Abstract
The extracellular signal-regulated kinase 1 and 2 (ERK1/2) mitogen-activated protein (MAP) kinase signaling pathway plays an important role in the proliferative response of mammalian cells to mitogens. However, the individual contribution of the isoforms ERK1 and ERK2 to cell proliferation control is unclear. The two ERK isoforms have similar biochemical properties and recognize the same primary sequence determinants on substrates. On the other hand, analysis of mice lacking individual ERK genes suggests that ERK1 and ERK2 may have evolved unique functions. In this study, we used a robust genetic approach to analyze the individual functions of ERK1 and ERK2 in cell proliferation using genetically matched primary embryonic fibroblasts. We show that individual loss of either ERK1 or ERK2 slows down the proliferation rate of fibroblasts to an extent reflecting the expression level of the kinase. Moreover, RNA interference-mediated silencing of ERK1 or ERK2 expression in cells genetically disrupted for the other isoform similarly reduces cell proliferation. We generated fibroblasts genetically deficient in both Erk1 and Erk2. Combined loss of ERK1 and ERK2 resulted in a complete arrest of cell proliferation associated with G1 arrest and premature replicative senescence. Together, our findings provide compelling genetic evidence for a redundant role of ERK1 and ERK2 in promoting cell proliferation.
The extracellular signal-regulated kinase 1 (ERK1)/ERK2 (ERK1/2) mitogen-activated protein (MAP) kinase pathway is an evolutionarily conserved signaling module that plays a central role in the control of cell proliferation (9, 30, 38, 41, 48). This pathway is activated by a wide variety of growth and mitogenic factors acting through diverse families of cell surface receptors. Ligand binding to growth factor receptors typically leads to the activation of the small GTPase Ras, which allows the recruitment of the MAP kinase kinase kinase Raf to the membrane and mediates the sequential phosphorylation and activation of the Raf-MEK1/MEK2 (MEK1/2)-ERK1/2 protein kinase cascade. Once activated, ERK1/ERK2 phosphorylate numerous cytoplasmic and nuclear substrates, including protein kinases and phosphatases, signaling effectors, transcriptional regulators, and cytoskeletal proteins (48, 72). Signaling output from the ERK1/2 MAP kinase pathway is modulated by interactions with scaffolds and regulatory proteins, which regulate the localization, amplitude, and duration of the signal (13, 42, 49).
In contrast to invertebrates, which express a single gene for each component of the Raf-MEK-ERK pathway, mammals and most vertebrates analyzed to date express two isoforms of ERK. One important issue that remains largely unaddressed is whether ERK1 and ERK2 isoforms have evolved unique physiological functions or whether they are used interchangeably in a cell-type-specific manner to reach a sufficient threshold of ERK activity. ERK1 and ERK2 are encoded by distinct genes and display 83% amino acid identity overall (5, 6). The two enzymes have similar biochemical properties and are activated by MEK1/2 with comparable efficiencies (50). Also, the substrate primary sequence determinants recognized by the two ERK isoforms are similar (19). ERK1 and ERK2 are coexpressed in the vast majority of cell lines and tissues tested although their relative abundances are variable (4, 5). Notably, some specific regions of the adult mouse brain appear to express exclusively Erk1 or Erk2 mRNA, suggesting that a single ERK isoform can mediate cellular functions in these areas (11). With the exception of a few reports describing the preferential activation of a single isoform (47, 53), multiple studies have shown that ERK1 and ERK2 are coactivated in response to extracellular stimuli (30). Detailed kinetic analysis in fibroblasts has revealed that the two isoforms are coordinately regulated in response to serum (40).
Although the above observations suggest that ERK1 and ERK2 are functionally equivalent, there is evidence for nonredundant functions of the two protein kinases. Analysis of mouse models has revealed that disruption of the Erk2 gene leads to early embryonic lethality, despite the wide expression of ERK1 in early mouse embryos (20, 52, 70). No proliferation of polar trophectoderm cells is observed in Erk2 homozygous mutants (52). In a model of cardiac ischemic injury, loss of a single allele of Erk2 was found to increase myocardial cell apoptosis, leading to decreased cardiac output (32). Under these experimental conditions, complete inactivation of Erk1 had no significant impact on myocardial infarction area and cardiac function, despite comparable reduction of total ERK1/2 kinase activity. ERK1-deficient mice were shown to be resistant to the development of skin papillomas in a two-step skin carcinogenesis protocol (7).
In vitro studies have also suggested that ERK1 and ERK2 may exert distinct functions in certain cellular contexts. For example, silencing of ERK2 expression by RNA interference (RNAi) in C2C12 myoblasts inhibits myogenin expression and myoblast fusion while inactivation of ERK1 has little effect (31). Keratinocytes from Erk1−/− mice show a reduced proliferative response to mitogenic factors and impairment in c-Fos expression (7). Other studies reported that knockdown of ERK2 expression restrains hepatocyte cell division, whereas ERK1 silencing specifically improves long-term hepatocyte survival (15, 16). Most intriguingly, it has been reported that ablation of ERK1 in fibroblasts by either gene targeting or RNAi enhances ERK2 signaling and leads to enhanced cell proliferation (62). In contrast, knockdown of ERK2 expression almost completely inhibits cell proliferation. It was further shown that overexpression of ERK1 inhibits oncogenic Ras-stimulated proliferation of NIH 3T3 cells. These findings have led to the hypothesis of a competition model where ERK1 acts as a negative regulator of cell proliferation by antagonizing ERK2 signaling (34, 62). In a more recent study, the authors have proposed that these functional differences between ERK1 and ERK2 are accounted for by a unique domain located at the N terminus of ERK1, which slows down nuclear shuttling of the kinase (37).
The objective of this study was to examine the individual contributions of ERK1 and ERK2 to cell proliferation control using a robust genetic approach. To this end, we have generated mouse embryonic fibroblasts (MEFs) genetically deficient for Erk1 or Erk2 or for both isoforms in well-defined genetic backgrounds. We now provide strong genetic evidence that ERK1 and ERK2 are redundant, positive regulators of cell proliferation.
MATERIALS AND METHODS
Reagents, antibodies, and plasmids.
