Abstract
Maintenance of energy homeostasis is a fundamental requirement for organismal fitness: defective glucose homeostasis underlies numerous metabolic diseases and cancer. At the cellular level, the ability to sense and adapt to changes in intracellular glucose levels is an essential component of this strategy. The basic helix-loop-helix-leucine zipper (bHLHZip) transcription factor complex MondoA-Mlx plays a central role in the transcriptional response to intracellular glucose concentration. MondoA-Mlx complexes accumulate in the nucleus in response to high intracellular glucose concentrations and are required for 75% of glucose-induced transcription. We show here that, rather than simply controlling nuclear accumulation, glucose is required at two additional steps to stimulate the transcription activation function of MondoA-Mlx complexes. Following nuclear accumulation, glucose is required for MondoA-Mlx occupancy at target promoters. Next, glucose stimulates the recruitment of a histone H3 acetyltransferase to promoter-bound MondoA-Mlx to trigger activation of gene expression. Our experiments establish the mechanistic circuitry by which cells sense and respond transcriptionally to various intracellular glucose levels.
The ability to sense and respond to changing nutrient levels in the surrounding environment is a central requirement for all life (24). No more fundamental energy source exists than the six-carbon sugar, glucose. Defects in glucose metabolism underlie numerous heritable genetic diseases, Alzheimer's disease, diabetes, and cancer (10, 23, 28).
Two basic helix-loop-helix leucine zipper (bHLHZip) transcription factor complexes, MondoA-Mlx and ChREBP-Mlx, act as transcriptional biosensors of glucose flux (6, 25). ChREBP is expressed predominantly in liver and upregulates genes involved in the conversion of glucose to lipid for energy storage and cell growth (3, 12, 15, 26). MondoA is expressed predominantly in skeletal muscle and upregulates glycolytic target genes (22). MondoA-Mlx and ChREBP-Mlx appear to be responsible for the majority of glucose-dependent transcription in their largely nonoverlapping target tissues (2, 7, 16, 25).
MondoA-Mlx heterodimers shuttle between mitochondria and the nucleus, fostering communication between these essential organelles (22). In the presence of glucose, MondoA-Mlx accumulates in the nucleus, facilitating activation of gene expression (25). MondoA contains five N-terminal domains known as the Mondo conserved regions (MCRs), which regulate nuclear accumulation of the heterodimer (8). Amino acids 126 to 135 (LTKLFECMTL [underlining indicates hydrophobic amino acids]) within the MCRII domain of MondoA define a Crm1-dependent nuclear export sequence (NES), which follows the hydrophobic-rich consensus Φ-X3-Φ-X2-Φ-X-Φ (11). Point mutation of methionine 133 to alanine within the NES ablates nuclear export of MondoA (8); thus MondoA(M133A) is a useful tool to study NES-dependent function of the heterodimer. Whether high concentrations of intracellular glucose disrupt interaction between MCRII and Crm1, leading to MondoA-Mlx accumulation in the nucleus, is unknown.
Thioredoxin-interacting protein (TXNIP) is a direct and glucose-dependent target of MondoA (1, 25). TXNIP negatively regulates glucose uptake (13, 21), and, hence, defects in TXNIP expression or function may precede the onset of type 2 diabetes (5, 20). The glucose-dependent occupancy of MondoA-Mlx at TXNIP requires a double-E-box-like promoter element known as the carbohydrate response element (ChoRE) (18, 25). We have shown that TXNIP functions downstream of MondoA to negatively regulate glucose uptake when intracellular glucose concentration is exceedingly high (25).
Both glucose and the nonmetabolizable glucose analog 2-deoxyglucose (2DOG) promote nuclear accumulation of MondoA-Mlx. Our previous work demonstrates that phosphorylation of glucose by hexokinases to glucose-6-phosphate (G6P) is critical for nuclear accumulation of MondoA-Mlx (25). Two models may explain how G6P regulates nuclear accumulation of MondoA. First, the MondoA-Mlx heterodimer could reside at the mitochondria when G6P levels are low and translocate to the nucleus when G6P levels are high. Alternatively, MondoA-Mlx could shuttle between the mitochondria and nucleus in the presence or absence of G6P. The latter model predicts that G6P augments the nuclear accumulation of the heterodimer through increase of nuclear import, increase in promoter occupancy, and/or decrease of nuclear export.
MondoA-Mlx is a predominant regulator of glucose-induced transcription and, via its regulation of TXNIP, activates a negative feedback loop governing glucose uptake. We show here that rather than simply controlling nuclear accumulation of MondoA-Mlx, glucose regulates three steps—nuclear accumulation, promoter occupancy, and coactivator recruitment—leading to transcriptionally active heterocomplexes.
MATERIALS AND METHODS
Cloning and mutagenesis.
Plasmids encoding V5-tagged MondoA and Flag-tagged Mlx have been described (8, 13, 25). Homo sapiens MondoA was cloned into the pWZL-BLAST vector (AddGene) for retroviral expression in mouse embryonic fibroblasts (MEFs). Mutations in MondoA were generated using the QuikChange XL site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions. Mutation was confirmed by sequencing of the entire MondoA cDNA. Gal4-tagged Crm1 was a gift of Bryan Cullen.
Cell culture and transient transfection experiments.
Cells were passaged in Dulbecco modified Eagle medium (DMEM) with antibiotics and 10% bovine calf serum (L6 cells) or 10% fetal bovine serum (FBS) (MEFs). L6 myoblasts and MEFs were transfected using Lipofectamine 2000 at 5 μg DNA: 4-μl lipid ratios and a 1:100 dilution of lipid to Opti-MEM (Invitrogen). All MondoA transfections were performed in the presence of cotransfected Flag-Mlx. Equal expression levels of overexpressed wild-type (wt) and mutant MondoA were confirmed by Western blotting. Glucose starvations were performed by incubating cells overnight in glucose-free DMEM plus 2% FBS and antibiotics. Treatment of glucose-starved cells with glucose (25 mM or 20 mM when comparing directly to 2-DOG) or 2-DOG (20 mM) was performed as described previously (25). Leptomycin B (LMB) (Biomol) was added to the media of transfected cells at a final concentration of 10 ng/μl for 4 h. Luciferase reporter assays were performed as described previously (18), and data are displayed as the average and standard deviation of one representative experiment performed in triplicate.
