Abstract
Histone lysine methylation and CpG DNA methylation contribute to transcriptional regulation. We have shown previously that dimethylated and trimethylated forms of histone H3 at lysine 4 (H3K4me2 and H3K4me3) are primarily depleted from CpG-methylated DNA regions by using patch-methylated stable episomes (minichromosomes) in human cells. This effect on H3K4me2 is clearly not linked to the transcriptional activity in the methylated DNA region; however, transcriptional activity may play a role in the presence of H3K4me3. Here, we present clear evidence of the impact of transcriptional activity on the overall level of H3K4me3 in the coding region and the lack of impact on H3K4me2. Our data also demonstrate the influence of transcriptional activity on the distribution of H3K4me3 and H3K4me2, but not that of total H3, in the 5′ end of the coding region relative to the 3′ end. The nature of the promoter (viral or endogenous) affects H3K4me3 much more than it affects H3K4me2, suggesting a potential fundamental difference in the recruitment of methyltransferase for H3K4 trimethylation.
Development of microarray-based analysis of DNA from chromatin immunoprecipitation (ChIP-on-chip assay) allows the efficient correlation of histone modifications, DNA sequences, and gene expression. By utilizing these assays, H3K4me2 and H3K4me3 have been found to be enriched around the transcriptional start site of active promoters, where TFIID and RNA polymerase are also present (3, 6, 9, 20, 21), as well as in the coding sequences in human cells (16). H3K4me2 and H3K4me3 are found at 46% and 66%, respectively, of the start sites of genes on human chromosomes 21 and 22 (3). H3K4me3 are found at more than 90% of the RNA polymerase II (Pol II) binding regions (2). It has been shown that the loss of H3K4me3 decreases transcriptional activity and reduces TFIID at some promoters without the canonical TATA box in human cells, leading to the hypothesis that H3K4me3 may define the core promoter by either anchoring TFIID to the activated promoter or recruiting TFIID during promoter activation (27). It has also been proposed that H3K4me3 facilitates the efficiency of postinitiation processes during active transcription (25). There are multiple known lysine methyltransferases (KMTs) and demethylases (KDMs) involving the modification of specific lysine residues (for a summary, see reference 1). While H3K4me2 and H3K4me3 are generally associated with active transcription, it is possible that different KMTs and KDMs are recruited to different sites in the genome based on the nature of the promoter, on the level of activity, and on other local histone modifications.
In one study, the levels of both H3K4me2 and H3K4me3 were found to be 2- to 3-fold lower in the coding region of a transgene with CpG methylation, and the level of methylated H3K9 was found to be higher. In that study, depletion of RNA Pol II was also observed (15). However, a lack of correlation between gene expression, histone modification, and DNA methylation has been described for some imprinted genes (29). Although it is clear that DNA methylation and histone modification are two important factors in gene regulation, the causal role of these factors has been difficult to infer from studies that correlate distribution of the histones, transcription activity, DNA sequences, and DNA methylation status. The stable replicating episomal (minichromosomal) system closely resembles endogenous events for transcription and chromatin, based on our previous studies and those of others (5, 13, 18, 19). With increasing numbers of antibodies (Abs) available for various histone modifications, we are in a unique position to determine the mechanistic relationships between histone modifications, DNA methylation, and transcription by using the stable episomal system. Our recent study using patch-methylated episomes in human cells clearly demonstrates that DNA methylation dictates the presence of H3K4me2, regardless of transcriptional activity, and the level of H3K4me3 may be influenced by transcription (18). In the current study, we further explored whether and how transcriptional activity influences the presence and distribution of H3K4me2 and H3K4me3 in a coding region.
MATERIALS AND METHODS
Plasmids.
pCLH22, which contains the Rous sarcoma virus (RSV) long terminal repeat (LTR) promoter driving the luciferase reporter gene, has been described previously (7). pCLH29 is identical to pCLH22 except that the RSV LTR promoter is replaced by the human cytomegalovirus (hCMV) promoter (Fig. 1A). pAWLuc is generated by deleting the EBNA1 gene from pCLH22 (Fig. 1A). Plasmids pEF1A-Luc and pGSTP-Luc are similar to pCLH22 except that the RSVLTR promoter is replaced by the endogenous promoter of the EF1A and GSTP1 genes, respectively, from the human genome. Plasmids pEB-Luc and pCLH42 are also similar to pCLH22 except for addition of a 1.2-kb DNA fragment containing the promoter from the human EDNRB gene and a smaller 757-bp promoter fragment from the same gene, respectively. pOLucOriP, pOLucRLTR, and pOLucΔLTR have been described previously (14). pCLH22γI was generated by inserting the 866-bp intron 2 of the γ-globin gene from pJM101 (17), between the RSV LTR promoter and the luciferase gene coding region on pCLH22.
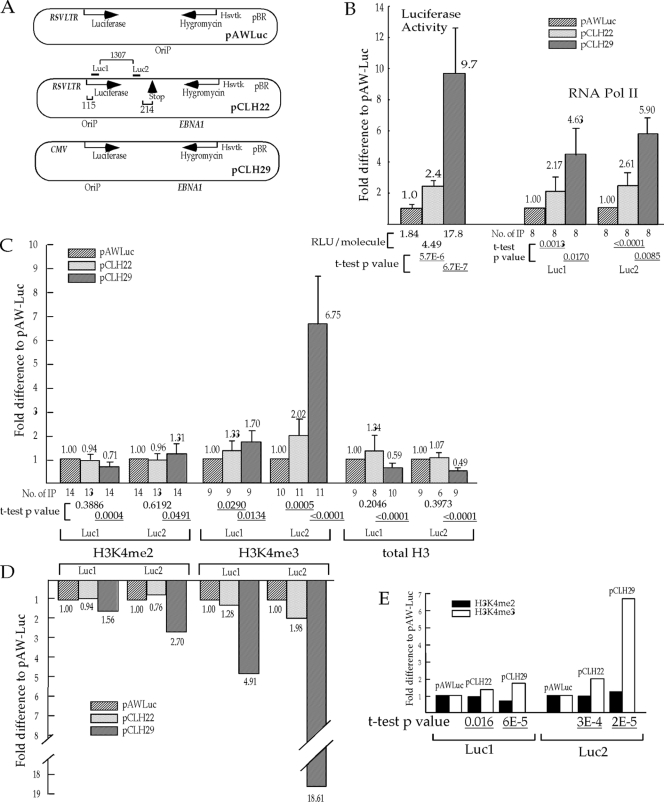
FIG. 1.
Transcriptional activity influences the level of H3K4me3 much more than the level of H3K4me2 in the coding region. (A) Illustration of the episomes. The two quantitative PCR (Q-PCR) amplicons, Luc1 and Luc2, are marked by bars above the diagram of pCLH22, and the distances (bp) from the transcription start site and the stop codon and between the two amplicons are indicated with brackets and numbers. (B) Relative transcriptional activity as assessed by the luciferase activity and the level of RNA Pol II on the episomes. The luciferase expression of each episome was normalized for the amount of episomal DNA harvested from transfected cells by Q-PCR analysis. Bars represent average relative luciferase expression of each episome from four different experiments with two duplicates (RLU/molecule listed under each relevant bar is calculated from luciferase assays on 6.6 × 106 to 5.7 × 107 molecules of episome in the cell sample) divided by the luciferase expression of pAWLuc. The relative levels of RNA Pol II in the Luc1 and the Luc2 regions of the episomes are shown as the fold difference from the RNA Pol II level of pAWLuc by normalizing the average %IP (calculated as described in Materials and Methods) of the region from each episome against the corresponding value from pAWLuc. (C) The relative levels of H3K4me2, H3K4me3, and total H3 in the Luc1 and the Luc2 regions of the episomes. The relative pull-down of each region was derived as described above for RNA Pol II as a ratio with the corresponding value for pAWLuc. For panels B and C, each bar, with accompanying values, represents the average relative pull-down from a range of 6 to 14 independent IP experiments as listed under each bar. The fold difference from the expression level of pAWLuc is displayed above each bar, and standard deviations are indicated by the error bars. A t test was carried out to test the hypothesis that no significant differences between pCLH29 and pAWLuc and between pCLH22 and pAWLuc could be detected by the assay. The two-tailed P values of the t test are listed under each bar, with the statistically significant P values of <0.05 underlined. (D) H3K4me2 and H3K4me3 levels normalized against the total H3 level. The histogram represents the relative level of each histone modification derived by dividing the average H3K4me2 or H3K4me3 level by the average total H3 level in each of the regions examined on each episome and then normalizing it against the pAWLuc value. Only IP experiments with total H3, H3K4me2, and H3K4me3 are included. The fold differences relative to the expression level of pAWLuc are displayed below each bar. (E) The change in the H3K4me3 level is significantly different from the change in the H3K4me2 level. The histogram shows the same parameters as the corresponding illustration in panel C, but these are grouped by episome instead of histone modification. A t test was carried out to test the hypothesis that there is no difference in the change of the H3K4me2 level and the change of the H3K4me3 level on pCLH22 or on pCLH29. The two-tailed P values of the t test are listed under each bar, with the statistically significant P values of <0.05 underlined. Hsvtk represents the thymidine kinase promoter from herpes simplex virus.
