Abstract
Despite their importance as agents of emerging disease, the time scale and evolutionary processes that shape the appearance of new viral species are largely unknown. To address these issues, we analyzed intra- and interspecific evolutionary processes in the Luteoviridae family of plant RNA viruses. Using the coat protein gene of 12 members of the family, we determined their phylogenetic relationships, rates of nucleotide substitution, times to common ancestry, and patterns of speciation. An associated multigene analysis enabled us to infer the nature of selection pressures and the genomic distribution of recombination events. Although rates of evolutionary change and selection pressures varied among genes and species and were lower in some overlapping gene regions, all fell within the range of those seen in animal RNA viruses. Recombination breakpoints were commonly observed at gene boundaries but less so within genes. Our molecular clock analysis suggested that the origin of the currently circulating Luteoviridae species occurred within the last 4 millennia, with intraspecific genetic diversity arising within the last few hundred years. Speciation within the Luteoviridae may therefore be associated with the expansion of agricultural systems. Finally, our phylogenetic analysis suggested that viral speciation events tended to occur within the same plant host species and country of origin, as expected if speciation is largely sympatric, rather than allopatric, in nature.
Although RNA viruses are the most common agents of emerging disease, key aspects of their evolution are still only partly understood. This is of both academic and practical importance, as virus evolution may compromise disease control strategies, including the rapid generation of genotypes that are able to evade host immune responses or of those that are resistant to antivirals or crop genetic resistance (20, 34, 47).
Most of our knowledge of the rapidity of RNA virus evolution comes from the study of animal viruses, for which estimates of rates of nucleotide substitution normally fall within 1 order of magnitude of 1 × 10−3 nucleotide substitutions per site per year (subs/site/year) and largely reflect the background mutation rate (10, 13, 29, 37, 53). Equivalent studies on plant RNA viruses have reported more heterogeneous rates. Early studies suggested that some plant RNA viruses evolved more slowly than RNA viruses that infect animals. For example, estimates of the nucleotide substitution rate in the range of ∼1 × 10−6 to 1 × 10−8 subs/site/year have been obtained for Turnip yellow mosaic virus (4, 23) and some tobamoviruses (21, 25). In contrast, more recent estimates using Bayesian coalescent methods applied to sequences with known dates of sampling and allowing for rate variation among lineages have reported substitution rates in the same range as those of animal RNA viruses (22, 63) and therefore suggest relatively high rates of mutation, as expected, given the intrinsically error-prone nature of RNA replication (15, 65). As well as the differences in how these rates are estimated, a reasonable biological explanation for such a diversity of rate estimates is that they are increased in the short term due to the presence of mutational polymorphisms but lower in the long term because any such deleterious mutations would have then been removed by purifying selection (17, 22). In particular, severe population bottlenecks at transmission would allow deleterious mutations to rise to a high frequency due to strong genetic drift. Such effects make it dangerous to extrapolate long-term rates of evolutionary change from the analysis of intraspecific sequence data (31). Differences in the strength of adaptive evolution could also cause rate heterogeneity, including such processes as competition for susceptible individuals and the colonization of new host species (22, 65).
Although there is a growing body of data on intraspecific evolutionary processes in plant RNA viruses, including rates of nucleotide substitution, there has been a general neglect of long-term evolutionary patterns, including the determinants of viral speciation. Exceptions are recent analyses of the Potyviridae and the Sobemovirus, which associated viral speciation with the development of agriculture (17, 24). Although RNA viruses reproduce asexually, it is informative to consider as analogies the two major forms of speciation used in studies of sexually reproducing eukaryotes: allopatric speciation, in which reproductive isolation follows geographic separation, and sympatric speciation, in which reproductive isolation occurs within an interbreeding population (67). In the context of RNA viruses, allopatric speciation can be thought of as the genetic diversification that occurs when viruses jump to new host species and thereafter evolve independently, as is commonly associated with the process of viral “emergence.” In contrast, sympatric speciation would occur when viruses diversify within a single host species, perhaps by exploiting different cell types (34). Despite the importance of these processes for our understanding of the macroevolution of RNA viruses, their respective roles are currently unknown.
To better understand the nature of long-term evolutionary processes in plant RNA viruses, we undertook an extensive molecular evolutionary analysis of the family Luteoviridae, a heterogeneous family of plant viruses divided into three genera, Luteovirus, Polerovirus, and Enamovirus, containing five, nine, and one classified species, respectively, as well as a number of unclassified species (18). The Luteoviridae possess positive-sense single-stranded RNA genomes of 5,600 to 6,000 nucleotides (nt). These genomes can harbor five or six open reading frames (ORFs). 5′-proximal partially overlapping ORF1 and -2 encode proteins P1 and P2, which are involved in virus replication. Low-frequency −1 ribosomal frameshifting in the overlapping region results in the P1-P2 fusion RNA-dependent RNA polymerase protein (RdRp). ORF3 encodes the coat protein (CP) and completely contains ORF4, which is not found in Enamovirus and is needed for virus movement in the plant (the movement protein, MP). ORF5, which is necessary for aphid transmission (6, 27, 49) and is also involved in virus movement (57) and Luteoviridae phloem limitation (58), is translated through in-frame read-through of the ORF3 stop codon, existing as a read-through domain (RTD) fused to the CP. Members of the genus Polerovirus have an extra ORF0 in the 5′ end of the genome partially overlapping ORF1. Its translation product (P0) acts as a repressor of the RNA-silencing plant defense response (44, 59). Finally, some Luteovirus species have an additional ORF6 with an unknown function in the 3′ end of the genome (18, 48, 70). As a consequence of this particular genomic organization, approximately one-third of the Polerovirus genome, and a smaller fraction in the Luteovirus genome, is composed of overlapping regions.
Due to their agronomic importance, gene sequence data, together with information on host range and geographical distribution, are available for a relatively large number of members of the family Luteoviridae. However, to date the only luteovirus for which rates of evolutionary change have been estimated is Barley yellow dwarf virus (BYDV). In this case, an analysis of substitution rates based on viral RNA extracted from herbarium specimens produced estimates of between 6.2 × 10−4 and 9.7 × 10−5 subs/site/year (43). Similarly, only one estimate of the point at which genetic diversity arose in the family Luteoviridae has been obtained, i.e., approximately 9,000 years ago, and therefore it is perhaps associated with the rise of agriculture (17). However, only a limited number of Luteoviridae species and sequences were included in this analysis. No studies have yet considered the mechanisms of speciation in the family Luteoviridae.
The family Luteoviridae also represents a useful data set to study two other evolutionary phenomena: the pattern and determinants of recombination, which appears to be commonplace within the family Luteoviridae (26, 49, 51, 70, 71), and the differing evolutionary dynamics in genes with overlapping reading frames. There are contrasting hypotheses as to why overlapping reading frames are so commonly used in RNA viruses. According to one view, gene overlapping maximizes the genetic information in smaller genomes (1, 39). Alternatively, it has been suggested that gene overlap generates mutational robustness (i.e., the ability to preserve phenotypes despite the genomic mutational load) at the population level (2, 16, 42). Under the latter hypothesis, gene overlapping generates hypersensitivity to deleterious mutations, as these affect more than one gene. Although this hypersensitivity reduces the capacity of each individual to buffer mutation effects, it represents a selective advantage for wild-type genotypes, which then bolsters robustness at the population level (16, 42). As a consequence of this elevated burden of deleterious mutation, RNA viruses with larger proportions of their genomes present as overlapping reading frames are expected to exhibit lower rates of nucleotide substitution (41, 50). Such a rate reduction has been observed in many animal DNA and RNA viruses (for example, see references 35, 36, 55, 77, and 78), although only a few studies have considered plant RNA viruses in this context (28, 61).
MATERIALS AND METHODS
Sequence data.
Available sequences representing the 15 members of the family Luteoviridae were compiled from GenBank. As temporal information is central to the analyses undertaken here, the year of isolation of each sequence was obtained either from GenBank or from the associated publications or was kindly provided by the relevant authors. Sequences from extensively passaged isolates in nonnatural hosts were excluded from the analyses. For a full list of the accession numbers, origins, and years of isolation of the sequence used, see Tables S1 to S9 in the supplemental material).
