Abstract
The induction of middle meiotic promoters is a key regulatory event in the life cycle of Saccharomyces cerevisiae that controls exit from prophase, meiosis, and spore formation. The Sum1 repressor and Ndt80 activator proteins control middle promoters by binding to overlapping DNA elements. NDT80 is controlled by a tightly regulated middle meiotic promoter through a positive autoregulatory loop and is repressed in vegetative cells by Sum1. It has previously been shown that the meiosis-specific kinase Ime2 promotes the removal of Sum1 from DNA. Here, we show that Sum1 is also regulated by the cyclin-dependent kinase, Cdk1. While sum1 phosphosite mutants that are insensitive to Cdk1 or Ime2 complete meiosis and form spores, a mutant that is insensitive to both Ime2 and Cdk1 (sum1-ci) blocks meiotic development in prophase with an ndt80Δ-like phenotype. Ectopic expression of NDT80 or mutation of a Sum1-binding element in the NDT80 promoter bypasses the sum1-ci block. Hst1 is a NAD+-dependent histone deacetylase that is linked to Sum1 by the Rfm1 tethering factor. Deletion of HST1 or RFM1 also bypasses the sum1-ci block. These results demonstrate that Sum1 functions as a key meiotic brake through the NDT80 promoter and that Cdk1 and Ime2 trigger exit from meiotic prophase by inhibiting the Sum1 transcriptional repression complex.
Gametogenesis is the process used by sexually reproducing organisms to produce haploid cells that are specialized for sexual fusion. During gametogenesis, precursor cells duplicate the genome, undergo crossing over, and then complete two rounds of chromosome segregation. The formation of haploids is coupled to differentiation programs that produce gametes. In the yeast Saccharomyces cerevisiae, the formation of haploids is coupled to spore formation. Spores can germinate and mate to regenerate diploids.
Sporulation in yeast is tightly regulated by a transcriptional cascade that controls the sequential induction of about 1,000 genes that can be broadly classified as early, middle, and late (5, 26). Early promoters are activated by the Ime1 transcription factor, which is produced in diploid cells in response to nutritional deprivation (the inducing signal for meiosis) (16, 21, 35). Early genes are expressed during premeiotic S phase and during prophase, when homologous chromosomes pair, synapse, and undergo reciprocal recombination. The expression of many early genes continues until pachytene, when homologous pairs of chromosomes are connected end to end by synaptonemal complexes (SCs).
Middle sporulation genes (MSGs) are activated by the Ndt80 transcription factor, which specifically recognizes DNA elements termed middle sporulation elements (MSEs) (6). NDT80 expression is IME1 dependent, and this may connect the early and middle phases of the transcriptional cascade. The NDT80 promoter is also activated in a positive autoregulatory loop by its own protein product. Thus, NDT80 is transcriptionally induced as a middle gene just prior to when most middle promoters are induced (23). The Ndt80-inducible gene product that promotes pachytene exit has been identified as the Cdc5 Polo-like kinase (32). Other Ndt80-inducible gene products include cyclins that promote the meiotic divisions and molecules such as Smk1 that control spore morphogenesis (5, 26). Thus, the Ndt80 regulon coordinately promotes pachytene exit, the meiotic divisions, and spore formation.
Exit from pachytene is a key decision point in meiotic development. In yeast, this transition is closely associated with the commitment point, after which the inducing signal (starvation) is no longer needed to complete the program (31). In addition, a checkpoint pathway that monitors recombination intermediates and defects in SC formation (the pachytene checkpoint) controls pachytene exit (13, 27). The pachytene checkpoint inhibits Ndt80 (6, 12, 34), and ndt80Δ cells block meiotic development at pachytene (40). These findings demonstrate that Ndt80 is a central component of the regulatory system that controls pachytene exit and commitment to meiotic development.
MSGs are repressed during vegetative growth by Sum1, a DNA-binding protein that specifically recognizes a subset of MSEs (39). Sum1 represses middle promoters by recruiting an NAD+-dependent histone deacetylase (Sir2 paralog), named Hst1, through a tethering factor, named Rfm1 (18). Sum1 also represses transcription in an Hst1-independent fashion. Sum1 represses NDT80, and it has been proposed that a regulated competition between the Sum1 repressor and the Ndt80 activator triggers the NDT80 positive autoregulatory loop (23). The DNA-binding domain of Ndt80 can displace the DNA-binding domain of Sum1 from MSE DNA (24). In addition, ectopic expression of Ndt80 can activate Sum1-repressible genes in vegetative cells (5). It is therefore likely that Ndt80 can competitively displace Sum1 from DNA in meiotic cells. However, Sum1 is displaced from DNA in ndt80Δ cells that have been transferred to sporulation medium (1). Thus, Sum1 can be removed from DNA in meiotic cells by a mechanism that does not require Ndt80 competition.
Ime2 is a meiosis-specific cyclin-dependent kinase (CDK)-like kinase that regulates multiple steps in meiotic development, including induction of the program, S phase, the nuclear divisions, and spore formation (15). It has been proposed that Ime2 collaborates with Cdk1 to regulate meiosis. One way that Ime2 regulates meiotic processes is to function in place of Cdk1. Thus, Ime2 is required for destruction of the Sic1 inhibitor of S phase in meiotic cells, while Cdk1 complexed with G1 cyclins (which inhibit meiotic development) targets Sic1 for destruction in mitotic cells (7-10). Another well-characterized Ime2 target is the Cdh1 substrate-bridging protein for the anaphase-promoting complex that is phosphorylated by both Ime2 and Cdk1 as meiosis is taking place (14). In this case, the differential sensitivities of the Ime2 and Cdk1 phosphoacceptor sites to phosphatases have been proposed to play a role in preventing S phase from taking place between meiosis I (MI) and MII.
It has previously been shown that Ime2 phosphorylates Sum1 on residue T306 and that a nonphosphorylatable Sum1-T306A protein (Sum1-i) is not removed from DNA in ndt80Δ cells (1, 22). However, in otherwise wild-type (NDT80) cells, Sum1-i does not block meiosis or spore formation. These findings suggest that while Ime2 promotes the Ndt80-independent removal of Sum1 from DNA, the removal of Sum1 is regulated through an additional mechanism.
