Abstract
Transcription of microRNAs (miRNAs) is thought to be regulated similarly to that of protein-coding genes. However, how miRNAs are regulated during the cell division cycle is not well understood. We have analyzed the transcription profiles of miRNAs in response to mitogenic stimulation in primary fibroblasts. About 33% of the miRNAs expressed in these cells are induced upon exit from quiescence. Many of these miRNAs are specifically induced by E2F1 or E2F3 during the G1/S transition and are repressed in E2F1/3-knockout cells. At least four miRNA clusters, let-7a-d, let-7i, mir-15b-16-2, and mir-106b-25, are direct targets of E2F1 and E2F3 during G1/S and are repressed in E2F1/3-null cells. Interestingly, these miRNAs do not contribute to E2F-dependent entry into S phase but rather inhibit the G1/S transition by targeting multiple cell cycle regulators and E2F targets. In fact, E2F1 expression results in a significant increase in S-phase entry and DNA damage in the absence of these microRNAs. Thus, E2F-induced miRNAs contribute to limiting the cellular responses to E2F activation, thus preventing replicative stress. Given the known function of E2F of inducing other oncogenic miRNAs, control of miRNAs by E2F is likely to play multiple roles in cell proliferation and in proliferative diseases such as cancer.
MicroRNAs (miRNAs) are small (∼23-nucleotide [nt]) regulatory RNA molecules that exert posttranscriptional control of specific target mRNAs (3). More than half of the human protein-coding genes appear to have been under selective pressure to maintain pairing of their 3′ untranslated regions (3′-UTRs) to miRNAs (5). This interaction is known to induce mRNA degradation or inhibition of translation through specific albeit imperfect base pairing (22, 24). Several target genes have been validated, indicating that each individual miRNA can target a few or, possibly, multiple genes. These small miRNAs therefore regulate fundamental cellular processes such as proliferation, differentiation, or apoptosis during development (65). In addition, deregulation of miRNAs is frequently observed in a wide range of diseases, including cancer (19, 21). The analysis of miRNA function and regulation in specific cellular processes has proven to be required for a full understanding of malignant transformation and to envision new therapeutic possibilities.
Tumor development is accompanied by a variety of genetic and epigenetic alterations in protein-coding genes and small, noncoding RNA genes. By regulating specific oncogenes or tumor suppressor molecules, miRNAs may have profound effects on tumor development. The first report linking miRNAs and cancer showed a frequent deletion of miR-15a and miR-16-1 in patients with B-cell chronic lymphocytic leukemia (13). Further analysis of miRNA expression signatures has suggested a frequent involvement of miRNAs in the initiation and progression of human tumors (12). Although the critical targets for most cancer-associated miRNAs are unknown, some relevant targets have been characterized in specific malignancies. The mir-15a-mir-16-1 (mir-15a-16-1) cluster may control proliferation, survival, and invasion regulators such as Bcl2, Wt1, Tab9B, and Mage83 (8, 19). The tumor suppressor let-7 miRNAs act as tumor suppressors in several pathologies, such as lung cancer, by modulating major oncogenes, such as Ras or Myc, among many others (60). miR-203 modulates T-cell and myeloid leukemias through the repression of the ABL oncogene and the BCR-ABL translocation product (11). In addition, cancer-associated miRNAs may regulate cell cycle progression at multiple levels by directly targeting proteins such as E2F transcription factors, cyclin-dependent kinases (CDKs), cyclins, and CDK inhibitors (10, 16, 19).
How miRNAs themselves are regulated is less understood. A few transcriptional networks control the expression of miRNAs that modulate the diverse pathways downstream of these networks. A few miRNAs are known to be induced by Myc or E2F transcription factors. Myc and E2F members are able to induce the mir-17-92 cluster that, in turn, controls E2F expression (1, 55, 71). E2F transcription factors are additionally modulated by several other miRNAs, such as members of the mir-106a-92 and mir-106b-25 clusters (paralogs to the mir-17-92 cluster) (57), miR-210 (30), miR-128 (74), miR-34 (66), or miR-20 (58). The oncogenic effect of Myc is not only exerted through the induction of the mir-17-92 cluster, since Myc also represses a significant number of antiproliferative miRNAs (14). Activation of the p53 tumor suppressor also results in induction or repression of several miRNAs. miR-34a is directly induced by p53, and it participates in the antiproliferative responses to p53 by repressing cell cycle regulators such as cyclin D1 and CDK6 (32). In addition, several miRNAs, including those carried by the mir-17-92, mir-106a-92, and mir-106b-25 paralog clusters, are downregulated by p53 in a E2F-dependent manner, leading to decreased proliferation and senescence (9, 57).
In the present study, we have examined the overall expression of miRNAs through the initial phases that drive the entry into the cell cycle in mammalian primary cells. A relevant percentage of miRNAs (about 30%) are induced after stimulation with serum and passage through the G1 phase of the cell cycle. Since E2F transcription factors control these stages of the cell cycle, we have characterized in detail the transcriptional control of several of these miRNAs by the activating E2F factors E2F1, -2, and -3. Some G1-induced miRNAs, such as the ones encoded by the mir-106b-25 or mir-15b-16-2 clusters or specific let-7 miRNAs, are induced by E2F1 or E2F3. In addition, these miRNAs are downregulated in E2F1-knockout or E2F3-knockdown cells, suggesting the functional relevance of these transcription factors in the control of these small RNAs. We have confirmed the direct binding of E2F factors to the promoter regions of four different miRNA clusters using chromatin immunoprecipitation (ChIP). Interestingly, the miRNAs expressed by these clusters inhibit entry into S phase and downregulate cell cycle regulatory genes, including E2F targets. These results suggest a role for E2F-induced miRNAs in limiting the cellular effects of E2F-dependent gene expression and in the prevention of replicative stress.
MATERIALS AND METHODS
Cell culture and transfections.
Mouse embryonic fibroblasts (MEFs) were isolated from wild-type mice, E2F1-null mice (23), or E2F2-null mice (53) using routine protocols and cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (28). For cell cycle entry analysis, these MEFs were stimulated with 10% FBS after 3 days in the presence of 0.1% FBS. This protocol usually results in a good synchronization in G0 (as detected by biochemical studies), although about 5% of cells seemed to be in S phase by DNA content analysis (28, 50, 51). For knockdown E2F3 expression, wild-type MEFs were trypsinized and infected with empty control or lentiviral vectors expressing small interfering RNAs against mouse E2F3, as described previously (34). Cells infected with virus were identified with green fluorescent protein (GFP) signaling by flow cytometry (∼90% of the transduced cells yielded a positive signal). Cells expressing the 4-hydroxytamoxifen (4-OHT)-inducible form of E2F1, E2F2, or E2F3 were kindly provided by K. Helin (52). Induction of E2F activity was achieved by treating cells with 300 nM 4-OHT, and miRNAs were analyzed after 8 h of treatment. For exogenous expression of miRNAs, miRNA genes were expressed in the pMirVec vector, as reported previously (70). For knockdown of endogenous miRNA expression, anti-miRNA inhibitors were purchased from Ambion and used by following the manufacturer's recommendations. MEFs were transfected with pMirVec plasmids or anti-miRNA inhibitors using Lipofectamine (Invitrogen).
Transcriptional profiles and target analysis.
Total RNA was prepared from cells using TRIzol reagent (Invitrogen). miRNA arrays were purchased from Invitrogen and processed according to the manufacturer's recommendations. Significantly deregulated microRNAs were computed using the TM4 package (61) and the limma package from Bioconductor (http://www.bioconductor.org). Clustering was performed using the self-organizing tree algorithm included in the TM4 package. Precomputed microRNA targets were obtained from miRBase targets database v5 (http://microrna.sanger.ac.uk/) or the EIMMo miRNA target prediction server (http://www.mirz.unibas.ch/ElMMo2/). Statistical significance was analyzed using the two-tailed Fisher exact test and Prism (GraphPad) software. Gene ontology was analyzed using the EIMMo server, the Ontologizer (http://compbio.charite.de/index.php/ontologizer2.html), or the FatiScan algorithm (Babelomics) (2).
