Abstract
Biogenesis of eukaryotic ribosomes requires a number of RNA helicases that drive molecular rearrangements at various points of the assembly pathway. While many ribosome synthesis factors are conserved among all eukaryotes, certain features of ribosome maturation, such as U8 snoRNA-assisted processing of the 5.8S and 28S rRNA precursors, are observed only in metazoan cells. Here, we identify the mammalian DEAD box helicase family member Ddx51 as a novel ribosome synthesis factor and an interacting partner of the nucleolar GTP-binding protein Nog1. Unlike any previously studied yeast helicases, Ddx51 is required for the formation of the 3′ end of 28S rRNA. Ddx51 binds to pre-60S subunit complexes and promotes displacement of U8 snoRNA from pre-rRNA, which is necessary for the removal of the 3′ external transcribed spacer from 28S rRNA and productive downstream processing. These data demonstrate the emergence of a novel factor that facilitates a pre-rRNA processing event specific for higher eukaryotes.
Synthesis of ribosomes is a highly complicated process that consumes a large amount of cellular resources and requires a large array of auxiliary factors. rRNAs, the main structural component of the ribosome, are transcribed as precursors (pre-rRNAs) that are processed to mature forms through cleavages, exonucleolytic trimming, and nucleotide modifications (34). Studies in the yeast Saccharomyces cerevisiae have led to the identification of more than 150 proteins acting in pre-rRNA processing and ribosome assembly (reviewed in references 14, 22, and 33). A variety of putative helicases, GTPases, and ATPases identified among ribosome synthesis factors are believed to facilitate extensive rearrangements in RNA and RNA-protein complexes occurring in the course of ribosome maturation (52). Functions of the majority of ribosome synthesis factors are still not completely understood. Based on sequence similarity, many yeast ribosome assembly factors have putative orthologs in higher eukaryotic species including humans (23). The overall pathway for ribosome maturation has also been conserved in eukaryotic evolution, but there are differences between organisms in the organization of the nucleolus (55) and pre-rRNA processing steps (12), pointing to the underlying mechanistic variations in this process.
In addition to protein factors, several dozen to hundreds of small nucleolar RNAs (snoRNAs) participate in eukaryotic ribosome maturation (reviewed in references 3, 16, and 31). The majority of snoRNAs act as guides for posttranscriptional nucleotide modifications in rRNA (2′-O-ribose methylation or pseudouridylation) that fine-tune ribosome performance (13, 30, 39). However, several snoRNAs, including U3, U14, and U17/snR30, are required for cleavages within pre-rRNA and therefore are essential for ribosome production and cell viability (2, 24, 28, 37, 42). Two additional snoRNAs, U8 and U22, participate in rRNA cleavages in vertebrates (44, 59), underscoring variability in the pre-rRNA processing machinery between different eukaryotes.
In mammals, the first cleavage in the newly synthesized 47S pre-rRNA transcript (Fig. 1A) occurs rapidly at site A′ in the 5′ external transcribed spacer (5′ETS) and is followed by processing at the 3′ end of 28S rRNA, termed site 6 (12, 21). The primary A′ cleavage, which is a prerequisite for all downstream processing steps in mammalian cells (29), requires U3 snoRNA (28). Extensive studies have shown that U3 base pairs with specific regions on the pre-rRNA transcript and directs cleavages in 5′ETS and internal transcribed spacer 1 (ITS1) as part of 18S rRNA formation (4, 8). U3 snoRNA is ubiquitous in eukaryotes and is necessary for the assembly of the large 80S to 90S processing complexes on nascent pre-rRNA transcripts (11, 19).
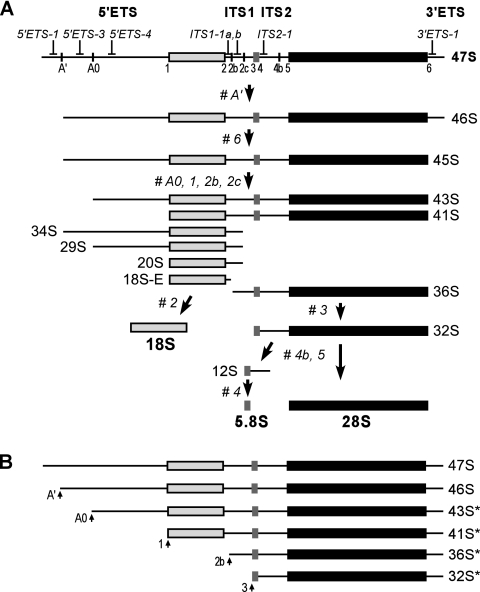
FIG. 1.
Processing of pre-rRNA in mouse cells. (A) Outline of the mammalian processing pathway. The primary transcript (47S pre-rRNA) is shown at the top. The main processing sites and oligonucleotide probes used for hybridization analysis are indicated. Processing sites termed 1 to 6 correspond to the ends of mature 18S, 5.8S, and 28S rRNAs. The external and internal transcribed spacers (ETS and ITS) contain additional cleavage sites designated A′, A0, 2b, 2c, and 4b. 47S pre-rRNA is normally short-lived and is rapidly cleaved at sites A′ and 6 to yield 45S. The order of subsequent cleavages within 5′ETS and ITS1 may vary, resulting in a series of alternate intermediates, of which the major ones are shown. The mature 18S is generated from 18S-E, and the mature 5.8S and 28S rRNAs are generated after processing of the ITS2 in 32S pre-rRNA. (B) 3′ETS-extended forms of rRNA precursors. In addition to the normal 47S and 46S pre-rRNAs, aberrant forms (labeled with asterisks) can be formed in cells with defective site 6 processing by cleavages at the indicated sites.
In comparison to 5′ETS processing, features of the processing step that removes the 3′ETS from the 3′ end of the 28S sequence in pre-rRNA (Fig. 1A) indicate that it occurs differently in lower and higher eukaryotes. In budding yeast, the corresponding cleavage is not known to require the assistance of any snoRNA (60). By contrast, studies in Xenopus laevis showed that processing at the 3′ end of 28S rRNA requires U8 snoRNA (44). Depletion of U8 or interfering with its binding to pre-rRNA led to the accumulation of abnormal 28S precursors in which 3′ETS extensions were not removed and inhibited the formation of mature 5.8S and 28S rRNAs (43, 45). Comparative analysis of vertebrate pre-rRNA sequences showed a conserved complementarity between U8 snoRNA and 28S rRNA and a less perfectly conserved complementarity between U8 and the 5.8S-ITS2 junction, suggesting a model in which this snoRNA acts as a chaperone assisting long-range pre-rRNA folding (41). U8 forms small nucleolar ribonucleoprotein (snoRNP) particles in which it associates with several common snoRNA-binding proteins such as fibrillarin (58), a complex of RNA-binding LSm proteins (57), and the decapping enzyme X29 (17), but few details are known about its activities in the context of preribosomal complexes. Here, we report the identification of a novel mammalian ribosome synthesis factor that functions in processing the 3′ end of 28S rRNA. Our results suggest that the DEAD box helicase family member Ddx51 assists in the displacement of U8 snoRNA from pre-rRNA during ribosome maturation in mouse fibroblasts, thereby acting to facilitate a processing step that occurs only in higher-eukaryote cells.