In this report, we show that Cdk1 negatively regulates Sum1 specifically in meiotic cells. We describe a Sum1 mutant that is insensitive to Cdk1 and Ime2 (sum1-ci) and show that this mutant blocks meiosis in prophase with a phenotype that is indistinguishable from that of ndt80Δ. Ectopic expression of NDT80 or mutation of an MSE in the NDT80 promoter that has previously been reported to interact with Sum1 bypasses the sum1-ci block. Moreover, hst1Δ and rfm1Δ mutants also bypass the block. These findings demonstrate that Cdk1 and Ime2 inhibit Sum1 at the NDT80 promoter in a pathway that involves Rfm1/Hst1. The data demonstrate that Sum1 is a key regulatory brake that must be inhibited before cells can exit from prophase and complete meiotic development.
MATERIALS AND METHODS
Yeast strains and plasmids.
All strains used in this study are listed in Table 1 and are derived from the rapidly sporulating SK1 strain background. To generate the sum1-c and sum1-ci alleles, we started with the SK1 form of SUM1 in plasmid pMES39 and sum1-i in plasmid pMES42 (1). All of the SP and TP minimal Cdk1 phosphoacceptor sites in the amino-terminal third of Sum1 were changed to AP by mutating codon 242 from TCA to GCA, codon 306 from ACT to GCT, codon 313 from ACA to GCA, codon 315 from ACA to GCA, and codon 318 from ACG to GCG using a QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA). In addition, a silent mutation was introduced at codon 353 to create an MluI restriction enzyme recognition site. To change the minimal Cdk1 phosphoacceptor sites in the remaining carboxy-terminal two-thirds of Sum1, an MluI/BspEI DNA fragment containing codons 353 to 834 of SUM1 was chemically synthesized (Genscript, Piscataway, NJ). This DNA fragment is identical to the wild-type (SK1 form) of SUM1 except for the following substitutions: mutation of codon 379 from TCT to GCT, of codon 409 from TCT to GCT, of codon 512 from TCT to GCT, of codon 616 from TCG to GCT, of codon 697 from ACC to GCC, of codon 738 from TCA to GCA, and of codon 817 from ACT to GCT. This mutant fragment was ligated into the MluI/BspEI sites of plasmids pMES65 and pMES69 to create pMES71 (sum1-ci) and pMES77 (sum1-c), respectively. The entire open reading frames (ORFs) of these plasmids were sequenced. Plasmids were linearized with HindIII prior to transformation, and integrated mutations were confirmed by sequencing DNA fragments generated by PCR. Strains of yeast that express estradiol-inducible NDT80 were generated by standard genetic methods and the ndt80::TRP1-PGAL1-NDT80 and ura3::PGPD1-GAL4(848)-ER::URA3 chromosomal insertion strains described by Benjamin et al. (3).
TABLE 1.
Yeast strains
| Strain | Genotypea |
|---|---|
| LNY150 | MATa/MATα leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 |
| MSY311 | sum1-i::URA3/sum1-i::URA3 cdc28-as1/cdc28-as1 SMK1-HA::KAN/SMK1-HA::KAN |
| MSY312 | cdc28-as1/cdc28-as1 SMK1-HA::KAN/SMK1-HA::KAN |
| MSY334 | SMK1-HA::KAN/SMK1-HA::KAN |
| MSY338 | sum1-ci::URA3/SUM1 SMK1-HA::KAN/SMK1-HA::KAN |
| MSY339 | sum1-ci::URA3/sum1-ci::URA3 SMK1-HA::KAN/SMK1-HA::KAN |
| MSY349 | sum1-c::URA3/sum1-c::URA3 SMK1-HA::KAN/SMK1-HA::KAN |
| MSY352 | sum1-i::URA3/sum1-i::URA 3SMK1-HA::KAN/SMK1-HA::KAN |
| MSY351 | ZIP1::GFP700/zip1::GFP700 ndt80Δ::LEU2/ndt80Δ::LEU2 |
| MSY362 | ZIP1::GFP700/zip1::GFP700 |
| MSY386 | zip1::GFP700/zip1::GFP700 sum1-ci::URA3/sum1-ci::URA3 |
| MSY350 | ndt80::TRP1-PGAL1-NDT80/ndt80::TRP1-PGAL1-NDT80 ura3::PGPD1-GAL4(848)-ER::URA3 |
| MSY385 | ndt80::TRP1-PGAL1-NDT80/ndt80::TRP1-PGAL1-NDT80 ura3::PGPD1-GAL4(848)-ER::URA3 sum1-ci::URA3/sum1-ci::URA3 |
| MSY390 | hst1Δ::KAN/hst1Δ::KAN sum1-ci::URA3/sum1-ci::URA3 |
| MSY391 | hst1Δ::KAN/hst1Δ::KAN |
| MSY392 | sum1-ci::URA3/sum1-ci::URA3 |
| MSY434 | sum1-ci::URA3/sum1-ci::URA3 TRP1-NDT80-trp1::hisG/TRP1-NDT80-trp1::hisG SMK1-HA::KAN/SMK1-HA::KAN |
| MSY428 | sum1-ci::URA3/sum1-ci::URA3 ndt80Δ::LEU2/ndt80Δ::LEU2 TRP1-NDT80-trp1::hisG/TRP1-NDT80-trp1::hisG |
| MSY426 | sum1-ci::URA3/sum1-ci::URA3 ndt80Δ::LEU2/ndt80Δ::LEU2 TRP1-NDT80-trp1::hisG/trp1::hisG |
| MSY438 | sum1-ci::URA3/sum1-ci::URA3 ndt80Δ::LEU2/ndt80Δ::LEU2 TRP1-ndt80-ΔM1-trp1::hisG/TRP1-ndt80-ΔM1-trp1::hisG |
| MSY444 | sum1-ci::URA3/sum1-ci::URA3 TRP1-ndt80-ΔM1-trp1::hisG/TRP1-ndt80-ΔM1-trp1::hisG SMK1-HA::KAN/SMK1-HA::KAN |
| MSY447 | rfm1Δ::KAN/rfm1Δ::KAN |
| MSY448 | rfm1Δ::KAN/rfm1Δ::KAN sum1-ci::URA3/sum1-ci::URA3 |
| MSY458 | ndt80Δ::LEU2/ndt80Δ::LEU2 |
| MSY460 | ndt80Δ::LEU2/ndt80Δ::LEU2 sum1-c::URA3/sum1-c::URA3 |
| EWY167 | sum1-ci::URA3/sum1Δ::KAN |
All strains are MATa/MATα diploids in the LNY150 (SK1) genetic background (35). The his4-N or his4-G allelic status of the strains has not been tested unless otherwise indicated.
