Abstract
Estrogen and progesterone are the defining hormones of normal female development, and both play critical roles in breast carcinogenesis. Cyclin D1 is a breast cancer oncogene whose amplification is linked to poor prognosis in estrogen and progesterone receptor-positive breast cancers. Here we report that cyclin D1 regulates progesterone receptor expression, consequently enhancing responses to estrogen and progesterone. Estrogen treatment of cyclin D1 transgenic mice increased progesterone receptor expression and induced mammary hyperplasias that were stimulated by progesterone and blocked by a progesterone antagonist. Progesterone receptor levels decreased in cyclin D1 knockout mice. Cyclin D1 regulated progesterone receptor expression through a novel estrogen- and cyclin D1-responsive enhancer in DNA encoding part of the 3′ untranslated region of the progesterone receptor gene. Small inhibitory RNAs for cyclin D1 decreased progesterone receptor expression and estrogen receptor binding to the 3′ enhancer region in human breast cancer cells. Since estrogen and progesterone regulate cyclin D1, our results suggest that cyclin D1's participation in a feed-forward loop could contribute to increased breast cancer risks associated with estrogen and progesterone combinations. Additionally, its regulation of the progesterone receptor identifies a novel role for cyclin D1 in ovarian hormone control of breast development and breast carcinogenesis.
We have long known that ovarian hormones control normal breast development (13). Likewise, the connection between ovarian hormones and breast cancer was first therapeutically exploited over 100 years ago (3). While estrogen and progesterone (Pgr) combine to regulate normal breast development during pregnancy, the Women's Health Initiative study of hormone replacement has clearly demonstrated an increased breast cancer risk when progestins are combined with estrogen (7). This result reinvigorated the view that combinations of progesterone with estrogen can be carcinogenic.
Cyclin D1 (CCND1) is a multifunctional G1-phase cyclin whose regulatory effects are particularly important in breast development and cancer (44). The cloning of cyclin D1 at 11q13 translocation breakpoints in parathyroid and mantle cell tumors was strong evidence of its oncogenic function, and evidence indicating that D1 was a G1 cyclin was a key finding that linked cancer to perturbations of the cell cycle. Extensive evidence links cyclin D1 to cyclin-dependent kinase regulation of the cell cycle (27), although a recent report demonstrates that it also acts in transcriptional control (4). 11q13 amplification has been repeatedly demonstrated in breast cancers (22) where CCND1 overexpression has been linked to estrogen and progesterone receptor (PR) status. This linkage was first attributed to cyclin D1's regulation by estrogen and progesterone (12, 37). In this model, CCND1 is proposed to contribute to poor treatment response of estrogen receptor (ER)-positive tumors by acting downstream to promote hormone agonist- and antagonist-independent proliferation (47).
The connection between poor patient outcome and cyclin D1 status in ER-positive breast cancers also led to explorations of a second potential explanation for cyclin D1-ER linkage. Identification of a potential ER coactivator interaction motif in the cyclin D1 protein suggested that CCND1 can interact with ER coactivators to activate estrogen receptor binding elements (ERE) in a ligand-independent manner (29, 52). Importantly, this direct activation of EREs by CCND1 was not inhibited by antiestrogens (52) and did not require an interaction between cyclin D1 and its associated cyclin-dependent kinases (CDK4/CDK6). This CDK4/CDK6 independence is potentially important, since CDK activity is not always increased in cyclin D1-associated breast cancers (45). Importantly, knock-in of CDK-independent cyclin D1 repairs a mammary developmental defect seen during pregnancy in cyclin D1 knockout mice, specifically confirming cyclin D1's potential CDK-independent functions (23).
A study employing cyclin D1 transgenic mice provided the first in vivo demonstration that cyclin D1 is an oncogene (46). These mice demonstrate extensive preneoplastic hyperplasias that precede tumor onset by months. Using laser-capture microdissection and microarray analysis, we identified genes that were upregulated in invasive tumors compared with normal or hyperplastic tissues (48). Remarkably, these genes were responsive to estrogen and responded to cyclin D1 in a CDK4/CDK6-independent manner, and the cyclin D1 and estrogen effects were additive. This result suggested that, as a third explanation for their association in breast cancer, cyclin D1 and the ER might share target genes. To investigate this possibility, we treated cyclin D1 transgenic mice with estrogen, causing hyperplasias that exhibited increased progesterone receptor expression. We now report that cyclin D1 and estrogen regulate progesterone receptor expression, which identifies a mechanism that may couple the effects of estrogen and progesterone in breast tissues and explain the tight association between cyclin D1 expression and hormone receptor status in breast cancer.
MATERIALS AND METHODS
Animals and histology.
Mouse mammary tumor virus (MMTV) cyclin D1 transgenic mice were maintained as previously described (46). Pellets containing 17β-estradiol (Innovative Research) (0.72 mg/pellet) (49), mifepristone (RU486) (Innovative Research) (35 mg/pellet) (34), and/or progesterone (Innovative Research) (10 mg/pellet) (40) were implanted into wild-type (WT) and cyclin D1 transgenic mice 60 days of age, and sham-treated animals were used as controls. This estradiol dose provides twice the physiologic levels found during estrus but 1/20th of the levels found during pregnancy (30). The progesterone dose is sufficient to regulate ductal development in ovariectomized mice (40). The mifepristone dose blocks tumor development in BRCA−/− mice (34). Whole mounts were prepared 2 months later by dissecting the mammary fat pad from the pelt, fixing in 10% buffered formalin, staining with carmine red, dehydrating in graded series of alcohol, and clearing with xylene. Eight animals each were used for the wild-type and cyclin D1 transgenic mouse groups or were subjected to sham or estradiol treatment. Morphometry was performed by counting structures in three photomicrographs of each of the 32 resulting whole-mount preparations. Numbers of branches (B), termini (T), and lobuloalveolar structures (LAS) were counted and totaled for each image. The means and standard errors for the numbers of branches, termini, and alveolar structures per image were then plotted. Four animals each were used for the experiments combining estradiol, mifepristone, and/or progesterone treatment. Mammary glands were fixed in 10% formaldehyde in phosphate-buffered saline (PBS) for routine hematoxylin and eosin staining histology. Additional mammary glands from the same mice were harvested for RNA and protein analysis. Cyclin D1 knockout mice (JAX 002537 mice; Jackson Laboratories) and matched controls were sacrificed at 8 weeks of age and after 12 days of pregnancy, and the mammary tissues were used for RNA and protein analysis.
RNA and protein expression analysis.
mRNA levels in cellular RNA isolated from the indicated cells or mouse tissues by the use of Trizol (Invitrogen) were analyzed. mRNA was reverse transcribed into cDNA and converted to double-stranded DNA (dsDNA); quantitative real-time PCRs (qRT-PCR) were performed as described previously (48). Table 1 presents primer and probe sequences.
TABLE 1.
Primers used for RNA analysisa
| mRNA | Species | Forward primer | Reverse primer | Probe |
|---|---|---|---|---|
| IER-3 | Mouse | GATTATGCGCTGGATCTTAAAGC | TGCCTTTGTTTCTTCGGACTGT | CCGGCGGCCTTCTAAACGC |
| ETV5 | Mouse | GCATTGTACTCTGATGAAAGGACTCTT | TTTACAACAAATAAATCTCTAGCCAGTTG | ATAGCTTTCTAATCTATACACAGTCT |
| Cyclin D1 | Mouse | TTTCTTGTAGCGGCCTGTTGT | CGGAACACTAGAACCTAACAGATTAAATG | ATTTGCTCTGAGTCTCCGGT |
| Cyclin D1 | Human | GGATGCTGGAGGTCTGCGA | AGAGGCCACGAACATGCAAG | AGGAGGTCTTCCCGCTGGCCATGAAC |
| Estrogen receptor | Mouse | CACATTTCGTATCAACTTTTGTATCCA | CTGCTACATAAGATTGTCTGTCATTCAA | ACATGCCTATTGCTGGGT |
| Progesterone receptor | Mouse | |||
| Cyclin A2 | Mouse | |||
| Progesterone receptor A and B isoforms together | Human | CCTCGGACACCTTGCCTGAA | CGCCAACAGAGTGTCCAAGAC | |
| Progesterone receptor B isoform | Human | TAGTGAGGGGGCAGTGGAAC | AGGAGGGGGTTTCGGGAATA | |
| Progesterone receptor A and B isoforms together | Mouse | GGTGGAGGTCGTACAAGCAT | GGATTTGCCACATGGTAAGG | |
| Progesterone receptor B isoform | Mouse | CGGAGAAGAACAGCAGACTC | CCCAAAGAGACACCAGGAAG |
The primers and probe used for the progesterone receptor analysis were from a TaqMan Mm00435628_m1 assay (Applied Biosystems, Carlsbad, CA); those used for the cyclin A2 analysis were from a TaqMan Mm00438066_m1 assay (Applied Biosystems, Carlsbad, CA).
For Western analyses, 50 μg of protein sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were incubated with rabbit polyclonal anti-cyclin D1 (48), anti-PR antibody (C-20; Santa Cruz, Santa Cruz, CA), anti-beta-catenin (BD Transduction Laboratories, San Jose, CA), anti-IER3 (Abcam Inc., Cambridge, MA), anti-etv5 (Abnova Corp., Taipei City, Taiwan), anti-mitogen-activated protein kinase (anti-MAPK) (K-23; Santa Cruz Biotechnology, Santa Cruz, CA), or antiactin (Chemicon International, Temecula, CA) as indicated. The secondary antibodies used were purchased from Santa Cruz or were those included in an enhanced chemiluminescence detection kit (Amersham).
Proliferating T47D cells were incubated in phenol red-free Dulbecco's modified Eagle's medium (DMEM) with 5% charcoal stripped serum for 3 days before transient transfections were performed. Cyclin D1 (Ambion) and scramble (Dharmacon D-1205-20) small interfering RNAs (siRNAs) were transfected using NeoFX (Applied Biosystems). To demonstrate the knockdown efficiency of the siRNAs, cells were harvested 48 h after transfection and subjected to Western blot analysis. We also stably reduced cyclin D1 expression in T47D cells by the use of a lentiviral construct expressing a short hairpin RNA (shRNA) targeted to CTCTAAGATGAAGGAGACCAT in exon 2 of cyclin D1 (RNAi consortium no. TRCN0000040040).
ZR-75-1, CAMA, HCC1428, BT483, and MCF 7 breast cancer cells were grown under the conditions indicated by the supplier (ATCC). They were also transiently transfected with scramble or small inhibitory RNA for cyclin D1 (siCCND1) siRNAs as described for the T47D cells. Western blot analysis and quantitative real-time PCRs (qRT-PCR) were performed as described above.