MATERIALS AND METHODS
Plasmids and antibodies.
The full-length coding sequence of Ddx51 was amplified by PCR from mouse cDNA. The amino acid sequence of the clone was identical to the GenBank sequence EDL20011.1. For stable transfections, the coding sequence for Ddx51 containing the N-terminal Myc tag was cloned in pX18, the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible vector that provides bicistronic expression of sequences of interest with the H-2Kk marker for magnetic selection of expressing cells (D. G. Pestov, unpublished data). Point mutations in the helicase domain were introduced using the QuikChange protocol (Stratagene). We used the 9E10 antibody (Santa Cruz) for the detection of Myc-tagged proteins, affinity-purified polyclonal antibodies to detect Nog1 (36) and Bop1 (50), and the MCA-38F3 monoclonal antibody (EnCor Biotechnology) to detect fibrillarin.
Interaction assays.
We used standard two-hybrid techniques (18) to screen a mouse cDNA library using LexA-Nog1G224A as bait and to verify the interactions. For the in vitro interaction assay, the coding sequence for Nog1, as a fusion with an N-terminal S tag, was cloned into the pCITE-4a vector (Novagen). The Myc-Ddx51 coding sequence was amplified by PCR with a T7 promoter sequence incorporated into the 5′ primer. Both proteins were synthesized by coupled transcription/translation (TnT T7 Quick system; Promega); reaction mixtures were mixed, incubated for 30 min at room temperature, and further incubated for 1 h with S-agarose affinity beads (Novagen) equilibrated with 10 mM Tris, pH 7.5, 65 mM KCl, 0.8 mM MgCl2, 0.001% Triton X-100, and protease inhibitors (Roche). Beads were washed three times with 500 μl of the same buffer, and bound proteins were eluted by boiling with SDS-PAGE loading buffer.
Cell culture.
Mouse LAP3 cells, an NIH 3T3-derived line that supports IPTG-inducible expression (46), were maintained and transfected using the calcium phosphate method as described previously (50). Cells cotransfected with pX18-Myc-Ddx51 and the selectable marker pPGK-puro were cultured with 1 μg/ml puromycin to obtain a pool of drug-resistant clones, from which cells inducibly expressing Myc-Ddx51 were magnetically selected using MACSelect beads (Miltenyi Biotec). To downregulate endogenous Ddx51, we performed transfections with a siGENOME small interfering RNA (siRNA) SMARTpool (Dharmacon) as described previously (49). Knockdowns were monitored with reverse transcription-PCR (RT-PCR) to amplify the endogenous Ddx51 mRNA using gene-specific primers.
Immunofluorescence.
Cells stably transfected with Myc-Ddx51 or green fluorescent protein (GFP)-Ddx51 were induced with 1 mM IPTG for 16 h, fixed with phosphate-buffered saline (PBS) containing 3% paraformaldehyde, and permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. After washes with PBS, cells were blocked in 5% goat serum for 30 min and incubated with primary antibodies for 1 h in PBS containing 1% goat serum, followed by detection with Alexa 488- or 568-conjugated secondary antibodies (Invitrogen). Cells were mounted in ProLong Gold (Invitrogen) and analyzed on a Zeiss Axio Observer Z1 microscope equipped with the ApoTome system.
Hybridizations.
Total RNA was isolated from cells with Trizol according to the manufacturer's protocol (Invitrogen), separated on a formaldehyde-agarose gel, transferred to a nylon membrane, and analyzed by hybridizations with 32P-labeled oligonucleotide probes (47). Probes for pre-rRNA hybridization were as described previously (49). Additional probes are as follows: ITS1-1a, 5′-ACGCCGCCGCTCCTCCACAGTCTCCCGTT-3′; U14, 5′-AGCACTTCTGGTGGAAACTACGAATGGTTT-3′; U3, 5′-CTCGTCCTCGTGGTTTCGGGTGCTCTACACA-3′; U8, 5′-AATCAGCAGATCATGCAAGCTCCAATCATC-3′.
Sucrose gradient analyses.
Nuclei were sonicated and fractionated on a 10% to 30% (wt/wt) sucrose gradient as detailed before (47), except that we used a slightly modified lysis buffer containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM NaF, 2 mM EDTA, 0.05% (vol/vol) Igepal CA-630, 1 mM dithiothreitol (DTT), protease inhibitors (Roche), and 50 U/ml RiboLock RNase inhibitor (Fermentas). For the isolation and sucrose gradient analysis of core preribosomal particles, we followed a previously published protocol (35).
To analyze deproteinized pre-rRNA complexes, we treated the extracted core preribosomes with 2 mg/ml proteinase K (Roche) and 1% SDS for 30 min at room temperature and verified protein degradation by SDS-PAGE. Deproteinized complexes were centrifuged through a 10 to 30% (wt/wt) sucrose gradient, prepared in 10 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 2 mM MgCl2, for 3.5 h at 36,000 rpm (160,000 × g) and 5°C in a SW41Ti rotor (Beckman).
To isolate RNA, we treated gradient fractions with 0.1 mg/ml of proteinase K-1% SDS-15 mM EDTA for 1 h at 42°C, extracted them with phenol-chloroform, and precipitated RNA with 1 volume of isopropanol. To isolate proteins, we added 10 μg of bovine serum albumin to each 1-ml fraction, followed by trichloroacetic acid to a 10% final concentration. After 1 h of incubation on ice, proteins were precipitated by centrifugation at 15,000 × g for 30 min at 4°C. The pellet was washed with 1 ml of ethanol and 1 ml of acetone, air dried, resuspended in 20 mM Tris-HCl (pH 6.8)-1 mM EDTA-50 U/ml RNase A (Worthington), incubated for 15 min at 37°C, and mixed with SDS-PAGE loading buffer for separation on a gel and Western blotting.
RESULTS
Ddx51 is a novel nucleolar protein that associates with Nog1 and preribosomes.