To determine long-term evolutionary patterns in the Luteoviridae, we focused our analysis on the CP gene, as this is the gene with the largest data set available. Species with more than 20 available CP sequences were retained for this analysis, which meant that we were able to utilize 11 out of the 15 classified taxa in this analysis, i.e., the three BYDV species (BYDV-MAV, BYDV-PAS, and BYDV-PAV), Soybean dwarf virus (SbDV), Beet chlorosis virus (BChV), Beet mild yellowing virus (BMYV), Cucurbit aphid-borne yellows virus (CABYV), Cereal yellow dwarf virus-RPV (CYDV-RPV [from here on CYDV]), Potato leafroll virus (PLRV), Sugarcane yellow leaf virus (ScYLV), and Turnip yellows virus (TuYV). Among the unclassified members of the family Luteoviridae, sufficient sequence data were available only for BYDV-GAV. No members of the genus Enamovirus were available for study. Because of the very large size of our complete luteovirus data set, which greatly inhibits computational tractability, 20 CP sequences were randomly chosen for each viral species. This resulted in a total of 240 CP sequences, with dates of isolation ranging from 1921 to 2008. Sequences were aligned according to the amino acid sequence, and three sets of data were generated: (i) CP, comprising the complete CP nucleotide sequence (636 nt); (ii) CP3′, including the 421 nt of the 3′ end of the CP, as the first 215 nt is the most divergent CP region among the members of the family Luteoviridae; and (iii) CPov, which includes 363 nt from the center of the CP sequence (nt 216 to 579) and allows us to consider a region that completely overlaps ORF4.
Phylogenetic analyses of these 12 Luteoviridae species were carried out individually, except for the four BYDV species, which were analyzed together. BYDV is currently classified into four species (BYDV-GAV, BYDV-MAV, BYDV-PAS, and BYDV-PAV) (18, 38). However, as these taxa are very similar, we clustered them in one group, as proposed previously (49), from here on referred to as BYDV. Protein-coding alignments were derived for the RdRp (ORF1+ORF2), CP (ORF3), and RTD (ORF5) genes of each species and the P0 (ORF0) gene of the Polerovirus species. In addition, the two ORFs that make up the RdRp were analyzed separately. As similar evolutionary rates were obtained for ORF1, ORF2, and ORF1+ORF2, only estimates for ORF1+ORF2 analyses are presented here. Unfortunately, the number of full-length genome sequences was insufficient for most of the species (i.e., <15 sequences), so that a reliable analysis could not be undertaken. For each data set, sequence alignments were obtained using MUSCLE 3.7 (14) and adjusted manually according to amino acid sequences using Se-Al (http://evolve.zoo.ox.ac.uk/).
For each species, sequence alignments were also obtained for the P0/RdRp and CP/MP overlapping regions and for the corresponding nonoverlapping fragments. Sequence alignments of the overlapping regions were adjusted according to the amino acid sequence of each of the two genes involved, thus generating two data sets for each overlapping region. Since no differences in nucleotide substitution rate were observed between these two data sets in any overlapping fragment, only those with the least statistical error are presented here.
Estimation of substitution rates and age of genetic diversity.
For each data set, rates of nucleotide substitution per site and the time to the most recent common ancestor (TMRCA) were estimated using the Bayesian Markov Cain Monte Carlo (MCMC) method available in the BEAST package (11). The best-fit model of nucleotide substitution in each case was determined using Modeltest 3.7 (60), and all data sets were subsequently run using the general time-reversible substitution model with invariant sites and a gamma distribution of among-site rate variation (GTR+I+Γ4), with three partitions into individual codon positions. These sequence data were analyzed using a relaxed (uncorrelated, log-normal) molecular clock (values for the coefficient of variation were always >0, indicative of non-clock-like evolution; see reference 12) and a Bayesian skyline model as a coalescent prior, as estimating demographic parameters was not the aim of this study. To gauge the robustness of these estimates, we repeated the analysis first by using a strict molecular clock and the simpler Hasegawa, Kishino, and Yano (HKY85) model of nucleotide substitution (30) and second by excluding putative intraspecific recombinant sequences (see below for a description of our recombination detection methodology). Estimation of amino acid substitution rates was performed using the Whelan and Goldman substitution model (76) utilizing the same clock and demographic parameters as described above. In all cases, the BEAST analyses were run until all relevant parameters converged, with 10% of the MCMC chains discarded as burn-in. Statistical confidence is represented by values for the 95% highest probability density (HPD).
Our analyses of substitution rates and TMRCAs for the family Luteoviridae as a whole using either nucleotide or amino acid sequences of the CP failed to produce stable estimates (results available on request). This is likely a function of rate variation among viral species (see Results) and the short time scale of sampling relative to the total depth of the tree. We therefore estimated these parameters for the family Luteoviridae as a whole by using an empirical prior distribution on the substitution rate that is based on the lowest mean value (6 × 10−4 subs/site/year) of the substitution rates estimated for each virus (Table 1). The lowest substitution rate was chosen to be as conservative as possible. We also assessed the possible effect of excessive multiple substitutions at single nucleotide sites by analyzing (i) the first and second codon positions and (ii) the second codon positions independently. Finally, maximum clade credibility trees, with Bayesian posterior probability values providing a measure of statistical support at each node, were also inferred using BEAST.
TABLE 1.
Nucleotide substitution rate and TMRCA estimates for Luteoviridae species
| Virusa | Date range | nb | Parameter | P0 gene | RdRp gene | CP gene | RTD gene |
|---|---|---|---|---|---|---|---|
| BYDV (L) | 1917-2008 | 0-48-335-41 | Sub ratec (HPD)d | 6.7 × 10−3 (2.6 × 10−3-1.2 × 10−2) | 1.5 × 10−3 (9.8 × 10−4-2.0 × 10−3) | 6.0 × 10−4 (5.2 × 10−6-1.6 × 10−3) | |
| TMRCAe (HPD) | 45 (130-13) | 247 (418-139) | 1,318f (2,009-363) | ||||
| SbDV (L) | 1990-2007 | 0-13-44-34 | Sub rate (HPD) | 2.6 × 10−3f (1.9 × 10−8-8.3 × 10−3) | 6.0 × 10−4 (2.6 × 10−4-9.9 × 10−4) | 2.3 × 10−3 (9.9 × 10−4-3.9 × 10−3) | |
| TMRCA (HPD) | 30f (2,000-26) | 96 (271-45) | 90 (263-37) | ||||
| BChV (P) | 1985-2005 | 15-21-21-5 | Sub rate (HPD) | 7.2 × 10−3 (2.7 × 10−4-1.1 × 10−2) | 1.6 × 10−3 (5.2 × 10−4-2.8 × 10−3) | 2.7 × 10−3 (2.6 × 10−4-4.2 × 10−3) | NDg |
| TMRCA (HPD) | 13 (14-12) | 26 (48-24) | 68 (121-24) | ND | |||
| BMYV (P) | 1980-2005 | 25-25-20-5 | Sub rate (HPD) | 7.7 × 10−4 (1.2 × 10−6-1.6 × 10−3) | 3.7 × 10−4 (3.4 × 10−5-7.3 × 10−4) | 4.9 × 10−5f (1.2 × 10−8-4.9 × 10−4) | ND |
| TMRCA (HPD) | 143 (289-19) | 88 (214-34) | 227f (2,000-34) | ND | |||
| CABYV (P) | 2003-2008 | 1-27-47-1 | Sub rate (HPD) | ND | 4.3 × 10−3 (2.2 × 10−4-9.4 × 10−3) | 3.5 × 10−2 (1.2 × 10−2-6.6 × 10−2) | ND |
| TMRCA (HPD) | ND | 233 (369-20) | 21 (22-20) | ND | |||
| CYDV (P) | 1925-2006 | 5-32-75-8 | Sub rate (HPD) | ND | 2.3 × 10−2 (1.1 × 10−3-5.3 × 10−2) | 7.4 × 10−4 (1.4 × 10−4-1.3 × 10−3) | 3.3 × 10−3 (5.5 × 10−4-5.9 × 10−3) |
| TMRCA (HPD) | ND | 42 (119-7) | 456 (746-85) | 56 (124-41) | |||
| PLRV (P) | 1974-2008 | 35-36-31-18 | Sub rate (HPD) | 3.4 × 10−4 (1.7 × 10−4-5.2 × 10−4) | 5.9 × 10−5f (2.6 × 10−9-1.7 × 10−4) | 6.2 × 10−4 (6.6 × 10−5-8.9 × 10−4) | 2.9 × 10−4f (1.1 × 10−9-1.4 × 10−3) |
| TMRCA (HPD) | 145 (369-98) | 684f (2,000-65) | 54 (232-46) | 2,000f (2,000-2,000) | |||
| ScYLV (P) | 1989-2008 | 59-104-80-31 | Sub rate (HPD) | 1.2 × 10−3 (2.5 × 10−4-2.4 × 10−3) | 1.4 × 10−4 (5.2 × 10−5-2.3 × 10−4) | 1.1 × 10−3 (6.8 × 10−4-1.5 × 10−3) | 3.7 × 10−5f (4.9 × 10−7-8.7 × 10−5) |
| TMRCA (HPD) | 31 (55-13) | 347 (704-144) | 26 (33-23) | 980f (2,009-96) | |||
| TuYV (P) | 1980-2006 | 15-48-34-1 | Sub rate (HPD) | 2.7 × 10−3 (5.9 × 10−4-4.9 × 10−3) | 1.0 × 10−3 (2.0 × 10−4-2.1 × 10−3) | 8.6 × 10−4 (3.7 × 10−4-1.4 × 10−3) | ND |
| TMRCA (HPD) | 39 (89-26) | 43 (219-33) | 96 (108-32) | ND |
Virus species. The genus is indicated in parentheses (L, Luteovirus; P, Polerovirus).