ChIP and electrophoretic mobility shift experiments.
We performed chromatin immunoprecipitation (ChIP) as previously described (48). We evaluated estrogen receptor genomic binding in T47D cells grown in phenol red-free DMEM with 5% charcoal-stripped serum for 3 days and treated with vehicle or 100 nM 17β-estradiol for 4 h. We performed chromatin immunoprecipitation by a method that we previously described (48). Cross-linking was performed by adding 37% formaldehyde to reach a final concentration of 1.5% for 15 min at 25°C with gentle agitation. The reaction was stopped by adding 0.125 M glycine for 5 min. Cells were scraped and collected by centrifugation; the pellet was washed twice with ice-cold PBS (120 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH 7.4) and twice with immunoprecipitation (IP) buffer (150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% NP-40, 50 mM Tris-HCl [pH 7.5], 0.5 mM dithiothreitol [DTT]) and resuspended with IP buffer supplemented with 0.5% SDS and protease inhibitor cocktail (Roche). Sonication was performed until the desired DNA fragment sizes were reached. The lysate was then subjected to centrifugation at 12,000 × g for 10 min. The supernatant was collected and diluted 5-fold with IP buffer. For immunoprecipitation, anti-ER antibody (HC-20) from Santa Cruz or an equal concentration of normal rabbit serum was added to the lysate and incubated at 4°C overnight with agitation. Protein A Sepharose beads (Amersham) were added to the mixture and incubated for 1 h. The beads were collected by centrifugation and washed twice with IP buffer, twice with high-salt IP buffer (500 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% NP-40, 50 mM Tris-HCl [pH 7.5], 0.5 mM DTT), twice with stringent wash buffer (10 mM Tris-HCl [pH 7.5], 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA), and twice with Tris-EDTA (TE) buffer (10 mM Tris-HCl, 0.1 mM EDTA). The bound fraction was eluted in elution buffer (TE buffer with 1% SDS) at 65°C for 10 min. Cross-links were reversed by incubating the eluate at 65°C for 6 h. Samples were treated with proteinase K at 45°C for 1 h and subjected to phenol-chloroform extraction and ethanol precipitation. The primer sets used in quantitative PCRs (qPCR) are listed in Table 2.
TABLE 2.
Primers used for chromatin immunoprecipitation and electrophoretic mobility shift experiments
| Position with respect to TSS | Amplified region | ChIP-PCR forward primer | ChIP-PCR reverse primer |
|---|---|---|---|
| −56 | −120 to 0 | CAGAATAACGGGTGGAAATG | TGGCCAGACAGCTTTCTAAC |
| 90 | 41 to 150 | TGTGCGTGTGGGTGGCATTC | CGAAGATCTCAGATCCCAGT |
| 326 | 276 to 375 | GAGAGCTTCACAGCATGCAC | CTTGAGTGGCTGCGGCTGCG |
| 580 | 521 to 640 | CGACAGCCACAGTTCCCCTG | GTGTTGAATGTGGCTGGACCG |
| 650 | 606 to 710 | GGTGGAGATCCCTCCGGTCC | CTCCAGGAGGAGGGAAAAGG |
| 745 | 701 to 790 | CCTCCTGGAGACGGGGGAGG | GCCCGCCACGTGGGGAGCCC |
| 1233 | 1181 to 1285 | CTTCCCGAAGATCCACCGGC | GGCAGCTGCCGTCCCGGAGC |
| 51200 | 51128 to 51307 | GAGTTATTTGATTTCTTCGTTG | GAAGATGGAAGCAGAACACC |
| 79800 | 79733 to 79903 | GTGTATTGTGTGTGATGCAAG | GTTACTTGAACTCACTCACTTG |
| 95585 | 95538 to 95660 | CTAGTCCCAGAATGTCAGCC | GCCTCTCCCATGTCTCCATG |
| EMSA probes | Designated | ||
| 95585 | 585 | CTGTATGTCATGCTGACCTTATTGTTAAACAC | |
| 95585 mutant | 585μ | CTGTAGATCTTGCGTCTATTATTGTTAAACAC | |
| 95585 core | 585core | CGTGTCATGCTGACCGA | |
| 95585 mutant core | 585μcore | GCAGCTTATGCTGATAG |
T47D cell nuclear extracts were obtained and analyzed using previously published methods (9). Gel shift activity for DNA binding proteins was determined using electrophoretic mobility shift assay (EMSA) binding buffer (20 mM HEPES [pH 7.5], 50 mM KCl, 7.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 0.5 mM DTT) and 2 ng/μl poly(dI-dC) as a nonspecific competitor. Complexes formed in binding buffer were resolved on a 4% nondenaturing polyacrylamide gel containing 0.5× Tris-borate-EDTA (TBE) buffer at 4°C. Double-stranded oligonucleotides were purified by polyacrylamide gel electrophoresis and labeled using [γ-32P]ATP and polynucleotide kinase. Each binding reaction mixture contained between 0.1 and 0.5 ng of labeled oligonucleotides. Competition experiments were performed using a 200-fold molar excess of unlabeled oligonucleotides. Oligonucleotide sequences are listed in Table 2.
Reporter constructs and transfections.
Transient transfections were performed using T47D cells for the majority of the reporter assays (but see Fig. 8, which shows the results of experiments in which we used MCF7 cells). The transient siCCND1 transfections were performed as described above. Cyclin D1 overexpression was accomplished using the plasmids pRC-CMV-cyclin D1 and pRC-CMV-mutant K112E as previously described (48). The plasmids hPRAB, hPRA, and hPRB were gifts from I. De Vivo (17). The 3× estrogen response element (ERE) luciferase plasmid was obtained from Addgene (14). The 1.3-kb nucleotide portion of the 3′ untranslated region (UTR) of the progesterone receptor gene (+94788 to +96092 relative to the B from the transcription start site [TSS]) was amplified by PCR using primers PR3F114788IF (GTCTGGATCCGTCGACGTCCTAAATTGCTTATCCTTACTTCACTAAG) and PR3R116092IF (AAGGGCATCGGTCGACTGGCAAAAAGCTTGAAACCACTGCTGTC) and cloned to SalI-digested pGL2 reporter vectors by the use of an In-Fusion cloning kit (Clontech). To insert the 3′ UTR in the opposite orientation, the same nucleotide portion was amplified using primers PR3F114788IV (AAGGGCATCGGTCGACGTCCTAAATTGCTTATCCTTACTTCACTAAG) and PR3R116092IV (GTCTGGATCCGTCGACTGGCAAAAAGCTTGAAACCACTGCTGTC) and similarly cloned into pGL2.
FIG. 8.
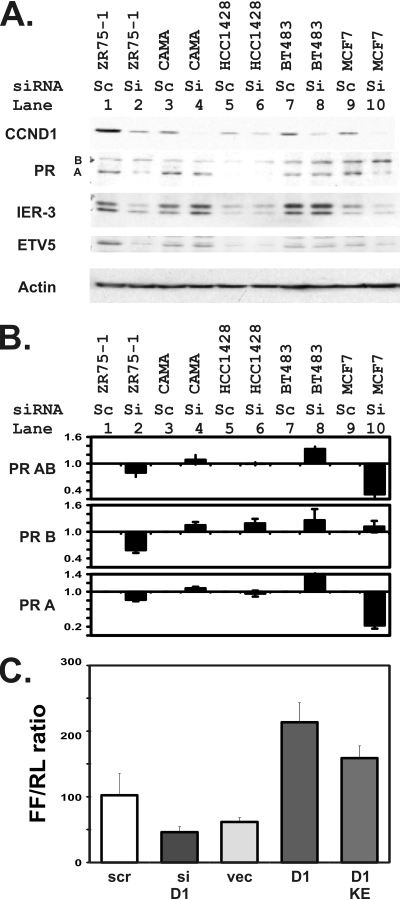
Progesterone receptor expression is blocked by a small inhibitory RNA for cyclin D1 in breast cancer cells in addition to the T47D cells used for the previous four figures, and decreases in cyclin D1 sensitize cell lines to both antiestrogen and antiprogesterone treatments. (A and B) Progesterone receptor expression responds to CCND1 knockdowns in several human breast cancer cell lines. (A) Immunoblot experiments were performed using the indicated gene products and 50 μg of lysates harvested 48 h after transfection as described for Fig. 4. Cyclin D1 levels differed between the cell lines, with those toward the left of panel A showing generally higher levels, as is consistent with the known results of 11q13 amplifications in ZR75-1 and CAMA cells. siCCND1 knocked down CCND1 protein levels in all cell lines tested. As we observed with respect to T47D cells, CCND1 knockdown decreased levels of PR isoform A in MCF7 and ZR75-1 cells. Similar losses of candidate CCND1-target genes were seen in parallel for IER3 and ETV5 in these cell lines. An actin loading control is shown. (B) The indicated human breast cancer cell lines were transfected with scramble (Sc) or siCCND1 (Si). Levels of isoform AB- and B-specific transcripts of the progesterone receptor were measured using equal amounts of reverse-transcribed cDNA via isoform-specific qRT-PCR. Isoform A levels were derived by subtracting B from AB levels. The means and standard errors for fold changes from scramble to siCCND1 transfectants for each of the indicated cell lines are indicated. (C) The 3′ UTR of the PR gene responds to cyclin D1 in MCF 7 cells. The pGL2P-UTR vector was transfected into MCF 7 cells together with a scramble control RNA (scr), siCCND1 (siD1), an empty vector control (vec), an expression plasmid for cyclin D1 (D1), and the plasmid expressing the K112E mutant of cyclin D1 (KE). Reporter gene expression measured as described above is plotted as the ratio of the firefly luciferase (FF) in the PR reporter to the CMV-renilla control reporter.
