [DOI] [PubMed] [Google Scholar]
- 52.Larson, R. A., D. Dai, V. T. Hosack, Y. Tan, T. C. Bolken, D. E. Hruby, and S. M. Amberg. 2008. Identification of a broad-spectrum arenavirus entry inhibitor. J. Virol. 82:10768-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, England.
- 54.Lenz, O., J. ter Meulen, H. Feldmann, H.-D. Klenk, and W. Garten. 2000. Identification of a novel consensus sequence at the cleavage site of the Lassa virus glycoprotein. J. Virol. 74:11418-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li, J., X. Chen, J. Huang, S. Jiang, and Y. H. Chen. 2009. Identification of critical antibody-binding sites in the HIV-1 gp41 six-helix bundle core as potential targets for HIV-1 fusion inhibitors. Immunobiology 214:51-60. [DOI] [PubMed] [Google Scholar]
- 56.Li, Z. N., B. J. Lee, W. A. Langley, K. C. Bradley, R. J. Russell, and D. A. Steinhauer. 2008. Length requirements for membrane fusion of influenza virus hemagglutinin peptide linkers to transmembrane or fusion peptide domains. J. Virol. 82:6337-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 58.Maiztegui, J. I., K. T. McKee Jr., J. G. Barrera Oro, L. H. Harrison, P. H. Gibbs, M. R. Feuillade, D. A. Enria, A. M. Briggiler, S. C. Levis, A. M. Ambrosio, N. A. Halsey, and C. J. Peters. 1998. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J. Infect. Dis. 177:277-283. [DOI] [PubMed] [Google Scholar]
- 59.McCormick, J. B., P. A. Webb, J. W. Krebs, K. M. Johnson, and E. S. Smith. 1987. A prospective study of the epidemiology and ecology of Lassa fever. J. Infect. Dis. 155:437-444. [DOI] [PubMed] [Google Scholar]
- 60.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mkrtchyan, S. R., R. M. Markosyan, M. T. Eadon, J. P. Moore, G. B. Melikyan, and F. S. Cohen. 2005. Ternary complex formation of human immunodeficiency virus type 1 Env, CD4, and chemokine receptor captured as an intermediate of membrane fusion. J. Virol. 79:11161-11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peters, C. J. 2002. Human infection with arenaviruses in the Americas. Curr. Top. Microbiol. Immunol. 262:65-74. [DOI] [PubMed] [Google Scholar]
- 64.Radoshitzky, S. R., J. Abraham, C. F. Spiropoulou, J. H. Kuhn, D. Nguyen, W. Li, J. Nagel, P. J. Schmidt, J. H. Nunberg, N. C. Andrews, M. Farzan, and H. Choe. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radoshitzky, S. R., J. H. Kuhn, C. F. Spiropoulou, C. G. Albariño, D. P. Nguyen, J. Salazar-Bravo, T. Dorfman, A. S. Lee, E. Wang, S. R. Ross, H. Choe, and M. Farzan. 2008. Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc. Natl. Acad. Sci. U. S. A. 105:2664-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reeves, J. D., S. A. Gallo, A. N., J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 68.Sanchez, A., D. Y. Pifat, R. H. Kenyon, P. C. J., J. B. McCormick, and M. P. Kiley. 1989. Junin virus monoclonal antibodies: characterization and cross-reactivity with other arenaviruses. J. Gen. Virol. 70:1125-1132. [DOI] [PubMed] [Google Scholar]
- 69.Saunders, A. A., J. P. Ting, J. Meisner, B. W. Neuman, M. Perez, J. C. de la Torre, and M. J. Buchmeier. 2007. Mapping the landscape of the lymphocytic choriomeningitis virus stable signal peptide reveals novel functional domains. J. Virol. 81:5649-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shu, W., H. Ji, and M. Lu. 1999. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry 38:5378-5385. [DOI] [PubMed] [Google Scholar]
- 71.Spiropoulou, C. F., S. Kunz, P. E. Rollin, K. P. Campbell, and M. B. A. Oldstone. 2002. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J. Virol. 76:5140-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 73.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U. S. A. 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.York, J., S. S. Agnihothram, V. Romanowski, and J. H. Nunberg. 2005. Genetic analysis of heptad-repeat regions in the G2 fusion subunit of the Junin arenavirus envelope glycoprotein. Virology 343:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.York, J., D. Dai, S. A. Amberg, and J. H. Nunberg. 2008. pH-induced activation of arenavirus membrane fusion is antagonized by small-molecule inhibitors. J. Virol. 82:10932-10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.York, J., and J. H. Nunberg. 2007. A novel zinc-binding domain is essential for formation of the functional Junín virus envelope glycoprotein complex. J. Virol. 81:13385-13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.York, J., and J. H. Nunberg. 2007. Distinct requirements for signal peptidase processing and function of the stable signal peptide (SSP) subunit in the Junin virus envelope glycoprotein. Virology 359:72-81. [DOI] [PubMed] [Google Scholar]
- 79.York, J., and J. H. Nunberg. 2009. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junin virus envelope glycoprotein GPC. J. Virol. 83:4121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.York, J., and J. H. Nunberg. 2006. Role of the stable signal peptide of the Junín arenavirus envelope glycoprotein in pH-dependent membrane fusion. J. Virol. 80:7775-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.York, J., V. Romanowski, M. Lu, and J. H. Nunberg. 2004. The signal peptide of the Junín arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J. Virol. 78:10783-10792. [DOI] [PMC free article] [PubMed] [Google Scholar]