Cell lines, transfection, and IPTG treatment of cells.
The calcium phosphate transfection method was used to transfect 293/EBNA1 (7) and 293lacI#18 (14) cells. All transfections were done at least in duplicate in each experiment, and the transfected cells were harvested for analyses at least twice in nearly all experiments to ensure that no changes occurred over time. In some experiments, IPTG (isopropyl-β-d-thiogalactopyranoside) was used to inhibit lacI binding to induce transcription of the luciferase gene on the episome. The cells were divided equally into two 100-mm plates when they become confluent in the 35-mm plate after transfection, and one plate was treated with 10 μM IPTG, and the other one was not treated (control).
Cell harvest and analyses.
As described previously, 1.25% of the cells were harvested for the luciferase assay, 2.5% of the cells were replated on a 100-mm-diameter tissue culture plate, and the remaining cells were harvested for a chromatin immunoprecipitation (ChIP) assay each time when transfected cells reached confluence. DNA quantitation was determined by real-time quantitative PCR (Q-PCR) using multiple TaqMan probe and primer sets for different regions of the episome. Luciferase activities were determined using a Monolight 2020 luminometer (Analytical Luminescence) and normalized by the quantity of plasmid DNA recovered from the same harvest as determined by Q-PCR. This ensures that the luciferase expressions from the same quantities of plasmids from different transfections were being fairly compared. ChIP assays were performed as described previously (18). After the cell suspension was sonicated and centrifuged, a 100-μl aliquot of soluble chromatin was used for each immunoprecipitation (IP) by using anti-dimethyl-histone H3 (lys4) (Upstate Biochemical), and a 300-μl aliquot of soluble chromatin was used for each IP by using the anti-trimethyl-histone H3 (lys4) (Upstate Biochemical), anti-histone H3 (against C terminus; AbCam and Active Motif), or anti-RNA Pol II (clone 8WG16; Covance). A 100-μl aliquot of soluble chromatin was reserved as the total chromatin fraction (TCF) without IP steps for quantitation. For each immunoprecipitation, 1 μg antibody was mixed with the soluble chromatin, 6 μg sheared salmon sperm DNA, and 20 μl protein G-Sepharose slurry (in radioimmunoprecipitation assay [RIPA] buffer) with rotary mixing at 4°C overnight. A 100-μl aliquot of soluble chromatin was processed without antibodies in parallel as a negative control (no Ab) for the immunoprecipitation experiments. The Sepharose beads were collected by centrifugation and washed, immunocomplexes were eluted, the formaldehyde cross-links were reversed, and the DNA was phenol-chloroform extracted and ethanol precipitated after proteinase K treatment as described previously (18). The TCF aliquot was processed through the same steps from the reverse cross-linking step as for the other immunoprecipitation samples. The final DNA pellet was dissolved in 50 μl Tris-EDTA (TE; pH 8.0) for Q-PCR analysis. Multiple independent immunoprecipitations for anti-H3K4me2, anti-H3K4me3, and anti-RNA Pol II were carried out for each experiment as indicated in the figure legends.
Q-PCR and statistical analysis.
Q-PCR was performed with a Bio-Rad iCycler using iQ Supermix (Bio-Rad) and 1 μl of each DNA sample. Fluorescently labeled TaqMan probes for six regions of plasmid pCLH22 were described previously (8, 18). All Q-PCRs were carried out using the same two-step program: 40 cycles of 95°C for 15 s and 60°C for 1 min. Within each 96-well plate of Q-PCR volumes, titrations of known amounts of pCLH22 DNA were included as positive controls and for quantitation. DNA from the TCF and ChIP sample immunoprecipitated without antibodies (no Ab) were included in each 96-well plate of Q-PCRs on ChIP samples immunoprecipitated with antibodies. All Q-PCRs were done in duplicate. The fraction of immunoprecipitated DNA (%IP) was calculated by dividing the amount of DNA from sample immunoprecipitated with antibodies (after the background amplification from the no-Ab control was subtracted) by the amount of DNA in the corresponding TCF sample. A two-sample t test assuming unequal variances was carried out to test the hypothesis that the levels of the modified histone or RNA Pol II of each of the episomes tested in the experiment and the episome with the lowest transcription level are not significantly different. The two-tailed P values of the t test are listed under each histogram in the figures.
RESULTS
Transcriptional activity has different impacts on the presence of H3K4me2 and the presence of H3K4me3 in the coding region.
In a previous study, we observed a tight correlation between CpG-methylated DNA and the absence of H3K4me2, and this correlation does not appear to be mediated through transcriptional activity in the region. In contrast, the transcriptional activity does influence the presence of H3K4me3 (18). Here, we used three stable episomes with a 10-fold range of luciferase reporter gene expression to investigate specifically whether transcriptional activity impacts the presence of H3K4me2 and the presence of H3K4me3 differently (Fig. 1A and B). These stable episomes were transfected into 293/EBNA1 cells individually, and the transfected cells were harvested serially over several weeks for luciferase expression measurement and ChIP analysis. Four independent transfections were carried out for each episome. In our study, ChIP assays for these experiments were followed by Q-PCR using two different probe and primer sets, for Luc1 and Luc2, which are located at the 5′ and 3′ ends of the coding region, respectively (Fig. 1A). The ChIP analysis using anti-RNA Pol II showed RNA Pol II levels in the Luc1 and Luc2 regions of pCLH22 and pCLH29, respectively, that were significantly higher than these levels for pAWLuc (Fig. 1B). These findings are consistent with the results of the luciferase assay. This demonstrates that the luciferase activity measurements closely correlate with the RNA Pol II level, which directly reflects transcriptional activity. The ChIP analysis using anti-H3K4me2 showed a <1.5-fold difference in the percent pull-down in the luciferase coding region for the three episomes, with pCLH29 being significantly different from pAWLuc (Fig. 1C). This finding further supports our inference from a previous study that the level of H3K4me2 is not markedly influenced by transcriptional activity in a consistent manner (18).
In contrast, a significantly increased H3K4me3 level in the luciferase coding region was detected on both pCLH22 and pCLH29 relative to the value for pAWLuc (Fig. 1C). Interestingly, the observed increase in the H3K4me3 level in the Luc2 region was larger than that in the Luc1 region on both pCLH22 and pCLH29 (Fig. 1C). The trend of these H3K4me3 increases is similar to that of RNA Pol II in both the Luc1 and Luc2 regions. These findings suggest that the level of H3K4me3 in the coding region may be influenced by the transcriptional activity, as reflected by the increase in luciferase expression and the increased level of RNA Pol II. The fact that the increased level of H3K4me3 is more prominent in the 3′ end of the coding region (1.8 kb downstream from the transcriptional start site) for both pCLH22 and pCLH29 strongly suggests that this is not a collateral effect of the increase in this modification at the proximal promoter region due to the strength of the promoter.