We identified Ddx51, a previously uncharacterized protein belonging to the DEAD box helicase family (10), in a two-hybrid screen for proteins interacting with the mammalian ribosome assembly factor Nog1. In yeast, Nog1p is required for the production of 60S ribosomal subunits (15, 26, 27, 48), and we showed previously that blocking the function of mouse Nog1 through a G224A mutation arrests preribosome maturation in mammalian cells (36). The Ddx51 clone isolated in the screen of a mouse cDNA library with the Nog1G224A bait interacted equally well with both the wild-type Nog1 and its mutant form (Fig. 2A). To verify the interaction, we synthesized Myc-tagged Ddx51 and S-tagged Nog1 by in vitro translation and coincubated these proteins together with S-agarose beads. Western blot analysis showed that Myc-Ddx51 was efficiently retained on the beads in the presence of S-tagged Nog1 (Fig. 2B, lane 3) but not alone (lane 5), consistent with the physical interaction between the two proteins.
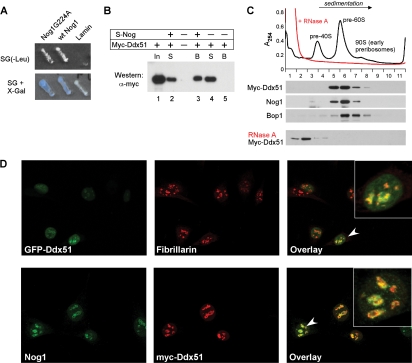
FIG. 2.
Ddx51 and Nog1 interact in a yeast-two hybrid system in vitro and colocalize in cells. (A) A two-hybrid interaction between Ddx51 (prey) and LexA-fused baits: wild-type Nog1, mutant Nog1G224A with an altered conformation of the GTP-binding domain, and the nuclear protein lamin (negative control). Growth on leucine-deficient medium and activation of the LacZ reporter were used to score the interaction. (B) In vitro binding of Nog1 and Ddx51. In vitro-transcribed and -translated S-tagged Nog1 and Myc-tagged Ddx51 were coincubated with the S-agarose affinity resin for 1 h at room temperature. After washes, bound proteins were eluted with the SDS-PAGE loading buffer and analyzed by Western blotting. In, input; S, supernatant; B, beads. (C) Ddx51 cofractionates with preribosomes. Purified nuclei from Myc-Ddx51-expressing cells were lysed by sonication and separated on a sucrose gradient. Positions of preribosomes are marked by peaks of absorbance at 254 nm. Gradient fractions were analyzed by Western blotting using antibodies against the Myc tag to detect Ddx51 and specific antibodies against ribosome assembly factors Nog1 and Bop1. RNase A treatment prior to loading on a gradient was used to disrupt preribosomes in the nuclear extract. (D) Ddx51 localizes predominantly to the nucleolus. Stably transfected GFP-Ddx51 cells (top row) were costained with an antibody against nucleolar marker fibrillarin followed by Alexa 594-conjugated secondary antibodies (red). Stably transfected Myc-Ddx51 cells (bottom row) were probed with anti-Myc and affinity-purified Nog1 antibodies. Alexa 488 (green) and 568 (red) secondary antibodies were used for the visualization of the endogenous Nog1 and Myc-Ddx51, respectively.
We next cloned the complete coding sequence of mouse Ddx51 with an N-terminal Myc tag into the mammalian expression vector pX18 and transfected mouse LAP3 cells (46) to obtain cells with stable, IPTG-inducible expression (see Materials and Methods for details). To evaluate whether Ddx51, like Nog1, might function in the biogenesis of new ribosomes, we purified nuclei from these cells, disrupted them by sonication, and separated nuclear components by sucrose gradient centrifugation. As shown in Fig. 2C, the tagged Ddx51 cosedimented with high-molecular-weight preribosome complexes that contained Nog1 and other assembly factors such as Bop1 (50, 51). Treatment of nuclear extracts with RNase A prior to loading on a gradient resulted in a shift of Myc-Ddx51 to top fractions, supporting the idea that it is physically associated with large ribonucleoprotein (RNP) particles (Fig. 2C, bottom). In further support for a role of Ddx51 in ribosome maturation, fluorescence microscopy showed that Ddx51 was localized to the nucleus and was especially abundant in the nucleolus (Fig. 2D). The nucleolar pattern was observed for the GFP-Ddx51 fusion by costaining for the nucleolar marker fibrillarin (Fig. 2D, top) and for the Myc-tagged Ddx51, which was costained with antibodies against endogenous Nog1 (Fig. 2D, bottom).
Ddx51 is required for processing the 3′ end of 28S rRNA.
To investigate the role of Ddx51 in ribosome biogenesis, we first analyzed pre-rRNA maturation in cells in which expression of the endogenous protein was knocked down using RNA interference (RNAi). RT-PCR confirmed efficient reduction of the Ddx51 mRNA levels after transfection with a specific siRNA oligonucleotide pool (Fig. 3A). Northern hybridizations of the cellular RNA with oligonucleotide probes complementary to different regions of the pre-rRNA transcript (Fig. 1A) showed that depletion of Ddx51 led to a slight decrease in the levels of multiple pre-rRNAs, including 34S, 32S, and 12S, but at the same time it increased levels of the early 47S to 45S precursors (Fig. 3B, lanes 1 and 2). This result suggested that, in the absence of Ddx51, the efficiency of an early pre-rRNA processing step may be reduced. Hybridization with probe 3′ETS-1 revealed an abnormal accumulation of precursors extended into the 3′ETS (47S and 46S, 41S*, 36S*, and 32S*), indicating that processing at site 6 downstream from 28S rRNA is inhibited (Fig. 1A and B). A strong accumulation of 3′ETS-extended pre-rRNAs was previously observed after expression of the dominant negative mutant Nog1G224A (36) and was also noticeable, albeit at a lower level, after siRNA-mediated downregulation of Nog1 (Fig. 3B, lane 3). Notably, Nog1 knockdown led to a strong accumulation of several other pre-rRNAs, such as 36S and 32S, and reduced 12S levels (Fig. 3B, lane 3), consistent with an essential role of this protein in ITS2 processing (36). The lack of similar defects after Ddx51 depletion indicates that this protein is not involved in late steps in the maturation of pre-28S rRNA. However, our results demonstrate that Ddx51 shares with Nog1 the ability to affect the early cleavage of the 3′ end of 28S rRNA at site 6.