Immunofluorescence.
Immunofluorescence and microscopy were conducted essentially as described previously (8). V5 tag localization was scored as predominantly cytoplasmic (Cyto), equal in the cytoplasm (C) and nucleus (N) (N = C), or predominantly nuclear (Nuc). Data are the average and standard deviation for two biological replicates, each scoring at least 50 cells from 10 random fields. Slides were blinded prior to scoring.
Western blotting.
Whole-cell lysates were prepared in ice-cold PBS plus 0.1% NP-40 and protease inhibitors. Western blotting using anti-MondoA at 1:500, anti-Mlx (affinity-purified rabbit polyclonal serum raised against TDDEDSDYHQEAYKESYK; Quality Controlled Biochemicals) at 1:500, anti-tubulin (Sigma) at 1:10,000, and anti-TXNIP at 1:1,000 (MBL International) was performed as described previously (25).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (25). Briefly, 3 × 106 cells were formaldehyde cross-linked, and isolated nuclei were sonicated four times for 25 s using a Sonicator Ultrasonic Processor XL (Misonix), with pulses of 0.9 s on/0.1 s off in 1 ml nuclei lysis buffer (50 mM Tris, 10 mM EDTA, 1% SDS plus protease inhibitors). Cleared lysates were diluted 1:10, precleared with 20-μl beads alone for 4 h, and immunoprecipitated overnight with 20-μl beads plus 5-μl antibodies against MondoA, acetylated histone H3 (Millipore), or trimethylated histone H3 lysine 4 (Active Motif). Coimmunoprecipitated DNA was purified and quantified by quantitative PCR (qPCR) with normalization to an off-target control region. TXNIP exon 1 primers flank the site of a nucleosome relevant to MondoA- and ChREBP-dependent regulation of TXNIP (4, 13). Rat and mouse primer sequences for the TXNIP E box, TXNIP exon 1, the ARRDC4 promoter, and PFKFB3 off target are available upon request. Data represent the average and standard error of the mean for two biological replicate experiments.
MondoA knockout (KO) and rescue.
C57BL/6 animals harboring MondoAlox/+ were intercrossed, and embryonic fibroblasts prepared at day 14 of embryogenesis (30). Following genotyping, MondoAlox/lox MEFs were infected with pBabePuroCre, resulting in 100% deletion of sequences between exons 12 and 15 of MondoA. These sequences encode the bHLHZip domain of MondoA, required for DNA binding and dimerization with Mlx. As such, their deletion completely abrogates MondoA's function as a transcriptional activator. A detailed description of our targeting of the MondoA allele will be described elsewhere. Knockout cells were rescued through retroviral expression of MondoA using the pWZL-BLAST vector and selected with blasticidin as directed by the manufacturer (Invitrogen).
Glucose uptake assays.
MEFs at subconfluent densities in 35-mm plates were washed with Krebs-Ringer-HEPES (KRH) buffer and then incubated for 10 min in 1 ml KRH containing 1 mM 2-deoxy-d-glucose and 1 μCi 2-[3H(G)]-deoxy-d-glucose (specific activity, 5 to 10 Ci/mmol; PerkinElmer). Glucose uptake was terminated by aspirating the incubation solution and washing cells with ice-cold KRH. Cells were solubilized with 0.5% NP-40 plus 0.5 M NaOH in H2O, and an aliquot of the lysate was assessed by scintillation counting for radiolabel incorporation. Assay results were normalized by determining radiolabel incorporation in control cells pretreated with cytochalasin B (10 μM; Sigma) and by normalization to protein content for each sample. The average and standard deviations of one representative experiment performed in quintuplicate are shown.
RESULTS
MondoA is a glucose-responsive regulator of TXNIP in skeletal muscle cells.
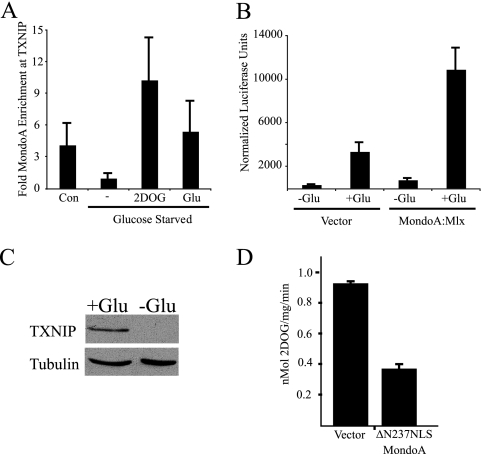
Our previous work with epithelial cells demonstrated that MondoA occupies the TXNIP promoter and regulates TXNIP expression in a glucose-dependent manner (13, 25). To determine whether the same regulatory circuitry exists in skeletal muscle cells, in which MondoA is most highly expressed (2), we examined TXNIP regulation in L6 myoblasts. MondoA occupancy of the TXNIP promoter was low in glucose-free media and increased by the addition of glucose or 2DOG (Fig. 1 A). In addition, the activity of a TXNIP-luciferase reporter was stimulated by the addition of glucose, and luciferase activity was further increased by ectopic expression of MondoA (Fig. 1B). TXNIP protein expression in L6 cells was strictly glucose dependent (Fig. 1C). Expression of a dominant active mutant of MondoA, ΔN237NLS MondoA, also decreased glucose uptake in L6 cells, similar to our observations with other cell types (Fig. 1D) (25). Therefore, MondoA's glucose-dependent regulation of TXNIP and its negative impact on glucose uptake extend to L6 myoblasts, suggesting that MondoA may be a ubiquitous regulator of the glucose-dependent transcriptional response.
FIG. 1.
