Abstract
Complex I (CI) is the largest enzyme of the mammalian mitochondrial respiratory chain. The biogenesis of the complex is a very complex process due to its large size and number of subunits (45 subunits). The situation is further complicated due to the fact that its subunits have a double genomic origin, as seven of them are encoded by the mitochondrial DNA. Understanding of the assembly process and characterization of the involved factors has advanced very much in the last years. However, until now, a key part of the process, that is, how and at which step the mitochondrially encoded CI subunits (ND subunits) are incorporated in the CI assembly process, was not known. Analyses of several mouse cell lines mutated for three ND subunits allowed us to determine the importance of each one for complex assembly/stability and that there are five different steps within the assembly pathway in which some mitochondrially encoded CI subunit is incorporated.
Complex I (CI) (NADH-ubiquinone oxidoreductase; EC 1.6.5.3) is one of the main electron entry points in the mitochondrial respiratory electron transport chain catalyzing the oxidation of NADH to reduce ubiquinone to ubiquinol (31, 39, 40), contributing to the proton motive force to synthesize ATP by the process called oxidative phosphorylation (OXPHOS).
CI assembly is a difficult problem to address due to the large size of the complex and its dual genomic nature, as 7 out of its 45 subunits are encoded by the mitochondrial DNA (mtDNA) (10, 11). Until very recently, mammalian CI assembly was explained using two different and apparently contradictory models. One model was proposed by following the time course of formation of CI intermediates in human cells in culture once mitochondrial protein synthesis had recovered after its inhibition by doxycycline (36). Based on these observations, human CI was proposed to be assembled through two different modules corresponding to the membrane and peripheral arms. The other model was proposed after analysis of a cohort of four CI-deficient patients in which seven putative assembly intermediates containing a combination of both peripheral- and membrane arm subunits were identified. Thus, an assembly pathway in which the peripheral- and membrane arm subassemblies came together before the completion of each of the arms was proposed (4). However, the most recent studies have refined the previous models and propose an overlapping view of the process. One study, by green fluorescent protein (GFP) tagging of the NDUFS3 subunit, identified six peripheral-arm intermediates. The second and third smaller NDUFS3-containing subassemblies were accumulated and could not advance into higher-molecular-mass species when mitochondrial protein synthesis was inhibited, thus determining the entry point of the mitochondrially encoded subunits in the CI assembly pathway (37). The most recent study analyzed the incorporation of the mitochondrial subunits in a time course to the fully assembled CI and, on the other hand, the incorporation of the nuclear subunits by importing them into isolated mitochondria (24). Although these two models differ in the order in which some subunits are incorporated, they agree on the general human CI assembly pathway, which takes place via evolutionarily conserved modular subassemblies (14, 25, 28, 37).
However, the specific entry points of all the mtDNA-encoded CI subunits (ND subunits) in the CI assembly pathway and their roles in the stability of the complex remained to be clarified. Structural studies related to mutations in the ND subunits in pathological cases have given some hints as to the importance of each of them for CI assembly/stability. In this case, defects in specific ND subunits do not have the same effect: ND1, ND4, and ND6 seem to be fundamental to CI assembly, while ND3 and ND5 are important for its activity but not for assembly. On the other hand, mutations in ND2 alter CI assembly, with abnormal intermediate accumulation (19).
In this article, we present new insights into the roles of the ND subunits by using mouse cells deficient for ND4, ND6, and a combination of ND6 and ND5. This study has allowed us to propose the five different entry points by which the mtDNA-encoded subunits are sequentially incorporated into the CI assembly pathway, completing the current view of the process. We conclude that ND4 and ND6 are required for the proper function and assembly of CI, although at different degrees due to their different entry points and roles in the CI assembly pathway.
MATERIALS AND METHODS
Cell lines and media.
All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL) supplemented with 5% fetal bovine serum (FBS) (Gibco BRL). Mouse cells without mtDNA (ρ°929) were generated by long-term growth of the L929 mouse cell line (ATCC CCL-1) in the presence of high concentrations of ethidium bromide (EtBr), as previously described (34). TmNIH 3T3 cells (control cells) were generated by transformation of ρ°929neo cells by cytoplast fusion, using NIH 3T3 fibroblasts as mitochondrial donors (26).
DNA analysis and mtDNA sequencing.
Total DNA was isolated from cells by digestion with proteinase K in TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5) containing 0.5% SDS and RNase A, purified by extraction with phenol-chloroform-isoamyl alcohol, and precipitated with ethanol. The complete mtDNA was amplified in 24 overlapping 800- to 1,000-bp-long PCR fragments using a multifunctional robot (Genesis 150 Tecan; Crailsheim) (20). Primers were designed using the reference sequence (NC_005089) (7). Assembly and identification of variations in the mitochondrial DNA were carried out using the Staden package (32). Quantification of the C insertion or deletion in mt-Nd6 was achieved by allele-specific termination primer extension (7). The positions in the mtDNA correspond to the reference mtDNA sequence of C57BL/6J (7).
Blue Native electrophoresis and in-gel activity assays.
Mitochondria were isolated from culture cell lines according to the method of Schägger (29), with some modifications (3). Digitonin-solubilized mitochondrial protein (100 μg) was run through Blue Native (BN) gradient gels (3 to 13%). After electrophoresis, the gels were further processed for in-gel activity assays (IGA), Western blotting (WB), two-dimensional (2D) SDS-PAGE, or 2D BN-PAGE (dodecylmaltoside [DDM]) analysis.
Complex I in-gel activity assays (CI-IGA) were performed after BN-PAGE by incubating the gels at room temperature in the presence of 0.1 M Tris-HCl, pH 7.4, 0.14 mM NADH, and 1 mg/ml of nitro-blue tetrazolium (NBT).
Strips from the first-dimension BN-PAGE were excised and used for 2D SDS-PAGE (41) or 2D BN-PAGE. Second-dimension BN-PAGE was similar to first-dimension BN-PAGE (gradient gels from 5 to 13%), and 0.02% DDM was added to the cathode buffer (27).