The NDT80 promoter plasmids used in this study were generated in the Segall laboratory from the set of pRS316(−505)ndt80 minigene plasmids described by Pak and Segall (23). To construct TRP1-selectable autonomous and integrating derivatives of these plasmids, the wild-type and MSE1Δ (M1Δ) NotI/SalI fragments (containing the promoter and entire open reading frame in each case) were subcloned into the same sites of pRS314 or pRS304, respectively, and the promoter mutations were verified by sequencing. The pRS304 derivatives were linearized with XbaI prior to transformation to direct integration to trp1 by homologous recombination.
Growth and sporulation.
Cells were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose), SD drop-out medium (0.67% Difco yeast nitrogen base without amino acids, 2% glucose and supplemented with nutrients essential for auxotrophic strains), or YEPA medium (1% yeast extract, 2% peptone, 2% potassium acetate). For sporulation, cells were grown overnight in liquid YEPA medium to a density no greater than 107 cells/ml. Cells were harvested and washed once in sporulation medium (2% potassium acetate, 10 μg/ml adenine, 5 μg/ml histidine, 30 μg/ml leucine, 7.5 μg/ml lysine, 10 μg/ml tryptophan, 5 μg/ml uracil) before being resuspended in sporulation medium at 4 × 107 cells/ml.
Western blotting.
Cellular extracts were prepared with the NaOH lysis-trichloroacetic acid precipitation procedure as previously described (29). Samples were resolved electrophoretically through 8% polyacrylamide gels, transferred onto Immobilon-P membranes (Millipore, Billerica, MA), and blocked at room temperature in phosphate-buffered saline (PBS)-Tween (PBST) and I-block (Tropix, Foster City, CA). Antibody staining was performed overnight at 4°C with monoclonal anti-hemagglutinin ([HA] 1:5,000; Covance), monoclonal anti-PSTAIR (1:20,000; Sigma), or rabbit anti-Ime2 (1:10,000; generously provided by Aaron Mitchell) that were visualized using an alkaline phosphatase-conjugated secondary antibody or rabbit anti-Ndt80 (1:50,000; generously provided by Kirsten Benjamin) that was visualized using horseradish peroxidase-conjugated secondary antibody.
Miscellaneous assays and procedures.
Meiosis was scored in at least 200 DAPI (4′,6′-diamidino-2-phenylindole)-stained cells for each sample as previously described (37). The microscopic analysis of ZIP1-GFP700 (in which green fluorescent protein [GFP] is inserted at position 700 of Zip1) was carried out on live cells visualized at a magnification of ×400 with a Leica microscope (Leica Microsystems GmbH, Wetzlar, Germany) using iVision software (North York, ON, Canada) as described previously (30, 38). To measure total Zip1-GFP fluorescence, 2 × 107 cells were washed once and resuspended in PBS, followed by the addition of formaldehyde to 4%. After a 10-min incubation at room temperature, cells were washed in PBS, and fluorescence was measured using a FLUOstar OPTIMA plate reader (BMG Labtech GmbH, Offenberg, Germany). The incorporation of dityrosine into the spore walls of cells was carried out as previously described using cells that had been sporulated in liquid and collected on a nitrocellulose filter using a fritted-glass funnel (19). The preparation of total RNA and Northern blot hybridization analyses were carried out as previously described using the hybridization probes described by Lindgren et al. (17).
RESULTS
Inhibition of Cdk1 prevents SMK1 expression specifically in sum1-i cells.
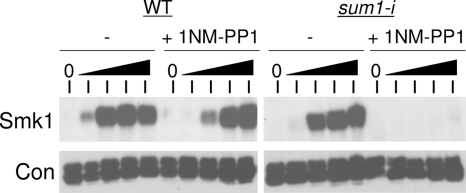
Ime2 functions in a meiosis-specific pathway to promote the removal of Sum1 from middle-meiotic promoters (1). However, a form of Sum1 that cannot be phosphorylated by Ime2 (Sum1-i) has only modest effects on meiosis. Ime2 and Cdk1 can coregulate meiotic substrates (14). One way to address the possibility that Sum1 is coregulated by Ime2 and Cdk1 is to assay Sum1 activity in a sum1-i strain in which Cdk1 has been inactivated. Inhibition of a form of Cdk1 that has been engineered to be sensitive to the ATP analog 4-amino-1-tert-butyl-3-(1-naphthylmethyl)phenylpyrazolo[3,4-d]pyrimidine ([1-NM-PP1] Cdk1-as1) blocks premeiotic S phase (3). However, Ndt80 is still produced in these blocked cells (but with a delay). It has also been shown that cells blocked in premeiotic S phase due to deletion of the S-phase promoting cyclins, Clb5/Clb6, express middle genes (33). Smk1 production is an excellent readout for Sum1 repression (1, 20, 25, 39). We assayed Smk1-HA in cdk1-as1 cells that had been treated with 1-NM-PP1. In SUM1 cells, 1-NM-PP1 delayed Smk1-HA expression by roughly 1.5 h (Fig. 1). In contrast, treatment of sum1-i cells with 1-NM-PP1 completely eliminated Smk1-HA expression at even the latest time point tested. One interpretation of these data is that Cdk1 and Ime2 negatively regulate Sum1 in meiotic cells.