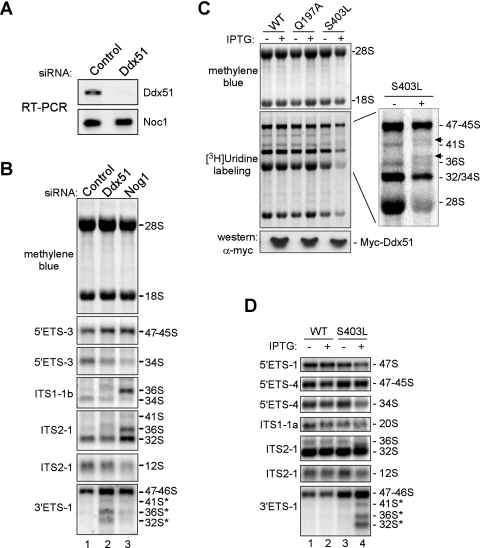
FIG. 3.
Ddx51 knockdown and expression of the dominant inhibitory mutant Ddx51S403L affect processing of pre-rRNA. (A) RT-PCR to assess downregulation of the Ddx51 mRNA 36 h after transfection with a Ddx51-specific siRNA oligonucleotide pool or control nontargeting siRNA. Efficiency of RT-PCR was verified by parallel amplification of an unrelated (Noc1) mRNA. (B) Northern analysis of pre-rRNA in cells transfected with control and Ddx51- and Nog1-targeting siRNAs. Equal amounts of total RNA (as verified by methylene blue staining of the blot) were hybridized with the indicated probes against different regions in pre-rRNA (see Fig. 1A for probe location). Positions of rRNAs and their precursors are indicated on the right. (C) Analysis of rRNA synthesis in cells inducibly expressing wild-type Ddx51 (WT) or the Q197A or S403L mutant by metabolic labeling with [3H]uridine. Labeled rRNA was separated on an agarose-formaldehyde gel, transferred to a membrane, stained with methylene blue, and detected by fluorography. The bottom panel shows Western blotting of protein lysates from the same cells. Each cell pool was either uninduced (−) or induced with IPTG (+). (D) Northern analysis of pre-rRNA processing defects caused by expression of Ddx51S403L. Unlabeled RNA isolated from the same cells as in panel C was separated on a gel and hybridized with pre-rRNA-specific probes.
The dominant negative phenotype of the helicase domain mutation S403L in Ddx51.
We next sought to assess the effect of a catalytically inactive Ddx51 on preribosome maturation. Compared to depletion, mutations that affect functionality of preribosome components but do not prevent their incorporation into preribosomes are less likely to distort the overall architecture of these complexes and thus are useful for studying specific activities (35). Proteins of the DEAD box family share a conserved core sequence of ∼350 to 400 amino acids that include well-defined motifs involved in ATP binding, hydrolysis, and helicase activities (10, 53). In the core helicase sequence of Ddx51, we introduced mutation Q197A into the Q motif, controlling ATP binding and hydrolysis (54), and mutation S403L into motif III, required for helicase activity (9). Both types of mutations were previously found to frequently create dominant negative phenotypes in yeast helicases (5, 20). We transfected mouse cells with constructs providing IPTG-inducible expression of the wild-type and mutant Ddx51 and isolated stably transfected cells. To assess the net effect of the mutants on rRNA synthesis, we metabolically labeled these cells with [3H]uridine. The Q197A mutant showed little effect in this test, whereas induction of the S403L mutant repressed production of new 28S and 18S rRNAs (Fig. 3C). Additional bands of rRNA precursors were also observed after induction of S403L (Fig. 3C, inset, arrows), similar to the previously studied mutants of pre-60S processing factors affecting cleavage between 28S and 3′ETS (35, 51).
To examine the effects of Ddx51S403L on pre-rRNA processing in more detail, we analyzed steady-state pre-rRNA levels by Northern hybridizations (Fig. 3D). Induction of the mutant protein strongly inhibited pre-rRNA processing at site 6, leading to a prominent accumulation of aberrant 3′ETS-extended pre-rRNA species (Fig. 3D, lane 4). Hybridization with the 3′ETS-1 probe also revealed a strong increase in the 47S/46S signal. This was entirely due to accumulation of 46S, as hybridization with probe 5′ETS-1 showed slightly reduced levels of the 47S pre-rRNA, which probably resulted from an inhibitory effect of abnormal processing on pre-rRNA transcription in these cells. The increase in the level of the normally short-lived 46S precursor implies that its processing is considerably slowed down by the Ddx51 mutant. The slow early processing, as well as the reduced transcription rate, likely accounts for diminished levels across the range of downstream intermediates including 34S, 20S, and 12S (Fig. 3D, lane 4) and the reduced production of mature rRNAs observed in metabolic labeling (Fig. 3C). The phenotypes of the Ddx51S403L mutant and knockdown of endogenous Ddx51 (Fig. 3B) thus provide consistent evidence that Ddx51 participates in the early maturation of the pre-rRNA transcript by facilitating processing at the 3′ end of 28S rRNA.
Defect in Ddx51 function inhibits 3′ETS removal but does not block ribosome assembly.
The unusually focused effects of Ddx51 on pre-rRNA maturation led us to examine the association of the wild-type and mutant Ddx51 with preribosomes in more detail. Previous studies showed that high-molecular-weight complexes that contain pre-rRNAs and tightly associated ribosome assembly factors can be isolated from nuclei of mammalian cells by lysis under high-salt conditions (0.5 M NaCl-0.05 M MgCl2) followed by extraction with EDTA-containing buffer (35, 61). Analysis of these “core” preribosomal particles (which form three major peaks at ∼30S, 50S, and 80S) (Fig. 4A) showed that Myc-Ddx51 was stably bound to preribosomes and resistant to high salt and chelation of divalent cations (Fig. 4A, left). Like the wild-type protein, Myc-Ddx51S403L peaked in fractions 4 and 5, containing Nog1, implying that this mutation does not compromise the incorporation of the mutant protein into preribosomes (Fig. 4A, center). In contrast to the dominant negative mutant Nog1G224A, which induced a distinct peak of stalled complexes on the UV absorbance profile (Fig. 4A, right, arrow), the UV profile of nuclear RNPs in Ddx51S403L-expressing cells did not show any significant abnormalities (Fig. 4A, center), indicating that inactivation of Ddx51 does not affect major structural transitions that occur during the maturation of preribosomal particles.
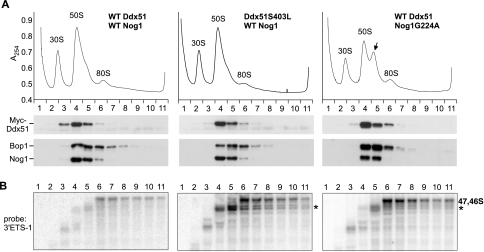
FIG. 4.