n, numbers of sequences of the genes (P0-RdRp-CP-RTD).
Sub rate, nucleotide substitution rate (subs/site/year).
95% HPD value.
TMRCA, years.
Unreliable estimate.
ND, no estimate determined due to a low number of sequences or a narrow date range.
Robustness of temporal signal.
To test the strength of the temporal signal in these data, essential to the accurate estimation of substitution rates, the BEAST analyses described above were repeated on data sets in which sampling times were randomized such that they lack any temporal structure. For computational tractability, this randomization was undertaken using the gene with the smallest number of taxa for each viral species. Runs for randomized data were repeated 10 times. The mean and 95% HPDs of the substitution rate estimates for the randomized data were then compared with those obtained from the real data; major differences in these estimates indicate the presence of temporal structure.
Detection of recombination.
We determined the occurrence of recombination within and between the RdRp and CP genes for each of the Luteoviridae species. In each case, sequences of different genes belonging to the same isolate were concatenated and recombination breakpoints were detected by using three different methods available in the RDP3 package (http://darwin.uvigo.es/rdp/rdp.html), i.e., RDP, GENECONV, and Bootscan, and employing the default parameters (45). To be as conservative as possible, only recombination signals detected by all of the methods were considered (P < 0.05). The lack of sufficient isolates with both the P0 and RTD genes prevented us from including these two genes in the analyses.
Analysis of selection pressures.
Selection pressures for each gene in each luteovirus species were measured as the mean number of nonsynonymous (dN)-to-synonymous (dS) nucleotide substitutions per site (dN/dS ratio) using the single-likelihood ancestor counting method implemented in the HYPHY package (40). In all cases, dN/dS ratio estimates were based on neighbor-joining trees inferred under the GTR substitution model, with 95% confidence intervals (CI) calculated assuming a χ2 distribution.
Modes of speciation.
To determine the respective roles of allopatric versus sympatric speciation, we assessed the strength of clustering by host species and country of sample origin within the interspecific Luteoviridae phylogeny. If allopatric speciation following host jumping were the dominant process, we would expect no significant association between phylogeny and host species, and perhaps between phylogeny and geography. In contrast, if sympatric speciation were the most important macroevolutionary process in these data, we would expect a significant association between phylogeny and both the host species and the country of sampling.
To undertake this analysis, we utilized the 240-sequence CPov Luteoviridae data set described above. However, to avoid biasing this analysis toward intraspecific evolutionary patterns, the data were subsampled such that only one sequence was chosen randomly for each species, unless a specific virus was located from multiple hosts or multiple countries, in which case one representative sequence was chosen for each host/country. This subsampling resulted in a data set of 89 isolates representing 45 different host species isolated from 32 different countries, on which we inferred BEAST trees as described above. We then employed the association index (AI) (75) and the parsimony score (PS) (19) to determine whether particular traits (host, country) are more strongly associated with the underlying phylogeny than expected by chance. These analyses were undertaken by using the BaTS method (54), which utilizes the posterior sample of trees produced by BEAST (with the first 10% removed as burn-in), thereby incorporating phylogenetic uncertainty. Null distributions for both statistics were generated by using 1,000 data replications.
RESULTS
Nucleotide substitution rates in Luteoviridae species.
We first estimated rates of nucleotide substitution within each luteovirus species (Table 1). Reliable estimates could not be obtained for the RTD in BChV, BWYV, CABYV, and TuYV and for the P0 in CABYV and CYDV due to insufficient data numbers. In addition, some rate estimates exhibited a very wide range of 95% HPD values, with lower values of ≤1 × 10−7, strongly suggesting that they are unreliable. Those data sets with very wide 95% HPD intervals under both models were excluded from further analysis. However, no major differences in evolutionary rates were observed when the data were reanalyzed using the HKY85 substitution model and a strict molecular clock. Similarly, the exclusion of putative intraspecific recombinants had no significant impact on rate estimates (results available on request).
Mean evolutionary rates across all of the genes in all of the species analyzed ranged over 2 orders of magnitude, from 3.5 × 10−2 to 1.4 × 10−4 subs/site/year (Table 1), with the highest rates recorded in CABYV and CYDV (at >1 × 10−2 subs/site/year). Substitution rates for the CP gene ranged from 3.5 × 10−2 to 6 × 10−4 subs/site/year. Importantly, mean evolutionary rates in amino acids covered the same range as those for nucleotides, from 1.1 × 10−2 to 4.1 × 10−4 subs/residue/year, indicating that our results are robust to the effect of multiple substitution (see Table S10 in the supplemental material). Examining rates across all of the genes revealed no significant variation in substitution rates between species for the P0 and RTD genes, although differences were found for the RdRp and CP genes. For example, in the RdRp, BYDV and CYDV showed significantly higher substitution rates (i.e., nonoverlapping HPD values) than BMYV, ScYLV, and TuYV.
Strength of temporal signal.
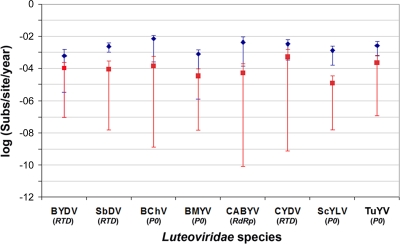
In all cases, estimates of the mean substitution rate differed by at least 1 order of magnitude between the real and randomized data sets (Fig. 1). More importantly, the 95% HPD values for all of the randomized controls excluded the mean substitution rates estimated for the real data, indicating that they are significantly different. In addition, the lower 95% HPD values in the randomized data sets differed by at least 3 orders of magnitude from the corresponding mean estimates, and all were equal to or lower than 1 × 10−7 and hence strongly indicative of insufficient temporal structure. Far narrower 95% HPD results were observed for the real data. Hence, the sequence data analyzed here contain sufficient temporal structure for reliable estimation.
FIG. 1.
Estimates of the rate of nucleotide substitution per site in the gene with the lowest number of sequences within each Luteoviridae species using the real (blue diamonds) and randomized (red squares) dates of isolation, with 95% HPD values shown. In each case, the gene used is shown in parentheses under the name of the species.
Nucleotide substitution rates and the effect of gene overlap.
Nucleotide substitution rates were estimated for the P0/RdRp and CP/MP overlapping regions, and their corresponding nonoverlapping fragments, for those species for which sufficient sequence data were available (Table 2). All PLRV rate estimates, as well as that for the CP/MP overlapping region of BMYV, exhibited a very wide range of 95% HPD values, indicating that there is insufficient temporal signal in these data for meaningful analysis.
TABLE 2.