5-Bromo-2-deoxyuridine (BrdU) was from Roche Diagnostics. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was from Calbiochem. Commercial antibodies were obtained from the following suppliers: anti-ERK1/2 CT from Upstate Biotechnology; anti-ERK2 from Zymed; anti-phospho-ERK1/2 (phosphorylation sites, Thr202/Tyr204), anti-phospho-p38 (Thr180/Tyr182), anti-phospho-Mnk1 (Thr197/202), anti-phospho-p90RSK (Thr573) (where RSK is ribosomal S6 kinase), anti-phospho-p90RSK (Ser380), and anti-phospho-MAPKAPK-2 (Thr334) from Cell Signaling Technology; anti-Mnk1 (G-19), anti-RSK1 (C-21), anti-c-Myc (9E10), anti-phospho-c-Myc (Thr58/Ser62), anti-p27 (C-19), and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; FL-335) from Santa Cruz Biotechnology; anti-α-tubulin (clone DM1A) from Sigma; anti-cyclin D1 (Ab-4) from Neomarkers; horseradish peroxidase (HRP)-conjugated goat anti-mouse and anti-rabbit IgG from Bio-Rad; and monoclonal anti-BrdU (clone 3D4) from BD Pharmingen.
The pLenti6/V5-large T lentiviral vector was constructed by subcloning the large T antigen sequence from pBabe-large T-puro (kindly provided by B. Thorens, University of Lausanne) into pLenti6/V5-DEST vector (Invitrogen). pMSCV-Cre-puro (where MSCV is mouse stem cell virus) was obtained from T. Hoang (Université de Montréal).
Maintenance, manipulation, and genotyping of mice.
Mice mutants for Erk1 (46) and Erk2 (52) were used to establish congenic strains by 10 successive generations of backcrossing to the CD-1 (Erk1 and Erk2 mice) and C57BL/6J (Erk1 mice) genetic backgrounds. FVB-GFP mice [FVB.Cg-Tg(ACTB-EGFP)B5Nagy/J; cells express green fluorescent protein (GFP)] were obtained from The Jackson Laboratory. Mice carrying a conditional Erk2 allele were generated by flanking exon 3 of Erk2 with loxP sites (to be reported elsewhere). Mice were maintained under specific-pathogen-free conditions according to the standards of the Canadian Council on Animal Care. All mice were housed in filter-topped isolator cages under a 12-h light-dark cycle.
Erk1+/− mice were intercrossed to generate Erk1−/− and Erk1+/+ embryo littermates used to derive MEFs. Erk2−/− and Erk2+/+ MEFs were derived from embryos generated by tetraploid aggregation (see below). For these experiments, superovulated Erk2+/− and FVB-GFP CD-1 females were mated with fertile Erk2+/− or wild-type CD-1 males, respectively. Pseudopregnant females used for chimeric embryo transfer were obtained by mating wild-type CD-1 females with vasectomized males. Embryo transfer was performed into pseudopregnant females at 2.5 days postcoitus (dpc). Erk1+/−; Erk2flox/flox mice were intercrossed to generate Erk1−/−; Erk2flox/flox and Erk1+/+; Erk2flox/flox embryo littermates that were used for isolation of MEFs.
For genotype screening, DNA was prepared from embryo tissue biopsy specimens. Genotyping of Erk1 null and wild-type littermates was determined by PCR using the following primers: 830, 5′-GAAGGAGCCAAGCTGCTATT-3′; 1118, 5′-AACGTGTGGCTACGTACTC-3′; and 1117, 5′-AGCAATGACCACATCTGCTA-3′. These primers amplify 330-bp and 600-bp fragments corresponding to the wild-type and mutant alleles of Erk1, respectively. Cycling conditions were as follows: 95°C for 30 s, 54°C for 45 s, and 72°C for 60 s for a total of 35 cycles. Erk2 genotyping was performed by PCR as described previously (52).
Tetraploid aggregation.
Tetraploid (4N) rescue experiments were performed essentially as described by Nagy and Rossant (43). Briefly, Erk2−/− morulae were obtained at the eight-cell stage by intercrossing CD-1 Erk2+/− mice. Wild-type embryos expressing GFP were collected at the two-cell stage from crosses of FVB-GFP male and CD-1 female mice and were used to produce 4N embryos by electro-fusion, followed by incubation in M16 medium for 24 h at 37°C in a humidified 5% CO2 incubator. Erk2−/− embryos at the eight-cell stage were then aggregated with two tetraploid wild-type embryos at the four-cell stage in a culture plate depression covered with M16 medium and mineral oil following removal of zona pellucida in acidic Tyrode's solution. Aggregates were incubated for 24 h, after which the chimeric embryos that developed successfully into blastocysts were transferred to uteri of pseudopregnant recipient females.
Cell culture, infections, and RNA interference (RNAi).
Erk1 and Erk2 mutant embryos were dissected at embryonic day 14.5 (E14.5), and MEFs were prepared as described previously (21). MEFs were routinely cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% newborn calf serum (NBCS) and antibiotics. The cells were made quiescent by incubation for 72 h in DMEM containing 0.1% NBCS. Erk1−/−; Erk2Δ/Δ MEFs were generated by Cre excision of the Erk2 floxed allele following infection (51) of Erk1−/−; Erk2flox/flox MEFs with the retroviral vector pMSCV-Cre-puro. Cre-expressing MEFs were cultured for 9 days in the presence of 2.5 μg/ml puromycin to completely ablate ERK2 expression. Erk1+/+; Erk2flox/flox MEFs infected with an empty pMSCV vector were used as controls in these experiments. For immortalization experiments, wild type, Erk1−/− and Erk2−/− MEFs were passaged according to a 3T3 protocol (59). Erk1−/−; Erk2flox/flox and Erk1+/+; Erk2Δ/Δ MEFs were infected (64) with pLenti6/V5-large T lentiviral vector to establish immortalized cell lines.
shRNA lentiviral infections of Erk1−/− or Erk2−/− MEFs were performed as described previously (64). The short hairpin RNA (shRNA) constructs for mouse Mapk1 (SHGLY-NM_011949), Mapk3 (SHGLY-NM_011952), and nontarget (NT) control (SHC002) were purchased from Sigma.
Kinase assays.
The enzymatic activity of ERK1 and ERK2 was measured by immune complex kinase assay using myelin basic protein (MBP) and [γ-32P]ATP as substrates (18). For immunoprecipitation of ERK1, cell lysates (∼150 μg of protein) were incubated for 3 h at 4°C with 3 μg of anti-ERK1/2 CT antibody (Upstate Biotechnology). ERK2 was immunoprecipitated by incubation for 2 h at 4°C with 2 μg of anti-ERK2 antibody (Zymed). Validation experiments confirmed that the anti-ERK1/2 CT antibody specifically precipitates the ERK1 isoform and that the anti-ERK2 antibody is specific to ERK2 (data not shown).