Analysis of proteins and pre-rRNA in core preribosomal particles by sucrose gradient centrifugation. (A) Preribosomes prepared from nuclei by high-salt-EDTA extraction as described in Materials and Methods were separated on a 10 to 30% sucrose gradient and fractionated with continuous monitoring of absorbance at 254 nm (A254). Proteins isolated from individual fractions were analyzed by Western blotting with anti-Myc antibody to detect transfected Myc-Ddx51 (wild type or mutant), while Nog1 and Bop1 were detected with specific affinity-purified antibodies. (Right) Myc-Ddx51 was coexpressed in cells with the dominant inhibitory mutant Nog1G224A. The arrow indicates stalled pre-60S particles accumulating as a result of defective Nog1 function. (B) Northern analysis of fractions corresponding to gradients in panel A reveals the presence of pre-rRNA species with unprocessed 3′ extensions. Asterisks indicate accumulating aberrant 3′ETS-extended precursors (Fig. 1B). The left and center panels were hybridized and exposed simultaneously using equally loaded blots (see Fig. S1A in the supplemental material) to ensure that the observed differences in 47S/46S signal intensity reflect actual changes of the levels of these intermediates in gradient fractions.
In electron microscopic studies of rodent preribosomes, complexes at early stages of maturation appear as loose filamentous structures and there is a gradual transition to more-compact, slower-sedimenting forms as pre-rRNAs within these complexes are processed (40). In a good agreement with this assessment, we found that preribosomes containing early precursors with unprocessed 3′ ends (47S and 46S) sedimented over a wide range of fractions in a sucrose gradient (Fig. 4B, left, fractions 6 to 11). By contrast, the bulk of later pre-60S complexes form a narrow peak in fractions 4 and 5. The transition from early to late complexes coincided with the removal of 3′ETS tails and the assembly of both Ddx51 and Nog1 (compare fractions 4 and 5 with 6 to 11 in Fig. 4A and B, left). Strikingly, in cells expressing Ddx51S403L, there was a marked accumulation of shorter 3′ETS-tailed species in both early (fractions 6 to 11) and late (fractions 4 and 5) particles (Fig. 4B, center, asterisk). Accumulation of these species was not observed in cells with the wild-type Ddx51 even after long exposure of the blot (see Fig. S1B in the supplemental material) and was not due to differences in loading (see Fig. S1A in the supplemental material). The sucrose gradient analysis performed with cells expressing Nog1G224A also showed a substantial increase in abnormal 3′ETS-tailed precursors (Fig. 4B, right). Ddx51 still cosedimented with preribosome complexes in these cells (Fig. 4A, right), indicating that the mutant Nog1 does not prevent Ddx51 association with preribosomes.
Together, these data indicate that, when site 6 processing is inhibited in preribosomes lacking proper Ddx51 or Nog1 activity, other cleavages in pre-rRNA can still occur, resulting in aberrant 3′ extended species (such as 32S* and 36S*) (Fig. 1B). The substantial increase in the 47S/46S pre-rRNA in these particles (Fig. 4B), however, indicates that these cleavages occur less efficiently than those in wild-type cells. The slow processing of the 47S/46S pre-rRNA can also explain the elevated steady-state levels of these normally short-lived intermediates observed in Northern hybridizations of total RNA (Fig. 3D, compare lanes 2 and 4).
Ddx51 affects association of U8 snoRNA with preribosomes.
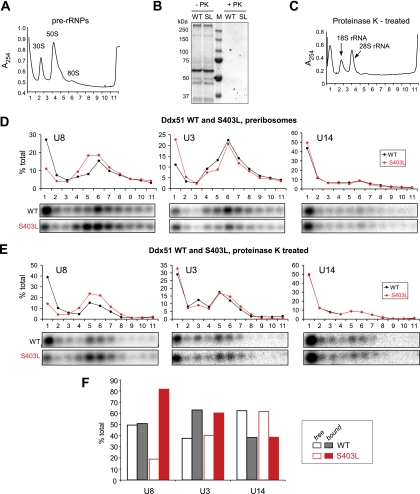
In previous studies in the Xenopus model system, depletion of U8 snoRNA and oligonucleotide-mediated interference with the displacement of U8 bound to pre-rRNA were shown to impair 3′ processing of 28S rRNA (44, 45). The highly conserved base complementarity between U8 and 28S rRNA in all studied species suggests that the mode of action of this snoRNA is conserved in evolution (41). To determine whether Ddx51 participates in U8 snoRNA-mediated processing events, we examined the association of U8 with preribosomes by Northern hybridizations of individual gradient fractions (Fig. 5A) using snoRNA-specific probes. In cells with wild-type Ddx51, U8 snoRNA formed two peaks on a gradient, in top fraction 1 and in fraction 6, containing 80S particles (Fig. 5D, WT). This agrees well with previous studies showing that most U8 in cells exists in complexes of approximately 10S and 80S, corresponding to free snoRNPs and preribosomes (58). Expression of Ddx51S403L resulted in marked changes in the U8 distribution pattern (Fig. 5D, S403L). First, the peak of U8 associated with 80S preribosomes in fraction 6 broadened to include fraction 5, indicating that this snoRNA persisted in late pre-60S particles. Second, the amount of free U8 in fraction 1 was reduced, likely as a result of the increased sequestration of U8 by preribosomes. In comparison, we found no similar changes in the distribution of two other snoRNAs required for pre-rRNA processing, U3 and U14 (Fig. 5D). We did find that the amount of free U3 at the top of the gradient increased after Ddx51S403L expression. However, since we do not observe defects in U3-mediated cleavages in these cells, we believe that this is likely a secondary effect caused by the slowed rate of early pre-rRNA processing, which increases the fraction of idling U3 snoRNPs.
FIG. 5.
Ddx51S403L inhibits the release of U8 snoRNA from pre-rRNA. (A) A UV absorbance profile of preribosomes separated on a 10 to 30% sucrose gradient (see Fig. 4A). Positions of major preribosome complexes are indicated by peaks of absorbance at 254 nm. (B) Treatment of preribosomes with proteinase K (PK) to obtain deproteinized complexes from cells expressing wild-type Myc-Ddx51 (WT) or Myc-Ddx51S403L (SL). Aliquots of the material taken before and after treatment were separated by SDS-PAGE, followed by Coomassie blue staining to verify protein degradation. (C) A typical absorbance profile of deproteinized pre-rRNA separated by sucrose gradient centrifugation (see Materials and Methods for details). (D) RNA isolated from individual fractions after separation of preribosomes on a gradient was analyzed by Northern hybridizations with U8, U3, and U14 snoRNA probes. The analysis was performed in parallel with cells expressing wild-type Ddx51 (WT) and Ddx51S403L (S403L). Hybridizations were quantified by phosphorimager analysis, and the data were plotted as the percentage of snoRNA in each fraction relative to the total in all 11 fractions. (E) Analysis of snoRNA association with deproteinized pre-rRNA complexes. RNA was reprobed on the same membrane consecutively to detect the indicated snoRNAs; the equivalent loading was verified by methylene blue staining (see Fig. S1D in the supplemental material). (F) The data obtained by quantification of the hybridization signal in panel E are presented as the ratios between “bound” (fractions 3 to 11) and “free” (fractions 1 and 2) snoRNAs.