It is possible that the observed differences in the levels of H3K4me3 are a reflection of the change in total H3 in the coding region due to the transcriptional activity. To test this, we carried out ChIP analysis by using antibodies against the C terminus of histone H3 to determine the levels of H3 in the luciferase coding region of these three episomes. We found that the total level of H3 is approximately 2-fold and significantly lower on pCLH29 than on pCLH22 or pAWLuc at both the Luc1 and Luc2 regions (Fig. 1C), indicating the possibility that the level of total H3 appears to change only when the transcriptional activity is extremely robust. The increase in H3K4me2 and H3K4me3 on pCLH29 compared with pAWLuc and pCLH22 is much more dramatic after normalization against the total H3 level (compare Fig. 1C and D).
To evaluate whether transcriptional activity has different impacts on the levels of H3K4me3 and H3K4me2 in the coding region, a t test was carried out to test the hypothesis that the change in the H3K4me2 level is the same as the change in the H3K4me3 level for each of the episomes, pCLH22 and pCLH29, from those levels on pAWLuc. It is clear, as shown in Fig. 1E, that the changes in the H3K4me3 levels were significantly different than the changes in the H3K4me2 levels in both Luc1 and Luc2 regions on pCLH22 (P values of 0.016 and 0.0003, respectively) as well as pCLH29 (P values of 0.00006 and 0.00002, respectively). Also, the levels of RNA Pol II correlate with the levels of H3K4me3 better than those of H3K4me2 (compare Fig. 1B and E). Taken together, these findings indicate that transcriptional activity through the coding region impacts the level of H3K4me3 differently and much more dramatically than it impacts H3K4me2 on these episomes.
Transcriptional activity from endogenous promoters impacts the level of H3K4me2 less than it impacts that of H3K4me3 in the coding region of the reporter gene.
The RSV LTR and hCMV promoters used for the experiments described above are viral promoters with very high transcriptional activity in 293/EBNA1 cells. We wondered whether endogenous human promoters covering a lower range of transcriptional activity would have the same impact on the presence of H3K4me2 and H3K4me3 in the coding region. Four stable episomes were generated by placing the promoters from the EDNRB (two versions), GSTP1, or EF1A gene immediately upstream of the luciferase reporter gene (Fig. 2A). After transfecting each episome individually into 293/EBNA1 cells, serial luciferase assays and ChIP experiments were carried out as described above over several weeks. Again, four independent transfections for each episome were done. The luciferase assays indicate that pCLH42, which harbors the 757-bp EDNRB promoter, and pEB-Luc, which has the 1.2-kb EDNRB promoter, have similarly low luciferase gene activities (Fig. 2B). The episomes harboring the GSTP1 promoter and the EF1A promoter have >2-fold and >400-fold higher luciferase activities than pCLH42, respectively (Fig. 2B). The percentages of RNA Pol II pull-down for pCLH42 and pEB-Luc were very low; therefore, the levels of RNA Pol II on the four episomes cannot be reliably compared. The levels of RNA Pol II in both the Luc1 and Luc2 regions of pEF1A-Luc are significantly higher than those of pGSTP-Luc (Fig. 2B).
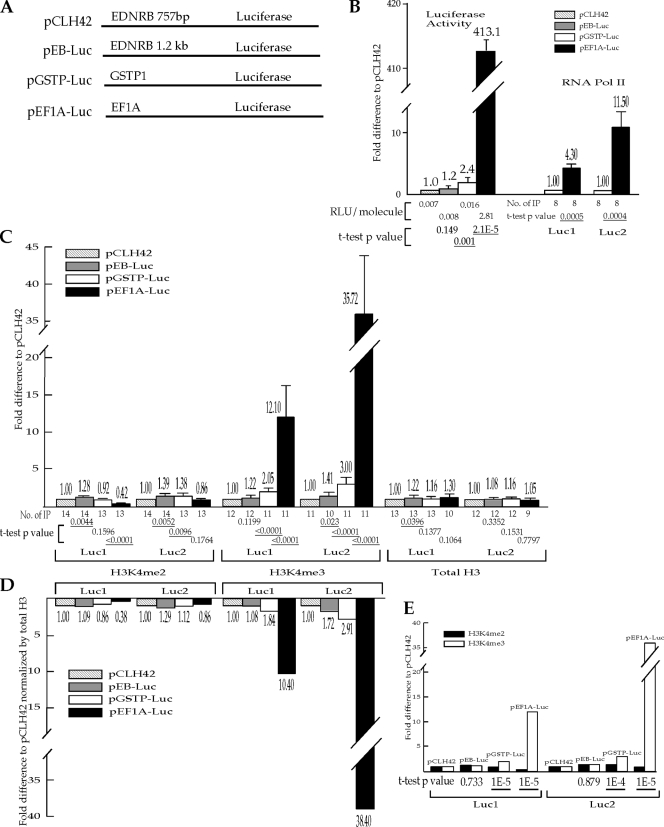
FIG. 2.
Transcriptional activity from endogenous promoters on the episomes influences the level of H3K4me3 much more than it influences the level of H3K4me2 in the coding region. (A) Illustration of the promoters on the episomes. These four episomes are identical to pCLH22 except for the promoters used upstream of the luciferase reporter gene as indicated. The two Q-PCR amplicons, Luc1 and Luc2, are the same as indicated in Fig. 1A. (B) Relative transcriptional activity as assessed by the luciferase activity and the level of RNA Pol II on the episomes. The relative reporter gene activity of each episome is calculated as described above but normalized against the corresponding values for pCLH42. The RLU/molecule listed under each relevant bar is calculated from luciferase activity measurements from 5 × 107 to 1.2 × 108 molecules of episome in the cell sample. The percentage of RNA Pol II pulldown was unreliably low for pCLH42 and pEB-Luc; therefore, the relative pulldown of RNA Pol II was derived by normalizing the average %IP of the region from pEF1A-Luc against the corresponding value from pGSTP-Luc. (C) The relative pulldowns of H3K4me2, H3K4me3, and total H3 in the Luc1 and Luc2 regions of the episomes. The relative pulldown of each region is shown as the fold difference from the value for pCLH42. For panels B and C, each bar, with the accompanying value, represents the average relative pulldown from a range of 8 to 14 independent IP experiments as listed under each bar. The fold difference from the value for pCLH42 is displayed on the top of each bar, and the standard deviation is indicated by an error bar. A t test was carried out to test the hypothesis that no significant difference between each episome and pCLH42 could be detected by the specific assay (only differences between pEF1A-Luc and pGSTP-Luc were tested for RNA Pol II pulldown). The two-tailed P values of the t test are listed under each bar, with the statistically significant P values of <0.05 underlined. (D) The average H3K4me2 and H3K4me3 levels normalized against the total H3 level. The histogram represents the level of each histone modification on each episome relative to that of pCLH42 after being normalized for the level of total H3 as described in the legend to Fig. 1. (E) The change in the H3K4me3 level is significantly different from the change in the H3K4me2 level. The histogram shows the same measurements as the corresponding illustration in panel C, but these are grouped by episome instead of histone modification. A t test was carried out to test the hypothesis that there is no difference in the change in H3K4me2 level and the change in the H3K4me3 level on pEF1A-Luc, pGSTP-Luc, or pEB-Luc. The two-tailed P values of the t test are listed under each bar, with the statistically significant P values of <0.05 underlined. The change in transcription on pEB-Luc compared with the change in transcription on pCLH42 as measured by luciferase activity was insignificant; therefore, the lack of a significantly different change in the H3K4me3 level relative to the change in the H3K4me2 level is expected.
The ChIP assay using anti-H3K4me2 antibody showed <2.4-fold differences in the levels of this histone modification at the Luc1 and Luc2 regions among all four episomes (Fig. 2C). The absolute percentages of pulldown of H3K4me2 at Luc1 and Luc2 on these four episomes are at levels similar to those of the episomes with viral promoters described above. The ChIP assay showed 12.1-fold and 35.7-fold increases in H3K4me3 in the Luc1 and Luc2 regions, respectively, on pEF1A-Luc compared with pCLH42 (Fig. 2C). A smaller increase in H3K4me3 was also detected in both of these regions on pGSTP-Luc compared with pCLH42 (Fig. 2C).