Cell proliferation and cell cycle analysis.
Cell proliferation was measured by the colorimetric MTT [3,4-(5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy phenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt] assay (10). Briefly, early-passage MEFs were plated in triplicate wells of 24-well plates (7,000 cells/well) and cultured in complete DMEM. Cell proliferation was determined at 24-h intervals by replacing the medium with 0.3 ml of DMEM containing 1 mg/ml MTT. After incubation of cells at 37°C for 1 h, the formazan product was solubilized in 500 μl of 2% glycine (0.1 M; pH 11) in dimethyl sulfoxide (DMSO). The absorbance was measured at 550 nm with reference at 620 nm. Manual cell counting was performed in parallel to MTT assays to confirm the results. The cells were plated as above, trypsinized daily, and counted with a hemacytometer. Essentially similar results were obtained by counting the cells (data not shown).
For cell cycle analysis, proliferating cells were pulsed with 10 μM BrdU for 1 h prior to harvesting. The cells were scraped in phosphate-buffered saline (PBS), fixed in cold 70% ethanol, and kept at −20°C until flow cytometry analysis. For the staining procedure, the cells were washed with blocking buffer (0.5% bovine serum albumin [BSA] in PBS), and DNA was denatured with 2 N HCl in PBS. The cells were washed again in blocking buffer and incubated for 2 min in 0.1 M sodium borate (pH 8.5) to neutralize any remaining HCl. After cells were washed with dilution buffer (blocking buffer with 0.5% Tween 20), they were incubated with anti-BrdU antibody (2.5 μg/ml) for 15 min at room temperature, followed by incubation with Alexa Fluor 350-conjugated anti-mouse IgG for another 15 min. The cells were then washed with PBS and incubated on ice for 30 min in propidium iodide (PI) buffer (0.1% sodium citrate, 50 μg/ml PI, and 0.2 mg/ml RNase) in the dark. Fluorescence was recorded on a LSRII cytometer, and the cell cycle distribution was determined using BD FACSDiva software.
Apoptosis assays.
Apoptotic cell death was evaluated by two-color annexin V staining. Early-passage MEFs were seeded at 275,000 cells/10-cm dish and grown for 3 days in complete DMEM. The cells were then harvested, washed twice with PBS, and resuspended in binding buffer (10 mM HEPES [pH 7.4], 140 mM NaCl, 2.5 mM CaCl2). Allophycocyanin (APC)-conjugated annexin V (BD Pharmingen) was added to the cells at a final dilution of 1:50. PI was added to a final concentration of 50 μg/ml. Cells were incubated for 30 min in the dark and analyzed by fluorescence-activated cell sorting (FACS) on an LSRII cytometer.
Replicative senescence analysis.
Early-passage MEFs were seeded at 33,600 cells/well in six-well plates. Three days after plating, the cells were washed in PBS and fixed for 5 min in 0.5% glutaraldehyde at room temperature. The cells were then incubated for 16 h at 37°C (no CO2) with fresh senescence-associated β-galactosidase (SA-β-Gal) stain solution: 1 mg/ml X-Gal (stock solution of 20 mg/ml in dimethylformamide), 40 mM citric acid, sodium phosphate buffer (pH 5.5), 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2 (12). After being washed in PBS, nuclei were stained with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) for 10 min, and the cells were viewed under phase contrast and UV fluorescence using a Leica DM IRB microscope. At least 200 cells were counted in random fields of three wells per MEF preparation.
Immunoblot analysis.
Cell lysis and immunoblot analysis were performed as described previously (55). Immunoblotting results were quantified by densitometry analysis using Multi Gauge software.
Real-time quantitative PCR analysis.
Total RNA was isolated with an RNeasy Mini Kit (Qiagen) and was reverse transcribed in a final volume of 100 μl using a High Capacity cDNA Archive Kit (Applied Biosystems) with random primers, as described by the manufacturer. Gene expression levels were determined using primer and probe sets from Applied Biosystems. PCRs were run using 2 μl of cDNA sample (20 to 50 ng), 5 μl of TaqMan Universal PCR Master Mix (Applied Biosystems), 0.5 μl of TaqMan Gene Expression Assay (20×), and 2.5 μl of water in a total volume of 10 μl. Real-time analysis of PCR product amplification was performed on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). All reactions were run in triplicate, and the average values were used for quantification. The mouse cyclophilin gene was used as endogenous control. The relative level of target gene expression was quantified using the ΔΔCT (where CT threshold cycle) method.
RESULTS
Loss of ERK1 slows down the proliferation rate of primary MEFs.
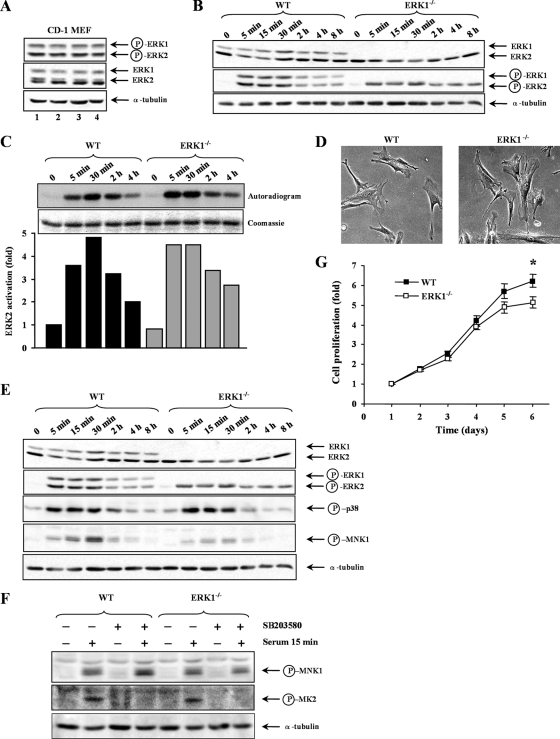
To rigorously test the role of ERK1 in the control of cell proliferation, we have prepared primary MEFs from E14.5 Erk1−/− embryos and wild-type control littermates. In order to control for the phenotypic variability associated with different genetic backgrounds, Erk1+/− mice in a mixed 129 × C57BL/6 background (46) were bred to a congenic CD-1 background (backcrossed to a CD-1 strain for more than 10 generations). Homozygous Erk1−/− mice in CD-1 background were born at expected Mendelian ratios, had normal size, and did not display any obvious anatomical abnormalities (data not shown). We estimated the ratio of ERK1 and ERK2 in embryonic fibroblasts from CD-1 mice by quantitatively measuring the phosphorylation of the two isoforms (epitope shared in ERK1 and ERK2) in serum-stimulated cells using a phospho-specific antibody (27). This analysis indicated that ERK1 and ERK2 are expressed in a stoichiometric ratio of 1:1.4 in these cells (Fig. 1A). Consistent with previous observations (16, 39, 45, 46), genetic ablation of ERK1 did not result in a compensatory increase in ERK2 expression (Fig. 1B). Also, serum addition induced similar kinetics of activation of ERK2 in ERK1-deficient fibroblasts and wild-type cells, as assessed by both activation loop phosphorylation immunoblotting (Fig. 1B) and immune complex kinase assay (Fig. 1C). Inactivation of the Erk1 gene had no detectable impact on the morphology or spreading of fibroblasts (Fig. 1D).