After electrophoresis, the gels were electroblotted onto Hybond-P polyvinylidene difluoride (PVDF) membranes (Amersham) and sequentially probed with specific antibodies against complex I (anti-NDUFA9, anti-NDUFB6, and anti-NDUFB8 from Molecular Probes and anti-NDUFS3 from MitoSciences), complex III2 (anti-Core2; Molecular Probes), and complex IV (anti-COI; Molecular Probes). Secondary antibodies used were peroxidase-conjugated anti-mouse antibodies (Invitrogen). The signal was generated using ECL Plus (Amersham). The sizes of the observed subcomplexes were determined by their relative migrations compared with those of the OXPHOS complexes (CI, 1,000 kDa; CII, 150 kDa; CIII2, 600 kDa; CIV, 240 kDa; CV, 750 kDa).
Pulse-chase experiments: mtDNA-encoded subunit protein labeling.
Labeling of mtDNA-encoded proteins was performed using [35S]methionine and [35S]cysteine (Expre35S35S Protein Labeling Mix; Perkin Elmer Life Sciences) in intact cells, as described previously (12, 16). Labeling in mitochondria isolated from mouse liver was performed as described previously (18).
Oxygen consumption measurements.
O2 consumption determinations in digitonin-permeabilized cells were carried out in an oxytherm Clark-type electrode (Hansatech) as previously described (23) with small modifications (7).
RESULTS
Generation and characterization of complex I mutant cell lines.
By exposure of mouse cells to a variety of chemical agents, we generated several lines carrying mutations in their mtDNA (8). To avoid biased results due to different alterations in the nucleus, transmitochondrial cybrids (a fusion between a cell and an enucleated cell) for the different mtDNA mutations in a homogeneous nuclear background were generated. The mutations described here were generated in two different mtDNA backgrounds, L929 (AJ489607) and NIH 3T3 (AY999076), by exposing the cells to the mutagen TMP.
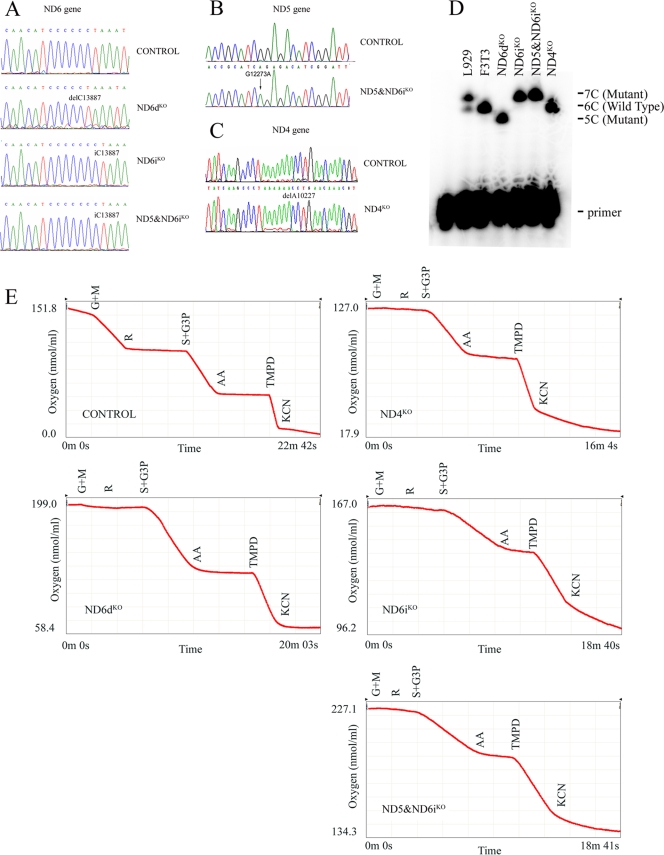
One of the three preexisting mutations found in the L929 cell mtDNA (1, 7) was a frameshift mutation (13887iC) that created a stop codon 51 to 53 bp downstream of a 6-C stretch within the mt-Nd6 gene, resulting in a 79-amino-acid truncated polypeptide. An ND6iKO mutant homoplasmic cell line (Fig. 1A and D) (1) could be isolated from the original heteroplasmic L929 cells.
FIG. 1.
Genetic and biochemical characterization of the ND mutants. (A to C) Chromatograms showing the homoplasmic mutations found in the mt-Nd6 gene (A), the mt-Nd5 gene (B), and the mt-Nd4 gene (C). (D) Allele-specific termination of a primer extension assay to confirm the homoplasmy of the insertion and deletion of a C in the ND6 mutants. (E) Polarographic measurements for the different cell lines employed in the study. Note the difference between the graph corresponding to the control and the rest. All the analyzed ND subunit mutants displayed undetectable NADH-linked substrate (glutamate plus malate) oxygen consumption, while the activities of CII plus CIII (succinate plus glycerol-3-P-linked oxygen consumption) and CIV (TMPD-linked oxygen consumption) were comparable to those of the control cells.
A new mutation in the mt-Nd5 gene induced by chemical mutagenesis of the L929 cell line allowed us to create a homoplasmic double-mutant cell line in CI by combination with the preexisting mt-Nd6 mutation (Fig. 1A, B, and D). The mt-Nd5 mutation, a G12275A transition, introduced a premature stop codon.
Two additional mutations were generated in the NIH 3T3 background. One was a deletion of one C (13887dC) in the mt-Nd6 gene—interestingly, at the same position as the naturally occurring L929 insertion—resulting in a 72-amino-acid-long truncated polypeptide (Fig. 1A and D, ND6dKO cell line; see Fig. S1 in the supplemental material) (26). The other was the deletion of an A in a track of 7 As at position 10227 in the mt-Nd4 gene. This mutation led to a truncated polypeptide of only 26 amino acids (Fig. 1C, ND4KO cell line).
In all cases, the truncated polypeptides were not detectable when the mtDNA-encoded polypeptides were specifically labeled (shown for the ND6dKO mutant in Fig. S1 in the supplemental material). Thus, all the cell lines employed in this study were knockouts for the mutated proteins.
The isolated CI mutant cybrids showed a dramatic loss of malate/glutamate-dependent O2 consumption, while the activities of complex III and complex IV were comparable to those of the controls (Fig. 1E).