Nucleotide substitution rate estimates for the overlapping and nonoverlapping regions of the P0 and RdRp genes, and the CP and MP genes of Luteoviridae species
| Virusa | Parameter | P0, nonoverlapping region | P0/RdRp |
CP, nonoverlapping region | CP/MP, overlapping region | |
|---|---|---|---|---|---|---|
| Overlapping region | Nonoverlapping region | |||||
| BYDV (L) | Length (nt) | 141 | 462 | |||
| Sub rateb (HPD)c | 1.9 × 10−2 (3.4 × 10−3-4.2 × 10−2) | 7.6 × 10−4 (4.6 × 10−4-1.1 × 10−3) | ||||
| SbDV (L) | Length (nt) | 33 | 570 | |||
| Sub rate (HPD) | 2.9 × 10−3 (9.4 × 10−5-7.1 × 10−3) | 5.9 × 10−4 (3.2 × 10−4-8.7 × 10−4) | ||||
| BChV (P) | Length (nt) | 136 | 612 | 2,834 | 81 | 528 |
| Sub rate (HPD) | 1.1 × 10−3 (9.2 × 10−4-2.1 × 10−2) | 5.2 × 10−3 (1.0 × 10−3-8.7 × 10−3) | 2.2 × 10−3 2.1 × 10−4-3.9 × 10−3) | NDd | 2.5 × 10−3 (3.2 × 10−4-4.5 × 10−3) | |
| BMYV (P) | Length (nt) | 124 | 596 | 2,649 | 81 | 528 |
| Sub rate (HPD) | 8.3 × 10−4 (1.1 × 10−4-1.6 × 10−3) | 4.3 × 10−4 (2.5 × 10−5-8.6 × 10−4) | ND | 6.0 × 10−3 (3.3 × 10−4-1.0 × 10−2) | 5.3 × 10−5e (4.4 × 10−8-1.5 × 10−4) | |
| CABYV (P) | Length (nt) | 121 | 595 | 2,568 | 28 | 575 |
| Sub rate (HPD) | ND | ND | ND | ND | 3.2 × 10−2 (6.2 × 10−3-6.7 × 10−2) | |
| CYDV (P) | Length (nt) | 151 | 619 | 2,616 | 153 | 462 |
| Sub rate (HPD) | ND | ND | ND | 1.9 × 10−2 (3.4 × 10−3-4.2 × 10−2) | 2.3 × 10−3 1.2 × 10−3-3.3 × 10−3) | |
| PLRV (P) | Length (nt) | 133 | 611 | 2,577 | 156 | 471 |
| Sub rate (HPD) | 1.3 × 10−4e (5.2 × 10−9-3.7 × 10−4) | 1.1 × 10−4e (1.9 × 10−9-2.9 × 10−4) | 5.4 × 10−5e (8.8 × 10−10-1.8 × 10−4) | 5.5 × 10−4 (2.2 × 10−4-9.2 × 10−4) | 1.8 × 10−4e (9.7 × 10−8-3.2 × 10−4) | |
| ScYLV (P) | Length (nt) | 175 | 596 | 2,616 | 138 | 453 |
| Sub rate (HPD) | 3.8 × 10−2 (1.0 × 10−3-8.6 × 10−2) | 4.7 × 10−4 (1.6 × 10−5-1.0 × 10−3) | 3.7 × 10−4 (1.9 × 10−4-5.9 × 10−3) | 2.3 × 10−3 (8.2 × 10−4-3.9 × 10−3) | 9.2 × 10−4 (4.1 × 10−4-1.4 × 10−3) | |
| TuYV (P) | Length (nt) | 142 | 608 | 2,499 | 81 | 528 |
| Sub rate (HPD) | 3.5 × 10−3 (6.6 × 10−4-6.7 × 10−3) | 4.8 × 10−3 (5.8 × 10−4-9.0 × 10−3) | 2.2 × 10−3 (8.5 × 10−4-5.3 × 10−3) | 2.6 × 10−3 (4.8 × 10−4-5.6 × 10−3) | 8.6 × 10−4 (4.2 × 10−4-1.3 × 10−3) | |
Virus species. The genus is indicated in parentheses (L, Luteovirus; P, Polerovirus).
Sub rate, nucleotide substitution rate (subs/site/year).
95% HPD value.
ND, not determined due to a lack of a sufficient number of sequences or too-short fragments.
Unreliable estimate.
For the P0/RdRp region, nucleotide substitution rates did not vary between the overlapping and nonoverlapping fragments within each virus species, with the exception of the P0 nonoverlapping fragment of ScYLV, which showed a higher rate than the corresponding overlapping fragment. In the case of CP/MP, the nonoverlapping region tended to exhibit higher evolutionary rates than the overlapping region in all of the Luteoviridae analyzed, this difference being significant in four of six species (Table 2). Thus, the theory that overlapping regions reduce the rate of evolutionary change holds for some, but not all, gene-species combinations in the Luteoviridae.
Nucleotide substitution rates at the family level.
Substitution rate estimates for the Luteoviridae as a whole—that is, at the interspecific level—using the CP gene resulted in unreliable estimates (i.e., a very wide range of 95% HPD values), irrespective of the substitution model or whether nucleotides or amino acids were utilized. We therefore estimated substitution rates using an empirical prior distribution on the mean substitution rate that conservatively reflects the lowest mean estimate of the substitution rate at the intraspecific level (i.e., 6 × 10−4 subs/site/year) and which effectively stabilizes rate estimates. The mean rates of nucleotide substitution obtained from these analyses were ∼4 × 10−4 subs/site/year (lowest HPD value = 2.6 × 10−4 subs/site/year) and hence lower than those seen in the majority of individual species (Table 3), perhaps reflecting the inclusion of transient deleterious mutations in the intraspecies comparisons (see Discussion). To remove any effect of site saturation at third codon positions, we repeated our analyses of the luteoviruses as a whole by using only the first and/or second codon positions of the CP. This resulted in estimates similar to those obtained by using all of the codon positions, again suggesting that our overall rate is robust (Table 3).
TABLE 3.
Nucleotide substitution rate and TMRCA estimates for the family Luteoviridae using different CP data sets and an empirical prior distribution of 6 × 10−4 subs/site/year
| Data seta | Mean sub rateb (range)c | Difference at codon position: |
Mean TMRCAe (range) | ||
|---|---|---|---|---|---|
| 1d | 2d | 3d | |||
| CP | 4.3 × 10−4 (2.6 × 10−4-6.1 × 10−4) | 0.99 | 0.51 | 1.50 | 1,732 (3,229 to 815) |
| CP3′ | 4.5 × 10−4 (2.8 × 10−4-6.2 × 10−4) | 0.70 | 0.43 | 1.87 | 1,761 (3,583 to 877) |
| CPov | 4.5 × 10−4 (2.7 × 10−4-6.3 × 10−4) | 0.78 | 0.47 | 1.75 | 2,007 (3,088 to 805) |
CP, complete coat protein nucleotide sequence (636 nt); CP3′, 421 nt of the 3′ end of the coat protein gene; CPov, 363 nt from the center of the coat protein gene (nt 216 to 579).
Sub rate, nucleotide substitution rate (subs/site/year).
The 95% HPD values are given.
Difference between the substitution rate and the overall mean substitution rate.
TMRCA, years.
Selection pressures in the Luteoviridae.
Our dN/dS ratio analysis revealed that although genes evolve under different pressures within and between species, all luteovirus genes are subject to purifying selection (dN/dS ratio of <1.0), with the exception of the MP gene (Table 4). Mean dN/dS ratios were also systematically lower for the RdRp and RTD genes (0.19 to 0.22) than for the other genes (0.31 to 1.18). In the case of the MP gene, the dN/dS ratios indicated purifying selection only for BYDV and CYDV (mean, ≤0.69; CI = 0.56 to 0.84), while the mean values for the remaining species were close to or greater than 1 (mean = 0.82 to 2.61; CI = 0.58 to 3.53), indicating that this gene is either evolving strictly neutrally or exhibits some localized positive selection.