To clone hPR AB-CER, primers PR3F115435IF and PR3R115672IF were used to amplify T47D genomic DNA. To clone hPR AB-REC, primers PR3F115435IV and PR3R115672IV were used. PCR fragments were inserted into the SalI site of hPR AB plasmid by the use of an In-Fusion cloning kit (Clontech). To construct hPR AB-3XCER, a primer PR-P1F- and primer PR3R115672IF-amplified fragment was similarly cloned to the hPR AB SalI site. The second copy of the central element-containing region (CER) was inserted in the same site by a primer PR-P2F- and primer PR3R115672IF-amplified fragment. This step was then repeated to introduce the third copy of CER. To move the tandem CERs to the proximal position upstream of hPR promoter, the 3XCER fragment was amplified from plasmid hPR AB-3XCER by primers IF-3xCERXhoF and IF-3xCERXhoR and cloned to the XhoI-linearized hPR AB plasmid by the use of an In-Fusion kit. hPRAB-CERμ1 and hPRAB-CERμ2 were mutated from hPRAB-CER by the use of a Stratagene QuikChange Lightning site-directed mutagenesis kit. All constructs were validated by sequencing. The primer sequences used for cloning were as follows: for PR3F115435IF, GTCTGGATCCGTCGACATTCCAAGGCAGAGCTCAGG; for PR3R115672IF, AAGGGCATCGGTCGACCTTAAAAATAATGCCTCTCCC; for PR3F115435IV, AAGGGCATCGGTCGACATTCCAAGGCAGAGCTCAGG; for PR3R115672IV, GTCTGGATCCGTCGACCTTAAAAATAATGCCTCTCCC; for PR-P1F, GTCTGGATCCGTCGAGACGTCATTCCAAGGCAGAGCTCAGG; for PR-P2F, TATTTTTAAGGTCGATGCATATTCCAAGGCAGAGCTCAGG; for IF-3xCERXhoF, CATTCAGAATCTCGAGCTGGATCCGTCGAGACGTC; and for IF-3xCERXhoR, GGCATTTCCACTCGAGGGCATCGGTCGACCTTAA.
Cells were transfected in 96-well plates with 200 ng of luciferase reporter plasmids by the use of Lipofectamine 2000 (Invitrogen) with 0.5 ng of pCMV-renilla as a control. Cells were treated with 100 nM estradiol 24 h after transfection and assayed using a dual-luciferase reporter assay system (Promega) after 24 h of treatment.
RESULTS
Estradiol treatment enhances mammary hyperplasia in cyclin D1 transgenic mice.
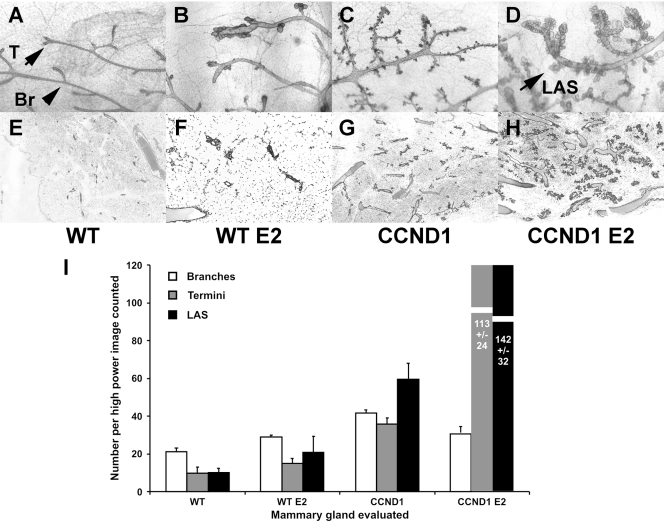
Since cyclin D1 is particularly associated with hormone receptor-positive cancers, we sought to determine whether constitutive cyclin D1 expression would accelerate the response to estrogen in an animal model. We stimulated mammary proliferation using wild-type and CCND1 transgenic mice and continuous-release 17β-estradiol (E2) pellets (49). We initiated treatment at 2 months of age and evaluated the proliferative response of mammary glands 2 months later using whole-mount and histologic assessments. Estradiol and cyclin D1 and their combination all caused different changes in mammary glands (Fig. 1). Ducts in untreated wild-type mice had a simple branching structure (Fig. 1A and B) that terminated in single end-bud structures (see letter T in Fig. 1A). Estradiol treatment led to dilated ducts and aberrant buds at the termini of ducts in wild-type mice (Fig. 1B and F). Cyclin D1 overexpression increased levels of both side branching and terminal acinar structures in the transgenic mice (Fig. 1C and G). The combination of estradiol treatment with cyclin D1 overexpression caused more marked changes. First, ducts were dilated and showed multiple abnormally enlarged buds that were reminiscent of normal alveolar structures (see “LAS” in Fig. 1D; also see Fig. 1H). Second, multiples of such alveolar structures were found along the sides of ducts, whereas no such structures were found in similar locations in E2-treated wild-type mice, and those found in untreated CCND1 mice were small. These effects combined to produce extensive alveolar hyperplasia in E2-treated CCND1 transgenic mice. We analyzed whole mounts and histology using a total of eight mice from each of the four groups and performed morphometric analyses as described in Materials and Methods. Numbers of LAS in E2-CCND1 mice increased at least 10-fold over LAS in the wild-type mice and 5.8-fold over LAS in E2-treated mice (P < 0.001) (Fig. 1I).
FIG. 1.
Alveolar structure numbers increase in mammary tissues constitutively expressing cyclin D1 after treatment with estrogen. Continuous release estradiol pellets were implanted subcutaneously into wild-type mice (B and F) and cyclin D1 transgenic mice (D and H) at 2 months of age as described in Materials and Methods. Control mice (A and E) were subjected to sham operations at the same time as control cyclin D1 transgenic mice (C and G). Mammary glands were harvested 2 months later for analysis using whole-mount preparations (A to D) and for routine hematoxylin and eosin staining histology (E to H). (A to D) Whole-mount photomicrographs (×10). (A) Wild-type mice (WT); (B) estradiol (E2)-treated WT mice; (C) transgenic MMTV-CCND1 mice (CCND1); (D) MMTV-CCND1 mice treated with estradiol (CCND1 E2). (E to H) Histologic photomicrographs (×40). (E) Wild-type mice (WT); (F) estradiol (E2)-treated WT mice; (G) transgenic MMTV-CCND1 mice (CCND1); (H) MMTV-CCND1 mice treated with estradiol (CCND1 E2). (I) Morphometric counts. We harvested whole-mount preparations for eight mice in each of the four groups (wild-type sham, wild-type E2, CCND1 sham, and CCND1 E2). Morphometric analyses were then performed by counting structures in three photomicrographs of each of the 32 whole-mount preparations. Numbers of branches, termini, and lobuloalveolar structures (LAS) were counted on coded photographs and totaled for each image. The means and standard errors for the numbers of branches, termini, and alveolar structures per image are shown.
Estradiol treatment increases progesterone receptor expression and signaling in cyclin D1 transgenic mice.
We next assessed expression of cyclin D1, the ovarian hormone receptors, and several target genes to identify genes whose altered expression accompanied these histologic changes. Using species-specific primers, we found that levels of murine CCND1 mRNA increased in response to transgene treatment or to E2 treatment but that combining the two treatments did not cause additive increases (Fig. 2 A). Since the human cyclin D1 transgene was not present in the wild-type mice, we further evaluated the increase in its mRNA value with respect to the murine wild-type value and found that the transgenic CCND1 value equally increased in the absence and presence of the E2 stimulus, as expected, since the MMTV element does not respond to the presence of estrogen (32). Thus, estradiol was not acting simply to induce expression of the CCND1 transgene.
FIG. 2.
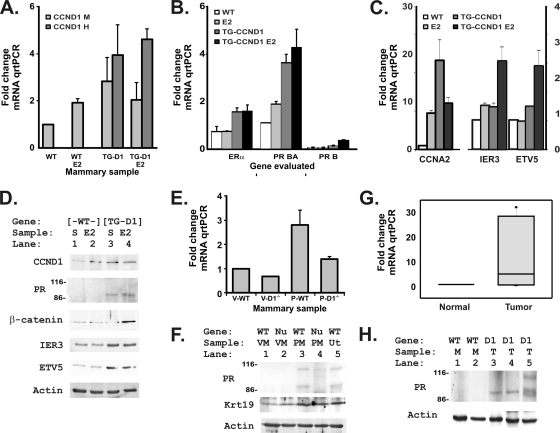
Estradiol increases progesterone receptor expression in mammary tissues of mice constitutively expressing cyclin D1. (A) Levels of both murine CCND1 mRNA (CCND1 M) and the human CCND1 mRNA that was used in our transgene (CCND1 H) were assessed by TaqMan-based quantitative real-time PCRs (qRT-PCR) to evaluate cDNAs reverse transcribed from 5 μg of total RNA harvested from each of the indicated specimens 2 months after hormonal treatments. We measured mRNA and cDNA amounts at each step in the analyses and used equal amounts of mRNA and cDNAs for each tissue and gene evaluated for each qRT-PCR. Murine cyclin D1 (CCND1 M) and human cyclin D1 (CCND1 H) levels were determined and plotted as fold changes in the threshold value for quantitation, setting the untreated wild-type (WT) mammary sample to a value of 1 and plotting fold changes for each of the other samples. The results for wild-type (WT), E2-treated wild-type (WT E2), CCND1 transgenic (TG-D1), and E2-treated CCND1 transgenic (TG-D1 E2) mice are indicated. The means and standard errors for 3 samples are shown. Human CCND1 was undetectable in wild-type mice, and the murine CCND1 primer-probe set did not detect human CCND1. (B) Estrogen receptor (ER) and progesterone receptor (PR) mRNAs were evaluated with TaqMan as described for panel A, using the specific primers and probes listed in Materials and Methods. The PR BA mRNA is 300 nucleotides longer than the PR A transcript. PR B isoform-specific primers were used to differentiate expression of this additional 5′ sequence (34). (C) Changes in cyclin A2 (CCNA2) expression were evaluated in the same tissues to detect possible CDK- and E2F-dependent effects. Increases for immediate-early response 3 (IER3) and ets variant 5 (ETV5), two candidate CCND1-target mRNAs, were evaluated in the same tissues. (D) Mammary protein lysates (50 μg) were probed for CCND1 (by using an antibody that detects both human and mouse cyclin D1), the progesterone receptor (PR), β-catenin, and the two candidate CCND1-target genes for which antibodies could detect the murine protein in mammary tissues. Size markers show that numbers of the PR A isoform are increased in the cyclin D1 transgenic mice, consistent with reports that PR A is the dominant PR isoform except during the later stages of pregnancy (1). An actin loading control is included. Wild-type (WT) and cyclin D1 transgenic (TG-D1) genotypes are indicated. Treatment groups were subjected to sham treatment (S) or treated with estradiol pellets (E2). (E) Progesterone receptor mRNA levels were measured in mammary tissues from virgin (V) and 12-day-pregnant (P), wild-type (WT), and CCND1−/− (D1−/−) mice. The mRNAs were isolated and measured as described above for panel A. Values are plotted as means and standard deviations for two glands of each developmental stage and phenotype. (F) Protein lysates from mammary glands of virgin (VM), 12-day-pregnant (PM), wild-type (WT), and CCND1−/− (Nu) mice were probed and compared with those of a wild-type uterine sample (Ut). In this case, both 114-kDa and 94-kDa PR bands are seen for the pregnant tissues, consistent with the reported expression of PR B and A during pregnancy (1). The membrane was then probed for keratin 18, a luminal mammary cell marker (2), to show that the PR changes were not due simply to changes in tissue composition. An actin loading control is shown. (G) Progesterone receptor mRNA was measured in mammary lysates from normal mammary tissues (Normal) and tumors induced by the MMTV-cyclin D1 transgene (Tumor). The mRNAs were isolated and measured as described above for panel A. (H) Lysates harvested from normal mammary glands (M) and cyclin D1-transgene-induced tumors (T) were probed for PR protein and an actin loading control.