Isolated mutant cell lines display defective complex I activity and assembly.
Up to now, most studies of CI assembly were obtained from BN-PAGE of mitochondrial cell fractions obtained using mild detergents, such as DDM (4, 35-37) or Triton X-100 (24, 33). However, if the detergent digitonin is used to solubilize the mitochondrial membranes, the interactions between the different respiratory chain complexes in the supercomplexes are maintained (3), permitting better resolution of the subassemblies.
To investigate the CI assembly state, digitonin-solubilized mitochondrial samples from the CI mutants were run through Blue Native gels, and then CI-IGA and WB, followed by immunodetection of the NDUFB6 subunit, were performed (Fig. 2A and B).
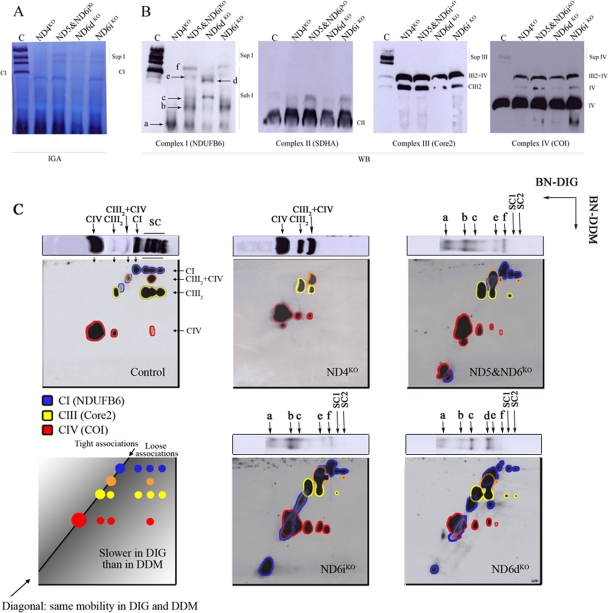
FIG. 2.
Detection of complex I-related species. (A) CI as a “monomer” and associated into supercomplexes was detected in 1D Blue Native gels in digitonin-solubilized mitochondria from control and mutant cells by CI-IGA after 2 h of development of the reaction. (B) WB and immunodetection with specific antibodies against the different respiratory chain complexes after 1D BNGE. Complex I was visualized using a specific antibody against the NDUFB6 subunit that detected mainly the monomeric CI and the CI-containing supercomplexes in the control samples and several CI subcomplexes in the mutant cell lines. (C) Digital composite of the immunodetections for complexes I (blue), III (yellow), and IV (red) performed after 1D BNGE in digitonin-solubilized mitochondria from control and mutant cells, followed by 2D BNGE run with the addition of DDM to the cathode buffer. The association of complexes III and IV is indicated in orange. See the text for a detailed explanation and Fig. S3 in the supplemental material for the original images used to make the composite. The NDUFB6-containing subcomplexes found in the mutant cells are named alphabetically (a to f); higher-molecular-mass associations of CI are indicated as SC1 (supercomplex 1) and SC2. The correspondence of the 2D DDM signals to those observed in the 1D gels (B) is shown.
When CI-IGA was performed in the control cell line, the band corresponding to the isolated CI and five additional bands corresponding to the CI-containing supercomplexes were detected (3, 30). The signal from isolated CI was absent in all mutants. However, active high-molecular-mass associations were found in ND5&ND6iKO, ND6dKO, and ND6iKO but not in the ND4KO cell line. Although these signals were faint after a 2-hour incubation, they could be easily visualized if the reaction was allowed to proceed overnight (see Fig. S2 in the supplemental material).
By WB analyses using an antibody against the membrane arm 17-kDa subunit (NDUFB6), it was observed that isolated monomeric CI was absent in all of the mutant cell lines, and the higher-molecular-mass associations observed in the CI-IGA were not detected in the ND5&ND6iKO, ND6dKO, and ND6iKO cell lines (Fig. 2A and B). Nevertheless, NDUFB6 was present in several subassemblies in the mutant cells that were prominent in the mutant ND6 cell lines, and most of them were absent in ND4KO. The aforementioned subcomplexes were named from smaller to bigger in alphabetical order (a to e). The accumulation of these subassemblies could be due to an assembly defect and also to a disturbance of the stability of the complex, as most of these intermediates are absent in control cells (35).
Supercomplexes in complex I mutant cell lines.
Based on the idea that supercomplexes are preexisting entities that can be efficiently separated into the individual respiratory complexes, we performed 1D BN-PAGE with the digitonin-solubilized mitochondria, followed by 2D BN-PAGE, adding DDM to the cathode buffer (27, 30). The complexes from the 1D BN-PAGE that retained their masses after the 2D BN-PAGE were found on a diagonal (Fig. 2C, diagram of tight associations). On the other hand, the supercomplexes that were dissociated into the individual complexes, that is, complexes with lower electrophoretic mobility when solubilized with digitonin than in the presence of DDM, were detected below the diagonal (Fig. 2C, diagram of loose associations). By immunodetection of specific subunits of CI (NDUFB6), complex III2 (Core2), and complex IV (COI), it was possible to distinguish the different complexes included in the supercomplexes in all the studied cell lines (Fig. 2C; see Fig. S3 in the supplemental material).
In control cells, the complexes on the diagonal (with the same mobility in digitonin and DDM) were identified as monomeric complex I (CI), dimeric complex III (CIII2), monomeric complex IV (CIV), and the III2- IV supercomplex (CIII2-IV) that was obtained in the digitonin solubilization and only partially dissociated by DDM (2). Two CI subassemblies that do not interact with any other complex could also be identified on the diagonal. On the other hand, when the signals detected below the diagonal were examined, direct interactions between complexes I, III2, and IV in the supercomplexes were clearly identified, because they were dissociated by DDM in the second dimension. Notice that two kinds of supercomplex III2-IV with different electrophoretic mobilities were found, one on the diagonal (i.e., no interaction with any other complex) and the other, located below the diagonal, resulting from direct interaction between CI, CIII2, and CIV.