Reverse transcription PCR (RT-PCR) analysis was carried out using SuperScript retrotranscriptase (Invitrogen). PCR products were obtained after 25 cycles of amplification, with an annealing temperature of 60 to 65°C. The following oligonucleotides were used: E2F1-RT-Fw, 5′-AGGCTGGATCTGGAGACTGA-3′; E2F1-RT-Rv, 5′-GAGTCCTCCGAAAGCAGTTG-3′; E2F2-RT-Fw, 5′-CCGCCACCACCTACTACACT-3′; E2F2-RT-Rv, 5′-CTGCCTACCCACTGGATGTT-3′; E2F3-RT-Fw, 5′-TGCAGTCTGTCTGAGGATGG-3′; and E2F3-RT-Rv, 5′-GGGTCTGTGTGTTTCCGTCT-3′. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as the normalization control. For quantitative analysis of miRNA expression, total RNA was amplified using oligonucleotides specific for different miRNAs (TaqMan microRNA assay kit; Applied Biosystems, Foster City, CA), according to the manufacturer's instructions, in an Applied Biosystems 7900HT fast real-time PCR apparatus. Amplification of Sno202 was used for normalization. The data analysis was done using the Sequence Detection Systems 2.2.2 program (Applied Biosystems, Foster City, CA).
Chromatin binding and immunoprecipitation.
Putative E2F recognition sites were analyzed using ConSite (http://consite.genereg.net/), and miRNA or miRNA clusters with putative E2F sites above the recommended threshold were selected. For chromatin binding analyses, chromatin was isolated as described previously (34) from a pool of fibroblasts derived from five embryos. A total of 100 to 120 μg of precleared chromatin was incubated overnight at 4°C with 4 to 5 μg of each of the following antibodies: SV40Tag (sc-147), E2F1 (sc-193), E2F2 (sc-633), E2F3 (sc-878), and E2F3 (sc-879) (all from Santa Cruz Biotechnology). Note that the polyclonal antibody sc-193 is thought to cross-react with E2F3 (29). Samples were then incubated with protein A-Sepharose at 4°C for 2 h. Immune complexes were recovered and washed as reported previously (34). The elution of the immune complexes was carried out with a buffer containing 0.1 M NaHCO3 and 1% sodium dodecyl sulfate (SDS). Cross-linking was reversed by addition of NaCl to a final concentration of 200 mM, followed by an overnight incubation at 65°C, and RNA was removed with 10 μg of RNase A. Proteins were digested with 80 μg of proteinase K at 42°C for 2 h, and the DNA was extracted with phenol-chloroform and ethanol precipitation. DNA was amplified using primers complementary to the CDK1 promoter as well as to specific sequences upstream of the different miRNA or miRNA clusters analyzed. These sequences were selected after analysis with the ConSite software. Oligonucleotide sequences are shown in the supplemental material. Quantification of immunoprecipitate-enriched DNA sequences was performed by real-time PCR with the Power SYBR green PCR master mix and the Applied Biosystems 7000 Sequence Detection System, and analysis was done with the Sequence Detection System 1.1 software. Samples were analyzed in triplicate. Data are represented as percentages of the input, calculated by the equation, 2ΔCT × 20, where the change in threshold cycle (ΔCT) is determined by CTdilution of the input − CTdilution of the IP sample and “20” refers to the input being 20% of the chromatin amount exposed to immunoprecipitation (IP).
Immunofluorescence.
For high-throughput microscopy studies, cells were grown on mClear 96-well dishes (Greiner Bio-One) and analyzer with a BD Pathway 855 bioimager (Becton Dickinson) essentially as described previously (54). 5-Ethynyl-2′-deoxyuridine (EdU) was detected using the Click-iT imaging kit (Invitrogen), whereas phosphorylation of H2AX (γH2AX) was detected using specific antibodies (Millipore), and secondary antibodies were conjugated with Alexa 488 or 594 (Invitrogen). Image acquisition was performed at room temperature using a 40× objective under nonsaturating exposure conditions. These images were processed using the AttoVision software (BD Biosciences) and Prism (GraphPad) for statistical purposes using Student's t test. To study bromodeoxyuridine (BrdU) incorporation, cells were treated with short pulses (30 min) of 10 mM BrdU. BrdU was detected using a specific antibody coupled to fluorescein isothiocyanate (FITC; Roche), counterstained with propidium iodide, and analyzed by cell cytometry (Becton Dickinson).
Protein analysis.
Protein lysates were obtained as reported previously (11). Proteins were transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA) and probed with antibodies against the following proteins: γH2AX (Millipore), cyclin E (Abcam), cyclin B1 (Chemicon), Cdk7 (Santa Cruz Biotechnology), MCM5 (a gift from Juan Méndez, CNIO), and cyclin D1, cyclin D2, Cdk7, E2F1, E2F2, and E2F3 (all from Santa Cruz Biotechnology). In addition, anti-α-tubulin antibody (Sigma) was used as a loading control. After being washed, blots were incubated with the appropriate secondary antibodies coupled to Alexa Fluor 680 or 800 (Invitrogen). Subsequently, membranes were scanned using the Odyssey infrared imaging system (LI-COR Biosciences).
RESULTS
miRNA expression during the early phases of the cell cycle.
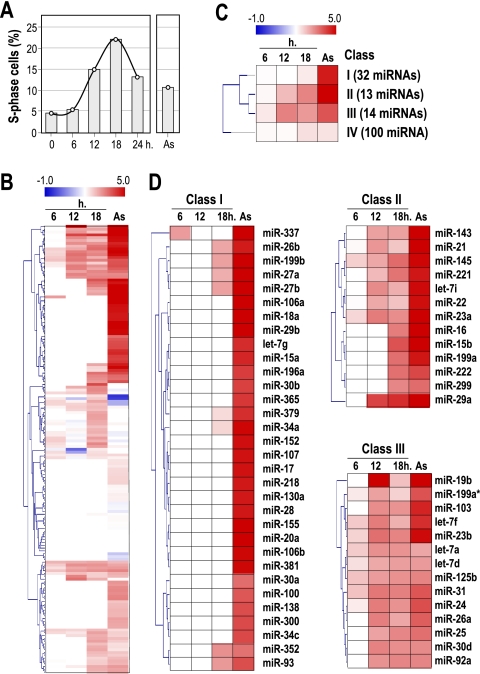
To investigate the expression of miRNAs during the early phases of the cell division cycle in normal cells, we used early-passage wild-type mouse embryonic fibroblasts (MEFs). MEFs were serum starved for 72 h and stimulated by addition of 10% fetal bovine serum. As reported previously (28), these wild-type cells peak in S phase at approximately 18 to 20 h after stimulation with serum (Fig. 1 A), and the cell cycle is completed in about 24 h. The miRNA expression profiles were analyzed before (0 h) and several time points (6 h, 12 h, and 18 h) after serum addition to analyze the expression of miRNAs during cell cycle entry. Asynchronously growing primary MEFs were also analyzed and compared to serum-starved cells. Under these conditions, 159 microRNAs display significant levels of expression in primary MEFs. The rest of the miRNA probes in the array (233 murine probes; see Materials and Methods) do not detect significant signals, suggesting that these miRNAs are not expressed in early-passage MEFs. As depicted in Fig. 1B, mitogenic stimulation with serum results in a wide upregulation of a significant number of miRNAs. No miRNA was consistently downregulated during cell cycle entry in this assay.
FIG. 1.
Transcriptional profile of miRNAs during the cell cycle. (A) Primary mouse embryonic fibroblasts (MEFs) were arrested by serum starvation, and entry into the cell cycle was stimulated by the addition of serum. These MEFs peak in S phase at about 18 h after stimulation with serum, as detected by DNA content analysis. (B) Expression profile of miRNAs during the G1/S transition and in asynchronously growing cells (As). Data are normalized versus the levels of expression in serum-starved cells (G0). Red indicates overexpression, and blue indicates downregulation compared to that of G0 cells. (C) Clustering of miRNAs (SOTA analysis) by their expression profile. Class I contains miRNAs that are upregulated in asynchronously growing cells, whereas miRNAs that are not significantly modulated by stimulation with serum are included in class IV. Classes II and III contain miRNAs upregulated 6 to 12 h after cell cycle entry by serum stimulation. (D) Expression profiles of miRNAs belonging to classes I, II, and III during cell cycle entry.
A classification of these expression profiles (self-organizing tree algorithm [SOTA]) indicates the presence of different clusters of miRNA expression during cell cycle entry (Fig. 1C; see also Fig. S1 and S2 in the supplemental material). Class I (32 miRNAs) corresponds to miRNAs that significantly accumulate in asynchronously proliferating cells but whose expression does not significantly increase during the early phases of the cell cycle, suggesting that these miRNAs are either accumulated after several cell cycles or induced by secondary pathways in proliferating cells (Fig. 1D). Class II (13 miRNAs) is formed of miRNAs whose expression levels show significant induction during G1, which is maintained or further increased in proliferating cells. Class III (14 miRNAs) also includes miRNAs that were partially and transiently induced 6 to 18 h after serum stimulation but whose expression levels, in most cases, are not further induced in asynchronously proliferating cells (Fig. 1D). Class IV (100 miRNAs) groups the rest of the miRNAs that are expressed in MEFs but whose levels of expression did not significantly change in proliferating versus quiescent cells (see Fig. S2 in the supplemental material). This classification suggests that both class II and class III miRNAs are specifically induced during the initial G1/S progression in exiting from quiescence.