FIG. 1.
Disruption of Erk1 gene attenuates the proliferation rate of primary MEFs in a CD-1 background. (A) Relative expression of ERK1 and ERK2 isoforms in CD-1 MEFs. Total lysates from exponentially proliferating CD-1 MEFs (n = 4) were analyzed by immunoblotting using antibodies specific for phosphorylated (P) ERK1/ERK2, total ERK1/ERK2, and α-tubulin. (B) MEFs derived from Erk1−/− embryos (ERK1−/−) or wild-type control littermates (WT) were made quiescent and then stimulated with 10% NBCS for the indicated times. Total lysates were analyzed by immunoblotting. (C) Kinetics of ERK2 activity. ERK1−/− or WT MEFs were treated as in panel B. Cell lysates were prepared, and the phosphotransferase activity of endogenous ERK2 was measured by immune complex kinase assay using MBP and [γ-32P]ATP as substrates. The top panel shows an autoradiogram and Coomassie staining of MBP. The lower panel is a bar histogram showing the quantification of 32P incorporation into MBP. (D) Morphology of ERK1−/− and WT MEFs. (E) ERK1−/− or WT MEFs were treated as in panel B. The expression and phosphorylation of Mnk1 and p38 were analyzed by immunoblotting. (F) ERK1−/− or WT MEFs were made quiescent, pretreated with SB203580 (10 μM) or DMSO (0.1%) for 30 min, and then stimulated with 10% NBCS for 15 min. Total lysates were analyzed by immunoblotting using antibodies specific for phosphorylated MNK1, phosphorylated MK2, and α-tubulin. (G) Proliferation rates of P3 MEFs prepared from Erk1−/− or wild-type littermate embryos were measured by the MTT assay. Values are expressed as fold increase in cell number and correspond to the mean ± standard error of the mean of five independent MEF preparations. The data are representative of three different experiments. *, P < 0.05.
We examined the specific contribution of ERK1 to the phosphorylation of known target substrates of the ERK1/2 pathway. For these experiments, the cells were made quiescent and then stimulated with 10% serum for different times. The phosphorylation of the protein kinase Mnk1 (17, 65) was monitored by immunoblot analysis with a phospho-specific antibody. Addition of serum to wild-type MEFs markedly increased the phosphorylation of Mnk1 on Thr 197/202, sites known to be phosphorylated by the MAP kinases ERK1/2 and p38 (66). Interestingly, loss of ERK1 resulted in a sizeable attenuation of the phosphorylation of Mnk1 in these cells (Fig. 1E). No change in the activating phosphorylation of p38 was observed under these conditions (Fig. 1E). Furthermore, treatment with the p38 inhibitor SB203580 did not affect the serum-stimulated phosphorylation of Mnk1, confirming that the effect of serum is mediated through the ERK1/2 pathway (Fig. 1F). We also probed the phosphorylation of c-Myc, RSK1, and Elk-1 using phospho-specific antibodies but observed no difference between Erk1−/− and control MEFs (data not shown).
We analyzed the proliferation of ERK1-deficient and control MEFs at primary passage 3 (P3). The proliferation rate of Erk1−/− MEFs was modestly but significantly reduced compared to wild-type littermate control cells (Fig. 1G). The difference in proliferation became more apparent after 4 days in culture, possibly reflecting the cumulative effect of a small cellular defect. Similar results were obtained when cells were grown in a lower (2.5%) serum concentration (data not shown). These results indicate that ERK1 is a positive regulator of cell proliferation.
The hypothesis by Vantaggiato et al. (62) that ERK1 acts as a negative regulator of cell proliferation was based on results obtained with MEFs derived from Erk1−/− mice enriched in a C57BL/6 background. To determine whether the genetic background of the mice might be responsible for the discrepancy in our results, we analyzed the proliferation rate of MEFs isolated from Erk1 mutant mice bred to a congenic C57BL/6J background. These cells express ERK1 and ERK2 in a ratio of 1:1.9 (Fig. 2A). Similarly, inactivation of Erk1 did not affect total ERK2 expression or ERK2-activating phosphorylation (Fig. 2B). Analysis of cell proliferation revealed that loss of ERK1 has no statistically significant impact on the proliferation rate of C57BL/6J embryonic fibroblasts although a slight reduction in the number of Erk1−/− cells was observed at days 5 and 6 (Fig. 2C). These findings are in agreement with the original observation by Pagès et al. (46) that MEFs derived from Erk1−/− mice in a mixed 129 × C57BL/6 background proliferate at the same rate as wild-type cells. Thus, although the genetic background may influence the impact of ERK1 loss on the proliferation of fibroblasts, our results do not support the idea that ERK1 negatively regulates cell proliferation.
FIG. 2.
Cell proliferation analysis of Erk1−/− primary MEFs in C57BL/6J background. (A) Relative expression of ERK1 and ERK2 isoforms in C57BL/6J MEFs. (B) MEFs derived from ERK1−/− or wild-type (WT) control littermate embryos were made quiescent and then stimulated with 10% NBCS for 5 min. The expression and phosphorylation of ERK1/ERK2 were analyzed by immunoblotting. (C) Proliferation curves of MEFs prepared from Erk1−/− or wild-type littermate embryos in a C57BL/6J background. Values correspond to the mean ± standard error of the mean of six independent MEF preparations. The data are representative of three different experiments.