In the ND4KO cell line, no NDUFB6-containing subcomplexes were detectable, and neither was the fully assembled free CI or CI-containing supercomplex. In addition, supercomplex III2-IV was highly accumulated, as was also observed in the 1D BN-PAGE (Fig. 2B).
In the ND6dKO cell line, the CI subassemblies already observed in the digitonin 1D PAGE (Fig. 2B) could be clearly identified. Analysis by this two-dimensional electrophoretic technique, using DDM in the second dimension, resolved subassembly “d,” which showed lower electrophoretic mobility than the complete CI in the 1D BN-PAGE and had the same electrophoretic mobility as subassembly “c” in the 2D DDM, which suggests that the two subassemblies could be the same (Fig. 2B and C). One could generally assume that the assembly intermediates with lower electrophoretic mobility in 1D BN-PAGE belong to a more advanced assembly step. Instead, in view of these results, we can reason that subassembly “d” is really subassembly “c” associated with some additional factors. Alternatively, subassembly “d” could represent the dimerization of subassembly “c.”
Very interestingly, this kind of 2D analysis permitted the identification of the components of the two supercomplexes, previously detected by CI-IGA, in the ND6 mutants. The band “SC1” is composed of CI and CIII2, and the band “SC2” is composed of CI, CIII2, and CIV. ND6iKO and the ND5&ND6iKO cell lines show patterns similar to that of ND6dKO, although subassembly “d” is absent. Moreover, the “CI” that formed part of these “supercomplexes” had the same electrophoretic mobility in BN-PAGE (DDM) as the complete CI (see Fig. S3 in the supplemental material). This result suggests that, in the absence of ND6, assembly may be continued to an almost complete CI, although in a very small proportion compared with the wild type.
Nuclear-encoded proteins contained in the complex I subcomplexes.
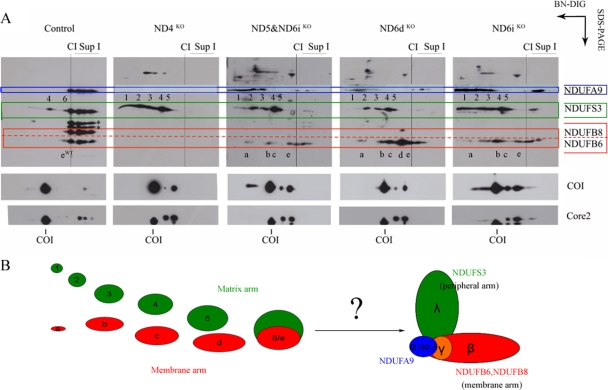
In order to further analyze the compositions of the NDUFB6-containing subassemblies, second-dimension denaturing gel electrophoresis (2D SDS-PAGE) from each CI mutant cell sample and the control cell line (Fig. 3) was carried out. The Western blots were immunodetected using antibodies against four CI subunits, each of them corresponding to a different part of the CI topology (25, 38) (Fig. 3B).
FIG. 3.
Characterization of the peripheral-arm and membrane arm CI subcomplexes. (A) Complex I was immunodetected by Western blot analysis after Blue Native gel electrophoresis in digitonin-solubilized mitochondria (BN-DIG) and 2D denaturing tricine-SDS-PAGE from the control and the mutant cells. Several antibodies against subunits belonging to different parts of the CI topology were used, as indicated. In the control cells, all the subunits were present in fully assembled CI and in the supercomplexes. In the mutant cells, the presence of these nuclear-encoded subunits in several subassemblies was characterized. The membrane arm NDUFB6-containing subcomplexes were named alphabetically (a to e, or eWT in the case of the subcomplex detected in the control cells), and the peripheral-arm NDUFS3-containing subcomplexes were named numerically (numbers 1 to 5) according to reference 37. Antibodies against complex III and complex IV were also used to detect their presence in the analyzed cell lines. (B) Schematic representation of the detected membrane arm (red) and peripheral-arm (green) subassemblies that come together to form the whole CI, in which each topological fraction is represented by a different color.
The NDUFS3 subunit is normally found in 6 different subassemblies (37), Four of them (numbers 1 to 4) were readily detected in all CI mutant cell lines (Fig. 3A), and the last one (number 6) was absent in the mutants and was identified in control cells showing a molecular mass near that of the complete complex. On the other hand, the membrane arm subcomplexes, defined by the NDUFB6 antibody (alphabetical nomenclature), are different from those containing NDUFS3 (peripheral arm), except for the last subassembly prior to the formation of the entire complex in the control cells (subcomplexes 6 and eWT). These results indicated that these proteins belong to different subassemblies, in accordance with the current idea that CI subunits are incorporated into discrete intermediates, which are then combined during assembly in a semisequential manner (38).
In control cells, free complex I and the CI-containing supercomplexes can be easily recognized using the tested antibodies. In the case of the NDUFA9 antibody, two additional signals, corresponding to unknown lower-molecular-mass polypeptides showing the typical pattern of the CI proteins, were observed (asterisks in Fig. 3A). The critical role of ND4 in CI assembly had been previously postulated by immunocapture studies (9, 22). In line with this idea, no signals for the NDUFB8 and NDUFB6 subunits were observed in the ND4KO cell line, but the immunoreactivity against NDUFS3 revealed four subcomplexes (Fig. 3A). This suggests that the membrane arm is not assembled in the absence of ND4 but that the mutation still allows the formation of the peripheral-arm subcomplexes.
After the ND6dKO and ND6iKO mutants were analyzed, the conclusion was that the absence of ND6 does not impair the formation of the peripheral- and hydrophobic-arm subcomplexes and that it is not necessary for the formation of some high-molecular-mass associations (Fig. 3A).
The study of the ND5&ND6iKO cell line revealed that, in the absence of both ND5 and ND6, the formation of subassemblies of the peripheral and hydrophobic arms was still allowed. This result underlines the importance of ND5 for CI stability and/or activity rather than for assembly, as reported previously (6, 21). Essentially the same membrane arm (NDUFB6-containing) and peripheral-arm (NDUFS3-containing) assembly intermediates are found in the ND5&ND6iKO and ND6iKO cell lines, indicating that the entry of ND5 must be successive to ND6 (Fig. 3A).