MondoA is regulated by glucose in L6 myoblast cells. (A) TXNIP promoter occupancy by MondoA in L6 cells grown in complete media (Con) or glucose-starved overnight and treated for 3 h with glucose-free media (−), 20 mM 2-deoxyglucose (2DOG), or 20 mM glucose (Glu). (B) Activity of a TXNIP-luciferase reporter plasmid in L6 cells cotransfected with expression plasmids as indicated, starved overnight for glucose, and incubated for 7 h in the absence or presence of 25 mM glucose. (C) TXNIP protein expression in whole-cell lysates from L6 cells treated overnight in the absence or presence of 25 mM glucose. (D) Glucose uptake in L6 cells infected with retroviral vector alone or ΔN237NLS MondoA.
Glucose regulates nuclear export of MondoA-Mlx heterodimers.
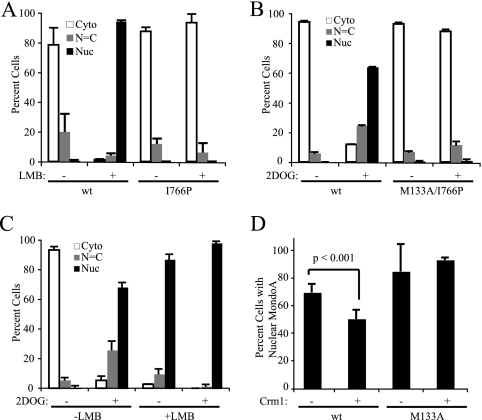
Dimerization with Mlx is required for MondoA nuclear accumulation, but the mechanism is unknown (25). It is possible that MondoA is subject to increased rates of nuclear export unless it is dimerized with Mlx. Alternatively, dimerization between MondoA and Mlx could be required for nuclear import. The first model predicts that inhibiting nuclear export will result in nuclear accumulation of MondoA regardless of dimerization status. The second model predicts that only MondoA-Mlx heterodimers should accumulate in the nucleus following blockade of nuclear export. To distinguish between these models, we blocked nuclear export of wt MondoA or MondoA(I766P), a leucine zipper mutant unable to heterodimerize with Mlx (25). MondoA(I766P) was highly cytoplasmic in the presence of the nuclear export inhibitor LMB, which blocks Crm1 function, whereas wt MondoA was almost 100% nuclear in the presence of LMB (Fig. 2 A). Similarly, the NES/dimerization double mutant MondoA(M133A/I766P) was cytoplasmic in the presence of 2DOG treatment, whereas wt MondoA accumulated in the nucleus in response to 2DOG as expected (Fig. 2B). Since blockade of nuclear export did not lead to nuclear accumulation of MondoA(I766P), we conclude that MondoA-Mlx heterodimerization is an obligate step required for MondoA nuclear entry.
FIG. 2.
Nuclear accumulation of MondoA is regulated by Mlx binding and glucose. (A to C) Quantified subcellular localization of V5 tag in L6 cells transfected with the indicated MondoA-V5 constructs and treated as indicated. (A) wt MondoA or MondoA(I766P) treated for 4 h with leptomycin B (LMB). (B) wt MondoA or MondoA(M133A/I766P), starved overnight for glucose and treated for 3 h in the presence or absence of 2DOG. (C) wt MondoA, starved overnight for glucose and treated for 4 h in the presence or absence of 2DOG and LMB. (D) wt MondoA or MondoA(M133A) and Gal4 vector alone or Gal4-Crm1, starved overnight for glucose and treated for 3 h with 2DOG. Only cells with predominantly nuclear MondoA were scored.
High-glucose conditions lead to nuclear accumulation of MondoA-Mlx, but under low-glucose conditions, it is unclear whether MondoA-Mlx is held at the mitochondria or shuttles between the mitochondria and the nucleus. LMB trapped nearly 100% of MondoA in the nucleus in the absence of glucose or 2DOG treatment (Fig. 2A and C), supporting the argument that MondoA-Mlx heterocomplexes shuttle between the mitochondria and nucleus in low-glucose conditions. Therefore, the shuttling of MondoA-Mlx in low-glucose conditions is likely a function of decreased nuclear import, decreased DNA binding, and/or increased nuclear export of the heterodimer relative to high-glucose conditions.
Previous studies of ChREBP showed that glucose increases the rate of nuclear import (6), but the role of glucose in nuclear export has not been addressed. If high intracellular glucose concentrations disrupt a nuclear export complex containing Crm1 and MondoA-Mlx, then overexpression of Crm1 should preserve such interactions even under high-glucose conditions, leading to increased nuclear export of MondoA-Mlx. Consistent with this hypothesis, Crm1 overexpression significantly decreased the 2DOG-induced nuclear accumulation of wt MondoA. Nuclear accumulation of the MondoA(M133A) NES mutant was unchanged by overexpression of Crm1; therefore this effect was specific to the NES function of MCRII (Fig. 2D). These data suggest that glucose-dependent nuclear accumulation of MondoA-Mlx heterodimers involves a blockade of nuclear export.
The basic region of MondoA contributes to nuclear accumulation.
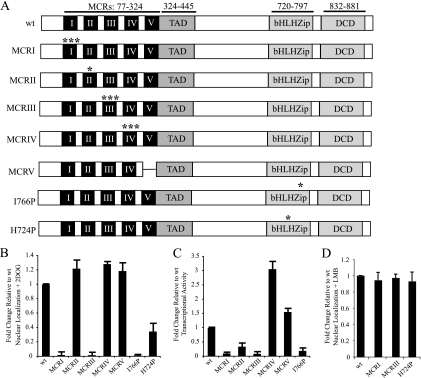
Glucose decreases nuclear export of MondoA-Mlx, which increases nuclear accumulation of the heterodimer. Glucose also increases occupancy of MondoA-Mlx at target promoters such as TXNIP. To test whether DNA binding is important for glucose-dependent nuclear accumulation of MondoA-Mlx, we made a single histidine-to-proline mutation in the basic region of MondoA, MondoA(H724P) (Fig. 3 A), and measured its nuclear localization in L6 cells. Mutations analogous to MondoA(H724P) are sufficient to ablate DNA binding in vitro for closely related bHLHZip proteins (2, 9, 19).