We then evaluated hormone receptor mRNAs and found that ER-alpha mRNA levels were only minimally affected by either estradiol treatment or transgene expression (Fig. 2B). Instead, progesterone receptor mRNA levels increased in response to the presence of both estradiol and cyclin D1 and especially increased in the E2-treated CCND1 mice to a level that was 4-fold above the wild-type value (Fig. 2B [PR]; P = 0.001). The PR mRNA is transcribed as a full-length BA transcript and a 300-nucleotide-shorter A transcript. The B-specific extension can be identified using PCR primers specific for its unique 5′ region. As previously reported (1), PR expression in these nonpregnant mammary samples was nearly entirely the PR A transcript. Cyclin A2 (CCNA2) was evaluated as a potential CDK4/E2F target gene, and its expression increased 20-fold in level in response to CCND1 expression (Fig. 2C). The combination of estradiol with CCND1 did not further increase CCNA2 numbers. We also evaluated expression of two candidate CCND1 targets that were found to be associated with tumor invasion in our previous study (48) (Fig. 2C). Expression of both the immediate-early response 3 (IER3) and ets variant 5 (ETV5) mRNAs increased in response to the combination of cyclin D1 and estrogen, along with that of the increased progesterone receptor mRNA. Since IER3 is a known progesterone target (35), the ETV5 and IER3 increases may have resulted as a response to the cyclin D1-induced increase in PR rather than as a direct effect of CCND1 and E2 on regulation.
To confirm the mRNA results, we performed immunoblot analyses for the progesterone receptor as well as for several progesterone receptor and cyclin D1 target genes (19, 36) (Fig. 2D). Progesterone receptor protein levels generally paralleled mRNA changes, and only the 94-kDa PR A isoform was seen, as previously described (1). PR levels clearly increased in the cyclin D1-expressing transgenic tissues. β-Catenin, a known target of PR function, increased in level only in the E2-CCND1 mice. This increase in β-catenin levels, together with the increased PR levels, suggested that the E2-CCND1 combination enhanced both PR expression and PR function (16). Indeed, the histology of the E2-CCND1 mice resembles that seen in PR overexpression (41) and in increased β-catenin signaling (26). Both the IER3 and ETV5 protein levels increased in response to the CCND1 transgene (Fig. 2D), although increases in IER3 and ETV5 protein numbers did not fully match the mRNA increases, suggesting that additional posttranscriptional mechanisms may limit additional estrogen-induced expression. Taken together, our results suggest that combining E2 with constitutive CCND1 expression causes a hyperplastic abnormality in virgin mice and that that abnormality is accompanied by increased PR and PR signaling in those mice.
Loss of cyclin D1 in knockout mice causes mammary growth failure during pregnancy, when progesterone receptor signaling is most critical for normal reproductive development (43). To determine whether PR expression is decreased in CCND1 knockout mammary glands, we measured PR mRNA and protein levels in glands harvested from virgin wild-type and CCND1−/− mice at 8 weeks of age and in glands harvested from wild-type and CCND1−/− mice at 12 days of pregnancy (Fig. 2E and F). Loss of cyclin D1 led to decreased PR mRNA levels in both stages. PR protein changes paralleled the changes in mRNA, and, as expected, we observed a PR B isoform band at 114 kDa in pregnant tissues. The results seen with a keratin 19 blot experiment suggested that the observed changes were not merely the result of changes in luminal cell composition in the knockout tissues. PR expression was increased in tumors in the cyclin D1 transgenic mice (Fig. 2G and H).
Estrogen-enhanced mammary hyperplasia is further regulated by progesterone agonists and antagonists in cyclin D1 transgenic mice.
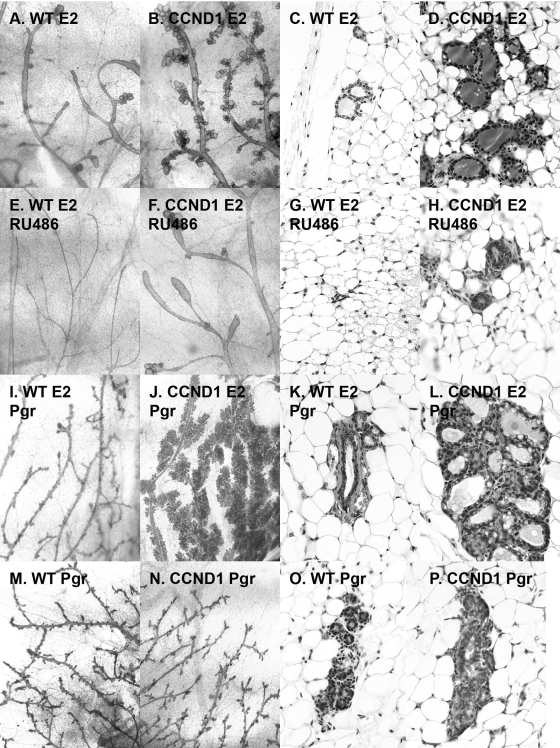
Given this evidence for increased PR signaling and the phenotypic changes that were consistent with a shift toward enhanced PR function in estrogen-treated cyclin D1 transgenic mice, we next sought to determine whether progesterone (Pgr) agonists or antagonists would enhance or block effects of the E2-CCND1 combination in virgin mice (Fig. 3). First, mifepristone (RU486) blocked much of the effect of E2 on CCND1 mice (Fig. 3F and H versus Fig. 3B and D). Importantly, the addition of progesterone pellets to E2 and CCND1 markedly stimulated the lobuloalveolar development beyond that achieved by E2 treatment and clearly increased hyperplasia compared to the results seen with E2-Pgr-treated wild-type mice (compare Fig. 3J and L to Fig. 3I and K). In contrast, treatment of wild-type and CCND1 mice with progesterone alone revealed few differences (Fig. 3M and O and Fig. 3N and P, respectively). We repeated these experiments using mice subjected to ovariectomy at 7 weeks of age. The histologic changes that accompanied estrogen and progesterone treatment of wild-type and cyclin D1 transgenic mice were not affected by ovariectomy, suggesting that these phenotypic changes were not dependent on endogenous ovarian hormone production (not shown).
FIG. 3.
The increased estrogen-induced lobuloalveolar structures in mammary tissues constitutively expressing cyclin D1 were blocked by a progesterone antagonist, enhanced when progestins were added to estrogen, and did not appear in response to progestins alone. (A to H) Mifepristone blocks estrogen effects in cyclin D1-expressing mice. The effect of treatment using continuous-release estradiol pellets was compared to the effect of treatment using estrogen plus continuous-release mifepristone (RU486) in wild-type mice (A and C versus E and G) and MMTV-cyclin D1 mice (B and D versus F and H) 2 months after pellet implantation at 2 months of age. Mammary glands were harvested for analysis using whole-mount preparations (×10) (A, B, E, and F) and for routine hematoxylin and eosin staining histology (×40) (C, D, G, and H). (I to P) Progestins enhance estrogen's effects on mammary tissues constitutively expressing cyclin D1. The effect of the combination of progesterone pellets with continuous-release estradiol pellets was compared to the effect of progesterone pellets alone in wild-type mice (I and K versus M and O) and MMTV-cyclin D1 mice (J and L versus N and P) 2 months after pellet implantation. Mammary glands were harvested from mice 4 months of age for analysis by whole-mount preparations (×10) (I, J, M, and N) and for routine hematoxylin and eosin staining histology (×40) (K, L, O, and P).
Identification of a 3′ estrogen receptor binding element in the progesterone receptor gene that responds to cyclin D1 and estrogen.
To determine whether the regulatory effects of CCND1 and estrogen on PR expression could also be found in human cells, we tested the effects of a small inhibitory RNA for cyclin D1 (siCCND1) on PR expression in T47D breast cells. Progesterone receptor mRNA levels decreased 60% ± 1% in response to siCCND1 compared to those seen with scramble siRNA-transfected control cells. This decrease was seen primarily in the shorter PR A isoform analyzed using isoform-specific primers. Western blot experiments confirmed that siCCND1 transfections decreased both cyclin D1 and PR levels (Fig. 4 A), and image analysis confirmed a 2-fold decrease in cyclin D1 protein levels. We then evaluated the effect of this PR change on IER3 expression and found that it also decreased in response to decreased CCND1.
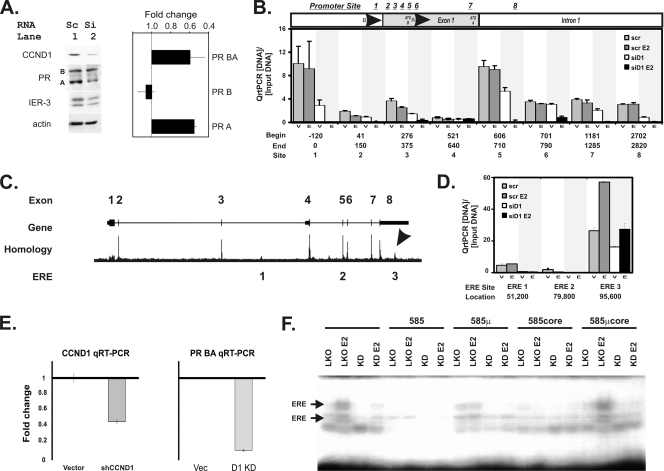
FIG. 4.