We also carried out ChIP analysis to determine the total H3 level in the luciferase coding region of these four episomes. The total H3 levels were within a 1.3-fold range in both the Luc1 and Luc2 regions for all four episomes, indicating that level of total H3 does not change significantly by the transcriptional activity on these episomes (Fig. 2C). Therefore, the difference in H3K4me2 and H3K4me3 on these episomes after normalization for the H3 level remained similar to the difference without the normalization (compare Fig. 2C and D).
Again, we used a t test to evaluate whether transcriptional activity has a differential impact on the levels of H3K4me3 and H3K4me2 in the coding regions of these four episomes. Excluding pEB-Luc, which has only a 1.2-fold increase in luciferase activity relative to pCLH42, the hypothesis that the change in the H3K4me2 level is the same as the change in the H3K4me3 level for each of the episomes relative to the levels on pCLH42 was clearly rejected (Fig. 2E). The change in the H3K4me3 level was significantly different from the change in the H3K4me2 level in both Luc1 and Luc2 regions on pGSTP-Luc (P values of 0.00001 and 0.0001, respectively) as well as pEF1A-Luc (P values of 0.00001 for both regions). These findings are consistent with the inference that transcriptional activity dictates the level of H3K4me3 in the coding region but has less influence on the level of H3K4me2 in the coding region. It also appears that the effects of transcriptional activity on H3K4me3 in the coding region are similar for viral promoters and endogenous promoters.
Transcriptional activity does not change the distribution of total H3, but it influences the distributions of H3K4me2 and H3K4me3 along the coding region of the reporter gene.
The transcriptional activity seems to have similar impacts on the levels of H3 modification on the episomes with viral promoters and those with endogenous promoters. We decided to assess whether transcriptional activity influences the presence of total H3, H3K4me2, and H3K4me3 in the coding region on the episomes with viral promoters and those with endogenous promoters in the same manner by using pCLH42 as the baseline for normalization to compare all seven episomes. The dramatic 2,000-fold range of reporter expression from these seven episomes was not caused by a small denominator, because the absolute luciferase readings were approximately 1.4 × 105 relative light units (RLU) and 8.0 × 107 RLU from 4 × 104 transfected cells harboring pCLH42 and pCLH29, respectively, before being normalized for episomal DNA in the cells. The viral promoters are much stronger than the endogenous promoters studied, and the EF1α promoter is the only endogenous promoter that is as strong as the viral promoters in 293/EBNA1 cells (Fig. 3A). The levels of RNA Pol II in the coding region of the reporter gene correlate well with the transcriptional activity of the promoter measured by the luciferase assay (compare the left and right sides of the vertical line in Fig. 3A). The level of RNA Pol II on the 5′ end (Luc1 region) is higher than that on the 3′ end (Luc2 region) of the coding region for all five episomes with reliable measurements of RNA Pol II in the coding region (compare the filled and unfilled bars on the right side of Fig. 3A).
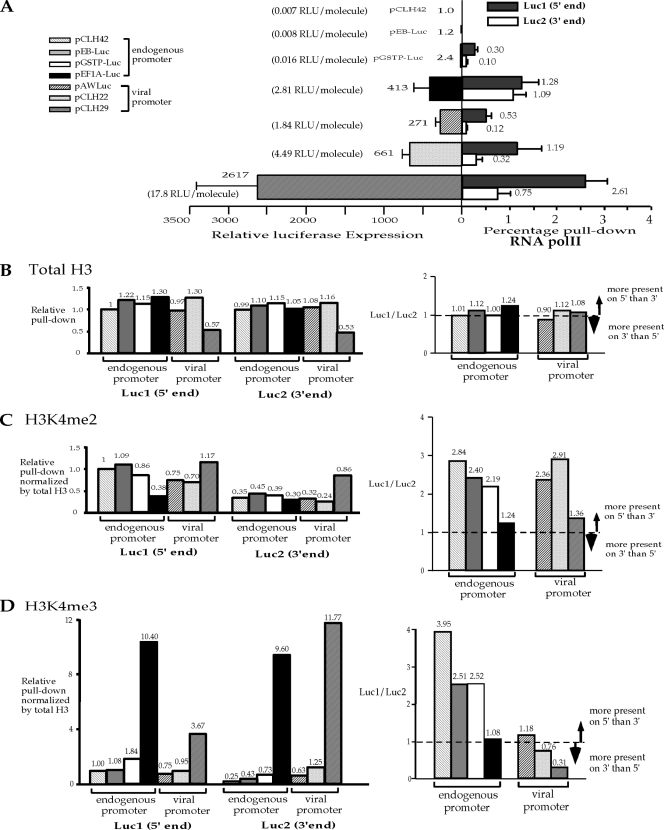
FIG. 3.
Transcriptional activity affects the distributions of H3K4me2 and H3K4me3 in the coding region. (A) Summary of relative activities of the luciferase reporter gene on all seven episomes and the RNA Pol II pulldowns from five of the seven episomes shown in Fig. 1 and 2. Bars on the left side represent average relative luciferase gene activities after normalization against the pCLH42 value. Bars on the right side represent average percent pulldowns of RNA Pol II in each of the two regions, Luc1 and Luc2. Standard deviations are indicated by the error bars. (B) Transcriptional activity does not have a dramatic impact on total H3 levels or distribution in the reporter coding region. (C) Transcriptional activity changes the distribution of H3K4me2 in the reporter coding region. (D) Transcriptional activity and the promoter influence the distribution of H3K4me3 in the reporter coding region. For panels B, C, and D, the histograms on the left side represent the specific histone levels in two regions (Luc1 and Luc2) of the reporter coding region from each of the seven episomes normalized by the corresponding value in the Luc1 region of pCLH42. The histograms on the right side represent the ratios of specific histone at the 5′ end to specific histone at the 3′ end of the reporter coding region from each of the seven episomes to reflect the distributions of the specific histone at the two ends of the coding region.
The levels of total H3 do not vary much for all the episomes, with the exception of pCLH29, which has approximately 2-fold less H3 in both Luc1 and Luc2 regions compared with the others (Fig. 3B, left side). It is possible that the appreciable change in total H3 level in the coding region occurs only when transcriptional activity is extremely high. This strongly suggests that the total H3 level in the coding region is not influenced by transcription activity, at least over this 600-fold range. Furthermore, the levels of total H3 at the 5′ end and the 3′ end of the coding region are similar, indicating an even distribution of total H3 throughout the coding region during active transcription (Fig. 3B, right side).
The H3K4me2 levels of the seven episomes are within a 3-fold range for both the 5′ end and the 3′ end of the coding region (Fig. 3C). The levels of H3K4me2 do not correlate with transcriptional activity on all of the episomes either as a group or when separated into endogenous and viral promoter groups (Fig. 3C). The ratio of the H3K4me2 level in the Luc1 region (5′ end of the coding region) to that in the Luc2 region (3′ end of the coding region) was determined in order to evaluate H3K4me2 distribution along the coding region. If this 5′-to-3′ ratio is greater than 1, then it would indicate that a higher level of H3K4me2 is detected at the 5′ end than at the 3′ end of the coding region. Conversely, it would indicate that H3K4me2 is present at a higher level at the 3′ end than at the 5′ end of the coding region if the 5′-to-3′ ratio is less than 1. Further deviation of the 5′-to-3′ ratio from 1 would indicate larger differences in the H3K4me2 modification levels between the 5′ and 3′ ends of the reporter gene. All seven episomes have a 5′/3′ ratio greater than 1 for the H3K4me2 level (Fig. 3C), indicating a consistently higher level of H2K4me2 at the 5′ end of the coding region than at the 3′ end, regardless of transcriptional activity or the nature of the promoter. In the endogenous promoter group, the 5′-to-3′ ratio of H3K4me2 showed slight decreases as the reporter gene expression increased (Fig. 3C). However, the 5′/3′ ratio of H3K4me2 does not show the same trend in the viral promoter group. It is clear that a stronger promoter does not lead to an increased H3K4me2 level in the coding region either immediately downstream from the promoter (Luc1 region) or at the 3′ end (Luc2 region). These findings suggest that the nature of the promoter and transcriptional activity have limited impact on the level of H3K4me2 modification in the coding region, but they may shift the distribution of H3K4me2 slightly along the coding region.