FIG. 3.
Glucose-dependent activities of MondoA require the MCR and bHLHZip domains. (A) Asterisks denote point mutations in MondoA MCRI (H78A/H81A/H88A), MCRII (M133A), MCRIII (I166A/W167A/R168A), MCRIV (Y210D/W211D/K212D), or the bHLHZip domain (I766P or H724P). MCRV is a deletion of amino acids 282 to 324. DCD, dimerization and cytoplasmic localization domain. (B to D) Localization or activity of MondoA-V5 mutants in L6 cells, normalized to wt. (B) Nuclear localization of the indicated mutants, with 2DOG treatment. (C) TXNIP luciferase activity of the indicated mutants, with glucose treatment. (D) Nuclear localization of the indicated mutants, with LMB treatment.
Following 2DOG treatment, MondoA(H724P) displayed reduced nuclear accumulation relative to wt MondoA (Fig. 3B). Following LMB treatment, this mutant phenocopied the nuclear accumulation of wt MondoA (Fig. 3D). Thus, MondoA(H724P) can enter the nucleus, suggesting that its impaired nuclear accumulation stems from a defect in DNA binding. Therefore, in addition to blocking nuclear export, these data suggest that glucose may also contribute to nuclear accumulation of MondoA-Mlx by increasing the promoter dwell time of the heterocomplex.
The Mondo conserved regions of MondoA control subcellular localization and TXNIP activation.
To determine whether the MCR domains of MondoA contribute to nuclear accumulation and TXNIP activation, we mutated each of the five MCRs individually. Single or triple point mutations in highly conserved residues of MCRs I to IV, known or predicted to disrupt the function of each domain, were generated (8). Due to the lower level of conservation of MCRV, a deletion was used to remove this domain (Fig. 3A).
The MCR mutants of MondoA fell into three functional classes: (i) cytoplasmic localization and impaired transcriptional activity, (ii) nuclear localization and enhanced transcriptional activity, and (iii) nuclear localization and moderately impaired transcriptional activity. The MCRI and MCRIII point mutants comprised class I and were cytoplasmic plus or minus 2DOG treatment (Fig. 3B and data not shown). Consistent with their lack of nuclear accumulation, these mutants did not activate the TXNIP reporter in response to glucose (Fig. 3C). Importantly, these mutants accumulated in the nucleus to an extent similar to that of wt MondoA, suggesting that their cytoplasmic localization was due to a defect in nuclear retention (Fig. 3D). Class II mutants, including the MCRIV triple point mutant and the MCRV deletion mutant, were highly nuclear in the presence or absence of 2DOG (Fig. 3B and data not shown). Consistent with their enhanced nuclear accumulation, these mutants were significantly more active than wt on the TXNIP reporter in response to glucose (Fig. 3C).
Class III comprised only the MondoA(M133A) point mutant in MCRII. Despite strong nuclear accumulation consistent with its NES mutation (Fig. 3B), MondoA(M133A) was only about 50% as transcriptionally active as the wt (Fig. 3C). Thus, MCRII of MondoA not only regulates nuclear export of MondoA-Mlx but also is necessary for full transcriptional activation at targets such as TXNIP.
MCRII regulates MondoA localization, TXNIP activation, and glucose uptake.
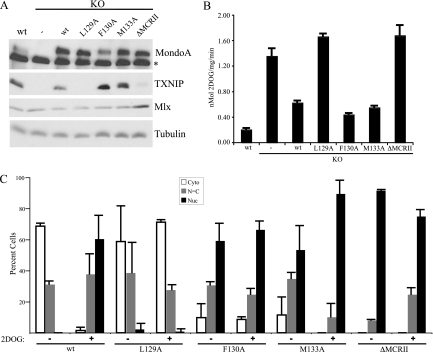
MCRII is implicated in nuclear accumulation and activation of TXNIP (Fig. 3). To investigate MCRII function further, we devised a knockout/rescue scheme to express wt MondoA or MCRII mutants of MondoA in a null background. Mouse embryonic fibroblasts (MEFs) were generated with exons 12 to 15 in the 3′ region of the MondoA gene flanked by loxP sites. Retroviral expression of Cre recombinase deleted these exons, resulting in a destabilized N-terminal fragment of MondoA unable to heterodimerize with Mlx or bind DNA. Cre-expressing MEFs were devoid of full-length MondoA and TXNIP protein (Fig. 4 A). Consistent with a loss of TXNIP, these cells displayed elevated glucose uptake (Fig. 4B) (25). As expected, rescue of the KO MEFs with retroviral expression of MondoA restored TXNIP expression and reduced glucose uptake (Fig. 4A and B). Perturbations in MondoA expression did not affect levels of Mlx protein (Fig. 4A).
FIG. 4.
MCRII controls localization and activity of MondoA-Mlx. MEFs from control mice (wt), MondoA KO, and KO cells rescued with retrovirally expressed wt MondoA (KO + wt) or MCRII mutants (KO + L129A mutant, KO + F130A mutant, KO + M133A mutant, and KO + ΔMCRII mutant) were measured for protein expression with the indicated antibodies (A), glucose uptake (B), and subcellular localization of MondoA in the presence or absence of 2DOG (C). Asterisk indicates nonspecific background band.
To better understand how MCRII regulates MondoA localization and transcriptional activity, we designed two additional mutants based on the consensus NES (Φ-X3-Φ-X2-Φ-X-Φ) of MCRII and introduced them into MondoA KO cells. L129 of MondoA does not contribute to this consensus, and thus, MondoA(L129A) should not disrupt NES function. This mutant was cytoplasmic in the absence of 2DOG and did not accumulate in the nucleus following 2DOG treatment (Fig. 4C). As expected, this constitutively cytoplasmic mutant failed to rescue TXNIP expression (Fig. 4A) and did not reduce glucose uptake (Fig. 4B). Since L129 of MondoA is dispensable for NES function, it is surprising that MondoA(L129A) does not accumulate in the nucleus following 2DOG treatment. This mutation may be hypermorphic for NES function or could drive cytoplasmic localization through a neomorphic mechanism.