TABLE 4.
dN/dS ratio estimates for genes of each Luteoviridae species
| Virusa | Mean dN/dS ratio (95% CI)b |
||||||
|---|---|---|---|---|---|---|---|
| P0 gene | RdRp gene | CP gene |
MP gene | RTD gene | |||
| Entire sequence | Overlapping region | Nonoverlapping region | |||||
| BYDV (L) | 0.09 (0.08-0.10) | 0.33 (0.31-0.36) | 0.58 (0.52-0.63) | 0.11 (0.09-0.13) | 0.64 (0.58-0.70) | 0.12 (0.11-0.13) | |
| SbDV (L) | 0.10 (0.08-0.11) | 0.25 (0.20-0.31) | 0.27 (0.19-0.34) | 0.08 (0.03-0.21) | 1.01 (0.85-1.19) | 0.08 (0.07-0.10) | |
| BChV (P) | 0.70 (0.48-0.97) | 0.25 (0.19-0.31) | 0.47 (0.36-0.60) | 0.55 (0.42-0.70) | 0.05 (0.00-0.24) | 0.99 (0.88-1.24) | NDc |
| BMYV (P) | 0.52 (0.42-0.64) | 0.41 (0.34-0.49) | 0.12 (0.07-0.21) | 0.14 (0.08-0.24) | 0.01 (0.00-0.12) | 2.61 (1.88-3.53) | ND |
| CABYV (P) | ND | 0.20 (0.18-0.22) | 0.26 (0.23-0.31) | 0.24 (0.20-0.28) | 0.42 (0.24-0.63) | 1.60 (1.24-3.04) | ND |
| CYDV (P) | ND | 0.12 (0.09-0.15) | 0.27 (0.22-0.32) | 0.53 (0.43-0.64) | 0.04 (0.03-0.06) | 0.69 (0.56-0.84) | 0.18 (0.15-0.22) |
| PLRV (P) | 0.36 (0.29-0.44) | 0.31 (0.27-0.34) | 0.33 (0.26-0.42) | 0.46 (0.35-0.59) | 0.18 (0.11-0.28) | 1.27 (1.01-1.57) | 0.37 (0.30-0.42) |
| ScYLV (P) | 0.49 (0.39-0.61) | 0.23 (0.21-0.26) | 0.35 (0.26-0.46) | 0.38 (0.26-0.53) | 0.31 (0.18-0.50) | 0.82 (0.58-1.10) | 0.20 (0.17-0.24) |
| TuYV (P) | 0.43 (0.36-0.52) | 0.32 (0.27-0.38) | 0.39 (0.31-0.48) | 0.44 (0.34-0.55) | 0.20 (0.10-0.36) | 1.00 (0.81-1.23) | ND |
| All | 0.50 | 0.22 | 0.31 | 0.40 | 0.16 | 1.18 | 0.19 |
Virus species. The genus is indicated in parentheses (L, Luteovirus; P, Polerovirus).
The 95% CIs shown are based on the assumption of a χ2 distribution.
ND, not determined due to lack of sufficient number of sequences.
We also investigated the nature of selection pressures in regions of gene overlap. Interestingly, we observed no significant difference in the dN/dS ratio between overlapping and nonoverlapping fragments of the CP gene (which overlaps the MP gene) in five of the nine species analyzed. In the remaining species, the dN/dS ratio was higher for the overlapping than for the nonoverlapping region (mean = 0.46 to 0.58 versus 0.04 to 0.18; CI = 0.35 to 0.70 versus 0.00 to 0.28, respectively), indicative of increased purifying selection in the latter (Table 4). Unfortunately, dN/dS ratios for overlapping and nonoverlapping fragments of the P0 and the RdRp genes could only be obtained for PLRV and ScYLV due to a lack of sequence data on the remaining species. In these two species, the dN/dS ratio was also higher in the overlapping than in the nonoverlapping regions of both genes (mean = 0.41 to 0.67 versus 0.03 to 0.13; CI = 0.35 to 0.95 versus 0.00 to 0.18; not shown in Table 4).
Phylogenetic relationships and age of the family Luteoviridae.
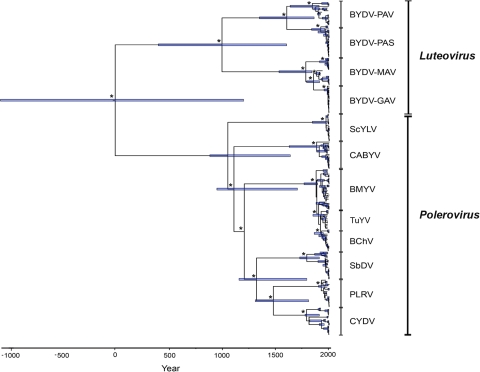
The maximum clade credibility tree for the CPov data set reveals that the members of the family Luteoviridae can be divided into two clusters largely corresponding to the Luteovirus and Polerovirus genera (very similar trees and TMRCAs were observed for all CP data sets) (Fig. 2). The Luteovirus cluster contained the BYDV species in isolation. All of the remaining species fell into the Polerovirus group, although support for the position of ScYLV was very low (posterior probability = 0.15 to 0.16). As noted previously (18), SbDV, the other virus classified within the genus Luteovirus, in fact clusters with the Polerovirus group.
FIG. 2.
Maximum clade credibility phylogeny of the family Luteoviridae based on the CPov data set using 20 sequences per species and an empirical prior distribution on the substitution rate. Branch tip times reflect the times of viral sampling. The tree is automatically rooted through the use of a relaxed molecular clock, and the total depth of the tree is the TMRCA of the family Luteoviridae. Asterisks indicate nodes with posterior probabilities of ≥0.90. Horizontal blue bars denote 95% HPD intervals for the age of each node. Vertical black bars delimit sequence clusters for each virus genus and species.
To determine the time scale of this evolutionary history, we estimated TMRCAs for each gene-virus species combination (Table 1). Most of these estimates indicated that the sampled genetic diversity arose no earlier than the second half of the 19th century. The only major differences in TMRCA estimates among genes were observed for BYDV and CYDV, where the RdRp gene diverged significantly later than the CP gene (mean estimates of 42 and 45 years for the RdRp gene versus 456 and 247 years for the CP gene), while the opposite pattern was observed for CABYV and ScYLV (TMRCAs of 233 and 347 versus 21 and 26 years, for the RdRp and CP genes, respectively). Importantly, no significant differences in TMRCA values were found in parallel analyses using the amino acid sequences (see Table S10 in the supplemental material). Hence, all estimates suggest that the sampled diversity within the Luteoviridae species arose during the last 500 years.
Our mean estimate for the TMRCA of the members of the family Luteoviridae sampled as a whole using the empirical prior distribution on the substitution rate was approximately 2,000 years (1,732 to 2,007 years), with upper 95% HPD values, representing the oldest credible age, of 3,088 to 3,583 years (Table 4). Similarly, mean estimates of TMRCAs for the Luteovirus and Polerovirus genera were 1,010 and 897 years, respectively (95% HPD = 403 to 1,606 and 300 to 1,051 years; Fig. 2). Finally, mean TMRCAs estimated for the different species within each genus were also very similar, ranging from the end of the 19th century to the beginning of the 20th century, and comparable to the results obtained for each species individually. Interestingly, BYDV retains the earliest genetic diversity, with coalescent events between 1 and 7 centuries before the other species (mean = 1,010 years ago; 95% HPD = 403 to 1,606 years ago). However, differentiation into the various BYDV “species” did not occur before the 18th century.
Recombination in the family Luteoviridae.
The TMRCA differences between genes noted above were also reflected in the topological incongruence of the RdRp and CP gene trees. Specifically, phylogenetic analyses of BYDV, CABYV, CYDV, and ScYLV revealed that the clustering of specific viral isolates differed between the RdRp and CP genes. In particular, the CP gene phylogeny divided isolates of CYDV and ScYLV into three clusters, as previously described (62, 74), while analyses of the RdRp gene resulted in only two clusters (see Fig. S1 and S2 in the supplemental material). Overall, these results are suggestive of recombination between the RdRp and CP genes in a number of Luteoviridae species.
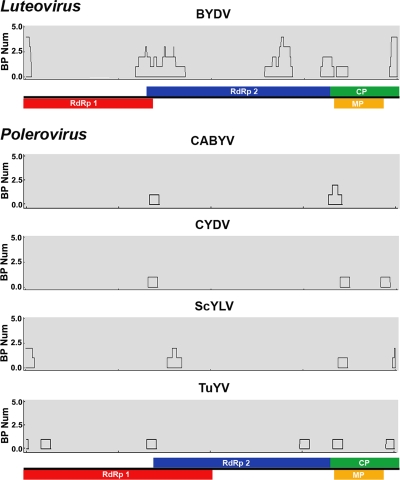
To better characterize the frequency and distribution of recombination events, we analyzed the concatenated RdRp and CP gene sequences of all of the species in the family by using three different recombination detection methods. This analysis revealed clusters of recombination breakpoints—putative recombination “hot spots”—in BYDV, CABYV, CYDV, ScYLV, and TuYV, although not in the remaining species (Fig. 3). Notably, the region comprising the end of the RdRp gene and the beginning of the CP gene and the overlapping region between RdRp ORF1 and -2 exhibited a concentration of breakpoints in these five species. Recombination signals were also detected in the 5′ end of the RdRp gene and the 3′ end of the CP gene in BYDV, ScYLV, and TuYV, which suggests that recombination may also occur in the boundaries between the P0 and RdRp genes and between the CP and RTD genes. Although most putative recombination events were observed at gene boundaries, we also observed recombinant breakpoints within the RdRp gene of BYDV and TuYV and in the CP gene of CYDV (Fig. 3).