As a validation for the expression profiles, we compared the expression of miRNAs expressed from the same transcript. Many miRNAs belonging to the same cluster display similar patterns of expression. For instance, three mature let-7 miRNAs, let-7a, let-7f, and let-7d (let-7a-7d transcript) are expressed as a transcriptional cluster, and all of them map to class III (Fig. 1 and Fig. 2). Similarly, miR-221 and miR-222, which are expressed in the same mir-221-222 cluster, or miR-143 and miR-145 (mir-143-145 cluster) belong to class II and display similar expression profiles. The expression of all clusters in which most members of the cluster are included in class II or class III is represented in Fig. 2. Clusters in which most members of the cluster were not included in class II or class III (e.g., the mir-17-92 cluster) were not further considered in this analysis (see Discussion). On the other hand, there is also a good correlation in nonexpressed miRNAs belonging to the same cluster, confirming that these clusters are not significantly expressed in primary MEFs under the conditions tested (data not shown).
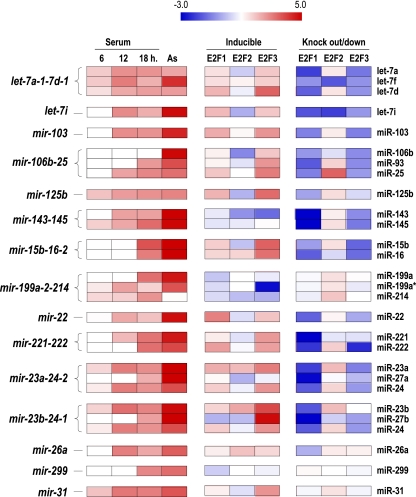
FIG. 2.
Control of miRNA expression by E2F transcription factors. This figure summarizes the expression profiles of class II and III miRNAs or miRNA clusters upon acute expression of E2F transcription factors in E2F-inducible cells or in E2F1-3-knockout/knockdown cells. miRNAs are organized by expression clusters. Their expression during stimulation with serum is shown in the first group of columns. The middle columns summarize their expression upon tamoxifen-inducible expression of E2F1, E2F2, or E2F3 estrogen receptor fusion proteins normalized versus noninduced cells. The columns on the right summarize the expression profiles in E2F1- or E2F2-knockout cells or in E2F3-knockdown cells normalized versus wild-type cells. In general, these data suggest a consistent control of several of these miRNA clusters mostly by E2F1 and E2F3 (upregulated in the middle group of columns and downregulated in the group of columns on the right).
miRNA expression in cells overexpressing or lacking E2F factors.
Class II and class III miRNAs are expressed in mid-G1 and the G1/S transition, which significantly depends on the inactivation of the retinoblastoma pathway and transcriptional induction by the E2F transcription factors (49, 67). In fact, several of the miRNA genes or clusters, including those in these categories, present putative E2F sites in their promoter sequences (see below). To analyze the expression of miRNAs upon specific activation of E2F factors, we used stable cell lines expressing E2F1, E2F2, or E2F3 factors fused to the estrogen receptor (ER) (52). These three E2F family members are thought to be responsible for the induction of genes during cell cycle progression (18, 67). These inducible cell lines express inactive E2F1, E2F2, or E2F3 proteins that are activated upon treatment with 4-hydroxytamoxifen (4-OHT) (52; data not shown). We then analyzed miRNA expression profiles in E2F-inducible cell lines 8 h after addition of 4-OHT by following the established protocol (52). As shown in Fig. 2, many class II or class III miRNAs are induced by individual or multiple E2F factors. For instance, all components of the let-7a-7d cluster are induced by E2F1 or E2F3 but not by E2F2. Similar results are found for other class II and class III miRNAs that, in general, are specifically induced by either E2F1 or E2F3. Some individual miRNAs such as miR-299 or other miRNA clusters, such as mir-143-145 or mir-199a-2-214, are not induced by any of these E2F factors in this assay.
We further tested the specificity of the E2F-dependent induction of class II or class III miRNAs by analyzing their expression profiles in primary MEFs derived from E2F1-deficient (23) or E2F2-deficient (53) mice. To obtain E2F3-deficient cells, we infected wild-type MEFs with a lentiviral vector expressing validated small hairpin RNAs against murine E2F3 (34). As shown in Fig. 2, many class II and class III miRNAs or miRNA clusters, such as let-7a-7d, let-7i, mir-106b-25, and mir-23a/b-24, are downregulated in E2F1- or E2F3-deficient MEFs. Others, such as mir-143-145 or mir-199a-2-214, also display significant downregulation in E2F-deficient cells, despite the lack of acute induction in E2F-inducible cell lines. These differences suggest that some clusters may accumulate during several cell cycles but are not significantly induced within the early exit from quiescence. Interestingly, several miRNAs displayed increased levels in E2F2-null MEFs (Fig. 2). It has been recently demonstrated that most E2F target-coding genes are upregulated in E2F2-null lymphocytes or MEFs, thus suggesting that E2F2 may have repressor functions (34, 53). All together, these results indicate that E2F1 and E2F3 transcription factors induce specific miRNAs or miRNA clusters during entry into the cell cycle.
E2F factors occupy the promoter regions of multiple microRNAs.
We analyzed putative E2F sites in miRNAs upregulated in E2F-inducible cells and downregulated in E2F-deficient cells using the ConSite algorithm (see Materials and Methods). Many of these miRNAs and miRNA clusters display putative E2F-recognition sites (above the recommended score) around their promoter areas (Fig. 3). As a test for the ability of E2F proteins to directly regulate the expression of miRNAs induced during G1, we asked whether E2F may bind directly to the promoters of these transcripts in serum-starved cells (0 h) or 18 h after serum stimulation in primary MEFs. We selected eight miRNA transcripts (let-7a-7d, let-7i, mir-106b-25, mir-15b-16-2, mir-22, mir-221-222, mir-23a-24-2, and mir-23b-24-1) whose expression was found to be modulated by E2F factors in the previous assays and who scored significantly for the predicted E2F recognition sites in their promoter sequences (Fig. 3). We also tested additional transcripts (mir-15a-16-1, mir-199a-1, and mir-143-145) whose expression patterns did not correlate properly with E2F activity. All three E2F factors, E2F1, E2F2, and E2F3, are expressed in primary MEFs, as detected by real-time RT-PCR (Fig. 4 A) and immunodetection with specific antibodies (Fig. 4B). We then performed chromatin immunoprecipitation (ChIP) followed by PCR with specific oligonucleotides to detect the predicted E2F binding within the promoters, as indicated in Fig. 3. Cdk1 promoter sequences were used as a positive control since the Cdk1 promoter is known to be directly regulated by E2F factors. The β-actin promoter was used as a negative control, since this promoter lacks E2F binding sites. In addition, as a control for specificity, we showed that an irrelevant antibody (anti-simian virus 40 [SV40] large T antigen) was unable to immunoprecipitate any of the various E2F target sequences in these experiments (Fig. 4C).
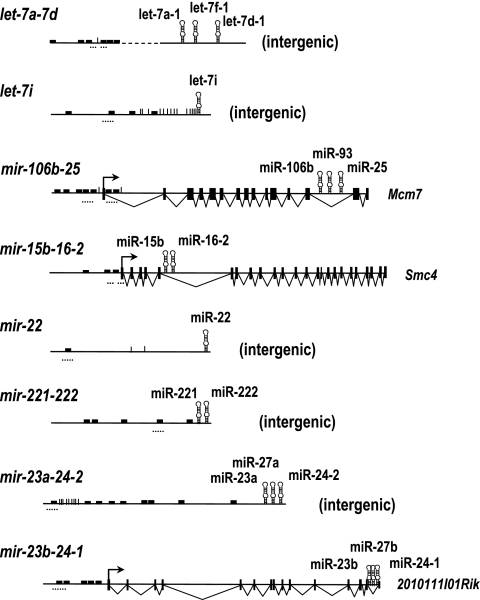
FIG. 3.
E2F sites in miRNA or miRNA clusters with differential expression in E2F-inducible or E2F-deficient cells. The structure of the mouse DNA loci expressing the different miRNAs is shown. let-7a-d, let-7i, mir-22, mir-221-222, and mir-23a-24-2 are intergenic transcriptional units, whereas mir-106b-25, mir-15b-16-2, and mir-23b-24-1 are expressed within the murine Mcm7, Smc4, and 2010111l01Rik transcripts, respectively. The predicted E2F recognition sites are indicated by small filled boxes, whereas the transcription start sites are indicated by vertical lines. The DNA regions analyzed by ChIP analysis are shown by small dotted horizontal lines.