FIG. 1.
Inhibition of Cdk1 prevents expression of Smk1 in sum1-i cells. cdk1-as1 SMK1-HA SUM1 (WT) or cdk1-as1 SMK1-HA sum1-i (sum1-i) cells were transferred to sporulation medium containing either 5 μM 1-NM-PP1 (+) to inhibit Cdk1-as1 or vehicle alone (−). Samples were withdrawn at 0, 5, 6.5, 8, and 9.5 h postinduction, and protein extracts were analyzed by immunoblotting using an HA antibody (Smk1) or a PSTAIR antiserum, which detects the constitutively expressed Cdk1 and Pho85 proteins as a loading control (Con).
A form of Sum1 that is insensitive to Ime2 and Cdk1 blocks MSG expression and meiosis.
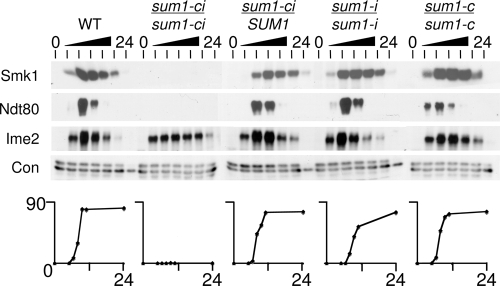
There are 11 minimal Cdk1 phosphoacceptor site occurrences (SP or TP) in Sum1. We changed all the minimal Cdk1 consensus sites in Sum1 to AP to generate a form of Sum1 that is insensitive to Cdk1 (sum1-c). The same substitutions were introduced into the sum1-i background to generate a form of Sum1 that is insensitive to both Cdk1 and Ime2 (sum1-ci). Consistent with the 1-NM-PP1 experiments, the sum1-ci substitutions completely prevented removal of Sum1-dependent repression as assayed by Smk1-HA expression (Fig. 2). Importantly, sum1-ci also prevented meiosis (less than 2.5% meiosis in sum1-ci diploids compared to >80% meiosis in wild-type cells). Moreover, Ndt80 was undetectable in sum1-ci cells. sum1-ci does not prevent induction of the program since Ime2, which is expressed as an early gene product, is expressed on schedule in the mutant. sum1-ci is recessive since the block to meiosis and Smk1-HA and Ndt80 induction were not observed in a sum1-ci/SUM1 heterozygote. The sum1-c and the sum1-i mutants had only modest effects on the expression of Smk1-HA, Ime2, and Ndt80, and these mutant cells completed meiosis at a rate and frequency that are comparable to those seen in the wild type. These findings show that the Ime2 phosphoacceptor site at T306 is essential for meiotic development when all 11 potential phosphoacceptor sites for Cdk1 have been eliminated. These data taken together with the inhibitor studies presented in Fig. 1 suggest that Cdk1 negatively regulates Sum1 in a pathway that is redundant with Ime2. However, it remains possible that Cdk1 exerts its effect on Sum1 through other protein kinases that phosphorylate SP/TP sites (such as mitogen-activated protein kinases [MAPKs]). It is also possible that protein kinases in addition to Cdk1 and Ime2 downregulate Sum1 through one or more of the 11 sites that have been mutated in sum1-c.
FIG. 2.
NDT80 and middle genes are not induced in a sum1-ci mutant. Cells of the indicated genotype were transferred to sporulation medium; collected at 0, 5, 6.5, 8, 9.5, 11, and 24 h postinduction; and analyzed for the completion of meiosis (lower panel). Protein extracts prepared from the same cells were analyzed by immunoblotting (upper) using antibodies for the indicated proteins. Con, control.
sum1-c is defective in the NDT80-independent pathway for removing Sum1 repression.
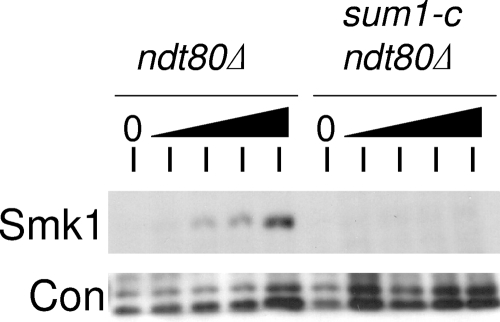
We previously demonstrated that the Sum1-i protein is not removed from chromatin in the absence of NDT80 (1). Thus, Smk1-HA is not derepressed when sum1-i ndt80Δ cells are transferred to sporulation medium. To test whether a sum1-c mutant is also defective in removing Sum1 repression in ndt80Δ cells, Smk1-HA was monitored in an ndt80Δ sum1-c strain. While Smk1-HA was expressed starting between 5 and 6.5 h in the ndt80Δ strain (around the time that exit from prophase occurs in wild-type cells), it is not expressed in the ndt80Δ sum1-c strain even at the latest time point tested (Fig. 3). Thus, sum1-c, similar to sum1-i, is defective in the NDT80-independent pathway for removing Sum1 repression. These findings suggest that Ime2 or Cdk1 can promote the removal of Sum1 repression in the NDT80-independent pathway.
FIG. 3.
sum1-c is defective in the NDT80-independent pathway for removing Sum1 repression. The indicated diploid strains were transferred to sporulation medium; collected at 0, 5, 6.5, 8, and 9.5 h; and analyzed by immunoblotting using an HA antibody (Smk1) or a PSTAIR antiserum as a loading control (Con).
sum1-ci and ndt80Δ have similar phenotypes.