A defect in Ddx51 function inhibits the release of U8 snoRNA base paired to pre-rRNA.
The above results suggest that U8 may be trapped in preribosomes in Ddx51S403L-expressing cells. This could occur through interactions with proteins in these particles or through direct base pairing with pre-rRNA. To distinguish between these possibilities, we treated nuclear lysates with proteinase K, which destroys protein components of preribosomal RNPs (Fig. 5B), and performed sucrose gradient centrifugation with the resulting “naked” RNA (Fig. 5C). Many snoRNAs are capable of extensive base pairing with pre-rRNA and remain stably bound after proteinase K treatment (32, 56). Northern blot analysis of the deproteinized complexes showed that a significant amount of U8 snoRNA was retained in rapidly sedimenting fractions (Fig. 5E), indicating that it remained base paired to pre-rRNAs after proteins were removed. Expression of the Ddx51S403L mutant dramatically changed the ratio between pre-rRNA-bound U8 and free U8 (Fig. 5E, F), implying that the lack of Ddx51 function affected the displacement of U8 directly from pre-rRNA. Control hybridizations of the same membrane did not show changes in the association of U3 and U14 snoRNA with pre-rRNA (Fig. 5E, F). We also verified the equivalent loading of gradient fractions by methylene blue staining of membranes prior to hybridization (see Fig. S1D in the supplemental material). Thus, we conclude that the activity of Ddx51 specifically influences the release of the U8 snoRNA base paired with pre-rRNA.
Inhibiting Nog1 function mimics the effects of Ddx51S403L on U8 snoRNA.
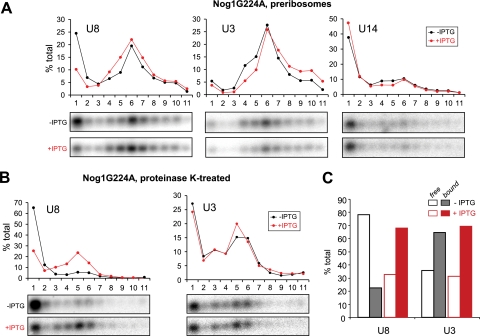
Given the similarity between Nog1 and Ddx51 in their effects on 3′ETS cleavage in preribosomes (Fig. 4B), we asked whether the two proteins might also overlap in their effects on U8. Northern analysis of preribosomes separated on a sucrose gradient from cells expressing Nog1G224A revealed a reduction of free U8 at the top of the gradient and the concomitant increase in U8 associated with preribosomes in fractions 4 to 7 (Fig. 6A), which were similar to the effects of Ddx51S403L. Likewise, a significant increase in the amount of U8 directly bound to pre-rRNA was evident in deproteinized complexes (Fig. 6B and C), indicating reduced displacement of U8 base paired with pre-rRNA. These results suggest that, in addition to the physical interaction with Nog1 (Fig. 2A and B), there is also functional linkage between Ddx51 and Nog1 in the preribosomal particle environment. Further studies probing the interaction between these two factors should help us better understand how U8 snoRNA-mediated events in ribosome maturation are controlled in higher eukaryotic cells.
FIG. 6.
Dominant negative Nog1G224A inhibits the release of U8 snoRNA from pre-rRNA. (A) Analysis of snoRNAs associated with preribosomes was performed as in Fig. 5D. Cell line D1411 (36), providing IPTG-inducible expression of Nog1G224A, was analyzed without and with IPTG induction. (B) snoRNA association with protein-free pre-rRNAs after proteinase K treatment in the uninduced and induced Nog1G224A-expressing cells. Analysis was performed as for Fig. 5E. The membrane was hybridized with U8 and then reprobed with a U3 probe. Equivalent loading of RNA from induced and uninduced cells was also verified by methylene blue staining of the blot (see Fig. S1D in the supplemental material). (C) The data obtained by quantification of the hybridization signal in panel B are presented as the ratios between “bound” (fractions 3 to 11) and “free” (fractions 1 and 2) snoRNAs.
DISCUSSION
Our knowledge of ribosome synthesis in eukaryotes is based largely on studies performed in the yeast S. cerevisiae. The insights gleaned from this organism are usually applicable to a wide range of eukaryotic species due to high conservation of ribosome components, homology between many assembly factors, and the overall similarity of pre-rRNA processing pathways. In some instances, however, pre-rRNA processing steps differ in lower and higher eukaryotes. One of the better-known examples is the use of additional snoRNAs, such as U8 and U22, in pre-rRNA processing in vertebrates (44, 59). Because of the relative paucity of data for higher eukaryotes, the molecular machinery that underlies the differences in ribosome maturation pathways is still very poorly understood. In this study, we characterized the novel DEAD box protein Ddx51, which plays a role in ribosome maturation in mammalian cells and functions in a pre-rRNA processing step that is mechanistically distinct from the corresponding step in yeast ribosome biogenesis. The evidence obtained by knocking down endogenous Ddx51 in mouse cells and expression of its dominant negative mutant is consistent with this factor playing a role in processing the 3′ end of 28S rRNA. Moreover, we found that expression of the Ddx51 mutant predicted to lack catalytic activity significantly increases the amount of U8 snoRNA base paired with pre-rRNA within preribosomal RNPs, suggesting that Ddx51 acts to promote U8 release during preribosome assembly.
Proteins of the DEAD box and related families of RNA helicases are found from bacteria to humans and participate in a multitude of cellular processes including translation initiation, pre-mRNA splicing, ribosome biogenesis, and RNA degradation (reviewed in references 6, 9, and 10). All these proteins contain several conserved motifs involved in binding and hydrolysis of nucleoside triphosphates and are predicted to function by promoting rearrangements of RNA molecules and RNA-protein complexes (25, 53). A systematic analysis of the putative helicases in S. cerevisiae showed that at least 20 of them are involved in ribosome biogenesis (5, 6, 20). Several of these proteins have been implicated in facilitating the binding and release of snoRNAs from pre-rRNA: Dbp4p is required for the release of U14 (32), Rok1 promotes the release of snR30 (7), and Has1 is required for U3 and U14 snoRNA release (38). Our analysis of Ddx51 indicates that this previously uncharacterized member of the family also acts by potentiating snoRNA release from preribosomes. Curiously, the target of Ddx51 is U8, a snoRNA found in higher eukaryotes but absent in S. cerevisiae, implying that Ddx51 has no functional counterpart in budding yeast. Consistent with this notion, we find no significant similarity between the N-terminal region in Ddx51, which excludes the conserved helicase core, and those of any of the DEAD box helicases encoded in the yeast genome (data not shown). Thus, it is possible that the function of Ddx51 has evolved in higher eukaryotes to specifically facilitate the U8 snoRNA-dependent step in the biogenesis of their ribosomes.