FIG. 3.
Recombination breakpoints in the RdRp and CP genes of members of the family Luteoviridae. Black boxes indicate the positions of clusters of recombination breakpoints. RdRp1 (red) and RdRp2 (blue) correspond to ORF1 and -2 encoding the RdRp, CP (green) indicates the CP gene, and MP (yellow) indicates the MP gene. BP Num is the number of recombination breakpoints in a 200-nt window.
Patterns of speciation.
Both the AI and PS statistics revealed a significant association between the interspecies phylogeny of the Luteoviridae and the plant host each virus species naturally infects (P < 0.001); hence, virus species that infect the same hosts tend to cluster together on the tree. Similarly, a statistically significant association was observed between the luteovirus phylogeny and the country of sampling (P < 0.001 in both tests), such that virus species that infect plant hosts in the same geographic region tend to cluster together. Together, these data suggest that viral speciation tends to occur within the same host species and in the same geographic area, which is compatible with genetic diversification through sympatric speciation.
DISCUSSION
Luteovirus evolutionary dynamics.
Although there was some gene- and species-specific variation, our large-scale comparative analysis revealed that members of the family Luteoviridae generally experience rates of nucleotide substitution ranging from 10−3 to 10−4 subs/site/year, comparable to those observed in animal RNA viruses (13, 37) and some other plant RNA viruses (22, 63). Most cases of gene-specific rate variation seem to reflect differences in selection pressure across the luteovirus genome, particularly as equivalent differences were found in the dN/dS ratio. A good example is provided by the RdRp and RTD genes, in which purifying selection was stronger (i.e., the dN/dS ratio was lower) for the Luteovirus species than for the Polerovirus species. In the case of the RdRp gene, this variation may reflect differences in the gene arrangement of the 5′ half of the genome. Specifically, Polerovirus species have an extra ORF0 encoding the P0 protein in the 5′-terminal region of the genome. Since the P0 gene is located in the −1 reading frame of the RdRp gene, synonymous nucleotide changes in the third codon position of the P0 gene result in amino acid changes in the overlapping region of the RdRp, increasing its overall dN/dS ratio. The analysis of the dN/dS ratio between the overlapping and nonoverlapping fragments of the RdRp supports this hypothesis. In the case of the RTD, which together with the CP is involved in specificity of vector-mediated transmission (6, 27, 49), as well as in long-distance movement through the phloem in a host-specific manner (57, 58), differences in the dN/dS ratio between the Luteovirus and Polerovirus genera may reflect differences in the vector species responsible for virus transmission and/or in the host range. Finally, the CP gene and the N-terminal region of RTD are highly conserved in BYDV and CYDV, which is suggestive of strong functional constraints (7, 27, 48). Thus, both the CP and RTD genes are expected to evolve at lower rates than other genes, which is in agreement with our observations.
The very high substitution rates observed for the CP gene of CABYV (at >1 × 10−2 subs/site/year) merit special attention, as they represent some of the highest rates observed to date in a plant RNA virus. However, such an elevated rate is likely to be a function of the fact that these sequences were only sampled over a very short time period (5 years). Sequences sampled over a short time scale tend to produce artificially inflated rate estimates, reflecting short-term mutation rates that include the circulation of transient deleterious mutations (i.e., polymorphisms), rather than more meaningful long-term rates of nucleotide substitution that measure evolutionary dynamics following the action of purifying selection (13). Indeed, that our substitution rate estimates are higher within (mean = 6 × 10−3 subs/site/year) than among (mean = 4.4 × 10−4 subs/site/year) luteovirus species suggests that transient deleterious mutations are a common component of intraspecific comparisons. It is clear that the time scale of sampling has a major impact on the reliability of substitution rate estimates.
Effect of gene overlap.
Although overlapping reading frames are predicted to result in a reduced rate of nucleotide substitution (41, 50), this effect was only apparent in some virus-gene combinations. In particular, although substitution rates were indeed generally higher in the nonoverlapping region of the CP gene than in the portion that overlaps the MP gene, no such rate differences were observed in the overlapping and nonoverlapping regions of the P0/RdRp genes. Again, this difference in evolutionary dynamics may reflect the specific structural-functional constraints of the genes in question. For example, the P0 gene contains sites important for RNA-silencing suppression and for viral replication (44, 56, 59), and mutations in these regions are expected to drastically affect virus fitness, which in turn constrain genetic diversity and hence impact on dN/dS ratio estimates (33, 68). Similarly, the putative catalytic sites for serine proteinase activity, the VPg domain, and the frameshift-inducing pseudoknot of poleroviruses are all located in the nonoverlapping region of the RdRp gene (8, 64, 73).
Recombination and modular evolution in the family Luteoviridae.
Luteoviruses are thought to be characterized by relatively frequent intra- and interspecific recombination (3, 26, 49, 51, 71). Indeed, we observed clusters of recombination breakpoints at the boundaries of the RdRp and CP genes of BYDV, CABYV, CYDV, ScYLV, and TuYV, which strongly suggest that these two genes have experienced different evolutionary histories in these species. This is supported by a marked incongruence between the RdRp and CP gene phylogenetic trees (see Fig. S1 and S2 in the supplemental material). Strikingly, between 66 and 100% of the recombination breakpoints detected here were located at gene boundaries, similar to results reported previously (51, 52, 72).
Such widespread intergenic recombination can also be considered a form of “modular evolution.” Modular evolution has been proposed as a major mechanism in generating genetic variability in animal and plant RNA viruses (5, 9). It has also been proposed that speciation in the family Luteoviridae may have resulted from gene module exchange with members of the Sobemovirus and Tombusvirus genera and between Luteovirus and Polerovirus species (3, 26, 49). However, despite the evident frequency of intergenic recombination, it is unclear whether the clustering of breakpoints at gene boundaries occurs because certain RdRp-CP-RTD combinations confer a selective advantage on particular host/vector combinations (46, 72) or because the recombination breakpoints that occur within genes are strongly injurious such that they are rapidly purged by purifying selection.
Macroevolution of luteoviruses.
Our time-structured phylogenetic analysis revealed three periods of evolutionary diversification in the family Luteoviridae (Fig. 2). First, on the basis of these data, the origin of the sampled Luteoviridae may have occurred within the last 4,000 years. Such a time scale of evolutionary change is rather shorter than that proposed for the Potyviridae at ∼6,500 years or even than that previously proposed for the Luteoviridae using Rice yellow mottle virus (a Sobemovirus) in isolation, both of which were linked to the rise of agriculture (17, 24). However, the probability distributions of these and our time estimates overlap, and difficulties in dating deep events in RNA virus evolution mean that all such conclusions should be drawn with caution (32). Indeed, it is noteworthy that we were unable to obtain reliable estimates of the substitution rate of the family Luteoviridae as a whole without using an empirical prior distribution and that our substitution rate estimates were higher than those for the family Potyviridae (∼1 × 10−4 subs/site/year) (24). Employing such lower rates would result in an evolutionary time scale for the luteoviruses that is highly comparable to that of the family Potyviridae and which similarly implies a role for early agriculture in viral speciation. It is also important to recall that the TMRCA estimates given here are for the sampled Luteoviridae species and so do not rule out the existence of extinct species of far greater antiquity, which might explain this difference from previous analyses. Finally, although recombination is known to influence estimates of divergence time (66), we observed no such adverse effects in our study.
The second key evolutionary period relates to the origin of the Luteovirus and Polerovirus genera, which we estimate took place no earlier than 1,500 years ago (although noting the same caveats as above). It is intriguing that both genera seemingly emerged at approximately the same time, although the ecological reasons (if any) underlying this are unclear. Finally, we observed that all individual luteovirus species appeared within the last 500 years. It is striking that the dates estimated for the majority of these speciation events fall into the same range as those of the Potyviridae and of other families of plant RNA viruses (17, 24, 43, 69). Given this overall similarity in evolutionary patterns, we hypothesize that the intensification of agriculture in the modern era, which resulted in an increase in cultivated plant populations, as well as the establishment of global communication networks, had a major effect on the extent and structure of genetic variation in multiple plant RNA viruses.