In contrast, the levels of H3K4me3 of the seven episomes vary dramatically both for the 5′ end and for the 3′ end of the coding region for pEF1A-Luc and pCLH29, which are the episomes with the highest transcriptional activity in the endogenous promoter and the viral promoter groups, respectively (Fig. 3D). Interestingly, the levels of H3K4me3 are higher on the episomes with endogenous promoters than episomes with viral promoters of comparable or higher transcriptional activity. For example, pEF1A-Luc has nearly 3-fold more H3K4me3 than pCLH29 at the 5′ end of the coding region (10.40 compared with 3.67; Fig. 3D); however, the luciferase expression from pEF1A-Luc is 6.3-fold lower than pCLH29 (413 compared with 2,617; Fig. 3A), and there is 2-fold less RNA Pol II at the 5′ end of pEF1A-Luc than at that of pCLH29 (1.28 compared with 2.61). Similarly, there is roughly 2-fold more H3K4me3 at the 5′ end of the coding region on pGSTP-Luc than on pCLH22 (1.84 compared with 0.95; Fig. 3D); and the luciferase expression from pGSTP-Luc is more than 100-fold lower than pCLH22 (2.4 compare with 661; Fig. 3A), and the RNA Pol II is essentially not detectable on pGSTP-Luc (at least 10-fold lower than pCLH22). While the levels of H3K4me3 on these episomes do not correlate with transcriptional activity as an entire group, a trend of increased H3K4me3 levels with increased transcriptional activity both at the 5′ end and at the 3′ end of the coding region was observed in both the endogenous promoter and viral promoter groups (Fig. 3D). These findings strongly suggest that the level of H3K4me3 in the coding region is affected by the nature of the promoter and the transcriptional activity.
The 5′-to-3′ ratio of H3K4me3 was lower than 1 for two of the episomes with viral promoters, indicating a greater amount of H3K4me3 at the 3′ end of the coding region (Fig. 3D). A greater amount of H3K4me3 at the 5′ end of the coding region was seen for the other five episomes, as indicated by the 5′-to-3′ ratio being greater than 1 (Fig. 3D). A trend of decreasing 5′-to-3′ ratios with increasing transcriptional activity for the seven episomes altogether or within each of the two groups of the episomes (viral and endogenous) is apparent (Fig. 3D). These findings suggest that increased transcriptional activity may shift the distribution of H3K4me3 to the 3′ end of the coding region in addition to increasing the level of this modification throughout the coding region.
The transcriptional activity has a direct impact on the H3K4me3 level in the coding region of the reporter gene on the episome.
Although it is clear that transcriptional activity affects the level of H3K4me3 in the coding region, this effect should ideally be tested without changing the promoter or the promoter activity. We have previously generated a robust system using stable episomes with three copies of the lac operator (lacO) positioned between the RSV LTR promoter and the luciferase reporter gene to modulate transcription (14). It has been established previously that lacI expression in human cells inhibits luciferase expression from the stable episome described above, and this inhibition can be reversed by the addition of IPTG (a lacI inhibitor) into the culture medium (14). The change in luciferase expression on the episome in this system is most likely determined by transcription elongation, since the first lacO site is 107 bp downstream from the transcription start site. Two other episomes, pOLucRLTR (with the RSV LTR promoter in the reverse orientation) and pOLucΔLTR (with the RSV LTR promoter deleted), were used as low-transcription/no-transcription controls. Another episome, pCLH22γI, which has a γ-globin intron placed between the RSV LTR and the luciferase reporter gene, was used as a control for potential effects of intronic sequence in the assays. The luciferase expression from pOLucOriP is known to increase when the transfected 293E/lacI cells are treated with IPTG; however, the luciferase expression from 293/EBNA1 cells (no lacI) that harbor pOLucOriP does not change with IPTG treatment, as illustrated and as previously shown (Fig. 4A) (14). These aspects of episomes pOLucOriP, pOLucRLTR, and pOLucΔLTR have been demonstrated previously (14) and again in the current experiment.
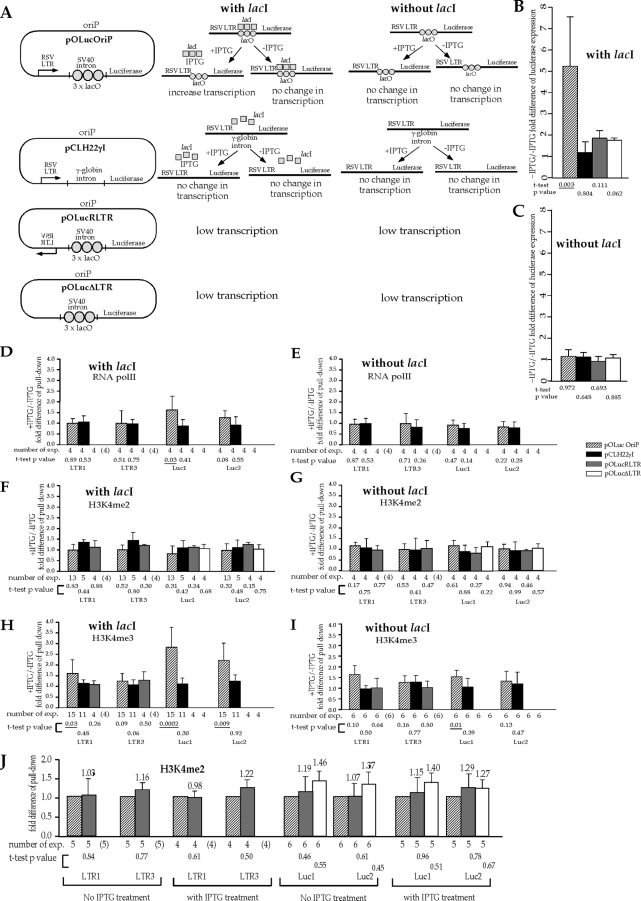
FIG. 4.