A deficiency in Ddx51 function in mammalian ribosome maturation is manifested primarily as the inhibition of pre-rRNA cleavage at site 6, separating 28S rRNA and the 3′ETS region (Fig. 3B to D). The elevated levels of 46S pre-rRNA in preribosomes (Fig. 4B) also indicate that cleavages separating the 18S and 28S branches of the pathway (Fig. 1A) are slowed down, but it is unclear whether this is simply due to inefficient early assembly in general or whether the cleavages in ITS1 and 3′ETS are partially coupled in mouse cells, similar to the situation in yeast (1). Overall, defects caused by mutant Ddx51 show a striking resemblance to the depletion of U8 snoRNA in the Xenopus system: an accumulation of abnormal 28S precursors extended at the 3′ end, inhibition of the formation of mature 5.8S and 28S rRNAs, and decreased levels of 18S due to partial inhibition of processing in ITS1 (43, 44).
According to the existing model, U8 snoRNA binds to newly transcribed pre-rRNA and facilitates its proper folding but later needs to be displaced for the 3′ cleavage of 28S and downstream processing to occur (43). Our study indicates that dissociation of U8 from pre-rRNA is not spontaneous but requires, or is enhanced by, Ddx51. The S403L mutation in the conserved helicase motif III in Ddx51 traps U8 in the pre-rRNA-bound state (Fig. 5D to F) and leads to the accumulation of particles containing pre-rRNAs that are internally cleaved in ITS1 but still possess unprocessed 3′ETS tails (Fig. 4B). These data are most easily interpreted as Ddx51 acting directly on U8, although we cannot exclude the possibility that Ddx51 might affect some other structural rearrangement in the preribosome particle, which causes U8 displacement indirectly. The same caveat applies to a number of other putative helicases proposed to act in snoRNA release and underscores the importance of developing methods to map intermolecular interactions within preribosomes with high precision.
Consistent with its having a role in ribosome biogenesis, Ddx51 is predominantly nucleolar (Fig. 2D) and cosediments with high-molecular-weight RNPs containing ribosome synthesis factors (Fig. 2C). In addition, we observe tight association of Ddx51 with salt-resistant core preribosomes (Fig. 4A), suggesting that the protein is stably integrated into these complexes. Unfortunately, we were unable to address the association with preribosomes in this study more directly because efficient immunoprecipitation of Myc-Ddx51 was found to require partially denaturing conditions, which disrupted its interactions (L. Srivastava and D. G. Pestov, unpublished observations). This is reminiscent of mammalian Nog1, which is also refractory to immunoprecipitation when it is incorporated into preribosomes (36), suggesting that Ddx51 might be similarly buried inside these large RNPs, which limits epitope accessibility on this protein.
The interaction of Ddx51 with the preribosome-associated G protein Nog1 in vitro and in the two-hybrid system (Fig. 2A and B) points to their cooperation in mammalian ribosome maturation, further supported by the overlapping processing defects caused by their depletion and dominant-inhibitory mutants (Fig. 3B, 4B). Nog1 is a conserved protein essential for the maturation of 60S ribosomal subunits and is found in a broad range of pre-60S complexes in S. cerevisiae (15, 26, 27, 48), but the exact role of this protein is still enigmatic. In our previous studies, we found that a mutation predicted to restrict conformational flexibility in Nog1 caused multiple defects in ribosome maturation including blocked cleavages in ITS2, accumulation of stalled 32S pre-rRNA-containing complexes, and inhibition of 3′ETS processing (36). Similar defects were also observed with certain mutations in the pre-60S ribosome assembly factors Bop1 and Pes1 (35, 51). Our results from this study suggest a possible explanation for the linkage between pre-60S ribosome factors and 3′ETS processing. In particular, the finding that U8 dissociation from pre-rRNA is inhibited by mutant Nog1 raises the possibility that U8 release might depend on the assembly of the proper pre-60S processing complex on a nascent transcript. Since preribosomes containing Nog1G224A are misassembled and unable to progress through maturation, as evidenced by the accumulation of stalled particles in fraction 5 (Fig. 4A, right), they might be in the wrong conformation to stimulate the Ddx51 activity required for effective U8 release. In this model, Ddx51 would not only provide mechanical assistance to displace U8 but also serve as part of a checkpoint enforcing the optimal order of events during ribosome assembly.
Supplementary Material
Acknowledgments
We thank Natalia Shcherbik for helpful comments on the manuscript.
This work was supported by National Institutes of Health grant GM074091 to D.G.P.
Footnotes
Published ahead of print on 19 April 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allmang, C., and D. Tollervey. 1998. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 278:67-78. [DOI] [PubMed] [Google Scholar]
- 2.Atzorn, V., P. Fragapane, and T. Kiss. 2004. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 24:1769-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachellerie, J. P., J. Cavaillé, and A. Hüttenhofer. 2002. The expanding snoRNA world. Biochimie 84:775-790. [DOI] [PubMed] [Google Scholar]
- 4.Beltrame, M., and D. Tollervey. 1992. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 11:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein, K. A., S. Granneman, A. V. Lee, S. Manickam, and S. J. Baserga. 2006. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis. Mol. Cell. Biol. 26:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleichert, F., and S. J. Baserga. 2007. The long unwinding road of RNA helicases. Mol. Cell 27:339-352. [DOI] [PubMed] [Google Scholar]
- 7.Bohnsack, M. T., M. Kos, and D. Tollervey. 2008. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 9:1230-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borovjagin, A. V., and S. A. Gerbi. 2005. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 33:4995-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordin, O., J. Banroques, N. K. Tanner, and P. Linder. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17-37. [DOI] [PubMed] [Google Scholar]
- 10.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 11.Dragon, F., J. E. G. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichler, D. C., and N. Craig. 1994. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49:197-239. [DOI] [PubMed] [Google Scholar]
- 13.Esguerra, J., J. Warringer, and A. Blomberg. 2008. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA 14:649-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 15.Fuentes, J., K. Datta, S. Sullivan, A. Walker, and J. Maddock. 2007. In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol. Genet. Genomics 278:105-123. [DOI] [PubMed] [Google Scholar]
- 16.Gerbi, S. A., A. V. Borovjagin, M. Ezrokhi, and T. S. Lange. 2001. Ribosome biogenesis: role of small nucleolar RNA in maturation of eukaryotic rRNA. Cold Spring Harb. Symp. Quant. Biol. 66:575-590. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh, T., B. Peterson, N. Tomasevic, and B. A. Peculis. 2004. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol. Cell 13:817-828. [DOI] [PubMed] [Google Scholar]
- 18.Golemis, E. A., and R. Brent. 1997. Searching for interacting proteins with the two-hybrid system III, p. 43-72. In P. L. Bartel and S. Fields (ed.), The yeast two-hybrid system. Oxford University Press, New York, NY.