ERK2-deficient primary MEFs proliferate more slowly.
Mice homozygous for a null Erk2 mutation die in utero around E6.5 from a defect in trophoblast development (52, 70), thereby precluding the isolation of embryonic fibroblasts. To circumvent the problem associated with the early embryonic lethality of ERK2-deficient embryos, we have aggregated wild-type tetraploid (4N) embryos with diploid embryos derived from CD-1 Erk2+/− intercrosses (Erk2+/+, Erk2+/−, and Erk2−/−). In these chimeras, the wild-type tetraploid cells contribute exclusively to extraembryonic structures (placental trophoblast cells and endoderm of the yolk sac), whereas diploid cells contribute to the embryo proper (58). Among the 36 embryos recovered at E13.5, we identified two living embryos that were of Erk2−/− genotype (Fig. 3). In agreement with a defect in trophoblast development, mutant Erk2−/− embryos showed a very strong contribution of wild-type 4N cells to placenta and yolk sac as revealed by GFP detection (Fig. 3A). Embryonic fibroblasts were prepared from the two Erk2−/− embryos and their control littermates. Since the MEFs are derived from embryos in a CD-1 background, this allows a genetically matched comparison with CD-1 Erk1 mutant cells.
FIG. 3.
Rescue of Erk2−/− mice from early embryonic lethality by tetraploid aggregation. Each eight-cell-stage embryo isolated from intercrosses of CD-1 Erk2+/− mice was aggregated with two tetraploid four-cell-stage wild-type embryos in vitro. After development to the blastocyst stage, chimeric embryos were transferred into the uteri of pseudopregnant females. (A) Macroscopic views of wild-type and Erk2−/− embryos at E13.5 (upper panels) and contribution of wild-type GFP-positive cells to the placenta and yolk sac endoderm (bottom panels). (B) Genotyping analysis of embryos obtained from tetraploid aggregation experiments. (C) Number and genotype of embryos obtained by tetraploid aggregation rescue experiments at E13.5.
We examined the consequence of the genetic disruption of the Erk2 gene on the expression and activity of ERK1. Loss of ERK2 did not lead to a change in ERK1 expression in embryonic fibroblasts (Fig. 4A). However, the absence of ERK2 resulted in a more sustained activation of ERK1 that was observed in cells stimulated to reenter the cell cycle (Fig. 4A and B). We also examined the specific involvement of ERK2 in the phosphorylation of Mnk1 and other ERK1/2 substrates. Analogous to ERK1, the loss of ERK2 translated into a significant decrease in the serum-stimulated phosphorylation of Mnk1 (Fig. 4C). These results indicate that activation of both ERK1 and ERK2 isoforms is required for optimal phosphorylation of specific target substrates in fibroblasts.
FIG. 4.
Disruption of Erk2 slows down the proliferation of primary MEFs in a CD-1 background. MEFs derived from ERK2−/− or wild-type (WT) control littermates were made quiescent and then stimulated with 10% NBCS for the indicated times. (A) The expression and phosphorylation of ERK1/ERK2 were analyzed by immunoblotting. (B) Kinetics of ERK1 activity. The phosphotransferase activity of endogenous ERK1 was measured by immune complex kinase assay as described in the legend of Fig. 1. (C) The expression and phosphorylation of Mnk1 and p38 were analyzed by immunoblotting. (D) Morphology of ERK2−/− and wild-type MEFs. (E) Proliferation curves of P3 MEFs prepared from Erk2−/− or wild-type littermate embryos obtained by tetraploid aggregation experiments. Values correspond to the mean ± standard error of the mean of two independent MEF preparations. The data are representative of three different experiments. *, P < 0.05.
The morphology of Erk2−/− MEFs was indistinguishable from that of wild-type control cells (Fig. 4D). We then asked whether the loss of ERK2 affects the rate of cellular proliferation. Erk2−/− MEFs clearly proliferated at a lower rate than wild-type littermate controls, and this difference was already visible after 2 days in culture (Fig. 4E). A similar proliferation defect was observed at lower serum concentrations (data not shown). We conclude from these results that ERK1 and ERK2 isoforms both act as positive regulators of cell proliferation.
RNAi-mediated silencing of ERK1 or ERK2 expression in the absence of the other isoform similarly reduces cell proliferation.
To further address the individual roles of ERK1 and ERK2 in cell proliferation, we next evaluated the impact of knocking down the expression of either isoform in cells genetically disrupted for the other isoform. To this end, primary Erk1−/− or Erk2−/− MEFs were infected with vesicular stomatitis virus (VSV)-pseudotyped lentiviral vectors expressing shRNAs targeting the mouse Erk2 or Erk1 gene, respectively. Populations of infected cells were selected with puromycin for 3 days prior to analysis. Immunoblot analysis confirmed that both shRNAs efficiently reduced the protein level and activating phosphorylation of their target ERK compared to nontarget control shRNA (Fig. 5A). Although complete elimination of ERK2 in Erk1−/− cells or ERK1 in Erk2−/− cells was not possible by this approach, lowering the level of either remaining ERK isoform resulted in a significant decrease in the rate of MEF proliferation (Fig. 5B). Together, these results indicate that the proliferation potential of fibroblasts is dependent on the total expression/activity of ERK1/2, demonstrating that ERK1 and ERK2 exert redundant functions in cell proliferation.
FIG. 5.
RNAi silencing of ERK1 or ERK2 restrains the proliferation of MEFs expressing a single ERK isoform. Erk1−/− or Erk2−/− MEFs were infected with lentiviruses encoding shRNAs to the ERK2 or ERK1 gene, respectively, and populations of transduced cells were selected with puromycin. After 72 h, the cells were replated at low density (day 0) to measure the rate of cell proliferation. A nontarget shRNA (NT) was used as a control. (A) The expression of ERK1/ERK2 and phospho-ERK1/2 was analyzed by immunoblotting at day 3. (B) Cell proliferation was measured by the MTT assay. Values are expressed as fold increase in cell number and correspond to the mean ± standard error of the mean of triplicate determinations. The data are representative of three different experiments.
Genetic inactivation of Erk1 and Erk2 results in proliferation arrest of MEFs.