In contrast to MondoA(L129A), MondoA(F130A) is predicted to disrupt the NES similarly to the MondoA(M133A) mutant. Consistent with this, both MondoA(M133A) and MondoA(F130A) localized constitutively to the nucleus, drove increased expression of TXNIP, and led to reduced cellular glucose uptake relative to wt MondoA (Fig. 4A, B, and C).
Next, we deleted the entire 12-amino acid domain [MondoA(ΔMCRII)]. MondoA(ΔMCRII) was highly nuclear in both the presence and absence of 2DOG (Fig. 4C). Unlike MondoA(F130A) and MondoA(M133A), MondoA(ΔMCRII) failed to rescue TXNIP expression (Fig. 4A) or block glucose uptake (Fig. 4B). Though deletion of MCRII results in nuclear accumulation of MondoA, the finding that MondoA(ΔMCRII) cannot activate TXNIP indicates that glucose-dependent nuclear accumulation of MondoA-Mlx is not sufficient for MondoA-Mlx transcriptional activity. Similar findings have been reported for ChREBP (6).
MCRII regulates glucose-dependent promoter occupancy independent of transcriptional activation.
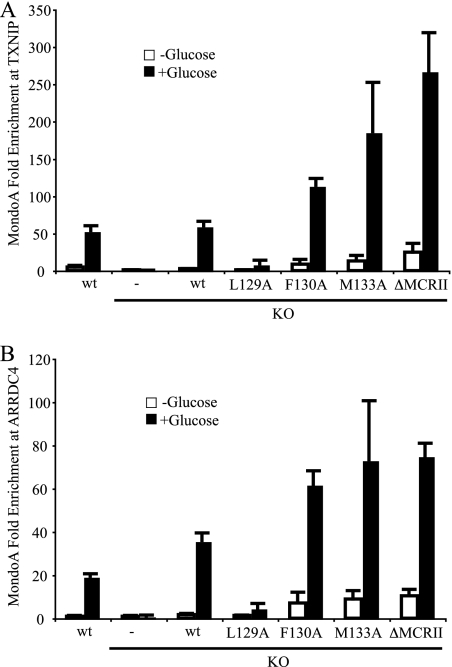
One explanation for the separable nuclear localization and transcriptional activity of MondoA is that the MCRII mutants may fail to occupy target promoters. We used chromatin immunoprecipitation (ChIP) to determine whether mutations in MCRII affected promoter occupancy by MondoA. As expected, wt MondoA in parental MEFs occupied the promoter of TXNIP in a robust and glucose-dependent manner, and occupancy was completely absent in KO cells (Fig. 5 A).
FIG. 5.
Nuclear accumulation of MondoA is not sufficient for promoter occupancy. (A) MondoA occupancy at the TXNIP promoter E box in wt MondoA, KO, and KO + rescue MEFs treated overnight in the presence or absence of glucose. (B) Experiments were performed as described for panel A except for the use of ARRDC4 promoter primers.
Rescue of KO cells with wt MondoA led to restoration of robust and glucose-dependent MondoA occupancy at the TXNIP promoter (Fig. 5A). As expected, MondoA(L129A) did not occupy the TXNIP promoter plus or minus glucose, while MondoA(F130A), MondoA(M133A), and MondoA(ΔMCRII) displayed enhanced occupancy of the TXNIP promoter (Fig. 5A). Importantly, TXNIP promoter occupancy by MondoA(F130A), MondoA(M133A), and MondoA(ΔMCRII) remained glucose dependent, even though each mutant accumulated in the nucleus in the presence or absence of glucose (Fig. 4C). Thus, nuclear accumulation of MondoA is uncoupled from promoter occupancy. Furthermore, each MCRII mutant occupied the TXNIP promoter more that the wild type, suggesting that CRM1 or another MCRII-bound factor inhibits promoter binding.
To demonstrate that these results were not specific to TXNIP, a second direct target of MondoA was examined. Previous work has shown that expression of the TXNIP paralogue ARRDC4 is MondoA dependent (25). MondoA occupied the ARRDC4 promoter in a glucose-dependent manner, similar to its occupancy of TXNIP (Fig. 5B). MCRII mutants of MondoA also occupied the ARRDC4 promoter similarly to the TXNIP promoter, demonstrating that MondoA-dependent activity at TXNIP is not a promoter-specific phenomenon.
MCRII recruits an H3-specific histone acetyltransferase to TXNIP.
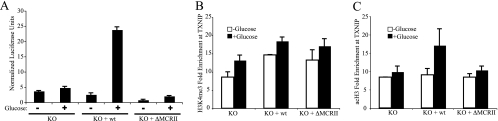
The MondoA(ΔMCRII) mutant occupies the TXNIP promoter but is unable to activate expression of TXNIP, suggesting an additional role for MCRII in coactivator recruitment. To investigate this further, we conducted TXNIP reporter assays. MondoA KO cells displayed baseline levels of TXNIP reporter activity in the presence or absence of glucose, and rescue with wt MondoA restored glucose-dependent TXNIP expression. As expected, MondoA(ΔMCRII) was unable to rescue TXNIP luciferase expression levels in MondoA KO cells (Fig. 6 A). Since no intrinsic chromatin modifying activity is predicted for the MCRII domain of MondoA (our unpublished sequence analysis), these data suggest that MCRII must be required to recruit a coactivator(s) to target promoters. To investigate MCRII-dependent recruitment of coactivators, we used ChIP to look for activating histone marks at the TXNIP promoter in MondoA KO cells or KO cells expressing wt MondoA or MondoA(ΔMCRII).
FIG. 6.
Glucose-dependent activation of TXNIP involves MondoA MCRII-dependent acetylation of histone H3. MondoA KO, KO + wt MondoA, and KO + MondoA(ΔMCRII) MEFs incubated overnight in the presence or absence of glucose and assayed for TXNIP luciferase activity (A) and H3K4me3 (B) or acetylated histone H3 (C) modifications at TXNIP exon 1.