Small interfering RNA (siRNA) knockdown of cyclin D1 blocks progesterone receptor expression and inhibits chromatin binding of the estrogen receptor to a novel 3′ estrogen response element in the progesterone receptor gene. (A) A small interfering RNA that knocks down cyclin D1 (siCCND1) decreases progesterone receptor expression. T47D breast cells were transfected with siCCND1 (Si) or a scramble control (Sc), and protein lysates were harvested 48 h later (left panel). We performed immunoblot analyses using 50 μg of protein lysates for the indicated genes. Immunoblots for cyclin D1 (CCND1), the progesterone receptor (PR) isoforms A and B, the immediate-early response gene 3 (IER-3) that we had previously demonstrated to have responded to cyclin D1 and estrogen (48), and an actin loading control are shown. (Right panel) Isoform BA and B transcripts of the progesterone receptor were measured using equal amounts of reverse-transcribed cDNA via isoform-specific qRT-PCR. Isoform A levels were derived by subtracting B from AB levels. The mean and standard error fold changes from scramble to siCCND1 transfectants are shown. (B) Analysis of cis sequences that have been reported to be estrogen control elements in the progesterone receptor gene promoters. Sites in the progesterone promoter region where estrogen regulation of PR expression has been proposed to occur are indicated. Candidate sites include the following: site 1, Sp1 (39); site 2, an AP1 at +90 from the PR B transcription start site (TSS) (33); site 3, a G/A 326 GATA5 breast cancer risk polymorphism (17); site 4, an Sp1-ERE half-site (33); site 5, the PR isoform A promoter region; site 6, a second AP1 at +745 from the PR B TSS (38); site 7, an RNA polymerase II binding site (6); and site 8, a rat promoter site that was previously shown to have responded to E2 (20). The locations of the start (Begin) and ending (End) points for the PCR primers used in these studies are indicated, along with the numbering system (Site) that identifies each site. We performed ChIP-PCR for ER binding as previously described (48). Anti-ER ChIP-PCR signals are plotted as a fraction of the input DNA for each reaction, subtracting the minimal ChIP-PCR backgrounds obtained using a nonspecific control serum from the αER signal. We measured αER ChIP-PCR DNA levels in cells transfected with a scramble control (scr) or an siCCND1 (siD1) oligonucleotide. Transfectants were treated with vehicle control (V) or estradiol (E). At each site, the bars depict the αER signal for scramble plus vehicle (scr), scramble plus E2 (scr E2), siCCND1 plus vehicle (siD1), and siCCND1 plus E2 (siD1 E2). (C) Locations of three conserved, canonical estrogen receptor binding sequences in the progesterone receptor gene. The progesterone receptor gene structure is shown diagrammatically using the UCSC genome browser to plot its exons. A homology plot from the genome browser is shown; an arrow identifies the region in the 3′ UTR of the gene that is the most highly conserved part other than its exons. The locations of the three canonical estrogen receptor binding elements that are conserved across mammalian species are indicated (ERE 1, 2, and 3). (D) Chromatin immunoprecipitation (ChIP) identified a novel ER-binding site in the DNA encoding the 3′ untranslated region (UTR) of the PR gene whose ER binding is uniquely stimulated by estradiol and blocked by an inhibitory siCCND1 oligonucleotide. We performed ChIP-PCR for ER binding at the three ERE sites as described for panel C. Anti-ER ChIP-PCR signals are plotted as a fraction of the input DNA for each reaction, subtracting the ChIP-PCR backgrounds. We measured αER ChIP-PCR DNA in cells transfected with a scramble control (scr) or an siCCND1 (siD1) oligonucleotide. Transfectants were treated with vehicle control (V) or estradiol (E). For each site, the bars depict the αER signal for scramble plus vehicle (scr), scramble plus E2 (scr E2), siCCND1 plus vehicle (siD1), and siCCND1 plus E2 (siD1 E2). Three conserved, canonical estrogen binding elements identified in the UCSC genome browser were compared; these sites are indicated according to the nucleotide distance downstream of the transcription start site of the B isoform of the progesterone receptor. (E) Stable knockdown of cyclin D1 reduces PR mRNA levels. We stably reduced cyclin D1 expression in T47D cells by expressing a lentiviral shRNA construct in pooled transfectants selected using puromycin. Changes in cyclin D1 and progesterone receptor mRNAs are shown as fold changes in mRNA levels compared to those of pooled control cells containing the parent pLKO vector (Vec). We used qRT-PCR to monitor mRNA levels as described in Materials and Methods. (F) Stable knockdown of cyclin D1 reduces binding to the canonical estrogen receptor binding site in the 3′ ERE of the progesterone receptor gene. We used electrophoretic mobility shift assays to monitor protein binding to an oligonucleotide containing the estrogen response element sequences 95,585 nucleotides downstream of the PR transcription start site. Protein binding to the oligonucleotide probe was evaluated in lysates harvested from the vector control cells (LKO) in the absence and presence of estradiol (E2). Levels of binding to the radiolabeled probe were compared using lysates harvested from the cells expressing the cyclin D1 knockdown shRNA (KD) in the absence and presence of estradiol (E2). The specificity of the binding was evaluated using a 200-fold excess of unlabeled probe (585), unlabeled 585 oligonucleotide containing a mutation in the ERE (585μ), an unlabeled smaller oligonucleotide containing the core ERE sequence (585core), and an unlabeled core oligonucleotide containing the ERE-inactivating mutation (EREμcore).
Although previous investigations failed to discover PR promoter sequences that contained the canonical estrogen receptor binding sequence (agGtCAnnnTGaCct; uppercase indicates conserved sites) (6), several investigators identified alternative promoter elements that responded to estrogen, including Sp1 (39), AP1 at +90 from the PR B transcription start site (TSS) (33), a G/A GATA5 breast cancer risk polymorphism (17), an Sp1-ERE half-site (33), a second AP1 at +745 (38), an RNA polymerase II binding site (6), and a rat promoter site that responded to E2 (20). We used ChIP-PCR to directly evaluate effects of estradiol and CCND1 on estrogen receptor binding to these candidates (Fig. 4B). According to the results of our chromatin immunoprecipitation studies, none of these previously proposed alternative promoter elements showed increased estrogen receptor binding with estrogen stimulation. Two of the eight sites showed a significant decrease in estrogen receptor binding after cyclin D1 knockdown, and they were located at the core progesterone receptor promoter sites for the B isoform (centered at −60) and the A isoform (centered at +658).
The Genome Browser of the University of California at Santa Cruz (UCSC) identifies three highly conserved, canonical estrogen receptor binding sites ([agGtCAnnnTGaCct]) (ERE 1-ERE 3) in the progesterone receptor gene that are located at 51,200, 79,800, and 95,585 nucleotides downstream of the transcription start site for the B isoform of the progesterone receptor (Fig. 4C) (21). A recent genome-wide screening showed that only the site at +95585 binds the estrogen receptor in response to estrogen treatment (6). We used ChIP together with quantitative PCR (ChIP-PCR) to directly evaluate effects of estradiol and CCND1 on these candidate sites (Fig. 4D). We first confirmed that estradiol induced anti-ER-bound DNA only for the ERE 3 site, which also showed the highest levels of αER-specific binding. The siCCND1 small inhibitory RNA decreased ER binding at all three sites, but only the binding to the ERE 3 site was both stimulated by estrogen and blocked by siCCND1.
To confirm the ChIP experiment results, we stably expressed an shRNA that targets exon 2 of cyclin D1 in T47D cells. This shRNA knocked cyclin D1 mRNA levels down by 50%, which resulted in a decrease of PR mRNA to 10% of its levels in control cells (Fig. 4E). We then evaluated DNA-protein interactions across the 240 nucleotides spanning the ERE 3 region by the use of electrophoretic mobility shift assays (EMSA). We first compared the DNA binding activity of lysates harvested from the vector control to that of CCND1 shRNA cells for 13 overlapping oligonucleotides that spanned the entire region. Cyclin D1 knockdown significantly altered binding at the canonical nucleotide 95585 ERE site (Fig. 4F) without altering binding at other adjacent sites (not shown). Using estrogen stimulation and cold-competitor oligonucleotides, we confirmed the estrogen response and sequence specificity of this ERE site.
We cloned a 1.3-kb genomic segment containing ERE 3 into standard luciferase reporter gene vectors to determine whether it could enhance transcription. Using PCR cloning, we placed it downstream of luciferase in reporter vectors lacking a heterologous promoter (pGL2-Basic [pGL2N]) and containing a heterologous promoter (pGL2 promoter [pGL2P]). We transfected reporter constructs together with either a scramble control siRNA (scramble) or siCCND1 (Fig. 5A and B) into T47D cells. In addition, we cotransfected the reporter constructs with either a control expression vector or a CCND1-expressing vector (pCCND1) (Fig. 5C and D). We performed these experiments using cells treated with vehicle alone (Fig. 5A and C) and cells treated with estradiol (Fig. 5B and D). The PR 3′ UTR sequences did not significantly increase luciferase expression in the absence of promoter sequences (Fig. 5A [pGL2N-UTR versus pGL2N]). However, they caused a 6-fold increase in expression when combined with a heterologous promoter (Fig. 5A [pGL2P-UTR versus pGL2P]).
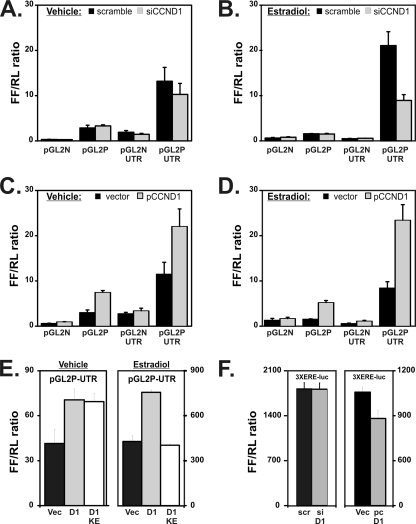
FIG. 5.