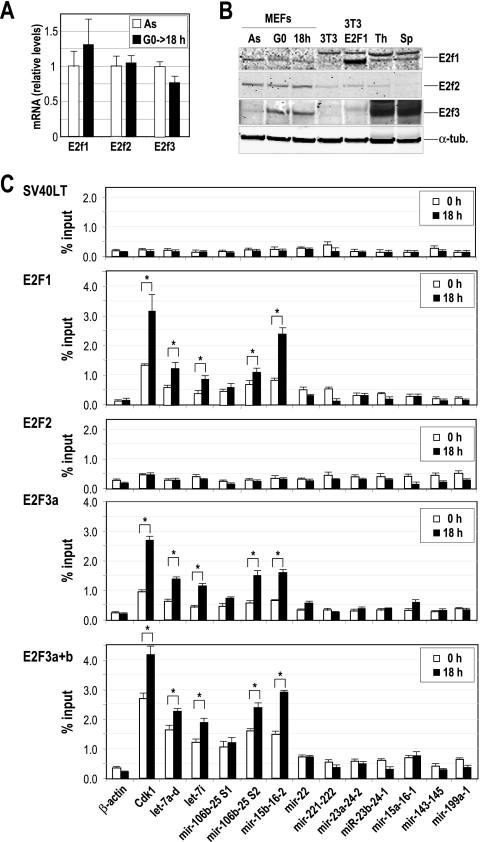
FIG. 4.
Chromatin immunoprecipitation of E2F transcription factors. (A) The relative expression of E2F factors is analyzed in primary MEFs by real-time PCR in asynchronously growing cells (As) or 18 h after serum stimulation of starved cells (G0→18 h). The mRNA levels were normalized versus the levels of GAPDH, and the mRNA level of E2F1 in As was considered to be 1. (B) Immunodetection of E2F1, E2F2, and E2F3 in As, serum-starved MEFs (G0) or G1/S primary MEFs (18 h after cell cycle entry). NIH 3T3 cells transfected with E2F1-expressing vectors or the parental cells were also used as a control. Thymus (Th) and spleens (Sp) from normal mice were also used as controls. α-Tubulin (α-tub.) was used as a loading control. (C) E2F1, E2F2, and E2F3 were immunoprecipitated from MEF protein lysates, and the predicted E2F binding sites of several miRNA clusters (dotted lines in Fig. 3) were amplified at t equals 0 h (serum starved) or 18 h after stimulation with serum. The promoter of Cdk1 was used as a positive control, whereas the β-actin promoter was used as a negative control. An unrelated antibody against the SV40 large T antigen (SV40LT) was used as a control for the IgG background at the specific miRNA promoters. Asterisks indicate significant differences (P < 0.01).
ChIP analyses carried out with wild-type MEFs revealed robust binding by E2F1 and E2F3 to several miRNA sequences, including those of the let-7a-7d, let-7i, mir-15b-16-2, and mir-106b-25 promoter sites (Fig. 4C). This interaction is significantly (P < 0.01) increased at 18 h, suggesting that these microRNAs are induced by E2F at the G1/S transition. We could not detect specific binding of E2F2 to any of the sequences tested, including that of Cdk1, suggesting that E2F2 is not bound to the promoter of these genes in primary MEFs. However, E2F2 was able to bind Cdk1 in parallel assays in lymphocytes as a control for the quality of the E2F2 antibody in these assays (34; data not shown). This analysis suggests a direct role for E2F1 and E2F3, but not E2F2, in the transcriptional induction of several miRNAs and miRNA clusters during G1 progression in primary fibroblasts.
Modulation of G1/S progression by E2F-induced miRNAs.
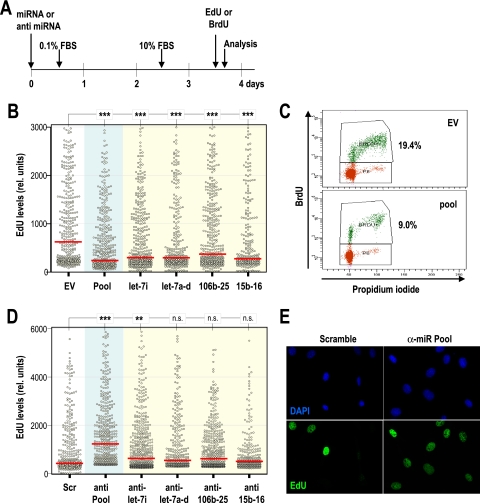
The induction of several miRNAs during G1 and their responsiveness to E2F transcription factors suggest that these miRNAs may induce G1/S progression or may contribute to the E2F-dependent transition to the S phase of the cell cycle. To test this possibility, we transfected MEFs with expression vectors carrying individual miRNAs or miRNA clusters induced by E2F1/3. Transfected cells were maintained in different concentrations of serum (0.1 to 5% FBS) to test whether miRNAs could favor entry into S phase under these suboptimal conditions. Surprisingly, most miRNAs tested did not favor cell cycle entry, but rather, they had negative consequences in S-phase entry (data not shown). To further analyze the effect of these miRNAs in cell cycle entry, MEFs were transfected with E2F-induced miRNAs, arrested in G0 during 48 h, and restimulated with serum, as indicated in Fig. 5 A. Cells were treated with a 4-h pulse of EdU and harvested 18 h after serum addition. To evaluate entry into S phase in individual cells, we then analyzed the distribution of EdU by high-throughput microscopy. As shown in Fig. 5B, the intensity of EdU was significantly lower in cells transfected with each of these miRNAs or with a pool containing the nine miRNAs induced by E2F (let-7i, let-7a, let-7f, let-7d, miR-106b, miR-93, miR-25, miR-15b, and miR-16-2). Similar results were obtained by analysis of BrdU (Fig. 5C) or thymidine (see Fig. S3 in the supplemental material) incorporation in similar assays. Expression of three other miRNAs or miRNA clusters that scored negative in the ChIP analysis (mir-22, mir-145-143, and mir-221-222) did not result in significant differences in S-phase entry (see Fig. S3 in the supplemental material).
FIG. 5.
E2F-induced miRNAs inhibit entry into S phase. (A) Schematic representation of the protocol followed to test the effect of microRNAs during cell cycle entry. Different miRNA clusters or anti-miRNA oligonucleotides were transfected in MEFs, and cells were arrested for 48 h without serum. Cells were harvested 18 h after serum stimulation and a 4-h pulse of EdU or BrdU. (B) Distribution of EdU intensity (relative units) per nucleus by high-throughput microscopy after transfection with individual miRNA clusters (yellow background) or a pool of the clusters (let-7i, let-7a-d, mir-15b-16-2, and mir-106b-25; blue background) that scored positive for direct interaction with E2F by ChIP analysis (Fig. 4). All individual clusters and the pool displayed significant inhibition of DNA replication in this assay. EV, empty vector. (C) Representative profiles of BrdU incorporation, as detected by cell cytometry, showing a significant reduction in progression into S phase after expression of the pool of miRNA clusters. (D) Distribution of EdU intensity per nucleus by high-throughput microscopy after transfection with anti-miRNA oligonucleotides against individual miRNA clusters (yellow background) or a pool of clusters (blue background). Scramble sequences (Scr) were used as a control. (E) Immunodetection of EdU (green) in MEFs transfected with scramble sequences or the pool of oligonucleotides against E2F-induced miRNAs (anti-miR pool). DNA is shown in blue after staining with DAPI (4′,6-diamidino-2-phenylindole). In panels B and D, red bars indicate median values. The statistical analysis was performed using Student's t test. ***, P < 0.001; **, P < 0.01; n.s., not significant (P > 0.05).
We next evaluated whether the absence of endogenous miRNAs may have a consequence in cell cycle entry in the presence of serum. Following use of the same experimental approach as that described in Fig. 5A, MEFs were transfected with anti-miR oligonucleotides that interfere with the expression of endogenous small RNAs (see Fig. S3 in the supplemental material). These cells were serum starved and restimulated with fresh serum, and the entry into S phase was monitored by EdU intake. As depicted in Fig. 5D and E, the elimination of the nine miRNAs encoded by the let-7i, let-7a-d, mir-106b-25, and mir-15b-16-2 clusters significantly promotes S-phase entry. The elimination of individual miRNAs has a partial or nonsignificant effect, suggesting that these miRNAs may cooperate in preventing S-phase entry. All together, these data suggest that E2F-induced miRNAs do not contribute to G1/S progression. Rather, their expression seems to have negative consequences in S-phase entry.
E2F-induced miRNAs prevent replicative stress.