Trimethylation of histone H3 lysine 4 (H3K4me3) was elevated above baseline levels by glucose in MondoA KO cells (Fig. 6B). Rescue with wt MondoA enhanced H3K4me3 levels in both the presence and absence of glucose, suggesting that MondoA promotes histone H3 lysine 4 trimethylation in a glucose-independent manner. Importantly, MondoA(ΔMCRII) also drove trimethylation of H3K4, suggesting that MCRII is not responsible for recruitment of the histone methyltransferase. It is surprising that MondoA can recruit a histone H3 lysine 4 methylase to the TXNIP promoter in the absence of glucose, despite weak but assayable occupancy of the TXNIP promoter by MondoA under these conditions. MondoA may “sample” target promoters in the absence of glucose, and this may be sufficient for certain histone modifications.
We next determined if MCRII of MondoA recruited histone acetyltransferase activity to the TXNIP promoter by measuring levels of acetylated histone H3 and H4. H3 acetylation at TXNIP was glucose dependent, consistent with our past results (Fig. 6C) (13). MondoA was required for the induction of H3 acetylation by glucose, demonstrating that MondoA recruits a histone H3 acetyltransferase to the TXNIP promoter in a glucose-dependent fashion. Levels of acetylated H3 at the TXNIP promoter were impaired with the MondoA(ΔMCRII) mutant, demonstrating that MCRII of MondoA was required for histone H3 acetylation at the TXNIP promoter. Levels of acetylated histone H4 at TXNIP required neither MondoA nor glucose (data not shown). These results suggest that the mechanism through which MCRII contributes to TXNIP activation involves glucose-dependent recruitment of a histone H3 acetyltransferase.
DISCUSSION
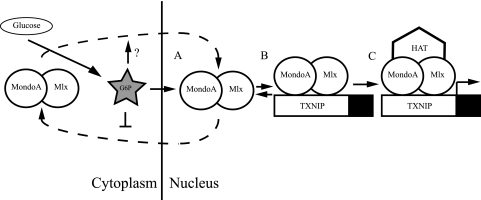
Proper utilization of glucose is central to all prokaryotic and eukaryotic life. In metazoans, MondoA is an essential transcriptional biosensor of intracellular glucose concentration. We propose that the primary glucose metabolite G6P regulates three fundamental aspects of MondoA function. (i) G6P promotes nuclear accumulation of MondoA by decreasing nuclear export, increasing promoter binding, and possibly through increasing nuclear import as shown for ChREBP (6). (ii) G6P enhances promoter occupancy of MondoA at targets such as TXNIP and ARRDC4. (iii) G6P directs recruitment of a histone H3 acetyltransferase to MondoA, thereby driving gene activation (Fig. 7).
FIG. 7.
Model of glucose-dependent regulation of MondoA. MondoA-Mlx heterodimers shuttle between cytoplasm and nucleus in the presence and absence of glucose (dashed lines). Intracellular glucose is metabolized to glucose-6-phosphate (G6P), which we propose as a MondoA signaling molecule that promotes nuclear accumulation (A), promoter occupancy (B), and recruitment of HAT coactivators (C) by MondoA-Mlx, in turn activating target genes, such as TXNIP.
Our data suggest that MondoA continually cycles between the cytoplasm and nucleus, monitoring intracellular glucose concentration. In low-glucose conditions, MondoA-Mlx cannot stably bind DNA and is immediately exported from the nucleus. In high glucose, MondoA-Mlx occupies target promoters, recruits transcriptional coactivators, and activates gene expression. How does G6P control three distinct steps of MondoA function? Regulation could proceed through a signaling cascade: G6P could invoke one or multiple signaling pathways to govern nuclear accumulation, DNA binding, and coactivator recruitment of MondoA. Alternatively, G6P could bind MondoA directly, regulating the activation steps by an integrated allosteric mechanism. These models are not mutually exclusive. However, the strong evolutionary conservation of the five MCRs suggests that they might bind a common metabolite directly. Thus, our data are most consistent with an allosteric regulatory model.
We propose a model in which G6P binding links the MCR and basic regions of MondoA, leading to increased nuclear accumulation and full transcriptional activity of the protein. The four tenets of this model are that (i) G6P dissociates Crm1 from MondoA-Mlx, (ii) the basic region of MondoA-Mlx binds DNA, (iii) promoter occupancy is further enhanced by a presumptive chromatin binding function of the MCRs, and (iv) MondoA-Mlx utilizes the MCR region to recruit a histone H3 acetyltransferase, facilitating glucose-dependent activation of targets such as TXNIP.
The first tenet of our model is that G6P dissociates Crm1 from the NES of MondoA and thus drives nuclear accumulation by restricting nuclear export. Due to the transient nature of Crm1-cargo binding, we have not been able to test this model directly. However, several experiments support this assertion. Overexpression of Crm1 reduced 2DOG-dependent nuclear accumulation of MondoA. Furthermore, 2DOG-dependent nuclear accumulation of MondoA is comparable to nuclear accumulation driven by LMB treatment. Finally, mutation of the NES located in MCRII of MondoA also results in constitutive nuclear localization, similar to LMB treatment. Thus, inhibition of nuclear export phenocopies 2DOG treatment, supporting the first tenet of our model. We propose that intracellular G6P levels act as a switch between Crm1-bound, cytoplasmic MondoA-Mlx and Crm1-unbound, promoter-bound MondoA-Mlx. The MondoA(H724P) DNA binding mutant displays a partial loss of 2DOG-induced nuclear accumulation. Therefore, we cannot rule out an additional contribution of DNA binding in the regulation of nuclear accumulation of MondoA-Mlx.