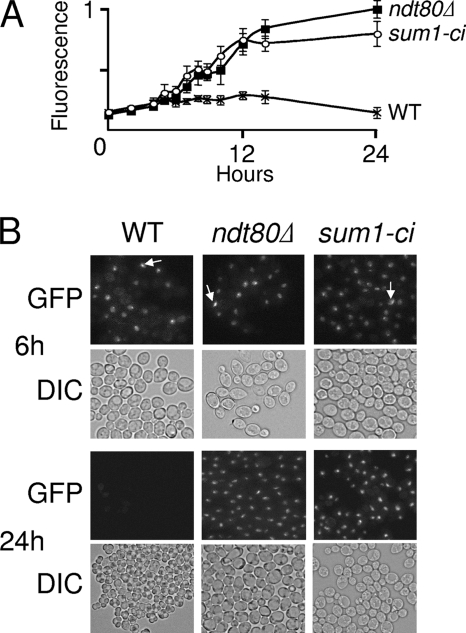
It has previously been shown that Sum1 can repress the NDT80 promoter (23, 39). The finding that Ndt80 is undetectable in a sum1-ci mutant suggests that Sum1-ci blocks meiosis by repressing NDT80. If this is the case, then sum1-ci should block cells with an ndt80Δ-like phenotype. We compared the fluorescence intensity of Zip1-GFP (a fluorescent marker for SC formation) (38) in sum1-ci, ndt80Δ, and wild-type cultures at various times after transfer to sporulation medium. Only a modest increase in fluorescence emission could be detected in wild-type cultures, likely reflecting the transient accumulation of SC and the rapid degradation of SC components that occur upon pachytene exit. In contrast, fluorescence increased throughout the course of the experiment in both the ndt80Δ and the sum1-ci cultures (Fig. 4A). Fluorescence microscopy revealed that Zip1-GFP accumulated in wild-type, sum1-ci, and ndt80Δ nuclei starting around 5 h postinduction (Fig. 4B). As previously described, in addition to the SC fluorescence, most of the fluorescent nuclei also contained a brightly staining complex (38). While the fractions of fluorescence-positive nuclei were comparable at early times (up until 6 h postinduction), at the 24-h time point 59% of the ndt80Δ nuclei and 61% of the sum1-ci nuclei fluoresced while only 0.5% of the wild-type nuclei fluoresced (Fig. 4B). Moreover, the frequencies of the brightly staining complexes in the fluorescent ndt80Δ and sum1-ci nuclei were identical at all time points. These data demonstrate that sum1-ci blocks meiotic development in prophase with a phenotype that is similar to that of ndt80Δ.
FIG. 4.
sum1-ci and ndt80Δ phenotypes are similar. (A) ZIP1-GFP (WT), ndt80Δ ZIP-GFP (ndt80Δ), and sum1-ci ZIP1-GFP (sum1-ci) cells were transferred to sporulation medium, collected at various times, and fixed, and relative fluorescence emission was monitored in a fluorometer (n = 4). (B) Live cells were analyzed by visible differential interference contrast (DIC) microscopy or by fluorescence microscopy for Zip1-GFP (GFP) at 6 and 24 h, as indicated. The brightly staining complex (arrows) was observed at a similar frequency in all fluorescence-positive cells.
Ectopic expression of NDT80 bypasses the sum1-ci block.
If sum1-ci blocks meiosis by repressing the NDT80 promoter, then the ectopic expression of NDT80 using a heterologous promoter might bypass the sum1-ci block. It has previously been shown that expression of NDT80 using the GAL1 promoter (pGAL-NDT80) can be induced using ß-estradiol in cells that have been engineered to express the Gal4 DNA-binding protein fused to the estrogen receptor (Gal4-ER) (3, 4). This system allows one to induce NDT80 around the time when NDT80 is normally expressed to a level that is comparable to that seen in wild-type cells. sum1-ci cells in the GAL4-ER pGAL-NDT80 background completed meiosis in a ß-estradiol-dependent fashion (Fig. 5 upper). Moreover, the fraction of ß-estradiol-treated cells that completed meiosis was only slightly lower in the sum1-ci background than in the SUM1 background. Spore formation in the SUM1 and sum1-ci ß-estradiol-treated cells was also comparable, as judged microscopically and using a semiquantitative fluorescence assay that detects incorporation of dityrosine into the outer layer of the spore wall (Fig. 5, lower panel). Moreover, the viability of enzymatically digested and microdissected spores formed by the ß-estradiol-treated sum1-ci cells (71%) was only modestly lower than the viability of the SUM1 spores (91%; compare to 100% viability for wild-type spores [for each strain, n = 144 analyzed spores]). These findings demonstrate that the ectopic expression of NDT80 can bypass the sum1-ci block to meiosis and spore formation and are consistent with the idea that sum1-ci blocks prophase exit and meiosis through the NDT80 promoter.
FIG. 5.
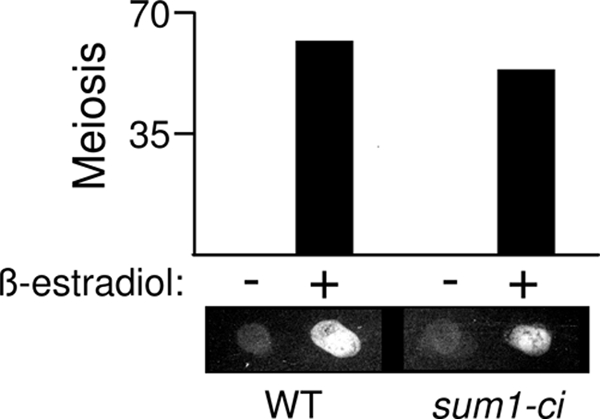
ß-Estradiol-inducible NDT80 bypasses the sum1-ci meiotic block. SUM1 GAL4-ER pGAL-NDT80 (WT) and sum1-ci GAL4-ER pGAL-NDT80 (sum1-ci) cells were transferred to sporulation medium, and 1 μM ß-estradiol (+) or vehicle alone (−) was added at 5 h to induce expression of NDT80. The completion of meiosis (MII) was monitored in three independent cultures at 24 h after cells were transferred to sporulation medium (error bars are too small to be visible). Equivalent numbers of cells were collected on a nitrocellulose filter and assayed for the presence of dityrosine, a component of the outer layer of the spore wall, by fluorescence.
Mutation of the Sum1-binding element in the NDT80 promoter bypasses the sum1-ci block.