Finally, we have, for the first time, been able to shed light on the patterns and processes of speciation within the family Luteoviridae. In particular, our observation of a statistically significant association between phylogeny and both host specificity and geographical origin suggests that many species of luteovirus may have arisen within the same host species and in a restricted geographic area, as is expected under a process of “sympatric” speciation. Such a predominance of sympatric speciation may be expected, given that both plants and the aphid vectors that transmit plant viruses clearly have a limited ability to move large geographic distances and that the anthropogenic factors that would assist allopatric speciation (such as increased transportation of crop plants) have generally occurred too recently to greatly influence speciation processes. Indeed, most luteovirus species are restricted to one plant family (27) and it has been previously suggested that specialization to increase within-host multiplication plays a major role in virus evolution (46). However, it is important to note that the presence of as-yet-unsampled Luteoviridae infecting different plant species may be biasing this analysis against the detection of allopatric processes. Similarly, it is clear that allopatric speciation can occur, particularly in the context of those plant species that are only infected by a single luteovirus, such as sugarcane infected by ScYLV (see reference 52). Despite these caveats, we believe that equivalent phylogenetic studies of speciation processes in other families of RNA viruses may prove to be equally informative.
Supplementary Material
Acknowledgments
This work was supported by Marie Curie fellowship PIOF-GA-2009-236470 to I.P. and NIH grant R01 GM080533 awarded to E.C.H.
We thank three reviewers for useful suggestions.
Footnotes
Published ahead of print on 7 April 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Barrell, B. G., G. M. Air, and C. A. Hutchison III. 1976. Overlapping genes in bacteriophage φX174. Nature 264:34-41. [DOI] [PubMed] [Google Scholar]
- 2.Belshaw, R., O. G. Pybus, and A. Rambaut. 2007. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 17:1496-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beuve, M., M. Stevens, H.-Y. Liu, W. M. Witermantel, S. Hauser, and O. Lemaire. 2008. Biological and molecular characterization of an American sugar beet-infecting Beet western yellows virus isolate. Plant Dis. 92:51-60. [DOI] [PubMed] [Google Scholar]
- 4.Blok, J., A. Mackenzie, P. Guy, and A. Gibbs. 1987. Nucleotide sequence comparisons of Turnip yellow mosaic virus isolates from Australia and Europe. Arch. Virol. 97:283-295. [DOI] [PubMed] [Google Scholar]
- 5.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-491. [DOI] [PubMed] [Google Scholar]
- 6.Brault, V., S. Périgon, C. Reinbold, M. Erdinger, D. Scheidecker, E. Herrbach, K. Richards, and V. Ziegler-Graff. 2005. The polerovirus minor capsid protein determines vector specificity and intestinal tropism in the aphid. J. Virol. 79:9685-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chay, C. A., U. B. Gunasinge, S. P. Dinesh-Kumar, W. A. Miller, and S. M. Gray. 1996. Aphid transmission and systemic plant infection determinants of Barley yellow dwarf Luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology 219:57-65. [DOI] [PubMed] [Google Scholar]
- 8.Cornish, P. V., M. Hennig, and D. P. Giedroc. 2005. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U. S. A. 102:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolja, V. V., and J. C. Carrington. 1992. Evolution of positive-strand RNA viruses. Semin. Virol. 3:315-326. [Google Scholar]
- 10.Drake, J. W., B. Charlesworth, D. Charlesworth, and J. F. Crow. 1998. Rates of spontaneous mutation. Genetics 148:1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drummond, A. J., S. Y. W. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy, S., L. A. Shackelton, and E. C. Holmes. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267-276. [DOI] [PubMed] [Google Scholar]
- 14.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elena, S. F., P. Agudelo-Romero, P. Carrasco, F. M. Codoner, S. Martin, C. Torres-Barcelo, and R. Sanjuán. 2008. Experimental evolution of plant RNA viruses. Heredity 100:478-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elena, S. F., and R. Sanjuán. 2005. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 79:11555-11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fargette, D., A. Pinel-Galzi, D. Sereme, S. Lacombe, E. Hebrard, O. Traore, and G. Konate. 2008. Diversification of Rice yellow mottle virus and related viruses spans the history of agriculture from the Neolithic to the present. PLoS Pathog. 4:e1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and B. A. Ball. 2005. Virus taxonomy: classification and nomenclature of viruses. 8th report of the International Committee on the Taxonomy of Viruses. Elsevier-Academic Press, San Diego, CA.
- 19.Fitch, W. N. 1971. Toward defining the course of evolution: minimal change for a specific tree topology. Syst. Zool. 20:406-416. [Google Scholar]
- 20.García-Arenal, F., and B. A. McDonald. 2003. An analysis of the durability of resistance to plant viruses. Phytopathology 93:941-952. [DOI] [PubMed] [Google Scholar]
- 21.Gibbs, A. J. 1980. How ancient are the tobamoviruses? Intervirology 14:101-108. [DOI] [PubMed] [Google Scholar]
- 22.Gibbs, A. J., D. Fargette, F. Garcia-Arenal, and M. J. Gibbs. 2010. Time—the emerging dimension of plant virus studies. J. Gen. Virol. 91:13-22. [DOI] [PubMed] [Google Scholar]
- 23.Gibbs, A. J., J. Blok, D. J. Coates, P. L. Guy, A. Mackenzie, and N. Pigram. 1986. Turnip yellow mosaic virus in an endemic Australian alpine Cardamine, p. 289-300. In B. A. Barlow (ed.), Flora and fauna of alpine Australasia; ages and origins. CSIRO, Collingwood, Victoria, Australia.
- 24.Gibbs, A. J., K. Ohshima, M. J. Phillips, and M. J. Gibbs. 2008. The prehistory of potyviruses: their initial radiation was during the dawn of agriculture. PLoS One 3:e2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbs, A. J., M. J. Gibbs, K. Ohshima, and F. Garcia-Arenal. 2008. More plant virus evolution; past present and future, p. 229-250. In E. Domingo, C. R. Parrish and J. J. Holland (ed.), Origin and evolution of viruses, 2nd ed. Academic Press, London, United Kingdom.
- 26.Gibbs, M. J. 1995. The luteovirus supergroup: rampant recombination and persistent partnerships, p. 351-368. In A. Gibbs, C. H. Calisher, and F. García-Arenal (ed.), Molecular basis of virus evolution. Cambridge University Press, Cambridge, United Kingdom.
- 27.Gray, S., and F. E. Gildow. 2003. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41:539-566. [DOI] [PubMed] [Google Scholar]
- 28.Guyader, S., and D. G. Ducray. 2002. Sequence analysis of Potato leafroll virus isolates reveals genetic stability, major evolutionary events and differential selection pressure between overlapping reading frame products. J. Gen. Virol. 83:1799-1807. [DOI] [PubMed] [Google Scholar]
- 29.Hanada, K., Y. Suzuki, and T. Gojobori. 2004. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol. Biol. Evol. 21:1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 31.Holmes, E. C. 2003. Patterns of intra- and inter-host nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 77:11296-11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes, E. C. 2003. Molecular clocks and the puzzle of RNA virus origins. J. Virol. 77:3893-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes, E. C. 2003. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 11:543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes, E. C. 2009. The evolution and emergence of RNA viruses. Oxford University Press, Oxford, United Kingdom.