We recently generated a conditional allele of Erk2 by flanking exon 3 with loxP sites (to be reported elsewhere). Mice carrying the floxed Erk2 gene were bred with Erk1−/− mice to generate Erk1+/−; Erk2flox/flox mice. These mice were then intercrossed to generate Erk1+/+; Erk2flox/flox and Erk1−/−; Erk2flox/flox embryos for the preparation of MEFs. To test the effect of the complete inactivation of both ERK1 and ERK2 isoforms, we infected Erk1−/−; Erk2flox/flox primary MEFs with a Cre recombinase expressing retroviral vector to generate Erk1−/−; Erk2Δ/Δ double knockout MEFs. Expression of Cre in Erk1−/−; Erk2flox/flox cells resulted in the complete elimination of ERK2 protein after 9 days (Fig. 6A). Moreover, no residual activating phosphorylation of ERK1/ERK2 could be detected by immunoblotting with a phospho-specific antibody (Fig. 6A). We also examined the phosphorylation of the ERK1/ERK2 substrates RSK1 and Mnk1. Combined loss of ERK1 and ERK2 resulted in the ablation of serum-induced phosphorylation of RSK1 on Thr574 and Ser380 (Fig. 6A). Direct phosphorylation of RSK on these residues is considered to be a bona fide readout of ERK1/ERK2 kinase activity in vivo (1). The phosphorylation of Mnk1 was also abrogated in double knockout MEFs (Fig. 6A).
FIG. 6.
Genetic inactivation of Erk1 and Erk2 genes arrests the proliferation of MEFs. (A) Erk1+/+; Erk2flox/flox and Erk1−/−; Erk2Δ/Δ MEFs were made quiescent and then stimulated with 10% NBCS for the indicated times. Total lysates were analyzed by immunoblotting using antibodies specific for phospho-ERK1/ERK2, total ERK1/ERK2, phospho-RSK (Thr573), phospho-RSK (Ser380), total RSK1, phospho-Mnk1, total Mnk1, and GAPDH. (B) Morphology of Erk1+/+; Erk2flox/flox and Erk1−/−; Erk2Δ/Δ MEFs. (C) Proliferation rates of Erk1+/+; Erk2flox/flox and Erk1−/−; Erk2Δ/Δ MEFs at P5 were measured by the MTT assay. Values are expressed as fold increase in cell number and correspond to the mean ± standard error of the mean of three independent experiments. *, P < 0.05.
The morphological appearance of Erk1−/−; Erk2Δ/Δ primary MEFs was similar to that of Erk1−/−; Erk2flox/flox cells except for the presence of a higher percentage of cells with a senescence-like morphology (Fig. 6B). We examined the impact of the loss of both ERK isoforms on the rate of cell proliferation. As shown in Fig. 6C, loss of ERK1 and ERK2 function resulted in a complete arrest of cell proliferation (Fig. 6C). It should be mentioned that Cre expression in MEFs and other cell lines was reported to induce DNA damage resulting in reduced proliferation (35). We also observed that retroviral Cre expression attenuates the proliferation of Erk1−/−; Erk2flox/flox cells, but this effect was variable, ranging from little to 25% inhibition, and could not account for the complete arrest of cell proliferation (data not shown).
Erk1−/− and Erk2−/− primary embryonic fibroblasts can be efficiently immortalized.
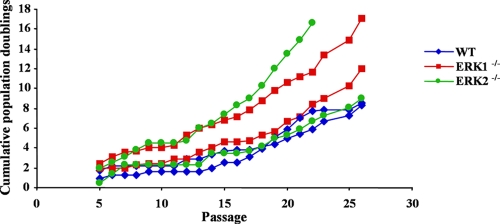
We wished to determine whether the loss of ERK1 and ERK2 isoforms affects the long-term proliferative capacity of MEFs. Wild-type, Erk1−/−, and Erk2−/− MEFs at P4 were serially passaged according to a 3T3-based protocol (59), and the cumulative population doublings of the cells were recorded at each passage. The proliferation rates of Erk1−/−, Erk2−/−, and control cells declined comparably with serial passage, and all individual MEF cultures entered into replicative arrest around passage 7 (Fig. 7). After this phase of proliferative decline, immortalized lines emerged from all MEF cultures. These results indicate that fibroblasts deficient in either ERK1 or ERK2 can be efficiently immortalized in culture.
FIG. 7.
Immortalization of Erk1−/− and Erk2−/− MEFs. MEF preparations derived from individual embryos were cultured according to a 3T3 protocol. The cells were counted, and the cumulative number of total population doublings was plotted at each passage.
We next tried to establish immortalized MEF lines deficient in both ERK isoforms. Since Erk1−/−; Erk2Δ/Δ MEFs fail to proliferate in culture and display morphological signs of senescence (Fig. 6), we decided to infect the cells with a lentiviral vector encoding the simian virus 40 (SV40) large T antigen to inactivate the retinoblastoma (Rb) and p53 pathways. Two strategies were used. Erk1−/−; Erk2flox/flox MEFs were infected first with the Cre-expressing retrovirus to excise Erk2 and then with the large T-encoding lentivirus. Alternatively, the cells were first infected with large T lentivirus prior to inactivation of the Erk2 gene. All attempts failed to establish cell lines of MEFs deficient in both ERK1 and ERK2.
Molecular basis of the proliferation defect observed upon loss of ERK1 and ERK2 in embryonic fibroblasts.