The second tenet of our model is that G6P is required for DNA binding by MondoA. MondoA(F130A), MondoA(M133A), and MondoA(ΔMCRII) are deficient in Crm1-dependent nuclear export, showing constitutive nuclear accumulation. The fact that these mutants occupy the TXNIP promoter only in the presence of glucose supports the argument that Crm1 binding does not directly inhibit DNA binding of MondoA-Mlx. Importantly, nuclear accumulation of MondoA-Mlx is not sufficient for DNA binding. Furthermore, each Crm1 binding mutant of MondoA displayed elevated TXNIP promoter occupancy relative to the wt, suggesting that G6P binding to the conserved MCR region, rather than Crm1 dissociation per se, unmasks the DNA binding function of MondoA-Mlx.
The third tenet of our model is that DNA-binding MondoA-Mlx heterodimers utilize the MCR region to stabilize MondoA at target genes, such as TXNIP and ARRDC4, in a G6P-dependent manner. Our mutagenesis data suggest a role for the MCR region in promoter occupancy by MondoA. However, MondoA(1-400), which contains the MCR region, lacks an identifiable DNA binding domain and cannot activate a TXNIP luciferase reporter (data not shown). These results suggest that the MCR region cannot interact with DNA independently. We propose that the basic region of MondoA is the prime determinant of DNA binding, while the MCR region enhances promoter occupancy by interacting with other chromatin factors, such as modified histones.
The final tenet of our model is that MCRII recruits a histone H3 acetyltransferase to MondoA-Mlx in a G6P-dependent manner. We hypothesize that G6P binding at MCRII drives coactivator recruitment by the adjacent transcriptional activation domain (TAD) of MondoA (2). Second-generation sequencing shows that a number of histone acetyltransferases (HATs) and histone deacetylases (HDACs) occupy promoters, such as TXNIP (29), suggesting that MondoA-Mlx may recruit numerous ancillary factors to activate transcription. Further, interaction between p300 and ChREBP has been demonstrated in solution (4). We are currently working to identify MondoA-bound coactivator(s) at target promoters. Building on our past results (13), we are also examining whether MondoA recruits corepressors to the TXNIP promoter in the presence of glutamine and resulting high-tricarboxylic acid (TCA) cycle intermediates.
In L6 cells, overexpressed MondoA(M133A) is roughly 50% as transcriptionally active as the wt. However, the same mutant expressed in MondoA KO MEFs is fully active. This may simply reflect a difference between these two cell types. However, it has been shown that ChREBP-Mlx multimerizes at target promoters, such as LPK and ACC (17). In L6 cells, the transcriptional activity of MondoA-Mlx may thus reflect the function of mixed hetero-oligomers. In contrast, the transcriptional activity of MondoA-Mlx complexes in MondoA KO MEFs is driven by pure wt or mutant homo-oligomers. The potential for dimeric or multimeric function of MondoA-Mlx will be an important consideration in functional studies of the heterocomplex.
MondoA and Mlx heterodimerize throughout their shuttling cycle (22), and we show that heterodimerization is required for nuclear import. We therefore propose that in addition to binding DNA, MondoA-Mlx heterodimerization acts as a checkpoint to exclude inactive monomers from nuclear entry. MondoA-Mlx dimerization is glucose independent (data not shown), yet Uyeda and Repa propose that glucose is required for ChREBP-Mlx dimerization and nuclear import. The key ChREBP residues identified for glucose-dependent heterodimerization of ChREBP-Mlx are not conserved in MondoA (27). Hence, some glucose response mechanisms of MondoA and ChREBP have apparently diverged.
Divergence between MondoA and ChREBP is especially evident in the MCRs. The MondoA(L129A) MCRII mutant used here is constitutively cytoplasmic and transcriptionally inactive, whereas the comparable ChREBP(L89A) mutant is constitutively nuclear and transcriptionally active (6). Deletion of MCRV from MondoA enhances its transcriptional activity, while MCRV deletions in ChREBP render the protein transcriptionally inert (14). Finally, point mutations within the MCR domains of MondoA affect its subcellular localization and transcriptional activity in different ways, whereas each MCR deletion in ChREBP abrogates its transcriptional activity (15). These differences may be due to varied spacing between the MCR domains among Mondo family members. The spacing between MCRII, MCRIII, and MCRIV is invariant across metazoans, suggesting that these MCRs function as a conserved structural module. However, MCRI and MCRV flank this MCRII/MCRIII/MCRIV module with linkers whose length and sequence vary between Mondo relatives, namely, MondoA and ChREBP. A full understanding of the regulatory differences between MondoA and ChREBP in their sensation of glucose will involve the dissection of the intramolecular interactions between MCRI, MCRII/MCRIII/MCRIV, and MCRV (25, 27).
In conclusion, we detail the multistage, glucose-dependent regulation of MondoA transcriptional activity. MondoA responds to the primary glycolytic metabolite G6P, which activates MondoA-Mlx heterodimers through a three-step regulatory cascade: nuclear accumulation, promoter occupancy, and coactivator recruitment. The MCR region of MondoA, namely, MCRII, plays a central role in this process. The best-characterized transcriptional targets of MondoA, TXNIP and ARRDC4, are activated through this pathway. The involvement of TXNIP and ARRDC4 in negative regulation of glucose uptake thus places MondoA at the center of a negative feedback loop controlling intracellular glucose uptake and glucose homeostasis.
Acknowledgments
We thank Laurie Jackson for technical assistance, Betty Leibold for critical review of the manuscript, the Graves and Cairns labs for reagents and advice, and members of the Ayer lab for helpful discussions and insights.
This work was supported by the Developmental Biology Training Grant T32 HD007491 (C.W.P.), National Institutes of Health grants GM55668 and GM60387 (D.E.A.), and funds from the Huntsman Cancer Foundation. DNA sequencing, oligonucleotide synthesis, and cell imaging facilities were supported by Cancer Center Support grant 2P30 CA42014.
Footnotes
Published ahead of print on 12 April 2010.