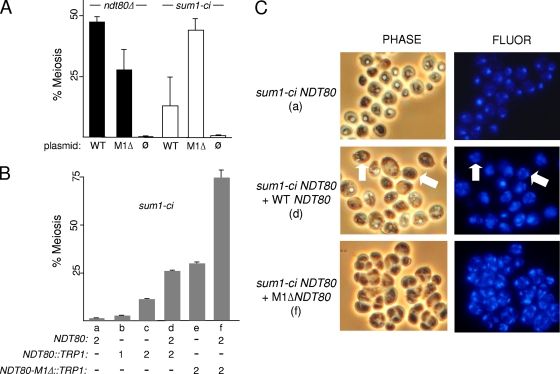
Pak and Segall previously demonstrated that the NDT80 promoter is regulated by multiple DNA elements including two URS1s that respond to Ime1, an MSE that responds to Ndt80 (named M2), and an MSE that responds to both Sum1 and Ndt80 (named M1) (23). In their study, the deletion of M1 reduced NDT80 expression (likely due to reduced Ndt80 binding) and also advanced the timing of NDT80 expression (likely due to reduced Sum1 binding). We compared the ability of wild-type and NDT80-M1Δ plasmids to complement the ndt80Δ strain and to bypass sum1-ci. M1Δ decreased the ability of the plasmid to complement the meiotic defect of the ndt80Δ strain (Fig. 6A, left). In contrast, M1Δ increased the ability of the plasmid to promote meiosis in the sum1-ci NDT80 strain (Fig. 6A right).
FIG. 6.
NDT80 promoter mutations bypass the sum1-ci block. (A) ndt80Δ or sum1-ci diploids harboring an NDT80 plasmid controlled by the wild-type promoter (WT), the promoter carrying a deletion of the MSE1 Sum1/Ndt80 binding site (M1Δ), or the plasmid lacking NDT80 (ø) were transferred to sporulation medium, and meiosis (MI) was scored at 24 h postinduction (n = 4). (B) sum1-ci diploids containing the indicated number of NDT80, NDT80::TRP1, or NDT80-M1Δ::TRP1 genes were analyzed as described for panel A (n = 3). (C) Strains a, d, and f from panel B were collected at 72 h postinduction, stained with DAPI, and analyzed using phase-contrast or fluorescence photomicrography. The arrows in the middle photomicrographs point to cells that have completed meiosis but have failed to form spores.
To further assess the significance of the Sum1 interaction with the NDT80 promoter, we tested integrating NDT80 plasmids with or without the M1Δ mutation for their ability to bypass sum1-ci. Integrating two copies of NDT80-M1Δ at TRP1 to generate a homozygous sum1-ci strain with two wild-type and two M1Δ copies of NDT80 caused substantially more bypass than integrating wild-type NDT80 (Fig. 6B, compare strains d and f). Moreover, spore walls could be observed by phase-contrast microscopy in the NDT80-M1Δ integrant while spore walls were not detected in the wild-type NDT80 integrant (Fig. 6C, compare photomicrographs of strains d and f). Further experiments with the spores formed in the NDT80-M1Δ integrant showed that they were not as resistant to enzymatic digestion as wild-type spores (resistance of the mutant to glusulase was less than 10% of the wild-type level). Nonetheless, tetrads could be identified in these digested samples, and 88% of the spores from these microdissected tetrads formed viable colonies (n = 128). Thus, NDT80-M1Δ efficiently bypasses the sum1-ci block to meiosis and partially bypasses the block to spore formation. The fact that high levels of meiosis and visible spore formation in the sum1-ci background require both the wild-type and M1Δ forms of NDT80 (Fig. 6 B, compare strains a, e, and f) suggests that M1 functions at two genetically separable steps in the NDT80 induction pathway: an initiation step (when Sum1 repression is removed) and an amplification step (when the positive autoregulatory loop is in force).
During the course of these experiments, we found that the wild-type NDT80 plasmid caused significant levels of sum1-ci bypass (either as an episome or when integrated into the genome, as shown in Fig. 6A and B). A single integrated copy of the wild-type NDT80 plasmid caused approximately the same fraction of cells to complete meiosis as two copies of NDT80 in its normal chromosomal context. We therefore infer that plasmid sequences adjacent to the NDT80 promoter can modestly activate transcription. Two copies of the integrated NDT80 plasmid caused a significant increase in meiosis, and the presence of all four copies of NDT80 caused an even greater increase (Fig. 6B, compare strains a to d). However, spores were not formed in any of these strains. These data demonstrate that increased NDT80 gene dosage can overcome the sum1-ci block to meiosis. Consistent with these results, meiosis was higher in diploids containing a single copy of sum1-ci (sum1-ci/sum1Δ NDT80/NDT80) (48.6% ± 2.0%) than in the corresponding homozygous sum1-ci strain (2.2% ± 0.3%). This finding is consistent with the low abundance of the Sum1 protein reported by other investigators (11). These data suggest that the balance between Sum1 and the NDT80 promoter can influence exit from meiotic prophase.
hst1Δ or rfm1Δ bypass the sum1-ci block.
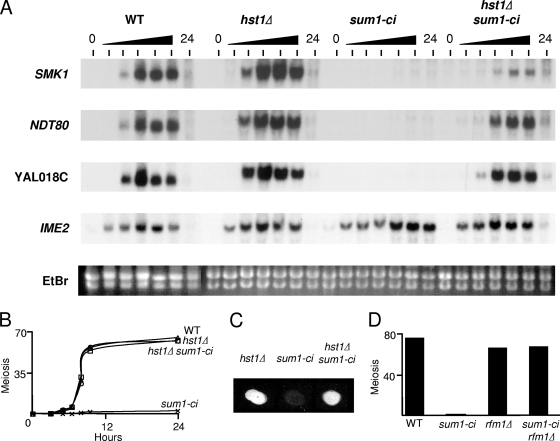
We previously demonstrated that the Hst1 histone deacetylase (Sir2 ortholog) is involved in an Ndt80-independent pathway for Sum1 removal from middle-meiotic promoters (1). hst1Δ cells complete meiosis and form spores (17, 39). We found that an hst1Δ sum1-ci double mutant completed meiosis, similar to the wild type (Fig. 7 B). Moreover, hst1Δ sum1-ci cells formed spores that appeared indistinguishable from wild-type cells as assayed by phase-contrast microscopy, and these spores were positive in the semiquantitative fluorescence assay for dityrosine incorporation (Fig. 7C). In addition, hst1Δ sum1-ci spores were mostly viable following enzymatic digestion/microdissection (viability of the hst1Δ sum1-ci, hst1Δ, and wild-type spores was 87%, 97%, and 100%, respectively [n = 144 spores analyzed for each strain]). Thus, hst1Δ efficiently bypasses the sum1-ci meiotic block. We also found that deletion of Rfm1, the tethering factor that links Hst1 to Sum1, bypassed the sum1-ci block (Fig. 7D).