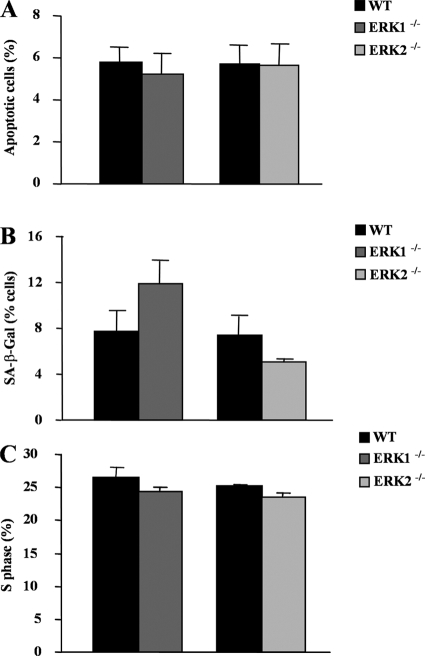
We carried out a series of experiments to investigate the nature of the proliferation defect observed in MEFs deficient in ERK1 or ERK2 or in both isoforms. The ERK1/2 signaling pathway is known to play a key role in the regulation of G1-to-S-phase progression (41). Activation of ERK1/ERK2 has also been associated with survival signaling in certain cell types (2). We first analyzed cells deficient in a single ERK isoform, which display a small proliferation defect. The extent of apoptosis in exponentially proliferating MEFs was measured by annexin V staining. As expected, the percentage of apoptotic cells was low in serum-containing medium. No significant difference was observed in the extent of apoptosis between the different genotypes (Fig. 8A). We also evaluated the percentage of cells entering into replicative senescence by monitoring SA-β-Gal activity in MEFs at P3. Consistent with other studies, the percentage of cells positive for SA-β-Gal at this early passage was under 15%, and no significant difference could be highlighted between Erk1−/− or Erk2−/− cells and their respective controls (Fig. 8B). These observations suggest that the proliferation defect of Erk1−/− and Erk2−/− mutant cells results from a defect in cell cycle progression. To address this question, we analyzed the cell cycle kinetics of asynchronously proliferating MEFs by flow cytometry with BrdU-PI staining. These experiments showed that the proportion of cells in S phase was slightly reduced by 8% in Erk1−/− MEFs and by 8% in Erk2−/− MEFs compared to levels in wild-type controls (Fig. 8C). Such small changes in cell cycle kinetics were expected, given the relatively small impact of the loss of a single ERK isoform on the proliferation of MEFs.
FIG. 8.
Impact of the loss of ERK1 or ERK2 on apoptosis, replicative senescence, and cell cycle progression of primary MEFs. Proliferating MEFs (P3) derived from CD-1 Erk1−/− or Erk2−/− mice or from their respective wild-type control littermate embryos were harvested at day 3 after plating. (A) Apoptosis was evaluated by two-color annexin V staining. Results are expressed as the mean ± standard error of the mean (n = 3). (B) Analysis of replicative senescence. Proliferating MEFs were fixed and stained for SA-β-Gal activity. Results are presented as the mean percentage ± standard error of the mean of SA-β-Gal-positive cells for wild-type (n = 4), ERK1−/− (n = 5), and ERK2−/− (n = 2) MEFs. (C) Cell cycle analysis. Asynchronously proliferating MEFs were pulsed with BrdU for 1 h prior to harvesting at day 3 after seeding. The cells were fixed and stained with PI, and the percentage of cells in S phase was determined by FACS analysis. Results are expressed as the mean ± standard error of the mean (n = 3).
We next characterized MEFs lacking the two ERK isoforms. The percentage of apoptotic cells was increased in Erk1−/−; Erk2Δ/Δ double knockout MEFs compared to control Erk1+/+; Erk2flox/flox cells (Fig. 9A). However, the extent of apoptosis remained low in serum-containing medium and could not account for the defect of proliferation. Strikingly, analysis of replicative senescence revealed that the percentage of SA-β-Gal-positive cells at early passage is about 3-fold higher in Erk1−/−; Erk2Δ/Δ MEFs than in control Erk1+/+; Erk2flox/flox cells (Fig. 9B). Expression of Cre alone had no significant effect on the induction of senescence (data not shown). This result suggests that replicative senescence limits the proliferation potential of cells lacking ERK1/2 activity. Flow cytometry analysis of the cell cycle demonstrated that combined loss of ERK1 and ERK2 completely blocks the progression of MEFs into S phase, with the resulting accumulation of cells in G1 phase (Fig. 9C). Thus, deficiency in ERK1/2 signaling leads to cell cycle arrest and premature entry into replicative senescence.
FIG. 9.
Combined loss of ERK1 and ERK2 impairs cell cycle progression and promotes replicative senescence. Proliferating Erk1+/+; Erk2flox/flox and Erk1−/−; Erk2Δ/Δ MEFs (P5) were harvested at day 3 after plating. (A) Apoptosis was measured by two-color annexin V staining. Results are expressed as the mean ± standard error of the mean (n = 3). (B) Replicative senescence was assayed by staining for SA-β-Gal activity. Bright-field and fluorescence (DAPI) photographs of representative fields are shown. The graph at right shows the quantification of SA-β-Gal activity. Results are presented as the mean percentage ± standard error of the mean of SA-β-Gal-positive cells (n = 3). (C) Cell cycle analysis. Asynchronously proliferating MEFs were pulsed with BrdU for 1 h prior to harvesting. The percentage of cells in S phase was determined by FACS analysis. Results are expressed as the mean ± standard error of the mean (n = 3). (D) Expression of cell cycle-regulatory genes. Erk1+/+; Erk2flox/flox and Erk1−/−; Erk2Δ/Δ MEFs were made quiescent and then stimulated with 10% NBCS for the indicated times. The expression of cell cycle genes was analyzed by quantitative PCR and is displayed in a heat map. Results represent the mean of two experiments. (E) Relative expression levels of the genes named in panel D. Gene expression levels were plotted as fold change over the value observed in Erk1+/+; Erk2flox/flox cells at time zero, which was arbitrarily set to 1. (F) Immunoblot analysis of cell cycle-regulatory genes. Results are representative of two experiments.
To further understand the molecular basis of the cell cycle defect of ERK1/ERK2-deficient fibroblasts, we monitored the expression of key regulators of G1-to-S-phase progression during mitogen-stimulated cell cycle reentry. We found that induction of the cell cycle regulatory genes encoding the transcription factors c-Myc, E2f1 and E2f3, the G1 cyclins cyclin D1, cyclin E1 and cyclin E2, and the replication licensing factor Mcm2 was completely abrogated in Erk1−/−; Erk2Δ/Δ double deficient MEFs compared to control Erk1+/+; Erk2flox/flox cells (Fig. 9D and E). In parallel, expression of the genes encoding the cyclin D1 transcriptional repressor Tob1 and the cyclin-dependent kinase (Cdk) inhibitors p21 and p27 was maintained at higher levels in ERK1/ERK2-deficient cells (Fig. 9D and E). Immunoblot analysis showed that cyclin D1 protein was barely detectable in ERK1/ERK2-deficient MEFs while p27 expression was stabilized and remained elevated during cell cycle progression (Fig. 9F). The induction of c-Myc expression and its phosphorylation on Thr58/Ser62 were also markedly impaired in the absence of ERK1 and ERK2.