Metabolic labeling and two-dimensional BN/SDS-PAGE analysis of CI assembly.
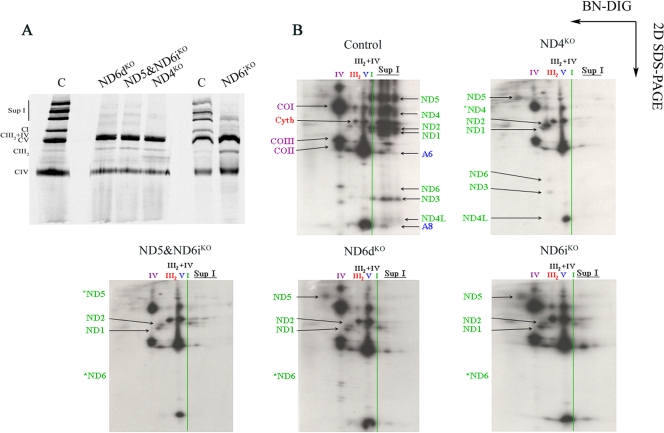
To elucidate which of the ND subunits are present in the detected subassemblies found in the mutants and, consequently, the order in which they are incorporated in the CI assembly process, the mtDNA-encoded subunits were selectively labeled in vivo using [35S]Met-Cys in the presence of cycloheximide, which blocks cytosolic protein synthesis. Then, the drug and the label were removed, and the incorporation of the labeled proteins into the fully assembled complexes and supercomplexes was followed after a 14-h chase, after which the labeled digitonin-solubilized mitochondria were analyzed by BN-PAGE (Fig. 4). In the 1D BN-PAGE (Fig. 4A), the typical pattern corresponding to the isolated CI and the five additional supercomplex bands was found in the control cells. The free CI signal was absent in all the mutant samples, and again, two bands corresponding to the formation of supercomplexes were found in ND5&ND6iKO, ND6dKO, and ND6iKO, but not in the ND4KO cell line (Fig. 4A).
FIG. 4.
Radiolabeling of the mtDNA-encoded subunits in vivo and their incorporation into the OXPHOS complexes and supercomplexes. Digitonin-solubilized samples from the wild-type and mutant cell lines were run through 1D Blue Native gels (A) and 2D denaturing tricine-SDS-PAGE (B) after a 2-h radioactive pulse and a 14-h chase. The signals from the different subunits were identified and localized in the positions corresponding to the fully assembled isolated complexes and associated into the supercomplexes or with the different subcomplexes: green, complex I subunits (NDL-6 and ND4L); red, complex III2 subunit cytochrome b (Cytb); purple, complex IV subunits COI, COII, and COIII (cytochrome c oxidase subunits I, II, and III); blue, complex V subunits A6 and A8 (ATPases 6 and 8) (see the text for detailed explanations).
The specific labeling of the mtDNA-encoded subunits could be resolved trough two-dimensional BN/SDS-PAGE (Fig. 4B). The obtained results indicated that ND1 and ND2 belong to two different subassembly modules, quite similar in molecular mass, which can be easily recognized in all CI mutant cell samples. The ND1-containing subcomplex corresponds to an ∼400-kDa subcomplex, and the one in which ND2 is detected corresponds to an ∼460-kDa subcomplex (25).
In the absence of ND4, the ND2-containing subassembly was readily accumulated, and ND6, ND3, and ND4L migrated to the same position, thus defining them as part of the same subcomplex. This analysis also confirmed the requirement for the ND4 subunit for the progression of the other subunits to higher-molecular-mass subcomplexes and proved the essential role of ND4 for CI assembly.
On the other hand, the absence of ND6 did not impair the progression of the ND2-containing membrane arm subassembly to molecular masses of 800 to 1,000 kDa. Previous reports suggested that ND6 is essential for CI assembly (5). However, we observed that although ND6 could play a crucial role in the maturation of the ND2 subassembly and its interaction with the ND1 subassembly, a residual amount of CI can progress to a nearly fully assembled state that associates into supercomplexes.
By comparing the mutant and control situations, we observed that to incorporate ND5 into CI, ND4 and ND6 need to be previously inserted. In line with these observations, ND5 has to be one of the last membrane arm subunits to be assembled.
Incorporation of the mtDNA-encoded subunits without a supply of nuclear-encoded proteins.
The time course assembly of the mtDNA-encoded subunits into the OXPHOS complexes was also analyzed in by in organello protein synthesis and pulse-chase experiments. Isolated mitochondria are capable of protein synthesis outside the whole cellular context, and the maximum amount of label was reached after 45 min under our incubation conditions (16, 17).
After the 40- to 45-min labeling pulse, mitochondria isolated from mouse liver were washed and further incubated in the presence of chloramphenicol to inhibit mitochondrial translation, and the mitochondria were prepared for BN-PAGE after different chase times. Thus, the progression of the labeled mitochondrial polypeptides was followed up to 3 h. This approach allowed us to study CI assembly without the external supply of nuclear-encoded structural subunits and assembly factors. The intermediates found when both types of solubilization were used, digitonin (Fig. 5A) and DDM (Fig. 5B), were mainly the same. The less labeled subunits were more easily detected when mitochondria were treated with DDM, probably because the detergent extracts more protein from the mitochondrial inner membrane than digitonin.
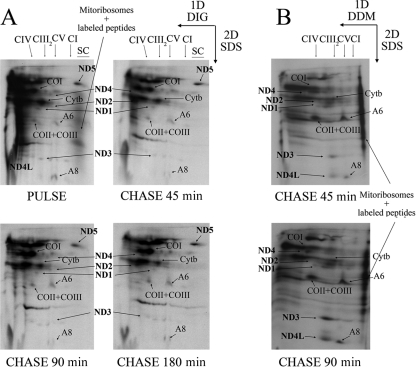
FIG. 5.