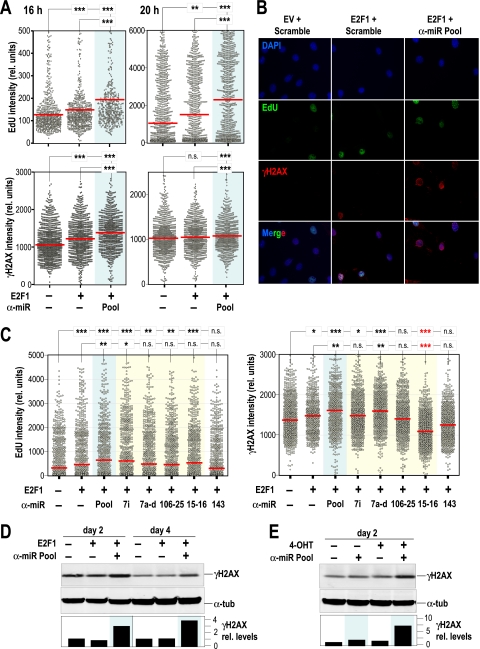
To further understand the relevance of miRNAs in E2F-dependent cell cycle entry, we transfected cells with E2F1 and tested the consequences of eliminating the endogenous E2F-induced miRNAs. E2F1 is known to promote S-phase entry, and its overexpression may lead to replicative stress (6). In fact, overexpression of E2F1 leads to increased incorporation of EdU at 16 h or 20 h after entry into the cell cycle. Interestingly, the effect of E2F1 is significantly more dramatic on cells devoid of E2F-induced miRNAs (Fig. 6 A and B). Furthermore, the unscheduled proliferation induced by E2F1 is accompanied by phosphorylation of H2AX (γH2AX), a widely used reporter of DNA damage and replicative stress. Importantly, the absence of E2F-induced miRNAs results in a significant increase in DNA damage, suggesting a protective role for E2F-induced miRNAs against excessive mitogenic signaling. The individual miRNA clusters seem to have a limited effect on preventing S-phase entry and DNA damage 18 h after transfection with E2F1 (Fig. 6C). However, all these E2F-induced miRNA clusters cooperate in protecting cells from the excessive mitogenic signaling caused by E2F1. The induction of DNA damage by E2F1 is not evident several days after transfection with E2F1-expressing vectors or after the treatment with tamoxifen in cells expressing the E2F1-ER constructs. However, the absence of E2F-induced miRNAs significantly increases the γH2AX signal 2 or 4 days after upregulation of E2F1 in both systems (Fig. 6D and E). Despite the significant effect on DNA damage signaling, the elimination of E2F-induced miRNAs does not result in a significant increase in apoptosis (see Fig. S4 in the supplemental material), suggesting possible independent roles of some of these miRNAs in apoptotic pathways. As an example, the specific elimination of miR-15 and miR-16 results in a significant increase in S-phase entry but, however, prevents the accumulation of γH2AX signals (Fig. 6C), suggesting additional targets apart from those involved in the G1/S transition. Thus, the elimination of E2F-induced miRNAs results in increased S-phase entry and DNA damage induced by E2F, although these defects do not provoke significant apoptosis, probably as a consequence of multiple signaling pathways modulated by these miRNAs.
FIG. 6.
E2F-induced miRNAs limit cell cycle progression and DNA damage induced by E2F. (A) Distribution of EdU or phosphorylated H2AX (γH2AX) intensity per nucleus in MEFs transfected with empty vector, a construct expressing E2F1, scramble oligonucleotides, or a pool of anti-miR oligonucleotides against the E2F-induced miRNAs (pool; blue background). Cells were transfected by following the protocol indicated in the legend to Fig. 5A and analyzed 16 or 20 h after serum stimulation. (B) Representative images of EdU (green) and γH2AX (red) in MEFs transfected with empty vector (EV), E2F1, scramble sequences, or the pool of oligonucleotides against E2F-induced miRNAs (anti-miR pool). DNA is shown in blue after staining with DAPI. (C) Distribution of EdU or phosphorylated H2AX (γH2AX) intensity per nucleus in MEFs transfected with E2F1 and anti-miR oligonucleotides against the individual miRNAs expressed in the let-7i, let-7a-d, mir-106b-25, or mir-15b-16-2 clusters (yellow background) or a pool of all these anti-miR oligonucleotides (pool; blue background). Cells were transfected by following the protocol indicated in the legend to Fig. 5A and analyzed 18 h after stimulation with serum. In panels A and C, red bars indicate median values. The statistical analysis was performed using Student's t test. ***, P < 0.001; **, P < 0.01; n.s., not significant (P > 0.05). Please note that anti-miRs against the mir-15b-16-2 cluster result in a significant and specific decrease in the γH2AX signal, suggesting specific targets involved in this pathway (red asterisks). (D) Immunodetection of γH2AX in protein lysates from MEFs 2 and 4 days after transfection with E2F1 and/or the anti-miR oligonucleotide pool. (E) Immunodetection of the indicated proteins in protein lysates from E2F1-ER cells 2 days after induction of E2F1 with 4-OHT. α-Tubulin was used as a loading control. The “−” symbols indicate empty vector or scramble sequences. α-tub, α-tubulin.
E2F-induced miRNAs significantly modulate cell cycle regulators and E2F-induced proteins.
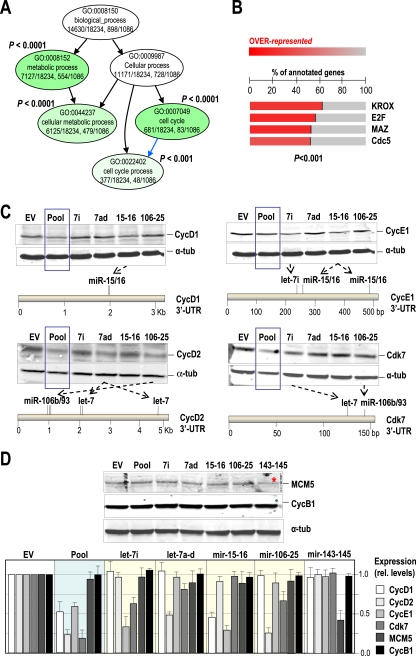
The fact that these E2F-induced miRNAs inhibit cell cycle progression suggests that they may participate to limit the induction of E2F targets. We therefore performed a bioinformatics analysis of the predicted targets of E2F-induced miRNAs. The combined predicted targets of let-7a, let-7f, let-7d, let-7i, miR-15b, miR-16-2, miR-106b, miR-93, and miR-25 were scored using miRBase or the EIMMo miRNA target prediction server and then analyzed for enrichment using specific gene ontology terms. We used very low P values (P < 0.001) to avoid inconsistent results due to the use of different databases or algorithms, as recently suggested (59). As shown in Fig. 7 A, this analysis indicates that E2F-induced miRNAs preferentially modulate targets involved in cellular processes such as cellular metabolism and the cell cycle (P < 0.0001). These targets include cyclin-dependent kinases (CDKs) or CDK-activating proteins, such as D-type and E-type G1 cyclins, CDC25 phosphatases, or even E2F factors like E2F1 (for a list of the specific targets included in these categories, see Table S1 in the supplemental material).
FIG. 7.
E2F-induced miRNAs target critical cell cycle regulators. (A) E2F-induced miRNA targets are enriched in cell cycle regulatory proteins. Only miRNA targets with P values of <0.001 (EIMMO algorithm) were selected for this analysis. Gene ontology (GO) analysis using the parent-child-union Bonferroni (Ontologizer) algorithm of predicted targets of these miRNAs shows a significant (adjusted P value < 0.001) enrichment in metabolic genes and cell cycle regulators. (B) Analysis of transcription factor recognition sites at the promoter region of the gene targets of E2F-induced miRNAs. A significant (adjusted P value < 0.001; FatiScan algorithm) enrichment is found for targets putatively regulated by the KROX, E2F, MAZ, or Cdc5 transcription factors. (C) Different miRNA clusters, the pool of E2F-induced miRNAs (blue box), or the empty vector (EV) were used to transfect primary MEFs, and the levels of cyclin D1, cyclin D2, cyclin E1, and Cdk7 were assessed by immunoblotting. α-Tubulin was used as a loading control. The positions of the predicted target sites for E2F-induced miRNAs are indicated in the 3′-UTRs of these genes. (D) Immunodetection of MCM5 and cyclin B1 after transfection with the indicated constructs. The red asterisk indicates that MCM5 is a predicted target of miR-145. In the histogram, protein levels from at least two different assays were normalized versus the levels of α-tubulin after expression of the indicated constructs.