Transcriptional activity has a direct impact on the presence of H3K4me3 in the reporter coding region. exp., experiments. (A) Illustration of the four episomes and the expected and tested effects of IPTG treatment on transcription of the luciferase reporter gene on these episomes in the cells with and without lacI expression. Binding of lacI to the lacO sites positioned between the promoter and the reporter coding region on pOLucOriP interferes with transcription of the luciferase reporter gene, and IPTG treatment would restore transcriptional activity of the luciferase gene on pOLucOriP in lacI-expressing cells but has no effect on pOLucOriP in cells without lacI expression. SV40, simian virus 40. (B) Effect of IPTG treatment on luciferase expression when the episomes are transfected into lacI-expressing cells. pOLucOriP is the only episome whose luciferase expression is significantly affected by the IPTG treatment as expected. (C) Lack of IPTG effect on luciferase expression when the episomes are transfected into cells that do not express lacI. In both panel B and panel C, the luciferase activity is normalized with the amount of episome in the transfected cells as measured by Q-PCR. The normalized luciferase activity of IPTG-treated cells is divided by that of untreated cells harboring the same episome to derive the fold difference of luciferase expression illustrated in the histogram. Each bar represents the average from four independent experiments, and the standard deviations are indicated by error bars. (D) Effect of IPTG treatment on RNA Pol II levels in the coding regions of the episomes in the lacI-expressing cells. (E) Effect of IPTG treatment on RNA Pol II levels in the coding regions of the episomes in cells with no lacI expression. In both panel D and panel E, pOLucRLTR and pOLucΔLTR have only background levels of transcription; therefore, there is no measurable DNA above the background in the Q-PCR of all four regions, reflecting the lack of RNA Pol II. (F) Effect of IPTG treatment on the H3K4me2 level in the coding regions of episomes in lacI-expressing cells. (G) Effect of IPTG treatment on the H3K4me2 level in the coding region of episomes in cells with no lacI expression. (H) Effect of IPTG treatment on the H3K4me3 level in the coding region of episomes in lacI-expressing cells. (I) Effect of IPTG treatment on the H3K4me3 level in the coding region of episomes in cells without lacI expression. In panels F, G, H, and I, there is no measurable DNA above the background for pOLucΔLTR in the Q-PCR of LTR1 and LTR3 regions because this episome does not have the RSV LTR. In panels H and I, there is no measurable DNA from pOLucRLTR and pOLucΔLTR from the H3K4me3 pulldown, reflecting the lack of H3K4me3 in these regions of the episome. The fold differences in pulldowns in panels D, E, F, G, H, and I were derived by dividing the percent pulldown from cells with IPTG treatment by the corresponding value from cells without IPTG treatment. Each bar represents the average of multiple independent pull-down experiments as indicated below the histogram. The number of experiments carried out is shown in parentheses when the DNA sequence was absent from the episome. The standard deviations are indicated by error bars. For panels B to I, a t test was carried out to test the hypothesis that the presence of RNA Pol II or the histone modification does not change with IPTG treatment. P values of the t test are listed under each bar, with statistically significant P values of <0.05 underlined. (J) H3K4me2 presence is not affected by transcriptional activity through the coding region. The levels of H3K4me3 on pOLucRLTR and pLucΔLTR in each of the four regions examined were normalized against that of pOLucOriP. There is no significant difference between pOLucRLTR and pOLucOriP or between pLucΔLTR and pOLucOriP, as indicated by the t test (P value listed under each bar), regardless of IPTG treatment in the lacI-expressing cells. Similar results were found in the cells that do not express lacI (data not shown).
The four episomes pOLucOriP, pCLH22γI, pOLucRLTR, and pOLucΔLTR were individually transfected into 293/EBNA1 cells and 293E/lacI cells. Cells from each transfection were divided into two culture dishes two to three days after transfection. One dish was treated with 10 μM of IPTG, and the other one was not treated. Cells were harvested for luciferase and ChIP assays when they were confluent, which was five to six days after the first split. Experiments with each episome were carried out four to six times. The only significant increase in luciferase expression (more than 5-fold) was observed when 293E/lacI cells harboring pOLucOriP were treated with IPTG (relative to when they were not treated) (Fig. 4B). We found little change in luciferase expression levels from all four episomes individually transfected into 293/EBNA1 cells, which do not express lacI, with or without IPTG treatment (Fig. 4C). These findings confirmed that IPTG treatment increases luciferase expression from lacI-expressing cells harboring episomes with lacO sites upstream of the luciferase coding region, and such treatment has no effect on luciferase expression from cells that do not express lacI. Neither lacI nor IPTG has any effect on luciferase expression from episomes without lacO sites upstream of the luciferase coding region (pCLH22γI), regardless of whether there is lacI expression in the cells. Also, placement of an intron between the promoter and the reporter gene sequence does not affect the luciferase expression when lacI and/or IPTG is present.
The ChIP assay using anti-RNA Pol II antibody showed a small but clear increase in the RNA Pol II level in both the Luc1 (t test P value of 0.03) and Luc2 (t test P value of 0.08) regions of pOLucOriP in lacI-expressing cells when they were treated with IPTG, while nearly no difference was detected in the LTR1 and LTR3 regions (Fig. 4D). The levels of RNA Pol II were very similar with and without IPTG treatment for all four DNA regions studied on pOLucOriP in 293/EBNA1 cells, which do not express lacI (Fig. 4E). Very little difference was observed in any of the four regions analyzed for pCLH22γI with and without IPTG treatment whether or not lacI was expressed in the cells (Fig. 4D and E), indicating that IPTG and lacI do not alter transcriptional activity of the reporter gene from the RSV LTR promoter. Q-PCR was carried out for all ChIP assays for all four regions, and there was no reliably detectable level of RNA Pol II on pOLucRLTR in all four regions or on pOLucΔLTR in the Luc1 and Luc2 regions (there is no LTR on this episome). This is consistent with the lack of transcription expected from these two episomes and the very low luciferase expression measured. Combined with the luciferase expression results, these results clearly indicate that IPTG treatment of lacI-expressing cells increases the transcription through the coding region while not affecting the activity at the promoter when lacO sites are placed between the promoter and the coding region of the reporter gene on the episome pOLucOriP.
The ChIP assay using anti-H3K4me2 antibody showed very little difference between the levels of H3K4me2 in all four regions of all four episomes with and without IPTG treatment in lacI-expressing cell lines (Fig. 4F). Similar results were observed in cells without lacI expression (Fig. 4G). These findings indicate that the presence of lacI and/or IPTG does not alter the H2K4me2 level in these four episomes. The similar levels of RNA Pol II and H3K4me2 in LTR1 and LTR3 regions with and without IPTG treatment (Fig. 4D to G) support the inference that the promoter is not influenced by IPTG treatment and the increased reporter gene activity. The similar H2K4me2 levels in the Luc1 and Luc2 regions, despite a clear increase in transcriptional activity on pOLucOriP in lacI-expressing cells with IPTG treatment, further support the conclusion that transcriptional activity does not influence the presence of H3K4me2 in the coding region.
In contrast, the ChIP assay using anti-H3K4me3 antibody showed a significant increase in H3K4me3 in the Luc1 and Luc2 regions on pOLucOriP in lacI-expressing cells with IPTG treatment (Fig. 4H). The levels of H3K4me3 in the LTR1 region of pOLucOriP in lacI-expressing cells and the Luc1 region of pOLucOriP in cells without lacI expression also showed statistically significant (P values of 0.03 and 0.01, respectively) increases upon IPTG treatment (Fig. 4H and I). No difference in H3K4me3 presence was detected in any regions on the other three episomes, regardless of the cells used or whether IPTG was present (Fig. 4H and I). Combined with results from the ChIP assay using anti-RNA Pol II antibody and the luciferase assay, these findings clearly show that transcriptional activity directly affects the level of H3K4me3 in the coding region.
The analyses above provide evidence that a 5-fold increase in transcriptional activity upon IPTG treatment primarily influences H3K4me3 in the coding region. The only difference among pOLucOriP, pOLucRLTR, and pOLucΔLTR is the lack of transcriptional activity from the RSV LTR promoter into the luciferase gene for the last two episomes. A comparison of pOLucOriP, pOLucRLTR, and pOLucΔLTR would allow the assessment of transcriptional activity over a much larger range for H3K4me2 and H3K4me3. Both pOLucΔLTR and pOLucRLTR have very low luciferase activities (0.0014 RLU/molecule and 0.0035 RLU/molecule, respectively, without IPTG treatment in lacI-expressing cells), nearly 80-fold lower than that of pOLucOriP. The H3K4me2 levels do not significantly differ in all four regions analyzed on these three episomes with or without IPTG treatment in lacI-expressing cells (Fig. 4J). It is important that the levels of H3K4me2 in each of the DNA regions analyzed are very similar for all three episomes (pOLucΔLTR does not have an RSV LTR). The same analysis cannot be carried out for H3K4me3, because it cannot be reliably measured on pOLucΔLTR and pOLucRLTR, as described above. However, the percent pull-down of H3K4me3 from these two episomes is at least 10-fold lower than that of pOLucOrip (and could be larger, but this determination is limited by the sensitivity of the assay). These findings further strengthen the conclusion that transcriptional activity does not influence H3K4me2 but affects the presence of H3K4me3 in the coding region.