Radiolabeling of the mtDNA-encoded subunits in organello and their incorporation into the OXPHOS complexes. Digitonin (A)- and DDM (B)-solubilized mitochondria isolated from mouse liver were run through 1D Blue Native gels and 2D denaturing tricine-SDS-PAGE after a 45-min radioactive pulse and the indicated chase times. Chase was performed in the presence of puromycin in the samples shown in panel A to eliminate the signal corresponding to the labeled peptides associated with the mitoribosomes. Newly synthesized mtDNA-encoded subunits could not be assembled into CI under these conditions; however, the same subcomplexes containing the ND subunits detected in the mutant cell lines (Fig. 4) were also formed in the isolated mitochondria. Moreover, mainly the same subcomplexes were detected independently of the detergent used to solubilize the mitochondrial membranes.
As can be seen in Fig. 5, the ND1 subunit was found in the ∼400-kDa subcomplex and ND2, ND3, and ND4L were incorporated into the ∼460-kDa intermediate right after the 40- to 45-min labeling pulse and remained accumulated mostly in the same subcomplexes during all the analyzed chase times. In addition, most ND4 was accumulated in a different assembly intermediate. The assembly of the whole CI and its association into supercomplexes could not be achieved under these conditions, at least during the almost 4-h-long experiments. As happened in the in vivo labeling, the signal for the ND6 subunit was very faint, and its localization within the subcomplexes was not clear. Under these conditions, labeled ND5 seemed to be quickly incorporated at positions corresponding to the fully assembled CI and supercomplexes, in agreement with the idea that ND5 is one of the last subunits to be incorporated. Also, by using this approach, we observed that ND2 and ND4 could progress into higher-molecular-mass positions, while most of the labeled ND1 remained in the ∼400-kDa intermediate at all times, in a fashion similar to what was observed in the ND6 mutant cell lines (see above). Moreover, the fact that the same subcomplexes are observed by in organello and in vivo labeling proves that they are bona fide assembly intermediates and not degradation products.
DISCUSSION
In the last few years, there has been great progress in the understanding of the very complicated CI assembly process. The study of the CI assembly state in CI-deficient human patients carrying mutations in different ND subunits has given some hints of the importance of each of them for the assembly/stability of the complex. In addition, the most recent assembly models define the possible entry point of the mitochondrially encoded CI subunits. However, the precise stage at which each ND subunit is incorporated and their importance for the correct progression of each step in the complex I assembly process was still not clear.
With this study, we have been able to elucidate the specific entry point of the mtDNA-encoded CI subunits and their presence within definite assembly intermediates (Fig. 6). Thus, the labeling of mtDNA-encoded proteins in four different CI mutant cell lines allowed us to determine the importance of specific subunits for the assembly/stability of the complex, as well as their hierarchy in the assembly process. The presence of the same ND subunit-containing assembly intermediates was further confirmed by the labeling of the mtDNA-encoded polypeptides in isolated mouse liver mitochondria.
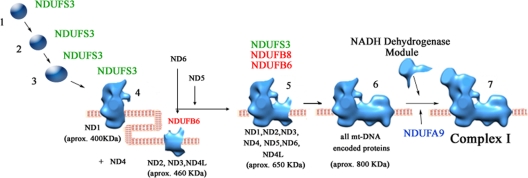
FIG. 6.
Complex I assembly model, including the ND subunit entry points determined in this study. Shown is a complex I assembly model for wild-type cells, based on the refined structure for CI shown by Clason et al. (13). The model unites previous knowledge and the new contributions from the present study.
The subassemblies that contained the ND subunits (Fig. 4 and 6) and NDUFS3 (Fig. 3) in the control and mutant systems were the same that had already been defined by using DDM, confirming their identities as true assembly intermediates and not degradation products or artifacts.
We have identified five entry points of the ND subunits in the CI assembly pathway constructed by several subassembly modules (Fig. 4 and 6). The specific labeling of the mtDNA-encoded subunits showed that ND1 and ND2 belong to two different subcomplexes that progress differently. ND1 joins peripheral-arm subunits, forming an ∼400-kDa intermediate that is anchored to the inner mitochondrial membrane (14, 37) step, in which the chaperones NDUFAF3 and NDUFAF4 are likely to be involved (28). On the other hand, the assembly factor CIA30 (NDUFAF1) would be involved in the initial assembly of the separate ND2-containing ∼460-kDa membrane subcomplex that, once it is mature, would finally join the ND1-containing subassembly to give rise to the complete CI. In accordance with this, the ∼460-kDa subcomplex was absent in CIA30-deficient human cells, but the ∼400-kDa ND1-containing subcomplex was accumulated (15). As can be seen in Fig. 4 and 6, in controls and in all the mutants, as well as in the in organello pulse-chase experiments (Fig. 5), ND2 was detected mainly in the ∼460-kDa subassembly, together with ND3 and ND4L. Therefore, we can define a first entry point for ND1 in the ∼400-kDa intermediate and a second entry point for ND2, ND3, and ND4L in the ∼460-kDa subcomplex (Fig. 6). In the absence of ND4, both ND1- and ND2-containing subassemblies were accumulated, as they could not go on into higher-molecular-mass positions. Thus, this defines a third entry point for ND4, confirming its importance for the progression of the membrane arm into the fully assembled CI. Furthermore, if ND4 is missing, NDUFB6 and NDUFB8 cannot be incorporated into the membrane arm assembly intermediates (Fig. 2 and 3) because the assembly process is impaired at early stages. In the absence of ND4, NDUFS3 is found in assembly intermediates 1 to 5, but not in the fully assembled complex I, supporting the idea of the assembly of the ND1-containing and membrane-anchored peripheral-arm fragment independently from the rest of the membrane arm. On the other hand, the absence of ND6 did not impair the progression of ND2 into higher-molecular-mass (800- to 1,000-kDa) subassemblies. There are two possible explanations for this observation: (i) the ND2 subassemblies are associated with some factors involved in subcomplex maturation or chaperoning or (ii) in the absence of ND6, a nonnatural pathway of CI assembly, in which a membrane arm subassembly is extended without the incorporation of the peripheral arm subunits, occurs. The labeling of the ND6 subunit is always faint, impairing its correct detection, but by analyzing this study's collection of ND6 mutants, it can be stated that ND6 is incorporated into the ∼460-kDa and successive ND2 subcomplexes but is not essential for their formation and that it is necessary for the incorporation of ND5, as the latter is accumulated at low-molecular-mass positions in the ND6dKO and ND6iKO cell lines. The incorporation of ND6 defines the fourth entry point of the mitochondrially encoded subunits and seems to facilitate bringing together the ND1-containing and the ND2-containing subcomplexes. In the ND6 mutants, NDUFB6 was found to be incorporated into the membrane arm subcomplexes (a to e) that are able to assemble up to high-molecular-mass positions. In this fashion, NDUFS3 is accumulated in subcomplexes 4 and 5 in all the mutants, in which it is proposed to interact with ND1 (37), forming subcomplex 4, the ND1-containing ∼400-kDa subcomplex. In our experiments, subcomplex 5 was shown to contain ND1 and NDUFS3 but not any of the other ND subunits (Fig. 3, 4, and 6). Thus, in the ND6 mutants, most of the membrane-anchored peripheral-arm NDUFS3-ND1-containing intermediates cannot get together with the NDUFB6-ND2-containing membrane arm intermediates.