In addition to the enrichment in cell cycle regulators, we also detected that the predicted targets for E2F-induced miRNAs display a highly significant (P < 0.001) enrichment for E2F-induced coding genes (Fig. 7B; see also Table S2 in the supplemental material). A similar level of enrichment was found for genes induced by additional transcription factors that may be involved in cell cycle progression, such as the Myc-associated zinc-finger transcription factor MAZ or genes containing KROX sequences which are recognized by early growth response C2H2-type zinc-finger proteins (EGR-1 to -4), which modulate genes involved in mitogenesis as well as differentiation (Fig. 7B).
To test the protein levels of some predicted or reported targets involved in cell cycle progression, cells were transfected with miRNAs and arrested in G0, and proteins were detected 18 h after serum stimulation. Some critical cell cycle regulators, such as D-type (D1 and D2) and E-type cyclins or the CDK-activating kinase CDK7, are indeed downregulated in MEFs overexpressing specific E2F-induced miRNAs or miRNA clusters in correlation with the presence of specific recognition sequences in their corresponding 3′-UTRs (Fig. 7C). Thus, cyclin D1 is downregulated by the pool of miRNAs, as well as specific overexpression of miR-15/16, as reported previously (47). Similar correlations are observed for cyclin D1, cyclin E1, and Cdk7. To discard possible general effects on cell cycle arrest, we also tested MCM5 and cyclin B1, two molecules whose protein levels correlate with cell proliferation. MCM5 is not targeted by any of these E2F-induced miRNAs, but it is a predicted target of miR-145. As represented in Fig. 7D, only the mir-143-145 cluster downregulates MCM5 in these assays. Similarly, cyclin B1, which is not a predicted target of any of the miRNAs studied in this work, does not change in these assays. Quantification of these results indicates that the pool of E2F-induced miRNAs is able to downregulate cyclins D1, D2, and E1 and Cdk7, but not MCM5 or cyclin B1. In addition, the effect of each the different miRNA clusters is target specific, and downregulation of specific proteins does not seem to simply represent overall proliferation defects. All together, these data suggest that miRNAs induced by E2F during the G1/S transition may help to limit the protein levels of critical cell cycle regulators, thus preventing excessive replication and DNA damage.
DISCUSSION
Transcription of miRNAs is regulated similarly to that of protein-coding genes, and these small noncoding RNAs are usually expressed by RNA polymerase II (33, 40). Although the induction of coding genes during cell cycle entry has been deeply studied, the response of miRNAs to serum stimulation is largely unknown. Our analysis in primary cells suggests that about 37% (59/159) of expressed miRNAs are induced upon stimulation with serum. About half of these induced miRNAs display a rapid expression after mitogenic signaling, suggesting a direct regulation during the early phases of the cells cycle similar to that observed for protein-coding genes (38).
Most mitogenic pathways that control cell cycle entry regulate the induction of D-type cyclins and the subsequent activation of CDKs, the catalytic activity required for G1/S progression (48). Active cyclin D-CDK4/6 complexes phosphorylate and inactivate the retinoblastoma protein (pRB). pRB is a general repressor of transcription by recruiting chromatin remodeling complexes to promoters and repressing their transcription. In fact, pRB directly binds E2F transcription factors, bringing the repressor machinery to gene promoters that contain E2F recognition sites. By inactivating pRB, cyclin-CDK complexes activate E2F and promote the expression of many genes required for DNA replication and mitosis (68). E2F family members have been divided into positive regulators of the cell cycle (activators; E2F1-3) and negative regulators of the cell cycle (repressors; E2F4-8) based on their transcriptional roles in vitro and conserved structural features (67, 68). The increased expression of E2F targets observed in E2F2-null cells has challenged this simplistic view by proposing that E2F2 may also have major repressor functions in several cell types, such as lymphocytes and fibroblasts (34, 53). Indeed, recent data suggest that not only E2F2 but also E2F1 and E2F3 may switch from activators to repressors depending on the differentiation stage of the cells and that these factors are not universally required for normal mammalian cell division (15, 17). It would be interesting to analyze to what extent E2F-induced miRNAs participate in this repressor function in differentiated cells.
Using E2F-inducible and E2F-deficient cells, we have identified in our analysis a set of miRNAs or miRNA clusters whose expression is positively modulated at least by E2F1 and E2F3. In addition, ChIP assays have identified direct binding of these transcription factors to the regulatory sequences of four genes, expressing the let-7a-7d, let-7i, mir-15b-16-2, or mir-106b-25 transcripts, during the G1/S transition. One of these E2F-induced miRNA clusters is expressed from the Mcm7 gene, a known target of E2F transcription factors (Fig. 3) (4). Indeed, approximately 50% of miRNAs are located in the introns of coding genes, and these intronic miRNAs are generally transcribed coincidentally with their host genes (62). All together, these data suggest that E2F1 and E2F3 induce at least let-7a-7d, let-7i, mir-15b-16-2, or mir-106b-25 during the early phases of the cell cycle.
Although we initially explored the possibility that these E2F-induced miRNAs could contribute to E2F-induced G1/S progression, our data (in agreement with published evidences) suggest that these miRNAs have rather negative consequences on S-phase entry and downregulate critical cell cycle regulators, such as cyclins or CDKs. Indeed, the predicted targets of these E2F-induced miRNAs are significantly enriched in E2F-target coding genes, suggesting that these miRNAs limit the cellular effects triggered by E2F. Thus, let-7 family members targeting multiple cell cycle regulators, including CDK4, CDK6, CDC25A, cyclin D1, cyclin D2, cyclin D3, and cyclin A (39, 43, 63, 72), are also able to directly downregulate E2F1 and E2F2 (7). Similarly, miR-15/16 miRNAs are known to regulate cell cycle progression by repressing critical cell cycle regulators, such as D- and E-type cyclins or CDKs, like CDK6 or CDK7 (20, 41, 42, 46, 47; our data). On the other hand, it has been reported that the miR-106b family regulates p21Cip1 and promotes cell cycle progression in specific cell lines (36). However, downregulation or p21Cip1 does not promote cell cycle progression in primary MEFs (56), and indeed, it may be important to support cyclin D-CDK4 activity in these primary cells (64).
These data indicate that the effect of let-7a-7d, let-7i, mir-15b-16-2, or mir-106b-25 limits cell cycle entry by E2F. It is tempting to speculate that these miRNAs may control the proper entry into S phase by limiting the excess of positive regulators of the cell cycle to avoid replicative stress in response to these transcription factors or to excessive mitogenic signaling. Indeed, it has been reported that overexpression of E2F is one of the major inductors of replicative stress and that the subsequent DNA damage response may lead to senescence in primary cells or early tumors (31). As a prediction, E2F-induced miRNAs may be linked to cellular senescence, as recently suggested for miR-15/16 and miR-106b/25 (45).
Since we have used restrictive criteria in the different steps of this identification, it is possible that additional miRNAs that are induced by serum may be direct or indirect targets of E2F. For instance, E2F factors, mostly E2F3, are known to directly induce the oncogenic mir-17-92 cluster (71). However, several mir-17-92 miRNAs, including miR-17, miR-18a, and miR-20a, map to class I in our analysis (Fig. 1), suggesting that induction of this cluster by E2F is not an early event in the exit from quiescence but rather an accumulative process in proliferating cells. Since two miRNAs expressed by this oncogenic cluster, miR-17-5p and miR-20a, also target E2F factors (55) or specific cyclins (73), these miRNAs may also act by modulating a feedback loop to control E2F activity upon continuous culture. Although we have not detected direct binding by ChIP assays during G0/G1, our results suggest that oncogenic miR-221 and miR-222 may also be induced by E2F factors in an accumulative manner, since they are dramatically reduced in E2F-null cells (Fig. 2). Continuous E2F-dependent induction of mir-221-222 and mir-17-92 may result in the elimination of the cell cycle inhibitors p21Cip1, p27Kip1, and p57Kip2 (25-27, 35, 44, 69), thus promoting maintained cell proliferation in these cells.
Entry into the cell cycle is one of the most critical phases of the cell cycle that is deregulated during tumor development (49). E2F-induced miRNAs may therefore have relevant roles in human cancer. Indeed, let-7 and miR-16 family members have multiple roles as tumor suppressors by targeting cell cycle regulators or critical oncogenes such as Ras or Myc (37, 41, 60). let-7i, in particular, is significantly reduced in ovarian cancer and has significant value as an independent prognostic factor (72). On the other hand, E2F factors induce relevant oncogene miRNAs, such as the mir-17-92 cluster (71) and perhaps mir-221-222. In this case, these miRNAs are not induced in exiting quiescence but rather accumulate with passages and may have critical roles by inhibiting cell cycle inhibitors of the CIP/KIP family. Thus, E2F factors induce different miRNAs with the capacity of either inhibiting (early-induced miRNA) or promoting (late-induced miRNA) cell cycle progression. These data are in agreement with the emerging picture of miRNA function in mammals, proposing that these noncoding genes not only participate in executive decisions but also perform much of the grunt work to micromanage protein output (5), in this case, by regulating the cell division cycle.