A 3′ estrogen-responsive enhancer in the progesterone receptor gene is regulated by cyclin D1. (A and B) Cloning of the 3′ ERE of the PR gene downstream of a basal promoter-luciferase construct activates transcription that responds to estrogen and cyclin D1. We cloned 1.3 kb of the PR 3′ UTR into luciferase reporter vectors. We performed reporter assays using T47D cells in the absence and presence of estradiol for 24 h. Knockdown efficiency for the siRNAs for these experiments is shown in Fig. 4A. Levels of firefly luciferase from the progesterone receptor plasmids are compared to those of an internal pCMV-renilla control (FF/RL ratio). Means ± standard errors of the results determined in three replicated experiments are shown. Reporter activity levels in cells transfected with a scrambled control siRNA (scramble) and an siCCND1 (siCCND1) in the presence of vehicle (A) or estradiol (B) were compared. Results for a pGL2-basic (pGL2N) vector with no promoter, a reporter containing a basal promoter (pGL2P), the basal vector containing the PR gene 3′ UTR (pGL2N-UTR), and the PR gene 3′ UTR region cloned in the promoter reporter (pGL2P-UTR) are shown. (C and D) Activity levels determined using the same reporters were further compared between cells transfected with an empty vector control (vector) or an expression plasmid for cyclin D1 (pCCND1) in the presence of vehicle (C) or estradiol (D). (E) The 3′ ERE of the PR gene is activated by cyclin D1 in a CDK4/CDK6-independent manner in the absence of estrogen and in a CDK4/CDK6-dependent manner in its presence. We evaluated the effect of wild-type cyclin D1 (D1) and the KE cyclin D1 mutant (D1 KE) on the 3′ UTR region by use of the PR gene 3′ UTR region cloned in the promoter reporter (pGL2P-UTR). Transfections were performed as described for panels C and D. We further compared cells transfected with vehicle-treated cells (left panel [Vehicle]) to cells treated with estrogen (right panel [Estradiol]). (F) Changes in cyclin D1 levels do not change levels of expression from a reporter plasmid containing a 3× isolated estrogen response element. Standard reporter vector containing a basal promoter activated by a 3× estrogen receptor binding element (14) was cotransfected with a scrambled control siRNA (scr) and an siCCND1 (siD1), and reporter activity in the presence of estradiol was determined (left panel). The same reporter was cotransfected with an empty vector control (Vec) or an expression plasmid for cyclin D1 (pc-D1 [right panel]).
Estradiol stimulated the PR UTR when combined with a heterologous basal promoter, taking estradiol's inhibition of the isolated promoter into consideration (compare Fig. 5A and B). This estradiol responsiveness of the PR UTR was best seen in the experiments using scramble control oligonucleotides and siCCND1 to determine whether decreased cyclin D1 could block the 3′ PR UTR's enhancing function (Fig. 5A and B). Importantly, siCCND1 decreased PR 3′ UTR-driven reporter activity to 78% of control levels in vehicle-treated control cells and to 42% of controls in estradiol-treated cells. Transfection of a cyclin D1 expression vector increased the reporter activity of the PR 3′ UTR (Fig. 5C and D). It doubled pGL2P-UTR activity in vehicle-treated cells and increased pGL2P-UTR reporter activity nearly 3-fold in the presence of estradiol treatment.
To determine whether these effects were related to cyclin D1-CDK interactions, we compared the effect of a wild-type cyclin D1 to that of a cyclin D1 K112E mutant that cannot interact with CDK4/CDK6 (15) (Fig. 5E). The KE mutant had effects equivalent to those of the wild-type cyclin D1 on the pGL2P-UTR reporter vector containing the 3′ UTR of the progesterone receptor in vehicle-treated cells. Interestingly, the KE mutation abrogated wild-type cyclin D1's 2-fold increase in activation of the pGL2P-UTR in estradiol-treated cells. Finally, a reporter construct containing a 3-fold repeat of an isolated estrogen receptor binding sequence (ERE) (14) did not respond in the same manner to the CCND1 expression plasmid or to siCCND1 (Fig. 5F). Taken together, these assays identify the 3′ UTR region of the PR gene as a unique cyclin D1- and estrogen-responsive element.
A 3′ estrogen/cyclin D1-responsive enhancer element interacts with the progesterone receptor A and B promoters.
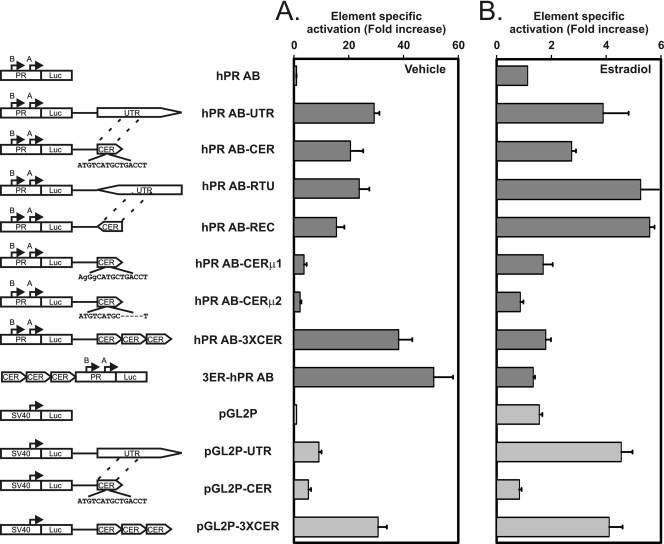
We further analyzed the 1.3-kb PR 3′ UTR region to determine whether it functions as an enhancer (Fig. 6). We first combined the 3′ UTR region with the progesterone receptor's own promoter region (17) and found that it activated reporter expression from the PR promoter 13-fold more potently than it activated the heterologous basal promoter in the pGL2 vector. We next found that its core 238 nucleotides that are conserved from lizards to humans are nearly as functional as the full 1.3-kb region when combined with the progesterone receptor's own promoter. The functions of both the full 1.3-kb UTR region and its core 238 nucleotides were independent of orientation, and increasing the core sequence copy number increased its function. The core 238-nucleotide element responded to estrogen in a position-independent manner. Finally, mutations of its ERE binding sequence (ATGTCATGCTGACCT) to AgGgCATGCTGACCT or ATGTCATGC-----T reduced its enhancement of promoter function and its estrogen response.
FIG. 6.
Definition of a core 238-nucleotide segment in DNA encoding part of the progesterone receptor 3′ UTR as a position- and orientation-independent enhancer of expression. To construct the plasmid diagrams, we obtained a luciferase (Luc) reporter vector containing the progesterone receptor promoter (PR) from the region spanning positions −711 to +822 (17), which is designated hPR AB in the figure. It contained both the PR isoform B transcription start site at 0 and the PR A isoform transcription start site at +751, as indicated in the diagrams of the various plasmids in the left column of the figure. We cloned the full 1.3-kb PR UTR region downstream of the luciferase in hPR AB in both orientations to produce hPR AB-UTR and hPR AB-RTU. The UTR orientation is indicated by the triangular point added to the rectangle in each diagram. We then cloned a 238-bp highly conserved central element-containing region (CER) from the 3′ UTR (positions 95435 to 95672 relative to the B form transcription start site) into the hPR AB plasmid in both orientations (producing hPR AB-CER and hPR AB-REC). We further introduced two mutations in the canonical ERE sequence in the CER portion of hPR AB-CER to interrupt estrogen receptor binding as predicted by previous studies (10) to make hPR AB-CERμ1 and hPR AB-CERμ2. Finally, we cloned a 3-fold repeat of the CER segment downstream and upstream of the hPR AB plasmid to evaluate the effects on gene expression of increasing its copy number. We compared these constructs to a smaller set of UTR and CER modifications of the pGL2 basal promoter reporter plasmid (pGL2P). (A) Element-specific activation compared to that seen with the parent hPR AB or pGL2P promoter vectors in the absence of estradiol. The indicated plasmids were transfected into T47D cells, and firefly luciferase expression was normalized to a cotransfected cytomegalovirus (CMV)-renilla luciferase construct. The activation effects of the individual elements were then plotted as the fold increases in expression from the element-containing plasmid compared to that of the promoter-only plasmids (hPR AB for the dark-gray columns and pGL2P for the light-gray columns). Values shown represent means ± standard errors of the results of three determinations for each experiment. The CER retains most of the 3′ UTR activity, and tandem repeats of the CER further stimulate the PR promoter. The indicated mutations in the core canonical estrogen response element abolish activation by the enhancer. (B) Element-specific stimulation of the 3′ UTR region of the progesterone receptor gene and its central element region by estradiol. The indicated plasmids were transfected as described for panel A, in the absence and presence of estradiol. The responses to estradiol of the individual elements were then plotted as the fold increases in expression from the element-containing plasmid between the vehicle-treated cells and cells treated with estradiol. Values shown represent means ± standard errors of the results of three determinations for each experiment.
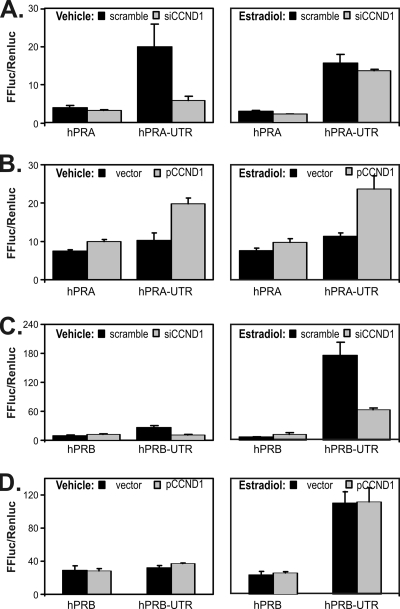
To evaluate cyclin D1's effects on PR isoform-specific promoters, we cloned the PR 3′ UTR enhancer into reporter constructs containing isolated PR A and B promoters (17) (Fig. 7). We repeated cotransfections with cyclin D1 and siCCND1. The PR A promoter is somewhat responsive to changes in CCND1 expression on its own, and its function and response to CCND1 are markedly increased by the addition of the PR 3′ UTR enhancer sequences (Fig. 7A and B). Using isoform-specific qRT-PCR, we also found that PR A mRNA decreases accounted for most of the response to siCCND1 (55% of levels in scramble-transfected controls). A strong reaction of PR A to cyclin D1 changes was seen in response to both siCCND1 and CCND1 overexpression. Interestingly, the PR B promoter was not responsive to CCND1 on its own, but siCCND1 blocked estrogen-induced increases when the PR B promoter was combined with the PR 3′ UTR enhancer (Fig. 7C and D). While the reporter gene result differs somewhat from the behavior of endogenous PR B (Fig. 4A), the added estrogen in these experiments likely accounts for the difference. The PR gene 3′ UTR enhancer therefore activates both PR isoform promoters, with the A isoform showing greater interaction with cyclin D1 and the B isoform being more responsive to estrogen.
FIG. 7.
The 3′ ERE of the PR gene differentially activates PR isoform-specific promoters, which respond to estrogen and cyclin D1. We combined the PR 3′ UTR (UTR) with PR reporters containing its individual A and B promoters. The 3′ UTR was cloned into PR A (hPRA) and PR B (hPRB) plasmids as indicated. We performed reporter assays in the absence (left panels) and presence (right panels) of estradiol as described in the previous figure legends. (A) Levels of activity induced by the hPRA and hPRA-UTR reporters were compared in the presence of a scrambled control siRNA (scramble) and in the presence of the siCCND1 oligonucleotide (siCCND1). (B) Levels of activity induced by the hPRA and hPRA-UTR reporters were compared in the absence (vector) and presence (pCCND1) of transfected cyclin D1. (C) Levels of activity induced by the hPRB and hPRB-UTR reporters were compared in the presence of a scrambled control siRNA (scramble) and in the presence of the siCCND1 oligonucleotide (siCCND1). (D) Levels of activity induced by the hPRB and hPRB-UTR reporters were compared in the absence (vector) and presence (pCCND1) of transfected cyclin D1. FFluc/Renluc, levels of firefly luciferase from the progesterone receptor plasmids compared to those of an internal pCMV-renilla control.