DISCUSSION
The stable episomal (minichromosomal) system described here is an ideal one for quantitative analysis of how zones of chromatin modification are influenced over a very wide range of transcriptional activity. In a previous study, we found that the level of H3K4me3 is not entirely correlated with the level of gene expression or RNA Pol II abundance and that transcription may play a contributory role in the level of H3K4me3 on the episome. We inferred that transcriptional activity might influence the H3K4me3 level more than that of H3K4me2. Using a series of stable episomes with viral or endogenous promoters upstream of the luciferase reporter gene, we specifically examined how transcriptional activity might impact H3K4me2 and H3K4me3 levels in the coding region. In this study, we demonstrated a clear association of transcriptional activity and H3K4me3 levels in the coding region that is distinct from H3K4me2 levels in the same regions. The levels of total H3 are similar in all cases except that the one episome with an extremely high level of transcription has approximately 2-fold less H3. We found that transcriptional activity influences the distribution of H3K4me2 and H3K4me3, while it does not change the distribution of total H3 in the coding region.
While promoters of different strengths allow us to analyze the same coding region with different transcriptional activities, it could potentially introduce other effects due to the nature of the promoters. However, this also gives us an opportunity to examine whether the nature of the promoter plays a role in the regulation of histone modification. On these stable episomes, a much higher level of H3K4me3 was observed in the coding region of the reporter gene downstream of endogenous promoters than that of viral promoters of comparable transcriptional activity, indicating that the nature of the promoter has a much greater impact on H3K4me3 than on H3K4me2 in the coding region. Utilizing episomes with lacO sites between the promoter and the coding region of the reporter, we further showed that transcriptional activity has a direct impact on the level of H3K4me3 in the coding region while not affecting H3K4me2 as dramatically in the same regions. These results suggest that transcription may impact the H3K4me3 level in the coding region through two different pathways, and there may be fundamental differences between how endogenous promoters and viral promoters interact with components of the transcriptional apparatus (see below).
H3 levels are stable over at least a 600-fold range in luciferase activity.
We observed similar levels of total H3 at the 5′ and 3′ ends of the coding region on each of these seven stable episomes across a 600-fold range in transcriptional activity with only a 2-fold reduction on the episome with 2,600-fold-higher luciferase activity. This finding suggests that H3 is most likely evenly distributed in the coding region during active transcription, regardless of the strength of transcriptional activity. This is consistent with the observation in Saccharomyces cerevisiae (yeast) that RNA Pol II occupancy is higher at the 5′ end than at the 3′ end of the coding region, while H3 occupancy does not vary greatly (23). In yeast, a partial loss of H3 and H4 tetramers from the coding regions was observed at the 4.2% of genes that are most actively transcribed at a rate of more than 30 mRNA per hour (11). It has also been reported that the H3 density in yeast decreases about 2-fold when a silent gene is activated, and a reduced histone density is detected in the coding region when RNA Pol II passes at a rate of at least once per minute (23). Transcriptional activity has been linked to H3 exchange in Drosophila and mouse cells (4, 24, 28), even though very little exchange of H3 and H4 was detected in HeLa cells stably expressing green fluorescent protein-tagged H2B, H3, and H4 (10).
In the current study, the levels of total H3 in the coding region of six of the seven stable episomes studied are very similar, even though the transcriptional activities span a 600-fold range. Interestingly, the level of H3 in the coding region of the episome with the highest transcriptional activity (2,600-fold higher than the lowest activity) is about 2-fold lower than those found in the other six episomes. It is likely that the change in H3 occupancy in the coding region can be clearly detected, within the limitations of the current assay, only when the transcriptional activity is extremely high. This finding would support the hypothesis that histone eviction and deposition occur at each passage of RNA Pol II, and the rate of RNA Pol II initiation determines the relative nucleosome occupancy in a transcribing region (23); this finding also indicates the potential similarity between human cells and yeast cells for histone displacement and transcription.
Limited effect of transcription on H3K4me2 levels.
We did not observe any consistent correlation of the RNA Pol II level and transcriptional activity with the H3K4me2 level in the current study. Although the level of H3K4me2 varied depending on the episome studied, there is no consistent trend associated with transcriptional activity. It is clear that a stronger promoter does not lead to an increased presence of H3K4me2 in the coding region either immediately downstream from the promoter (Luc1 region) or at the 3′ end (Luc2 region). Furthermore, control episomes lacking a promoter or having a promoter in the reverse orientation confirm that transcriptional activity does not have much impact on the H3K4me2 level in the coding region. We observed more H3K4me2 at the 5′ end than at the 3′ end of the coding region, but there is also a lack of correlation between the transcription level and the ratio of H3K4me2 at the 5′ end to H3K4me2 at the 3′ end. These findings support our previous hypothesis that, unlike DNA methylation, transcriptional activity does not play a major role in dictating H3K4me2 presence in the transcription zone on the episome. However, we cannot rule out the possibility that transcription activity may influence the level of H3K4me2 in the coding region through other factors.
H3K4me3 levels strongly correlate with transcriptional activity and possible mechanisms.
In contrast, a much better correlation between the levels of transcription and the presence of H3K4me3 was observed within the group of episomes with viral promoters and within those with endogenous promoters. It is interesting that the presence of H3K4me3 in the coding region is greater on the episomes with endogenous promoters than on the episomes with viral promoters at a similar or even lower level of transcriptional activity. For example, pEF1A-Luc expresses approximately 60% of the luciferase activity of pCLH22, while the H3K4me3 level on pEF1A-Luc is nearly 11-fold higher in the Luc1 region and >7-fold higher in the Luc2 region than in the respective regions on pCLH22. This is also true for other stable episomes with endogenous promoters compared with episomes that have viral promoters. This phenomenon was not observed for H3K4me2. This suggests that there is a fundamental difference between how endogenous and viral promoters function in human cells. Initiation of transcription from endogenous promoters appears to lead to a much more active recruitment of H3K4me3 modification, but not H3K4me2 modification, to the coding region. It would be interesting to know what factors contribute to the crucial difference for this H3K4me3 recruitment during transcription.
The influence of transcriptional activity on the distribution of H3K4me3 in the coding region is more consistent than that on H3K4me2. Higher transcriptional activity correlates with more H3K4me3 in the 3′ end than in the 5′ end of the coding region on the stable episome, regardless of the type of promoter used. Hence, the increase in H3K4me3 in the coding regions with very high transcriptional activity is not a collateral effect of the increased presence of this modification at the proximal promoter region (due to the strength of the promoter). Also, it appears that the effects of transcriptional activity on the H3K4me3 level in the coding region are similar for viral promoters and endogenous promoters, even though the nature of the promoter does play a crucial role in the level of H3K4me3 in the coding region, independent of the transcriptional activity. Increased transcription activity appears to shift the distribution of H3K4me3 to the 3′ end of the coding region in addition to increasing the presence of this modification throughout the coding region.
Using a system with lacI expression in human cells and lacO sites between the promoter and the coding region on the episome, we saw no appreciable change in H3K4me2 levels in the coding region, despite a clear >5-fold increase in transcriptional activity on the episome with IPTG treatment. This finding further supports the conclusion that transcriptional activity does not influence the presence of H3K4me2 in the coding region dramatically. In the same experiments, a modest increase in the H3K4me3 level in the coding region on the episome with IPTG treatment was observed. In this system, the only difference between stable episomes in untreated cells and cells treated with IPTG was the increased transcriptional activity through the luciferase coding region, since the same transfected cells were used for the assays. We did not detect much effect of IPTG on the episome when transfected into cells without lacI expression or on an episome without lacO sites. Therefore, the finding of increased H3K4me3 in the coding region with IPTG treatment strongly indicates that transcriptional activity directly affects the presence of H3K4me3 in the coding region. Also, the H3K4me3 levels on the control episomes without a promoter or with a promoter in the reverse orientation were reduced more than 10-fold to a level that is below the detection limit of the assay, while the levels of H3K4me2 did not show significant change. Our results in the current study provide strong evidence that transcriptional activity plays a much more important and direct role in dictating the presence of H3K4me3 in the coding region but has much less influence on the presence of H3K4me2 in that region.