REFERENCES
- 1.Alvarez, C. E. 2008. On the origins of arrestin and rhodopsin. BMC Evol. Biol. 8:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billin, A. N., A. L. Eilers, K. L. Coulter, J. S. Logan, and D. E. Ayer. 2000. MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol. Cell. Biol. 20:8845-8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess, S. C., K. Iizuka, N. H. Jeoung, R. A. Harris, Y. Kashiwaya, R. L. Veech, T. Kitazume, and K. Uyeda. 2008. Carbohydrate-response element-binding protein deletion alters substrate utilization producing an energy-deficient liver. J. Biol. Chem. 283:1670-1678. [DOI] [PubMed] [Google Scholar]
- 4.Cha-Molstad, H., G. Saxena, J. Chen, and A. Shalev. 2009. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. J. Biol. Chem. 284:16898-16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J. Q., S. T. Hui, F. M. Couto, I. N. Mungrue, D. B. Davis, A. D. Attie, A. J. Lusis, R. A. Davis, and A. Shalev. 2008. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB J. 22:3581-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, M. N., B. L. O'Callaghan, and H. C. Towle. 2008. Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity. J. Biol. Chem. 283:24029-24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Luis, O., M. C. Valero, and L. A. Jurado. 2000. WBSCR14, a putative transcription factor gene deleted in Williams-Beuren syndrome: complete characterisation of the human gene and the mouse ortholog. Eur. J. Hum. Genet. 8:215-222. [DOI] [PubMed] [Google Scholar]
- 8.Eilers, A. L., E. Sundwall, M. Lin, A. A. Sullivan, and D. E. Ayer. 2002. A novel heterodimerization domain, CRM1, and 14-3-3 control subcellular localization of the MondoA-Mlx heterocomplex. Mol. Cell. Biol. 22:8514-8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferré-D'Amaré, A. R., G. C. Prendergast, E. B. Ziff, and S. K. Burley. 1993. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363:38-45. [DOI] [PubMed] [Google Scholar]
- 10.Hunt, A., P. Schonknecht, M. Henze, U. Seidl, U. Haberkorn, and J. Schroder. 2007. Reduced cerebral glucose metabolism in patients at risk for Alzheimer's disease. Psychiatry Res. 155:147-154. [DOI] [PubMed] [Google Scholar]
- 11.Hutten, S., and R. H. Kehlenbach. 2007. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 17:193-201. [DOI] [PubMed] [Google Scholar]
- 12.Iizuka, K., and Y. Horikawa. 2008. Regulation of lipogenesis via BHLHB2/DEC1 and ChREBP feedback looping. Biochem. Biophys. Res. Commun. 374:95-100. [DOI] [PubMed] [Google Scholar]
- 13.Kaadige, M. R., R. E. Looper, S. Kamalanaadhan, and D. E. Ayer. 2009. Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 106:14878-14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, M. V., B. Chang, M. Imamura, N. Poungvarin, and L. Chan. 2006. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes 55:1179-1189. [DOI] [PubMed] [Google Scholar]
- 15.Li, M. V., W. Chen, N. Poungvarin, M. Imamura, and L. Chan. 2008. Glucose-mediated transactivation of carbohydrate response element-binding protein requires cooperative actions from Mondo conserved regions and essential trans-acting factor 14-3-3. Mol. Endocrinol. 22:1658-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma, L., L. N. Robinson, and H. C. Towle. 2006. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J. Biol. Chem. 281:28721-28730. [DOI] [PubMed] [Google Scholar]
- 17.Ma, L., Y. Y. Sham, K. J. Walters, and H. C. Towle. 2007. A critical role for the loop region of the basic helix-loop-helix/leucine zipper protein Mlx in DNA binding and glucose-regulated transcription. Nucleic Acids Res. 35:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minn, A. H., C. Hafele, and A. Shalev. 2005. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology 146:2397-2405. [DOI] [PubMed] [Google Scholar]
- 19.Nair, S. K., and S. K. Burley. 2003. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112:193-205. [DOI] [PubMed] [Google Scholar]
- 20.Parikh, H., E. Carlsson, W. A. Chutkow, L. E. Johansson, H. Storgaard, P. Poulsen, R. Saxena, C. Ladd, P. C. Schulze, M. J. Mazzini, C. B. Jensen, A. Krook, M. Bjornholm, H. Tornqvist, J. R. Zierath, M. Ridderstrale, D. Altshuler, R. T. Lee, A. Vaag, L. C. Groop, and V. K. Mootha. 2007. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 4:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patwari, P., W. A. Chutkow, K. Cummings, V. L. Verstraeten, J. Lammerding, E. R. Schreiter, and R. T. Lee. 2009. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J. Biol. Chem. 284:24996-25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sans, C. L., D. J. Satterwhite, C. A. Stoltzman, K. T. Breen, and D. E. Ayer. 2006. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol. Cell. Biol. 26:4863-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah, S., M. Iqbal, J. Karam, M. Salifu, and S. I. McFarlane. 2007. Oxidative stress, glucose metabolism, and the prevention of type 2 diabetes: pathophysiological insights. Antioxid. Redox Signal. 9:911-929. [DOI] [PubMed] [Google Scholar]
- 24.Smith, E., and H. J. Morowitz. 2004. Universality in intermediary metabolism. Proc. Natl. Acad. Sci. U. S. A. 101:13168-13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoltzman, C. A., C. W. Peterson, K. T. Breen, D. M. Muoio, A. N. Billin, and D. E. Ayer. 2008. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc. Natl. Acad. Sci. U. S. A. 105:6912-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsatsos, N. G., and H. C. Towle. 2006. Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem. Biophys. Res. Commun. 340:449-456. [DOI] [PubMed] [Google Scholar]
- 27.Uyeda, K., and J. J. Repa. 2006. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 4:107-110. [DOI] [PubMed] [Google Scholar]
- 28.Vander Heiden, M. G., L. C. Cantley, and C. B. Thompson. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, Z., C. Zang, K. Cui, D. E. Schones, A. Barski, W. Peng, and K. Zhao. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, S., G. Ying, Q. Wu, and M. R. Capecchi. 2007. Toward simpler and faster genome-wide mutagenesis in mice. Nat. Genet. 39:922-930. [DOI] [PubMed] [Google Scholar]