Cyclin D1 knockdown decreases progesterone receptor expression in additional breast cancer cells.
Having identified a direct link between CCND1 levels and PR expression, we sought to determine whether additional breast cancer cell lines exhibited this CCND1 control of PR (Fig. 8). siCCND1 transfection effectively reduced CCND1 protein levels in five additional cell lines (Fig. 8A), and PR was consequently reduced in two of them, namely, the ZR75-1 and MCF7 cell lines. We further found that IER3 and ETV5 levels also decreased in response to CCND1 and PR decreases in the same two cell lines. We used isoform-specific qRT-PCR to analyze PR isoform transcripts (Fig. 8B). We found that levels of the combined PR A-and-B transcript, predominately, those of PR A-specific mRNA, decreased in response to the siCCND1 knockdown in ZR75-1 and MCF7 cells. Finally, we evaluated the response of the 3′ UTR enhancer to cyclin D1 expression in MCF 7 cells (Fig. 8C). As described for the T47D cells, decreased cyclin D1 levels decreased the PR UTR enhancer activity, increased cyclin D1 levels increased the PR UTR enhancer activity, and the cyclin D1 K112E mutant remained active in these cells.
DISCUSSION
While studies of cyclin D1 have long established its primary role in cell cycle regulation, additional studies examining its effects on gene expression have suggested that it might have additional CDK4/CDK6-independent effects (29, 52). Since those previous studies initially relied on reporter assays, endogenous targets were not identified (51). By identifying genes associated with cyclin D1's effects on tumor invasion, we recently identified genes that responded to the combination of estrogen and cyclin D1 (48). Given that these genes responded to this combination, we treated cyclin D1 transgenic mice with estrogen. We now report that the combination of estrogen and constitutive cyclin D1 expression increased progesterone receptor expression, increased PR function, increased PR-target gene expression, and increased PR effects (Fig. 1 to 3).
The phenotypic changes accompanying cyclin D1 overexpression shown here, together with the previously reported phenotype of cyclin D1 knockout mice, support the idea that the PR is a cyclin D1 target. The histologic appearance of cyclin D1 transgenic mammary glands treated with estrogen (Fig. 1) strongly resembles glands in mice constitutively expressing additional PR (41, 42). The failure of mammary development during pregnancy in cyclin D1 null mice (43) is nearly identical to the loss of mammary development in PR null mice (28). In contrast, mammary glands in mice lacking the ER do not resemble glands in mice lacking cyclin D1 (11). Our data suggest that cyclin D1 may act both as a cell cycle mediator of ovarian hormone activity and as a developmental switch that increases in mammary glands to augment their effects on mammary development. This model implies an additional potential explanation for the decreased tumor formation seen in erbB2 and ras transgenic mice when they are cyclin D1 null (50), since the decreased tumorigenesis could result from loss of PR function and/or loss of cell cycle control. Notably, a progesterone receptor antagonist blocks tumorigenesis in BRCA mutant mice (34).
After years of studies of estrogen regulation of the PR gene, a clear estrogen-responsive enhancer has not yet been identified in its sequences (31). Various studies of PR promoter proximal elements particularly failed to identify canonical estrogen receptor binding elements. Here we show that a 238-nucleotide sequence in the 3′ UTR of the progesterone gene is a classic estrogen-responsive enhancer. It contains a canonical estrogen receptor binding element whose sequence is highly conserved. The homology plot in Fig. 4C shows that this element is the most highly conserved region of the progesterone receptor gene, excluding its coding exons. The estrogen receptor binds this sequence in an estrogen-dependent manner. In reporter experiments, it activates estrogen-regulated expression of both the PR promoter and a heterologous promoter. Its activation is independent of position and orientation and increases with increased copy numbers (Fig. 6).
Given the importance of progesterone receptor expression, exploring unique features of its enhancer is important to understand how ovarian hormones regulate breast development and carcinogenesis. We have shown using ChIP, siRNA knockdowns, and cyclin D1 overexpression studies that this enhancer is uniquely responsive to cyclin D1. Its response to cyclin D1 is independent of the presence of CDK4/CDK6 in the absence of estrogen, although its response is dependent on the presence of CDK4/CDK6 in the presence of estrogen (Fig. 5E). Estrogen and cyclin D1 additively stimulate its function, potentially explaining their combined in vivo effects on mammary development. In contrast, a reporter construct containing a 3-fold repeat of the canonical estrogen response element did not respond to cyclin D1 decreases or increases (Fig. 5F). Importantly, the estrogen receptor binding element was required for the PR enhancer's function both for promoter activation and for estrogen response (Fig. 6).
Our results provide the first in vivo identification of a gene containing an estrogen receptor binding element (ERE) that responds to cyclin D1 in the manner previously proposed (29, 52). On the other hand, we did not confirm that cyclin D1 could activate an isolated ERE (Fig. 5F), demonstrating that other elements in the PR enhancer are required for its cyclin D1 responsiveness. Our results suggest that cyclin D1 has at least two mechanisms responsible for activation of the PR enhancer. Further, we could not demonstrate direct binding of cyclin D1 to the PR enhancer in ChIP experiments, largely because available cyclin D1 antibodies immunoprecipitate several nonspecific bands. Given our results, the PR enhancer obviously contains additional DNA binding elements likely to contribute to its unique response to cyclin D1. Brown and colleagues have found that FOXA1 binding in combination with the ER defines enhancer-driven lineage-specific transcription for a number of ER-targeted enhancers (5). The PR 3′ UTR enhancer region lacks a FOXA1 binding site, although the results of another genome-wide ChIP scan previously suggested that FOXA1 can bind in this region (25). Additionally, there are no E2F sites in the PR enhancer, and PR is not known to be regulated through the cell cycle. Rather, a transcription regulatory element search (TRES) of the core region of this PR enhancer identified conserved sites for AP4, E47, Sox-5, and HOXA9 (18). Since the PR enhancer is now a well-characterized endogenous target sequence that is regulated by both increased and decreased cyclin D1 levels, identification of combinations of transcription factors involved in its regulation would be important to gain a better understanding of mechanisms explaining cyclin D1's role in gene expression control.
The role of progesterone in breast cancer pathogenesis has long been controversial (24). Progesterone's contribution to rapid signaling through the activity of mitogen-activated protein kinases is important in the deregulation of breast cancer cell proliferation (8). Hormone replacement therapies that combine estrogen with progesterone clearly demonstrate that these hormones together can promote breast carcinogenesis (7). We now show that cyclin D1's tight association with the ER and PR status of breast cancers may result from the combination of ovarian hormone control of cyclin D1's cell cycle effects together with a feed-forward regulation of PR expression by cyclin D1 and estrogen. This mechanism would provide a strong selective pressure for increasing cyclin D1 expression in hormone receptor-positive breast cancers. We further show that this regulation is encoded in a novel 238-nucleotide enhancer in the progesterone receptor gene. Identification of additional regulators of this enhancer should offer important insights into cyclin D1's CDK4/CDK6-independent transcriptional effects and could potentially identify therapeutic targets whose manipulation could inhibit the combined effects of cyclin D1 and estrogen in cases of breast cancer.
Acknowledgments
For this work, C. Yang, L. Chen, and E. V. Schmidt were supported by a grant from the National Cancer Institute of the National Institutes of Health (grant RO1 CA69069). C. Yang received additional support from grant R01 CA112021, and both E. V. Schmidt and L. Chen received support from grant U01 AI07033. C. Li was supported by the Harvard Breast Cancer SPORE grant P50 CA89393.
We gratefully acknowledge review of the histology results determined for our mice by Robert Cardiff of the University of California at Davis.
Footnotes
Published ahead of print on 19 April 2010.