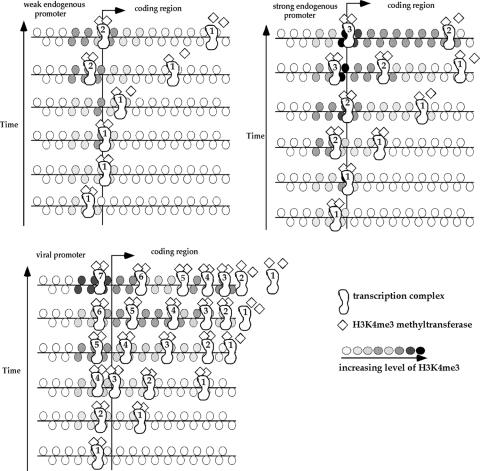
It has been shown in yeast that H3K4me3 methylation depends on the recruitment of the Set1 histone methyltransferase by RNA Pol II during elongation (with the involvement of RNA polymerase II-associated factor complex, histone H2B ubiquitination, and BUR kinase; for reviews, see references 12 and 26). It is likely that the tight association between transcriptional activity and the presence of H3K4me3 in the coding region observed in the current study in human cells is the result of a similar interaction of RNA Pol II and histone methyltransferase. A potential model (Fig. 5) is the transcription initiation complex recruiting H3K4me3 methyltransferase, which dissociates from the complex after a certain time interval. A stronger promoter would initiate more often, leading to a higher level of H3K4me3 at and around the promoter as well as the 5′ end of the coding region. The histone methyltransferase dissociates from the transcription complex as the complex moves toward the 3′ end, and it leads to the decrease in H3K4me3 at the end of the coding region. A weak promoter has fewer transcription complexes initiating and a lower level of H3K4me3 than a strong promoter. The first two steps of the transcription elongation process, promoter escape and promoter-proximal pausing, also play important rate-limiting roles in the process (for a review, see reference 22). These two steps may take longer intervals with decreasing promoter strength, and histone methyltransferase would dissociate from the transcription complex closer to the 5′ end of the coding region. Therefore, the overall H3K4me3 level decreases while the ratio of H3K4me3 at the 5′ end to H3K4me3 at the 3′ end of the coding region increases as the promoter strength decreases. The switch to a higher level of H3K4me3 at the 3′ end than at the 5′ end of the coding region occurs on the episomes with viral promoters having extremely high transcription activity. It is possible that transcription complexes pause at the end of the coding region before dissociation of histone methyltransferase when an extremely high number of transcription complexes is initiated from the viral promoters and leads to a greater H3K4me3 presence at the 3′ end of the coding region. This is consistent with our observation that the increase in RNA Pol II is slightly higher in the 3′ end of the coding region as the promoter strength increases on the episomes (Fig. 1B and 2B). In addition, it is likely that different H3K4me3 methyltransferases may be recruited to the transcription complex indirectly through other factors at different promoters. This may account for the overall level of H3K4me3 immediately downstream from viral promoters being lower than the level downstream from endogenous promoters with similar levels of transcriptional activity. Until all the factors involved in the transcription complex from each specific promoter are fully known and analyzed, our understanding of the interplay of histone modification, transcription, and promoter function remains incomplete. Our current study indicates that knowledge about the general distribution of various histone modifications is only the beginning of this understanding.
FIG. 5.
A proposed model for the influence of transcriptional activity on the H3K4me3 level and H3K4me3 distribution in the coding region. The overall increase in H3K4me3 with increased transcriptional activity in each of the viral and endogenous promoter groups may be a simple consequence of the increased number of transcriptional complexes moving through the coding region. If the dissociation time of the histone methyltransferase from the transcription complex is invariant, then with decreased transcription and increased promoter pause time, the histone methyltransferase will dissociate closer to the 5′ end (leading to the increased ratio of H3K4me3 at the 5′ end relative to that at the 3′ end). The accumulation of transcription complexes at the end of the coding region when an extremely high number of complexes is initiated may lead to the higher H3K4me3 level at the 3′ end of the coding region on the episomes with very strong viral promoters. Different histone methyltransferases recruited to the viral and endogenous promoters may account for the overall lower level of H3K4me3 in the coding region downstream from the viral promoter.
Footnotes
Published ahead of print on 19 April 2010.
REFERENCES
- 1.Allis, C. D., S. L. Berger, J. Cote, S. Dent., T. Jenuwien, T. Kouzarides, L. Pillus, D. Reinberg, Y. Shi, R. Shiekhattar, A. Shilatifard, J. Workman, and Y. Zhang. 2007. New nomenclature for chromatin-modifying enzymes. Cell 131:633-636. [DOI] [PubMed] [Google Scholar]
- 2.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. [DOI] [PubMed] [Google Scholar]
- 4.Daury, L., C. Chailleux, J. Bonvallet, and D. Trouche. 2006. Histone H3.3 deposition at E2F-regulated genes is linked to transcription. EMBO Rep. 7:66-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han, L., I. G. Lin, and C. L. Hsieh. 2001. Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol. Cell. Biol. 21:3416-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39:311-318. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh, C. L. 1994. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irvine, R. A., I. G. Lin, and C. L. Hsieh. 2002. DNA methylation has a local effect on transcription and histone acetylation. Mol. Cell. Biol. 22:6689-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, T. H., L. O. Barrera, M. Zheng, C. Qu, M. A. Singer, T. A. Richmond, Y. Wu, R. D. Green, and B. Ren. 2005. A high-resolution map of active promoters in the human genome. Nature 436:876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura, H., and P. R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 12.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128:707-719. [DOI] [PubMed] [Google Scholar]
- 13.Lin, I. G., and C. L. Hsieh. 2001. Chromosomal DNA demethylation specified by protein binding. EMBO Rep. 2:108-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, I. G., T. J. Tomzynski, Q. Ou, and C. L. Hsieh. 2000. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol. Cell. Biol. 20:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorincz, M. C., D. R. Dickerson, M. Schmitt, and M. Groudine. 2004. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 11:1068-1075. [DOI] [PubMed] [Google Scholar]
- 16.Miao, F., and R. Natarajan. 2005. Mapping global histone methylation patterns in the coding regions of human genes. Mol. Cell. Biol. 25:4650-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 18.Okitsu, C. Y., and C. L. Hsieh. 2007. DNA methylation dictates histone H3K4 methylation. Mol. Cell. Biol. 27:2746-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrowski, M., H. Richard-Foy, R. Wolford, D. Berard, and G. Hager. 1983. Glucocorticoid regulation of transcription at an amplified, episomal promoter. Mol. Cell. Biol. 3:2045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roh, T. Y., S. Cuddapah, K. Cui, and K. Zhao. 2006. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. U. S. A. 103:15782-15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roh, T. Y., S. Cuddapah, and K. Zhao. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders, A., L. J. Core, and J. T. Lis. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557-567. [DOI] [PubMed] [Google Scholar]
- 23.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz, B. E., and K. Ahmad. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sims, R. J., III, S. Millhouse, C. F. Chen, B. A. Lewis, H. Erdjument-Bromage, P. Tempst, J. L. Manley, and D. Reinberg. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suganuma, T., and J. L. Workman. 2008. Crosstalk among histone modifications. Cell 135:604-607. [DOI] [PubMed] [Google Scholar]
- 27.Vermeulen, M., K. W. Mulder, S. Denissov, W. W. Pijnappel, F. M. van Schaik, R. A. Varier, M. P. Baltissen, H. G. Stunnenberg, M. Mann, and H. T. Timmers. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131:58-69. [DOI] [PubMed] [Google Scholar]
- 28.Wirbelauer, C., O. Bell, and D. Schubeler. 2005. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 19:1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamasaki-Ishizaki, Y., T. Kayashima, C. K. Mapendano, H. Soejima, T. Ohta, H. Masuzaki, A. Kinoshita, T. Urano, K. I. Yoshiura, N. Matsumoto, T. Ishimaru, T. Mukai, N. Niikawa, and T. Kishino. 2007. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol. Cell. Biol. 27:732-742. [DOI] [PMC free article] [PubMed] [Google Scholar]