Supplementary Material
Acknowledgments
We thank Lucía Barrado and Susana Temiño for technical assistance, Jon Fernández and Olatz Zenarruzabeitia for help with generation of E2F3-knockdown fibroblasts, J. Méndez for providing MCM5 antibodies, K. Helin for kindly providing E2F-inducible cells and vectors, and Ignacio Pérez de Castro and Oscar Fernández-Capetillo for valuable help and discussions.
U.L. is a recipient of a Basque Government fellowship for graduate studies. This work was funded by grants from the Association International for Cancer Research (AICR 08-0188 to M.M.), Fundación Mutua Madrileña Automovilista and Fundación Ramón Areces (to M.M.), Basque Government Department of Industry (Etortek-IE06-178 to A.M.Z.), and the MICINN (SAF2006-09437 and SAF2009-11426 to J.F.-P., SAF2005-07930-C02-01 and SAF2009-12037 to A.M.Z., and SAF2006-05186 and SAF2009-07973 to M.M.). The CBM/UAM group is supported by the CIBERER network of the MICINN. The Cell Division and Cancer Group of the CNIO is supported by the OncoCycle Programme (grant S-BIO-0283-2006) from the Comunidad de Madrid. A.M.Z. and M.M. are supported by the OncoBIO Consolider-Ingenio 2010 Programme (grant CSD2007-00017) from the MICINN, Madrid, Spain.
Footnotes
Published ahead of print on 19 April 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aguda, B. D., Y. Kim, M. G. Piper-Hunter, A. Friedman, and C. B. Marsh. 2008. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc. Natl. Acad. Sci. U. S. A. 105:19678-19683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Shahrour, F., P. Minguez, J. M. Vaquerizas, L. Conde, and J. Dopazo. 2005. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 33:W460-W464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. [DOI] [PubMed] [Google Scholar]
- 4.Arata, Y., M. Fujita, K. Ohtani, S. Kijima, and J. Y. Kato. 2000. Cdk2-dependent and -independent pathways in E2F-mediated S phase induction. J. Biol. Chem. 275:6337-6345. [DOI] [PubMed] [Google Scholar]
- 5.Bartel, D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. [DOI] [PubMed] [Google Scholar]
- 7.Bhat-Nakshatri, P., G. Wang, N. R. Collins, M. J. Thomson, T. R. Geistlinger, J. S. Carroll, M. Brown, S. Hammond, E. F. Srour, Y. Liu, and H. Nakshatri. 2009. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 37:4850-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonci, D., V. Coppola, M. Musumeci, A. Addario, R. Giuffrida, L. Memeo, L. D'Urso, A. Pagliuca, M. Biffoni, C. Labbaye, M. Bartucci, G. Muto, C. Peschle, and R. De Maria. 2008. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 14:1271-1277. [DOI] [PubMed] [Google Scholar]
- 9.Brosh, R., R. Shalgi, A. Liran, G. Landan, K. Korotayev, G. H. Nguyen, E. Enerly, H. Johnsen, Y. Buganim, H. Solomon, I. Goldstein, S. Madar, N. Goldfinger, A. L. Borresen-Dale, D. Ginsberg, C. C. Harris, Y. Pilpel, M. Oren, and V. Rotter. 2008. p53-repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol. Syst. Biol. 4:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bueno, M. J., I. P. de Castro, and M. Malumbres. 2008. Control of cell proliferation pathways by microRNAs. Cell Cycle 7:3143-3148. [DOI] [PubMed] [Google Scholar]
- 11.Bueno, M. J., I. Perez de Castro, M. Gomez de Cedron, J. Santos, G. A. Calin, J. C. Cigudosa, C. M. Croce, J. Fernandez-Piqueras, and M. Malumbres. 2008. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13:496-506. [DOI] [PubMed] [Google Scholar]
- 12.Calin, G. A., and C. M. Croce. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6:857-866. [DOI] [PubMed] [Google Scholar]
- 13.Calin, G. A., C. D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, E. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, L. Rassenti, T. Kipps, M. Negrini, F. Bullrich, and C. M. Croce. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 99:15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, T. C., D. Yu, Y. S. Lee, E. A. Wentzel, D. E. Arking, K. M. West, C. V. Dang, A. Thomas-Tikhonenko, and J. T. Mendell. 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, D., M. Pacal, P. Wenzel, P. S. Knoepfler, G. Leone, and R. Bremner. 2009. Division and apoptosis of E2f-deficient retinal progenitors. Nature 462:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chivukula, R. R., and J. T. Mendell. 2008. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem. Sci. 33:474-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong, J. L., P. L. Wenzel, M. T. Saenz-Robles, V. Nair, A. Ferrey, J. P. Hagan, Y. M. Gomez, N. Sharma, H. Z. Chen, M. Ouseph, S. H. Wang, P. Trikha, B. Culp, L. Mezache, D. J. Winton, O. J. Sansom, D. Chen, R. Bremner, P. G. Cantalupo, M. L. Robinson, J. M. Pipas, and G. Leone. 2009. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 462:930-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen, J., P. Cloos, U. Toftegaard, D. Klinkenberg, A. P. Bracken, E. Trinh, M. Heeran, L. Di Stefano, and K. Helin. 2005. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 33:5458-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croce, C. M. 2009. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10:704-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshpande, A., A. Pastore, A. J. Deshpande, Y. Zimmermann, G. Hutter, M. Weinkauf, C. Buske, W. Hiddemann, and M. Dreyling. 2009. 3′UTR mediated regulation of the cyclin D1 proto-oncogene. Cell Cycle 8:3584-3592. [DOI] [PubMed] [Google Scholar]
- 21.Esquela-Kerscher, A., and F. J. Slack. 2006. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 6:259-269. [DOI] [PubMed] [Google Scholar]
- 22.Eulalio, A., E. Huntzinger, and E. Izaurralde. 2008. Getting to the root of miRNA-mediated gene silencing. Cell 132:9-14. [DOI] [PubMed] [Google Scholar]
- 23.Field, S. J., F. Y. Tsai, F. Kuo, A. M. Zubiaga, W. G. Kaelin, Jr., D. M. Livingston, S. H. Orkin, and M. E. Greenberg. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85:549-561. [DOI] [PubMed] [Google Scholar]
- 24.Filipowicz, W., S. N. Bhattacharyya, and N. Sonenberg. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9:102-114. [DOI] [PubMed] [Google Scholar]
- 25.Fontana, L., M. E. Fiori, S. Albini, L. Cifaldi, S. Giovinazzi, M. Forloni, R. Boldrini, A. Donfrancesco, V. Federici, P. Giacomini, C. Peschle, and D. Fruci. 2008. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One 3:e2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fornari, F., L. Gramantieri, M. Ferracin, A. Veronese, S. Sabbioni, G. A. Calin, G. L. Grazi, C. Giovannini, C. M. Croce, L. Bolondi, and M. Negrini. 2008. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27:5651-5661. [DOI] [PubMed] [Google Scholar]
- 27.Galardi, S., N. Mercatelli, E. Giorda, S. Massalini, G. V. Frajese, S. A. Ciafre, and M. G. Farace. 2007. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 282:23716-23724. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Higuera, I., E. Manchado, P. Dubus, M. Canamero, J. Mendez, S. Moreno, and M. Malumbres. 2008. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 10:802-811. [DOI] [PubMed] [Google Scholar]
- 29.Giangrande, P. H., T. C. Hallstrom, C. Tunyaplin, K. Calame, and J. R. Nevins. 2003. Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol. Cell. Biol. 23:3707-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giannakakis, A., R. Sandaltzopoulos, J. Greshock, S. Liang, J. Huang, K. Hasegawa, C. Li, A. O'Brien-Jenkins, D. Katsaros, B. L. Weber, C. Simon, G. Coukos, and L. Zhang. 2008. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 7:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halazonetis, T. D., V. G. Gorgoulis, and J. Bartek. 2008. An oncogene-induced DNA damage model for cancer development. Science 319:1352-1355. [DOI] [PubMed] [Google Scholar]
- 32.He, L., X. He, S. W. Lowe, and G. J. Hannon. 2007. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat. Rev. Cancer 7:819-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hobert, O. 2008. Gene regulation by transcription factors and microRNAs. Science 319:1785-1786. [DOI] [PubMed] [Google Scholar]