FIG. 7.
hst1Δ and rfm1Δ bypass the sum1-ci block to middle-phase gene expression and meiosis. (A) Cells of the indicated genotype were harvested at 0, 3, 5, 6.5, 8, 9.5, and 24 h. RNA was prepared, and levels of the indicated mRNAs were analyzed by Northern blot hybridization. EtBr, the ethidium bromide-stained gel before transfer. (B) The completion of MII was quantified by fluorescence microscopy. (C) Equivalent numbers of cells of the indicated genotypes were collected on a nitrocellulose filter, and incorporation of dityrosine into spore walls was analyzed using the fluorescence assay. (D) Cells of the indicated genotype were harvested at 24 h postinduction, and the completion of MII was quantified by fluorescence microscopy.
Northern blot hybridization analyses showed that the IME2 early meiosis-specific gene is induced with comparable kinetics in sum1-ci, hst1Δ, sum1-ci hst1Δ, and wild-type cells (Fig. 7A). Thus, sum1-ci and hst1Δ do not influence induction of meiosis under the conditions tested. However, while IME2 mRNA accumulation was transient in wild-type cells, IME2 mRNA levels remained high in sum1-ci cells at the longest time point tested (24 h). NDT80 and SMK1 middle gene mRNAs were undetectable in sum1-ci samples at any of the time points tested. Similarly, YAL018C, a middle gene that is repressed by Hst1 in vegetative cells (2, 18), was not induced in the sum1-ci cells (detection of the derepressed level of YAL018C mRNA in hst1Δ vegetative cells requires overexposure of the autoradiogram [1]). In contrast to the sum1-ci mutant, NDT80 is expressed in the hst1Δ sum1-ci double mutant with only a modest delay and to about the same levels seen in the wild-type and hst1Δ controls. Thus, hst1Δ efficiently bypasses sum1-ci repression at the NDT80 promoter. Moreover, YAL018C, which is activated by Ndt80 (5), is efficiently induced. Therefore, the Ndt80 that is produced in the double mutant is active. While SMK1 is also induced in the sum1-ci hst1Δ cells, it is induced at a lower level than in the wild type. We previously demonstrated that SMK1 expression can be reduced by 80% before spore formation defects are observed (cells may make 5-fold more Smk1 than is needed to produce spores under standard laboratory conditions) (36), and the reduction in SMK1 mRNA is therefore consistent with the ability of sum1-ci hst1Δ cells to form spores on schedule. These observations are consistent with Sum1's ability to repress the SMK1 promoter in an Hst1-independent fashion (17, 25) and indicate that middle promoters can be differentially influenced by Sum1-ci when active Ndt80 has been produced. The nearly wild-type rates of meiosis and spore formation observed in the sum1-ci hst1Δ background suggest that many (and perhaps most) middle promoters are expressed at substantial levels and with nearly wild-type kinetics in sum1-ci cells when HST1 has been deleted.
DISCUSSION
The conversion of middle-meiotic promoters from a repressed (Sum1-occupied) to an activated (Ndt80-occupied) state is a critical regulatory transition in the life cycle of S. cerevisiae that controls exit from prophase (at pachytene) and completion of the meiotic program. Sum1 and Ndt80 bind to overlapping segments of DNA, and it has been proposed that middle-promoter induction is triggered by competitive displacement (24). However, Sum1 can also be removed from DNA by a pathway that does not require Ndt80 (1). Ime2 promotes the removal of Sum1 from DNA in the NDT80-independent pathway. In the present study, we have shown that Cdk1, similar to Ime2, inhibits Sum1 (Fig. 1 and 3) and that a form of Sum1 that is insensitive to both Cdk1 and Ime2 (Sum1-ci) blocks NDT80 expression (Fig. 2) and prophase exit (Fig. 4). The sum1-ci block can be bypassed by the ectopic expression of NDT80, mutation of a Sum1-binding site in the NDT80 promoter, or deletion of HST1 or RFM1 (Fig. 5, 6, and 7). These findings indicate that the Sum1/Rfm1/Hst1 complex functions as a key regulatory brake at the NDT80 promoter and that the phosphorylation of Sum1 by either Cdk1 or Ime2 triggers removal of the brake, induction of NDT80 expression, and the completion of meiotic development. The ability of either Cdk1 or Ime2 to downregulate Sum1 may allow multiple inputs (e.g., dependency, checkpoint, and nutritional signals) to modulate the efficiency of prophase exit.
The MSE1/Sum1 interaction at NDT80 as a switch.