REFERENCES
- 1.Aupperlee, M. D., K. T. Smith, A. Kariagina, and S. Z. Haslam. 2005. Progesterone receptor isoforms A and B: temporal and spatial differences in expression during murine mammary gland development. Endocrinology 146:3577-3588. [DOI] [PubMed] [Google Scholar]
- 2.Bartek, J., J. Bartkova, and J. Taylor-Papadimitriou. 1990. Keratin 19 expression in the adult and developing human mammary gland. Histochem. J. 22:537-544. [DOI] [PubMed] [Google Scholar]
- 3.Beatson, G. T. 1896. On the treatment of inoperable cases of carcinoma of the mamma: suggestions for a new method of treatment, with illustrative cases. Lancet ii:104-107. [PMC free article] [PubMed] [Google Scholar]
- 4.Bienvenu, F., S. Jirawatnotai, J. E. Elias, C. A. Meyer, K. Mizeracka, A. Marson, G. M. Frampton, M. F. Cole, D. T. Odom, J. Odajima, Y. Geng, A. Zagozdzon, M. Jecrois, R. A. Young, X. S. Liu, C. L. Cepko, S. P. Gygi, and P. Sicinski. 2010. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature 463:374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38:1289-1297. [DOI] [PubMed] [Google Scholar]
- 7.Chlebowski, R. T., L. H. Kuller, R. L. Prentice, M. L. Stefanick, J. E. Manson, M. Gass, A. K. Aragaki, J. K. Ockene, D. S. Lane, G. E. Sarto, A. Rajkovic, R. Schenken, S. L. Hendrix, P. M. Ravdin, T. E. Rohan, S. Yasmeen, and G. Anderson. 2009. Breast cancer after use of estrogen plus progestin in postmenopausal women. N. Engl. J. Med. 360:573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel, A. R., M. Qiu, E. J. Faivre, J. H. Ostrander, A. Skildum, and C. A. Lange. 2007. Linkage of progestin and epidermal growth factor signaling: phosphorylation of progesterone receptors mediates transcriptional hypersensitivity and increased ligand-independent breast cancer cell growth. Steroids 72:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driscoll, M. D., G. Sathya, M. Muyan, C. M. Klinge, R. Hilf, and R. A. Bambara. 1998. Sequence requirements for estrogen receptor binding to estrogen response elements. J. Biol. Chem. 273:29321-29330. [DOI] [PubMed] [Google Scholar]
- 11.Feng, Y., D. Manka, K. U. Wagner, and S. A. Khan. 2007. Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc. Natl. Acad. Sci. U. S. A. 104:14718-14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groshong, S. D., G. I. Owen, B. Grimison, I. E. Schauer, M. C. Todd, T. A. Langan, R. A. Sclafani, C. A. Lange, and K. B. Horwitz. 1997. Biphasic regulation of breast cancer cell growth by progesterone: role of the cyclin-dependent kinase inhibitors, p21 and p27(Kip1). Mol. Endocrinol. 11:1593-1607. [DOI] [PubMed] [Google Scholar]
- 13.Halban, J. 1900. Ueber den Einfluss der Ovarien auf die Entwicklung des Genitales. Mschr. Geburtsh. Gynaek. 12:496-503. [Google Scholar]
- 14.Hall, J. M., and D. P. McDonnell. 1999. The estrogen receptor beta-isoform (ERbeta) of the human estrogen receptor modulates ERalpha transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology 140:5566-5578. [DOI] [PubMed] [Google Scholar]
- 15.Hinds, P. W., S. F. Dowdy, E. N. Eaton, A. Arnold, and R. A. Weinberg. 1994. Function of a human cyclin gene as an oncogene. Proc. Natl. Acad. Sci. U. S. A. 91:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiremath, M., J. P. Lydon, and P. Cowin. 2007. The pattern of beta-catenin responsiveness within the mammary gland is regulated by progesterone receptor. Development 134:3703-3712. [DOI] [PubMed] [Google Scholar]
- 17.Huggins, G. S., J. Y. Wong, S. E. Hankinson, and I. De Vivo. 2006. GATA5 activation of the progesterone receptor gene promoter in breast cancer cells is influenced by the +331G/A polymorphism. Cancer Res. 66:1384-1390. [DOI] [PubMed] [Google Scholar]
- 18.Katti, M. V., M. K. Sakharkar, P. K. Ranjekar, and V. S. Gupta. 2000. TRES: comparative promoter sequence analysis. Bioinformatics 16:739-740. [DOI] [PubMed] [Google Scholar]
- 19.Kester, H. A., B. M. van der Leede, P. T. van der Saag, and B. van der Burg. 1997. Novel progesterone target genes identified by an improved differential display technique suggest that progestin-induced growth inhibition of breast cancer cells coincides with enhancement of differentiation. J. Biol. Chem. 272:16637-16643. [DOI] [PubMed] [Google Scholar]
- 20.Kraus, W. L., M. M. Montano, and B. S. Katzenellenbogen. 1993. Cloning of the rat progesterone receptor gene 5′-region and identification of two functionally distinct promoters. Mol. Endocrinol. 7:1603-1616. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn, R. M., D. Karolchik, A. S. Zweig, T. Wang, K. E. Smith, K. R. Rosenbloom, B. Rhead, B. J. Raney, A. Pohl, M. Pheasant, L. Meyer, F. Hsu, A. S. Hinrichs, R. A. Harte, B. Giardine, P. Fujita, M. Diekhans, T. Dreszer, H. Clawson, G. P. Barber, D. Haussler, and W. J. Kent. 2009. The UCSC Genome Browser database: update 2009. Nucleic Acids Res. 37:D755-D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lammie, G. A., and G. Peters. 1991. Chromosome 11q13 abnormalities in human cancer. Cancer Cells 3:413-420. [PubMed] [Google Scholar]
- 23.Landis, M. W., B. S. Pawlyk, T. Li, P. Sicinski, and P. W. Hinds. 2006. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell 9:13-22. [DOI] [PubMed] [Google Scholar]
- 24.Lange, C. A. 2008. Challenges to defining a role for progesterone in breast cancer. Steroids 73:914-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupien, M., J. Eeckhoute, C. A. Meyer, Q. Wang, Y. Zhang, W. Li, J. S. Carroll, X. S. Liu, and M. Brown. 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132:958-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaelson, J. S., and P. Leder. 2001. beta-catenin is a downstream effector of Wnt-mediated tumorigenesis in the mammary gland. Oncogene 20:5093-5099. [DOI] [PubMed] [Google Scholar]
- 27.Motokura, T., T. Bloom, H. G. Kim, H. Juppner, J. V. Ruderman, H. M. Kronenberg, and A. Arnold. 1991. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature 350:512-515. [DOI] [PubMed] [Google Scholar]
- 28.Mulac-Jericevic, B., J. P. Lydon, F. J. DeMayo, and O. M. Conneely. 2003. Defective mammary gland morphogenesis in mice lacking the progesterone receptor B isoform. Proc. Natl. Acad. Sci. U. S. A. 100:9744-9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuman, E., M. H. Ladha, N. Lin, T. M. Upton, S. J. Miller, J. DiRenzo, R. G. Pestell, P. W. Hinds, S. F. Dowdy, M. Brown, and M. E. Ewen. 1997. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol. Cell. Biol. 17:5338-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Offner, H., K. Adlard, A. Zamora, and A. A. Vandenbark. 2000. Estrogen potentiates treatment with T-cell receptor protein of female mice with experimental encephalomyelitis. J. Clin. Invest. 105:1465-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Malley, B. W. 2005. A life-long search for the molecular pathways of steroid hormone action. Mol. Endocrinol. 19:1402-1411. [DOI] [PubMed] [Google Scholar]
- 32.Otten, A. D., M. M. Sanders, and G. S. McKnight. 1988. The MMTV LTR promoter is induced by progesterone and dihydrotestosterone but not by estrogen. Mol. Endocrinol. 2:143-147. [DOI] [PubMed] [Google Scholar]
- 33.Petz, L. N., and A. M. Nardulli. 2000. Sp1 binding sites and an estrogen response element half-site are involved in regulation of the human progesterone receptor A promoter. Mol. Endocrinol. 14:972-985. [DOI] [PubMed] [Google Scholar]
- 34.Poole, A. J., Y. Li, Y. Kim, S. C. Lin, W. H. Lee, and E. Y. Lee. 2006. Prevention of Brca1-mediated mammary tumorigenesis in mice by a progesterone antagonist. Science 314:1467-1470. [DOI] [PubMed] [Google Scholar]
- 35.Richer, J. K., B. M. Jacobsen, N. G. Manning, M. G. Abel, D. M. Wolf, and K. B. Horwitz. 2002. Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J. Biol. Chem. 277:5209-5218. [DOI] [PubMed] [Google Scholar]
- 36.Rider, V., K. Isuzugawa, M. Twarog, S. Jones, B. Cameron, K. Imakawa, and J. Fang. 2006. Progesterone initiates Wnt-beta-catenin signaling but estradiol is required for nuclear activation and synchronous proliferation of rat uterine stromal cells. J. Endocrinol. 191:537-548. [DOI] [PubMed] [Google Scholar]
- 37.Said, T. K., O. M. Conneely, D. Medina, B. W. O'Malley, and J. P. Lydon. 1997. Progesterone, in addition to estrogen, induces cyclin D1 expression in the murine mammary epithelial cell, in vivo. Endocrinology 138:3933-3939. [DOI] [PubMed] [Google Scholar]
- 38.Schultz, J. R., L. N. Petz, and A. M. Nardulli. 2005. Cell- and ligand-specific regulation of promoters containing activator protein-1 and Sp1 sites by estrogen receptors alpha and beta. J. Biol. Chem. 280:347-354. [DOI] [PubMed] [Google Scholar]
- 39.Schultz, J. R., L. N. Petz, and A. M. Nardulli. 2003. Estrogen receptor alpha and Sp1 regulate progesterone receptor gene expression. Mol. Cell Endocrinol. 201:165-175. [DOI] [PubMed] [Google Scholar]
- 40.Shi, H. Y., J. P. Lydon, and M. Zhang. 2004. Hormonal defect in maspin heterozygous mice reveals a role of progesterone in pubertal ductal development. Mol. Endocrinol. 18:2196-2207. [DOI] [PubMed] [Google Scholar]
- 41.Shyamala, G., X. Yang, R. D. Cardiff, and E. Dale. 2000. Impact of progesterone receptor on cell-fate decisions during mammary gland development. Proc. Natl. Acad. Sci. U. S. A. 97:3044-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shyamala, G., X. Yang, G. Silberstein, M. H. Barcellos-Hoff, and E. Dale. 1998. Transgenic mice carrying an imbalance in the native ratio of A to B forms of progesterone receptor exhibit developmental abnormalities in mammary glands. Proc. Natl. Acad. Sci. U. S. A. 95:696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 44.Sutherland, R. L., and E. A. Musgrove. 2004. Cyclins and breast cancer. J. Mammary Gland Biol. Neoplasia 9:95-104. [DOI] [PubMed] [Google Scholar]
- 45.Sweeney, K. J., A. Swarbrick, R. L. Sutherland, and E. A. Musgrove. 1998. Lack of relationship between CDK activity and G1 cyclin expression in breast cancer cells. Oncogene 16:2865-2878. [DOI] [PubMed] [Google Scholar]
- 46.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669-671. [DOI] [PubMed] [Google Scholar]
- 47.Wilcken, N. R., O. W. Prall, E. A. Musgrove, and R. L. Sutherland. 1997. Inducible overexpression of cyclin D1 in breast cancer cells reverses the growth-inhibitory effects of antiestrogens. Clin. Cancer Res. 3:849-854. [PubMed] [Google Scholar]
- 48.Yang, C., S. Trent, V. Ionescu-Tiba, L. Lan, T. Shioda, D. Sgroi, and E. V. Schmidt. 2006. Identification of cyclin D1- and estrogen-regulated genes contributing to breast carcinogenesis and progression. Cancer Res. 66:11649-11658. [DOI] [PubMed] [Google Scholar]
- 49.Yang, X., S. M. Edgerton, S. D. Kosanke, T. L. Mason, K. M. Alvarez, N. Liu, R. T. Chatterton, B. Liu, Q. Wang, A. Kim, S. Murthy, and A. D. Thor. 2003. Hormonal and dietary modulation of mammary carcinogenesis in mouse mammary tumor virus-c-erbB-2 transgenic mice. Cancer Res. 63:2425-2433. [PubMed] [Google Scholar]
- 50.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 51.Zwijsen, R. M., R. S. Buckle, E. M. Hijmans, C. J. Loomans, and R. Bernards. 1998. Ligand-independent recruitment of steroid receptor coactivators to estrogen receptor by cyclin D1. Genes Dev. 12:3488-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwijsen, R. M., E. Wientjens, R. Klompmaker, J. van der Sman, R. Bernards, and R. J. Michalides. 1997. CDK-independent activation of estrogen receptor by cyclin D1. Cell 88:405-415. [DOI] [PubMed] [Google Scholar]