- 19.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schäfer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 20.Granneman, S., K. A. Bernstein, F. Bleichert, and S. J. Baserga. 2006. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol. Cell. Biol. 26:1183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurney, T. J. 1985. Characterization of mouse 45S ribosomal RNA subspecies suggests that the first processing cleavage occurs 600 +/− 100 nucleotides from the 5′ end and the second 500 +/− 100 nucleotides from the 3′ end of a 13.9 kb precursor. Nucleic Acids Res. 13:4905-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henras, A. K., J. Soudet, M. Gérus, S. Lebaron, M. Caizergues-Ferrer, A. Mougin, and Y. Henry. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinsby, A. M., L. Kiemer, E. O. Karlberg, K. Lage, A. Fausbøll, A. S. Juncker, J. S. Andersen, M. Mann, and S. Brunak. 2006. A wiring of the human nucleolus. Mol. Cell 22:285-295. [DOI] [PubMed] [Google Scholar]
- 24.Hughes, J. M., and M. J. Ares. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10:4231-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291:121-125. [DOI] [PubMed] [Google Scholar]
- 26.Jensen, B. C., Q. Wang, C. T. Kifer, and M. Parsons. 2003. The NOG1 GTP-binding protein is required for biogenesis of the 60 S ribosomal subunit. J. Biol. Chem. 278:32204-32211. [DOI] [PubMed] [Google Scholar]
- 27.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kass, S., K. Tyc, J. A. Steitz, and B. Sollner-Webb. 1990. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60:897-908. [DOI] [PubMed] [Google Scholar]
- 29.Kass, S., N. Craig, and B. Sollner-Webb. 1987. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol. Cell. Biol. 7:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King, T. H., B. Liu, R. R. McCully, and M. J. Fournier. 2003. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11:425-435. [DOI] [PubMed] [Google Scholar]
- 31.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 32.Kos, M., and D. Tollervey. 2005. The Putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol. Cell 20:53-64. [DOI] [PubMed] [Google Scholar]
- 33.Kressler, D., E. Hurt, and J. Baβler. 30 October 2009, posting date. Driving ribosome assembly. Biochim. Biophys. Acta doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed]
- 34.Lafontaine, D. L., and D. Tollervey. 2001. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2:514-520. [DOI] [PubMed] [Google Scholar]
- 35.Lapik, Y. R., C. J. Fernandes, L. F. Lau, and D. G. Pestov. 2004. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell 15:17-29. [DOI] [PubMed] [Google Scholar]
- 36.Lapik, Y. R., J. M. Misra, L. F. Lau, and D. G. Pestov. 2007. Restricting conformational flexibility of the switch II region creates a dominant-inhibitory phenotype in Obg GTPase Nog1. Mol. Cell. Biol. 27:7735-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, H. D., J. Zagorski, and M. J. Fournier. 1990. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang, X., and M. J. Fournier. 2006. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol. Cell. Biol. 26:7437-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang, X., Q. Liu, and M. J. Fournier. 2007. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 28:965-977. [DOI] [PubMed] [Google Scholar]
- 40.Matsuura, S., T. Morimoto, Y. Tashiro, T. Higashinakagawa, and M. Muramatsu. 1974. Ultrastructural and biochemical studies on the precursor ribosomal particles isolated from rat liver nucleoli. J. Cell Biol. 63:629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michot, B., N. Joseph, S. Mazan, and J. P. Bachellerie. 1999. Evolutionarily conserved structural features in the ITS2 of mammalian pre-rRNAs and potential interactions with the snoRNA U8 detected by comparative analysis of new mouse sequences. Nucleic Acids Res. 27:2271-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrissey, J. P., and D. Tollervey. 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 13:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peculis, B. A. 1997. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol. 17:3702-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peculis, B. A., and J. A. Steitz. 1993. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73:1233-1245. [DOI] [PubMed] [Google Scholar]
- 45.Peculis, B. A., and J. A. Steitz. 1994. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 8:2241-2255. [DOI] [PubMed] [Google Scholar]
- 46.Pestov, D. G., and L. F. Lau. 1994. Genetic selection of growth-inhibitory sequences in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 91:12549-12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pestov, D. G., Y. R. Lapik, and L. F. Lau. 2008. Assays for ribosomal RNA processing and ribosome assembly. Curr. Protoc. Cell Biol., chapter 22, unit 22.11. [DOI] [PubMed]
- 48.Saveanu, C., A. Namane, P. Gleizes, A. Lebreton, J. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shcherbik, N., M. Wang, Y. R. Lapik, L. Srivastava, and D. G. Pestov. 2010. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 11:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 20:5516-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2002. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 277:29617-29625. [DOI] [PubMed] [Google Scholar]
- 52.Strunk, B. S., and K. Karbstein. 2009. Powering through ribosome assembly. RNA 15:2083-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251-262. [DOI] [PubMed] [Google Scholar]
- 54.Tanner, N. K., O. Cordin, J. Banroques, M. Doère, and P. Linder. 2003. The Q. motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11:127-138. [DOI] [PubMed] [Google Scholar]
- 55.Thiry, M., and D. L. J. Lafontaine. 2005. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 15:194-199. [DOI] [PubMed] [Google Scholar]
- 56.Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomasevic, N., and B. A. Peculis. 2002. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell. Biol. 22:4101-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyc, K., and J. A. Steitz. 1989. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tycowski, K. T., M. D. Shu, and J. A. Steitz. 1994. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science 266:1558-1561. [DOI] [PubMed] [Google Scholar]
- 60.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 61.Warner, J. R., and R. Soeiro. 1967. Nascent ribosomes from HeLa cells. Proc. Natl. Acad. Sci. U. S. A. 58:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.