- 34.Infante, A., U. Laresgoiti, J. Fernandez-Rueda, A. Fullaondo, J. Galan, R. Diaz-Uriarte, M. Malumbres, S. J. Field, and A. M. Zubiaga. 2008. E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle 7:3915-3927. [DOI] [PubMed] [Google Scholar]
- 35.Inomata, M., H. Tagawa, Y. M. Guo, Y. Kameoka, N. Takahashi, and K. Sawada. 2009. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood 113:396-402. [DOI] [PubMed] [Google Scholar]
- 36.Ivanovska, I., A. S. Ball, R. L. Diaz, J. F. Magnus, M. Kibukawa, J. M. Schelter, S. V. Kobayashi, L. Lim, J. Burchard, A. L. Jackson, P. S. Linsley, and M. A. Cleary. 2008. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 28:2167-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanovska, I., and M. A. Cleary. 2008. Combinatorial microRNAs: working together to make a difference. Cell Cycle 7:3137-3142. [DOI] [PubMed] [Google Scholar]
- 38.Iyer, V. R., M. B. Eisen, D. T. Ross, G. Schuler, T. Moore, J. C. Lee, J. M. Trent, L. M. Staudt, J. Hudson, Jr., M. S. Boguski, D. Lashkari, D. Shalon, D. Botstein, and P. O. Brown. 1999. The transcriptional program in the response of human fibroblasts to serum. Science 283:83-87. [DOI] [PubMed] [Google Scholar]
- 39.Johnson, C. D., A. Esquela-Kerscher, G. Stefani, M. Byrom, K. Kelnar, D. Ovcharenko, M. Wilson, X. Wang, J. Shelton, J. Shingara, L. Chin, D. Brown, and F. J. Slack. 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 67:7713-7722. [DOI] [PubMed] [Google Scholar]
- 40.Kim, V. N., J. Han, and M. C. Siomi. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126-139. [DOI] [PubMed] [Google Scholar]
- 41.Klein, U., M. Lia, M. Crespo, R. Siegel, Q. Shen, T. Mo, A. Ambesi-Impiombato, A. Califano, A. Migliazza, G. Bhagat, and R. Dalla-Favera. 2010. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17:28-40. [DOI] [PubMed] [Google Scholar]
- 42.Lee, S. O., T. Masyuk, P. Splinter, J. M. Banales, A. Masyuk, A. Stroope, and N. Larusso. 2008. MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J. Clin. Invest. 118:3714-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legesse-Miller, A., O. Elemento, S. J. Pfau, J. J. Forman, S. Tavazoie, and H. A. Coller. 2009. let-7 overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J. Biol. Chem. 284:6605-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.le Sage, C., R. Nagel, D. A. Egan, M. Schrier, E. Mesman, A. Mangiola, C. Anile, G. Maira, N. Mercatelli, S. A. Ciafre, M. G. Farace, and R. Agami. 2007. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 26:3699-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, G., C. Luna, J. Qiu, D. L. Epstein, and P. Gonzalez. 2009. Alterations in microRNA expression in stress-induced cellular senescence. Mech. Ageing Dev. 130:731-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linsley, P. S., J. Schelter, J. Burchard, M. Kibukawa, M. M. Martin, S. R. Bartz, J. M. Johnson, J. M. Cummins, C. K. Raymond, H. Dai, N. Chau, M. Cleary, A. L. Jackson, M. Carleton, and L. Lim. 2007. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 27:2240-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, Q., H. Fu, F. Sun, H. Zhang, Y. Tie, J. Zhu, R. Xing, Z. Sun, and X. Zheng. 2008. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 36:5391-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malumbres, M., and M. Barbacid. 2009. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9:153-166. [DOI] [PubMed] [Google Scholar]
- 49.Malumbres, M., and M. Barbacid. 2001. To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer 1:222-231. [DOI] [PubMed] [Google Scholar]
- 50.Malumbres, M., R. Sotillo, D. Santamaria, J. Galan, A. Cerezo, S. Ortega, P. Dubus, and M. Barbacid. 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118:493-504. [DOI] [PubMed] [Google Scholar]
- 51.Martin, A., J. Odajima, S. L. Hunt, P. Dubus, S. Ortega, M. Malumbres, and M. Barbacid. 2005. Cdk2 is dispensable for cell cycle inhibition and tumor suppression mediated by p27(Kip1) and p21(Cip1). Cancer Cell 7:591-598. [DOI] [PubMed] [Google Scholar]
- 52.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murga, M., O. Fernandez-Capetillo, S. J. Field, B. Moreno, L. R. Borlado, Y. Fujiwara, D. Balomenos, A. Vicario, A. C. Carrera, S. H. Orkin, M. E. Greenberg, and A. M. Zubiaga. 2001. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity 15:959-970. [DOI] [PubMed] [Google Scholar]
- 54.Murga, M., I. Jaco, Y. Fan, R. Soria, B. Martinez-Pastor, M. Cuadrado, S. M. Yang, M. A. Blasco, A. I. Skoultchi, and O. Fernandez-Capetillo. 2007. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 178:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839-843. [DOI] [PubMed] [Google Scholar]
- 56.Pantoja, C., and M. Serrano. 1999. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene 18:4974-4982. [DOI] [PubMed] [Google Scholar]
- 57.Petrocca, F., R. Visone, M. R. Onelli, M. H. Shah, M. S. Nicoloso, I. de Martino, D. Iliopoulos, E. Pilozzi, C. G. Liu, M. Negrini, L. Cavazzini, S. Volinia, H. Alder, L. P. Ruco, G. Baldassarre, C. M. Croce, and A. Vecchione. 2008. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13:272-286. [DOI] [PubMed] [Google Scholar]
- 58.Pickering, M. T., B. M. Stadler, and T. F. Kowalik. 2009. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene 28:140-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ritchie, W., S. Flamant, and J. E. Rasko. 2009. Predicting microRNA targets and functions: traps for the unwary. Nat. Methods 6:397-398. [DOI] [PubMed] [Google Scholar]
- 60.Roush, S., and F. J. Slack. 2008. The let-7 family of microRNAs. Trends Cell Biol. 18:505-516. [DOI] [PubMed] [Google Scholar]
- 61.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 62.Saini, H. K., S. Griffiths-Jones, and A. J. Enright. 2007. Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. U. S. A. 104:17719-17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schultz, J., P. Lorenz, G. Gross, S. Ibrahim, and M. Kunz. 2008. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 18:549-557. [DOI] [PubMed] [Google Scholar]
- 64.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 65.Stefani, G., and F. J. Slack. 2008. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9:219-230. [DOI] [PubMed] [Google Scholar]
- 66.Tazawa, H., N. Tsuchiya, M. Izumiya, and H. Nakagama. 2007. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. U. S. A. 104:15472-15477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 68.van den Heuvel, S., and N. J. Dyson. 2008. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9:713-724. [DOI] [PubMed] [Google Scholar]
- 69.Visone, R., L. Russo, P. Pallante, I. De Martino, A. Ferraro, V. Leone, E. Borbone, F. Petrocca, H. Alder, C. M. Croce, and A. Fusco. 2007. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr. Relat. Cancer 14:791-798. [DOI] [PubMed] [Google Scholar]
- 70.Voorhoeve, P. M., C. le Sage, M. Schrier, A. J. Gillis, H. Stoop, R. Nagel, Y. P. Liu, J. van Duijse, J. Drost, A. Griekspoor, E. Zlotorynski, N. Yabuta, G. De Vita, H. Nojima, L. H. Looijenga, and R. Agami. 2006. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124:1169-1181. [DOI] [PubMed] [Google Scholar]
- 71.Woods, K., J. M. Thomson, and S. M. Hammond. 2007. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 282:2130-2134. [DOI] [PubMed] [Google Scholar]
- 72.Yang, N., S. Kaur, S. Volinia, J. Greshock, H. Lassus, K. Hasegawa, S. Liang, A. Leminen, S. Deng, L. Smith, C. N. Johnstone, X. M. Chen, C. G. Liu, Q. Huang, D. Katsaros, G. A. Calin, B. L. Weber, R. Butzow, C. M. Croce, G. Coukos, and L. Zhang. 2008. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 68:10307-10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu, Z., C. Wang, M. Wang, Z. Li, M. C. Casimiro, M. Liu, K. Wu, J. Whittle, X. Ju, T. Hyslop, P. McCue, and R. G. Pestell. 2008. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell Biol. 182:509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, Y., T. Chao, R. Li, W. Liu, Y. Chen, X. Yan, Y. Gong, B. Yin, W. Liu, B. Qiang, J. Zhao, J. Yuan, and X. Peng. 2009. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J. Mol. Med. 87:43-51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.