ND5 is incorporated at a later step than ND6, as demonstrated by the fact that the subassemblies that are accumulated in the ND5&ND6iKO cell line are the same as in the ND6iKO mutants and because ND5 is accumulated at low-molecular-mass positions in the ND6 mutants, thus defining the ND subunit fifth entry point.
The pulse-chase assays performed by our group and others in control cell lines (3, 24) revealed that the ND subunits require quite long chase times for their assembly into complex I and supercomplexes. In the control systems, when the mtDNA subunits are labeled in vivo or in organello (Fig. 5) at shorter chase times, we get a picture similar to that in the long chase periods in the mutant cell lines. This indicates that in the mutants, the CI assembly process is stalled, and that the observed subassemblies are true assembly intermediates and not misassembled or partially degraded complexes.
Supplementary Material
Acknowledgments
We are very grateful to Leo G. J. Nijtmans and Rutger O. Vogel for critically reading the manuscript. We thank Santiago Morales for his technical assistance.
Our work was supported by the Spanish Ministry of Science (SAF2009-08007 and CSD2007-00020), the EU (EUMITOCOMBAT-LSHM-CT-2004-503116), a Marie Curie European Reintegration Grant (PERG04-GA-2008-239372), and Group of Excellence grant DGA (B55). E.P.-C. was supported by an FPI fellowship, E.F.-V. by a Juan de la Cierva postdoctoral grant, and M.P.B.-B. by a Ramon y Cajal postdoctoral grant, all three provided by the Spanish MICINN.
Footnotes
Published ahead of print on 12 April 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Acin-Pérez, R., M. P. Bayona-Bafaluy, M. Bueno, C. Machicado, P. Fernandez-Silva, A. Perez-Martos, J. Montoya, M. J. Lopez-Perez, J. Sancho, and J. A. Enriquez. 2003. An intragenic suppressor in the cytochrome c oxidase I gene of mouse mitochondrial DNA. Hum. Mol. Genet. 12:329-339. [DOI] [PubMed] [Google Scholar]
- 2.Acin-Pérez, R., M. P. Bayona-Bafaluy, P. Fernandez-Silva, R. Moreno-Loshuertos, A. Perez-Martos, C. Bruno, C. T. Moraes, and J. A. Enriquez. 2004. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acin-Pérez, R., P. Fernandez-Silva, M. L. Peleato, A. Perez-Martos, and J. A. Enriquez. 2008. Respiratory active mitochondrial supercomplexes. Mol. Cell 32:529-539. [DOI] [PubMed] [Google Scholar]
- 4.Antonicka, H., I. Ogilvie, T. Taivassalo, R. P. Anitori, R. G. Haller, J. Vissing, N. G. Kennaway, and E. A. Shoubridge. 2003. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 278:43081-43088. [DOI] [PubMed] [Google Scholar]
- 5.Bai, Y., and G. Attardi. 1998. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 17:4848-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai, Y., R. M. Shakeley, and G. Attardi. 2000. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell. Biol. 20:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayona-Bafaluy, M. P., R. Acin-Perez, J. C. Mullikin, J. S. Park, R. Moreno-Loshuertos, P. Hu, A. Perez-Martos, P. Fernandez-Silva, Y. Bai, and J. A. Enriquez. 2003. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 31:5349-5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayona-Bafaluy, M. P., N. Movilla, A. Perez-Martos, P. Fernandez-Silva, and J. A. Enriquez. 2008. Functional genetic analysis of the mammalian mitochondrial DNA encoded peptides: a mutagenesis approach. Methods Mol. Biol. 457:379-390. [DOI] [PubMed] [Google Scholar]
- 9.Bourges, I., C. Ramus, B. Mousson de Camaret, R. Beugnot, C. Remacle, P. Cardol, G. Hofhaus, and J. P. Issartel. 2004. Structural organization of mitochondrial human complex I: role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem. J. 383:491-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll, J., I. M. Fearnley, R. J. Shannon, J. Hirst, and J. E. Walker. 2003. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell Proteomics 2:117-126. [DOI] [PubMed] [Google Scholar]
- 11.Carroll, J., I. M. Fearnley, J. M. Skehel, R. J. Shannon, J. Hirst, and J. E. Walker. 2006. Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 281:32724-32727. [DOI] [PubMed] [Google Scholar]
- 12.Chomyn, A. 1996. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 264:197-211. [DOI] [PubMed] [Google Scholar]
- 13.Clason, T., T. Ruiz, H. Schagger, G. Peng, V. Zickermann, U. Brandt, H. Michel, and M. Radermacher. 2010. The structure of eukaryotic and prokaryotic complex I. J. Struct. Biol. 169:81-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieteren, C. E., P. H. Willems, R. O. Vogel, H. G. Swarts, J. Fransen, R. Roepman, G. Crienen, J. A. Smeitink, L. G. Nijtmans, and W. J. Koopman. 2008. Subunits of mitochondrial complex I exist as part of matrix- and membrane-associated subcomplexes in living cells. J. Biol.Chem. 283:34753-34761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunning, C. J., M. McKenzie, C. Sugiana, M. Lazarou, J. Silke, A. Connelly, J. M. Fletcher, D. M. Kirby, D. R. Thorburn, and M. T. Ryan. 2007. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 26:3227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Silva, P., R. Acin-Perez, E. Fernandez-Vizarra, A. Perez-Martos, and J. A. Enriquez. 2007. In vivo and in organello analyses of mitochondrial translation. Methods Cell Biol. 80:571-588. [DOI] [PubMed] [Google Scholar]