It has previously been shown that the NDT80 promoter is repressed in vegetative cells by a Sum1-responsive MSE (M1) and by upstream repression sequence 1 (URS1) elements (23). Thus, NDT80 is derepressed in vegetative cells only when both URS1 and MSE-mediated repression have been eliminated (39). URS1s are converted from repression to activation elements in an IME1-dependent fashion following meiotic induction (35). The URS1/MSE combinatorial control of the NDT80 promoter therefore generates a window during meiotic development (framed by the presence of active Ime1) when Sum1 is poised to function as a gatekeeper for NDT80 expression. The genetic interactions between NDT80-M1Δ and sum1-ci reported in this study (Fig. 6) are consistent with the idea that Sum1 bound to MSE1 functions as an NDT80 gatekeeper. We previously proposed that the phosphorylation of Sum1 by Ime2 downregulates the Hst1 histone deacetylase and suggested that this is an important step in the Ndt80-independent pathway for Sum1 removal (1). The ability of hst1Δ or rfm1Δ to bypass the sum1-ci meiotic block (Fig. 7) is consistent with this model. These data also suggest that Ime2 and Cdk1 can function redundantly to downregulate Hst1. Sum1 and Hst1 function in promoter-specific repression and not long-range silencing of chromatin in S. cerevisiae (28). The role of Hst1 in controlling meiotic commitment in yeast raises the possibility that the transient inhibition of sirtuins can trigger commitment to differentiation programs in other organisms by transiently altering the chromatin structure of specific promoters. Taken as a whole, the data suggest that the competition-based removal of Sum1 and the positive autoregulatory loop at NDT80 are coupled feed-forward mechanisms that give switch-like properties to this transition in wild-type cells. Negative regulation of repressors (e.g., Sum1) may be reinforced by competitive displacement of activator/macromolecule interactions (e.g., Ndt80/DNA) to give irreversible switch-like properties to transcriptional cascades in a variety of biological systems.
Acknowledgments
We thank Kirsten Benjamin, Aaron Mitchell, and Andrew Vershon for yeast strains and immunological reagents. We are especially indebted to Jacqueline Segall for the generous gift of the NDT80 plasmids used in this study.
M.E.S. gratefully acknowledges support of NRSA award T32-DK07705. This work was supported by NIH grant GM061817 and an institutional REA award to E.W.
Footnotes
Published ahead of print on 12 April 2010.
REFERENCES
- 1.Ahmed, N. T., D. Bungard, M. E. Shin, M. Moore, and E. Winter. 2009. The Ime2 CDK-like kinase enhances the disassociation of the Sum1 repressor from middle meiotic promoters. Mol. Cell. Biol. 29:4352-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedalov, A., M. Hirao, J. Posakony, M. Nelson, and J. A. Simon. 2003. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:7044-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, K. R., C. Zhang, K. M. Shokat, and I. Herskowitz. 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17:1524-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlile, T. M., and A. Amon. 2008. Meiosis I is established through division-specific translational control of a cyclin. Cell 133:280-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. (Erratum, 282: 1421.) [DOI] [PubMed] [Google Scholar]
- 6.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 7.Colomina, N., E. Gari, C. Gallego, E. Herrero, and M. Aldea. 1999. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J. 18:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshaies, R. J., and J. E. Ferrell, Jr. 2001. Multisite phosphorylation and the countdown to S phase. Cell 107:819-822. [DOI] [PubMed] [Google Scholar]
- 9.Dirick, L., T. Bohm, and K. Nasmyth. 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 11.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 12.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochwagen, A., and A. Amon. 2006. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr. Biol. 16:217-228. [DOI] [PubMed] [Google Scholar]
- 14.Holt, L. J., J. E. Hutti, L. C. Cantley, and D. O. Morgan. 2007. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol. Cell 25:689-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honigberg, S. M. 2004. Ime2p and Cdc28p: co-pilots driving meiotic development. J. Cell. Biochem. 92:1025-1033. [DOI] [PubMed] [Google Scholar]
- 16.Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano, S. Sagee, and G. Shenhar. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224:111-171. [DOI] [PubMed] [Google Scholar]
- 17.Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon, and E. Winter. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCord, R., M. Pierce, J. Xie, S. Wonkatal, C. Mickel, and A. K. Vershon. 2003. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald, C. M., K. F. Cooper, and E. Winter. 2005. The Ama1-directed APC/C ubiquitin ligase regulates the Smk1 MAPK during meiosis in yeast. Genetics 171:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald, C. M., M. Wagner, M. J. Dunham, M. E. Shin, N. T. Ahmed, and E. Winter. 2009. The Ras/cAMP pathway and the CDK-like kinase Ime2 regulate the MAPK Smk1 and spore morphogenesis in Saccharomyces cerevisiae. Genetics 181:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore, M., M. E. Shin, A. Bruning, K. Schindler, A. Vershon, and E. Winter. 2007. Arg-Pro-X-Ser/Thr is a consensus phosphoacceptor sequence for the meiosis-specific Ime2 protein kinase in Saccharomyces cerevisiae. Biochemistry 46:271-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pak, J., and J. Segall. 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6417-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierce, M., K. R. Benjamin, S. P. Montano, M. M. Georgiadis, E. Winter, and A. K. Vershon. 2003. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol. Cell. Biol. 23:4814-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierce, M., M. Wagner, J. Xie, V. Gailus-Durner, J. Six, A. K. Vershon, and E. Winter. 1998. Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Primig, M., R. Williams, E. Winzeler, G. Tevzadze, A. Conway, S. Hwang, R. Davis, and R. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 27.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 28.Rusche, L. N., and J. Rine. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaber, M., A. Lindgren, K. Schindler, D. Bungard, P. Kaldis, and E. Winter. 2002. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol. Cell. Biol. 22:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherthan, H., H. Wang, C. Adelfalk, E. J. White, C. Cowan, W. Z. Cande, and D. B. Kaback. 2007. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 104:16934-16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simchen, G. 2009. Commitment to meiosis: what determines the mode of division in budding yeast? Bioessays 31:169-177. [DOI] [PubMed] [Google Scholar]
- 32.Sourirajan, A., and M. Lichten. 2008. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 22:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart, D., and C. Wittenberg. 1998. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 12:2698-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tung, K. S., E. J. Hong, and G. S. Roeder. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. U. S. A. 97:12187-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vershon, A. K., and M. Pierce. 2000. Transcriptional regulation of meiosis in yeast. Curr. Opin. Cell Biol. 12:334-339. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, M., P. Briza, M. Pierce, and E. Winter. 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151:1327-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner, M., M. Pierce, and E. Winter. 1997. The CDK-activating kinase Cak1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16:1305-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White, E. J., C. Cowan, W. Z. Cande, and D. B. Kaback. 2004. In vivo analysis of synaptonemal complex formation during yeast meiosis. Genetics 167:51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]