- 35.Hughes, A. L., K. Westover, J. da Silva, D. H. O'Connor, and D. I. Watkins. 2001. Simultaneous positive and purifying selection on overlapping reading frames of the tat and vpr genes of simian immunodeficiency virus. J. Virol. 75:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes, A. L., and M. A. Hughes. 2005. Patterns of nucleotide difference in overlapping and non-overlapping reading frames of papillomavirus genomes. Virus Res. 113:81-88. [DOI] [PubMed] [Google Scholar]
- 37.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:156-165. [DOI] [PubMed] [Google Scholar]
- 38.Jin, Z., X. Wang, S. Chang, and G. Zhou. 2004. The complete nucleotide sequence and its organization of the genome of Barley yellow dwarf virus-GAV. Sci. China C Life Sci. 47:175-182. [DOI] [PubMed] [Google Scholar]
- 39.Jordan, I. K., B. A. Sutter IV, and M. A. McClure. 2000. Molecular evolution of the Paramyxoviridae and Rhabdoviridae multiple-protein-encoding P gene. Mol. Biol. Evol. 17:75-86. [DOI] [PubMed] [Google Scholar]
- 40.Kosakovsky Pond, S. L., and S. D. W. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 41.Krakauer, D. C. 2000. Stability and evolution of overlapping genes. Evolution 54:731-739. [DOI] [PubMed] [Google Scholar]
- 42.Krakauer, D. C., and J. B. Plotkin. 2002. Redundancy, antiredundancy, and the robustness of genomes. Proc. Natl. Acad. Sci. U. S. A. 99:1405-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malmstrom, C., R. Shu, E. W. Linton, L. A. Newton, and M. Cook. 2007. Barley yellow dwarf viruses (BYDVs) preserved in herbarium specimens illuminate historical disease ecology of invasive and native grasses. J. Ecol. 95:1153-1166. [Google Scholar]
- 44.Mangwende, T., M.-L. Wang, W. Borth, J. Hu, P. H. Moore, E. T. Mirkov, and H. H. Albert. 2009. The P0 gene of Sugarcane yellow leaf virus encodes an RNA silencing suppressor with unique activities. Virology 384:38-50. [DOI] [PubMed] [Google Scholar]
- 45.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 46.Martin, R. R., P. K. Keese, M. J. Young, P. M. Waterhouse, and W. L. Gerlach. 1990. Evolution and molecular biology of luteoviruses. Annu. Rev. Phytopathol. 28:341-363. [Google Scholar]
- 47.McDonald, B. A., and C. Linde. 2002. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124:163-180. [Google Scholar]
- 48.Miller, W. A., and L. Rasochova. 1997. Barley yellow dwarf viruses. Annu. Rev. Phytopathol. 35:167-190. [DOI] [PubMed] [Google Scholar]
- 49.Miller, W. A., S. Liu, and R. Beckett. 2002. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol. Plant Pathol. 3:177-183. [DOI] [PubMed] [Google Scholar]
- 50.Miyata, T., and T. Yasunaga. 1978. Evolution of overlapping genes. Nature 272:532-535. [DOI] [PubMed] [Google Scholar]
- 51.Moonan, F., J. Molina, and T. E. Mirkov. 2000. Sugarcane yellow leaf virus: an emerging virus that has evolved by recombination between luteoviral and poleroviral ancestors. Virology 269:156-171. [DOI] [PubMed] [Google Scholar]
- 52.Moonan, F., and T. E. Mirkov. 2002. Analyses of genotypic diversity among North, South, and Central American isolates of Sugarcane yellow leaf virus: evidence for Colombian origins and for intraspecific spatial phylogenetic variation. J. Virol. 76:1339-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moya, A., E. C. Holmes, and F. González-Candelas. 2004. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2:279-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parker, J., A. Rambaut, and O. G. Pybus. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8:239-246. [DOI] [PubMed] [Google Scholar]
- 55.Pavesi, A. 2006. Origin and evolution of overlapping genes in the family Microviridae. J. Gen. Virol. 87:1013-1017. [DOI] [PubMed] [Google Scholar]
- 56.Pazhouhandeh, M., M. Dieterle, K. Marrocco, E. Lechner, B. Berry, V. Brault, O. Hemmer, T. Kretsch, K. E. Richards, P. Genschik, and V. Zieger-Graff. 2006. F-box-like domain in the Polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. U. S. A. 103:1994-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peter, K. A., D. Liang, P. Palukaitis, and S. M. Gray. 2008. Small deletions in the Potato leafroll virus readthrough protein affect particle morphology, aphid transmission, virus movement and accumulation. J. Gen. Virol. 89(Pt. 8):2037-2045. [DOI] [PubMed] [Google Scholar]
- 58.Peter, K. A., F. Gildow, P. Palukaitis, and S. M. Gray. 2009. The C terminus of the polerovirus P5 readthrough domain limits virus infection to the phloem. J. Virol. 83:5419-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfeffer, S., P. Dunoyer, F. Heim, K. E. Richards, G. Jonard, and V. Ziegler-Graff. 2002. P0 of Beet western yellow virus is a suppressor of posttranscriptional gene silencing. J. Virol. 76:6815-6824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 61.Ranjith-Kumar, C. T., K. Gopinath, A. N. K. Jacob, V. Svrividhya, P. Elango, and S. H. Savithri. 1998. Genomic sequence of Physalis mottle virus and its evolutionary relationship with other tymoviruses. Arch. Virol. 143:1489-1500. [DOI] [PubMed] [Google Scholar]
- 62.Robertson, N. L., and R. French. 2007. Genetic structure in natural populations of barley/cereal yellow dwarf virus isolates from Alaska. Arch. Virol. 152:891-902. [DOI] [PubMed] [Google Scholar]
- 63.Roossinck, M., and A. Ali. 2007. Mechanisms of plant virus evolution and identification of genetic bottlenecks: impact on disease management, p. 109-124. In Z. K. Punja, S. H. De Boer, and S. Sanfaçon (ed.), Biotechnology and plant disease management. CABI Publishing, Wallingford, United Kingdom.
- 64.Sadowy, E., M. Juszczuk, C. David, B. Gronenborn, and M. D. Hulanicka. 2001. Mutational analysis of the proteinase function of Potato leafroll virus. J. Gen. Virol. 82:1517-1527. [DOI] [PubMed] [Google Scholar]
- 65.Sanjuán, R., P. Agudelo-Romero, and S. F. Elena. 2009. Upper-limit mutation rate estimation for plant RNA virus. Biol. Lett. 5:394-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schierup, M. H., and J. Hein. 2000. Recombination and the molecular clock. Mol. Biol. Evol. 17:1578-1579. [DOI] [PubMed] [Google Scholar]
- 67.Schluter, D. 2001. Ecology and the origin of species. Trends Ecol. Evol. 16:372-380. [DOI] [PubMed] [Google Scholar]
- 68.Simmonds, P., and D. B. Smith. 1999. Structural constraints on RNA virus evolution. J. Virol. 73:5787-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmons, H. E., E. C. Holmes, and A. G. Stephenson. 2008. Rapid evolutionary dynamics of Zucchini yellow mosaic virus. J. Gen. Virol. 89:1081-1085. [DOI] [PubMed] [Google Scholar]
- 70.Smith, H. G., and H. Barker. 1999. The Luteoviridae. CABI Publishing, Wallingford, United Kingdom.
- 71.Stevens, M., B. Freeman, H.-Y. Liu, E. Herrbach, and O. Lemaire. 2005. Beet polerovirus: close friends or distant relatives? Mol. Plant Pathol. 6:1-9. [DOI] [PubMed] [Google Scholar]
- 72.Terauchi, H., K. Honda, N. Yamagishi, S. Kanematsu, K. Ishiguro, and S. Hidaka. 2003. The N-terminal region of the readthrough domain is closely related to aphid vector specificity of Soybean dwarf virus. Phytopathology 93:1560-1564. [DOI] [PubMed] [Google Scholar]
- 73.van der Wilk, F., M. Verbeek, A. M. Dullemans, and J. F. van den Heuvel. 1997. The genome-linked protein of Potato leafroll virus is located downstream of the putative protease domain of the ORF1 product. Virology 234:300-303. [DOI] [PubMed] [Google Scholar]
- 74.Viswanathan, R., M. Balamuralikrishnan, and R. Karuppaiah. 2008. Identification of three genotypes of Sugarcane yellow leaf virus causing yellow leaf disease from India and their molecular characterization. Virus Genes 37:368-379. [DOI] [PubMed] [Google Scholar]
- 75.Wang, T. H., Y. K. Donaldson, R. P. Brettle, J. E. Bell, and P. Simmonds. 2001. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J. Virol. 75:11686-11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whelan, S., and N. Goldman. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18:691-699. [DOI] [PubMed] [Google Scholar]
- 77.Zaaijer, H. L., F. J. van Hemert, M. H. Koppelman, and V. V. Lukashov. 2007. Independent evolution of overlapping polymerase and surface protein genes of hepatitis B virus. J. Gen. Virol. 88:2137-2143. [DOI] [PubMed] [Google Scholar]
- 78.Zhao, X., K. M. McGirr, and G. C. Buehring. 2007. Potential evolutionary influences on overlapping reading frames in the bovine leukemia virus pXBL region. Genomics 89:502-511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.