- 17.Fernández-Vizarra, E., J. A. Enriquez, A. Perez-Martos, J. Montoya, and P. Fernandez-Silva. 2008. Mitochondrial gene expression is regulated at multiple levels and differentially in the heart and liver by thyroid hormones. Curr. Genet. 54:13-22. [DOI] [PubMed] [Google Scholar]
- 18.Fernández-Vizarra, E., G. Ferrin, A. Perez-Martos, P. Fernandez-Silva, M. Zeviani, and J. A. Enriquez. 2010. Isolation of mitochondria for biogenetical studies: an update. Mitochondrion 10:253-262. [DOI] [PubMed] [Google Scholar]
- 19.Fernández-Vizarra, E., V. Tiranti, and M. Zeviani. 2009. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta 1793:200-211. [DOI] [PubMed] [Google Scholar]
- 20.Gallardo, M. E., R. Moreno-Loshuertos, C. Lopez, M. Casqueiro, J. Silva, F. Bonilla, S. Rodriguez de Cordoba, and J. A. Enriquez. 2006. m.6267G>A: a recurrent mutation in the human mitochondrial DNA that reduces cytochrome c oxidase activity and is associated with tumors. Hum. Mutat. 27:575-582. [DOI] [PubMed] [Google Scholar]
- 21.Hofhaus, G., and G. Attardi. 1995. Efficient selection and characterization of mutants of a human cell line which are defective in mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase. Mol. Cell. Biol. 15:964-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofhaus, G., and G. Attardi. 1993. Lack of assembly of mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase and loss of enzyme activity in a human cell mutant lacking the mitochondrial ND4 gene product. EMBO J. 12:3043-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofhaus, G., R. M. Shakeley, and G. Attardi. 1996. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 264:476-483. [DOI] [PubMed] [Google Scholar]
- 24.Lazarou, M., M. McKenzie, A. Ohtake, D. R. Thorburn, and M. T. Ryan. 2007. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol. 27:4228-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazarou, M., D. R. Thorburn, M. T. Ryan, and M. McKenzie. 2009. Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta 1793:78-88. [DOI] [PubMed] [Google Scholar]
- 26.Moreno-Loshuertos, R., R. Acin-Perez, P. Fernandez-Silva, N. Movilla, A. Perez-Martos, S. Rodriguez de Cordoba, M. E. Gallardo, and J. A. Enriquez. 2006. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 38:1261-1268. [DOI] [PubMed] [Google Scholar]
- 27.Nübel, E., I. Wittig, S. Kerscher, U. Brandt, and H. Schagger. 2009. Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics 9:2408-2418. [DOI] [PubMed] [Google Scholar]
- 28.Saada, A., R. O. Vogel, S. J. Hoefs, M. A. van den Brand, H. J. Wessels, P. H. Willems, H. Venselaar, A. Shaag, F. Barghuti, O. Reish, M. Shohat, M. A. Huynen, J. A. Smeitink, L. P. van den Heuvel, and L. G. Nijtmans. 2009. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am. J. Hum. Genet. 84:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schägger, H. 1995. Native electrophoresis for isolation of mitochondrial oxidative phosphorylation protein complexes. Methods Enzymol. 260:190-202. [DOI] [PubMed] [Google Scholar]
- 30.Schägger, H., and K. Pfeiffer. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smeitink, J., R. Sengers, F. Trijbels, and L. van den Heuvel. 2001. Human NADH:ubiquinone oxidoreductase. J. Bioenerg. Biomembr. 33:259-266. [DOI] [PubMed] [Google Scholar]
- 32.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 33.Sugiana, C., D. J. Pagliarini, M. McKenzie, D. M. Kirby, R. Salemi, K. K. Abu-Amero, H. H. Dahl, W. M. Hutchison, K. A. Vascotto, S. M. Smith, R. F. Newbold, J. Christodoulou, S. Calvo, V. K. Mootha, M. T. Ryan, and D. R. Thorburn. 2008. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am. J. Hum. Genet. 83:468-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiranti, V., K. Hoertnagel, R. Carrozzo, C. Galimberti, M. Munaro, M. Granatiero, L. Zelante, P. Gasparini, R. Marzella, M. Rocchi, M. P. Bayona-Bafaluy, J. A. Enriquez, G. Uziel, E. Bertini, C. Dionisi-Vici, B. Franco, T. Meitinger, and M. Zeviani. 1998. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 63:1609-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ugalde, C., R. J. Janssen, L. P. van den Heuvel, J. A. Smeitink, and L. G. Nijtmans. 2004. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum. Mol. Genet. 13:659-667. [DOI] [PubMed] [Google Scholar]
- 36.Ugalde, C., R. Vogel, R. Huijbens, B. Van Den Heuvel, J. Smeitink, and L. Nijtmans. 2004. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum. Mol. Genet. 13:2461-2472. [DOI] [PubMed] [Google Scholar]
- 37.Vogel, R. O., C. E. Dieteren, L. P. van den Heuvel, P. H. Willems, J. A. Smeitink, W. J. Koopman, and L. G. Nijtmans. 2007. Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J. Biol. Chem. 282:7582-7590. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, R. O., J. A. Smeitink, and L. G. Nijtmans. 2007. Human mitochondrial complex I assembly: a dynamic and versatile process. Biochim. Biophys. Acta 1767:1215-1227. [DOI] [PubMed] [Google Scholar]
- 39.Walker, J. E. 1992. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25:253-324. [DOI] [PubMed] [Google Scholar]
- 40.Wikström, M. 1984. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 169:300-304. [DOI] [PubMed] [Google Scholar]
- 41.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue Native PAGE. Nat. Protoc. 1:418-428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.