DISCUSSION
Compelling biochemical, pharmacological, and genetic evidence have linked the ERK1/2 MAP kinase signaling pathway to the control of cell proliferation in fibroblasts and other cell types (reviewed in reference 41). However, the question remains whether the two structurally related isoforms ERK1 and ERK2 similarly contribute to cell proliferation. Early studies of MEFs prepared from Erk1−/− embryos did not reveal any impact of the loss of ERK1 on the rate of cell proliferation (3, 46). Impaired proliferation of ERK1-deficient thymocytes was reported initially (46), but this observation could not be reproduced in more recent studies (14, 45). Also, hepatocytes from wild-type and Erk1−/− mice were found to replicate with the same kinetics in the regenerating liver after partial hepatectomy and in primary culture in vitro (16). The only specialized cell type where ERK1 function was shown to be necessary for normal cell proliferation is keratinocytes (7). The early embryonic lethality of Erk2−/− mice has precluded the isolation of primary cells amenable to cell proliferation studies (52, 70). However, analysis of chimeric embryos has demonstrated that ERK2 functions cell-autonomously in the development of extra-embryonic cell lineages and is most likely required for the proliferation of polar trophectoderm cells (52). These studies in genetically deficient mice have suggested that ERK2 is more important than ERK1 for cell proliferation control. However, they do not conclusively resolve the question of whether the apparent requirement for ERK2 is due to its specific pattern of expression in mouse tissues or to genetic buffering of Erk1 loss or whether it is an indication of functional differences between ERK1 and ERK2 isoforms.
The individual role of ERK isoforms in cell proliferation has also been studied using RNA interference approaches, which allow for the partial inactivation of gene function in established cell lines and are less susceptible to long-term gene compensatory mechanisms. These studies have confirmed the important function of ERK2 in regulating cell proliferation. For example, silencing of ERK2 expression has been shown to slow down the proliferation of HeLa cells (33), ovarian cancer cells (74), C2C12 myoblasts (31), fibroblasts (62), and hepatocytes (16). In these studies, knockdown of ERK1 expression was also reported to inhibit the proliferation of HeLa cells and ovarian cancer cell lines (33, 74). Intriguingly, Vantaggiato et al. (62) have reported that ablation of ERK1 function in both mouse embryonic and NIH 3T3 fibroblasts by gene disruption or RNA interference enhances cell proliferation. This has led to the proposal that ERK2 drives cell proliferation, whereas ERK1 act as a negative regulator by interfering with ERK2 signaling (34, 62). During the course of our study, Lefloch et al. (28) also reported that silencing of ERK2 expression slows down the proliferation of NIH 3T3 fibroblasts, whereas reduction of ERK1 expression has no effect. Interestingly, these authors showed that silencing of ERK1 expression further reduces cell proliferation in cells with a limiting level of ERK2.
Here, we have used a robust genetic approach to analyze the individual functions of ERK1 and ERK2 in cell proliferation control using primary embryonic fibroblasts as a model. We prepared ERK1-deficient MEFs from Erk1 mutant mouse intercrosses and used a tetraploid aggregation procedure to rescue the early embryonic lethality of Erk2−/− mice and generate genetically matched ERK2-deficient cells. We show that loss of either Erk1 or Erk2 slows down the proliferation rate of early-passage fibroblasts. The decline in proliferation is more important in ERK2-deficient cells, consistent with the higher level of expression of ERK2 in these cells. Of note, we observed negligible functional compensation by the remaining ERK2 isoform in ERK1-deficient MEFs, as monitored by activating loop phosphorylation and kinase activity. However, in the case of ERK2-deficient MEFs, the kinase activity of ERK1 increased by 2- to 3-fold although this compensation was insufficient to rescue the proliferation defect. Again, this observation is in agreement with the relative expression levels of ERK1 and ERK2 isoforms. To further analyze the specific contribution of ERK1 and ERK2, we evaluated the impact of silencing the expression of ERK1 or ERK2 by RNA interference in cells genetically disrupted for the other isoform. Lowering the level of either remaining ERK isoform similarly resulted in a marked decrease in cell proliferation. Finally, we generated fibroblasts genetically deficient in both Erk1 and Erk2 by infecting Erk1−/−; Erk2flox/flox primary MEFs with a retroviral vector expressing the Cre recombinase. Combined loss of ERK1 and ERK2 function resulted in a complete arrest of cell proliferation. Altogether, these findings provide compelling genetic evidence for a redundant role of ERK1 and ERK2 in promoting cell proliferation. To further substantiate this idea, we have quantitatively measured the level of activating phosphorylation of ERK1/ERK2 (using a phospho-specific antibody that recognizes an identical epitope in the two isoforms) in the different MEF populations and plotted this value against their relative rates of cell proliferation. As shown in Fig. 10, the rate of MEF proliferation was strongly correlated (r2 = 0.90) with the total level of phosphorylated ERK1/ERK2, demonstrating that cell proliferation depends on the total activity of ERK1 and ERK2, which is dictated by the relative level of expression of the two isoforms. Our results confirm and extend the proposition of Lefloch et al. (28) by providing a genetic demonstration of the redundant function of ERK1 and ERK2 in cell proliferation.
FIG. 10.

The rate of cell proliferation is tightly correlated with the level of phosphorylated ERK1/ERK2 in MEFs. The extent of phospho-ERK1/2 staining was quantified at 5 min after NBCS stimulation and is expressed as a percentage of the level observed in control littermate MEFs (n ≥ 2). The rate of cell proliferation was estimated from the MTT assay at day 5 or 6 (last day of the experiment) and is expressed as a percentage of the proliferation measured in control MEFs (n = 3).
Our findings also disprove the model that ERK1 and ERK2 compete with each other and exert antagonistic effects on cell proliferation (34, 37, 62). This discrepancy cannot be explained by differences in genetic background, as we clearly show that loss of Erk1 does not increase the proliferation of MEFs in a C57BL/6 (used in the study of Vantaggiato et al. [62]) genetic background.
The total ablation of ERK1 and ERK2 leads to G1 arrest and accelerated replicative senescence of MEFs. The cell cycle arrest was associated with complete abrogation of the serum-stimulated induction of c-Myc, Cyclin D1, Cyclin E1/Cyclin E2, E2f1/E2f3, and Mcm2, genes that are critically involved in G1 progression and DNA replication, and with upregulation of the antiproliferative genes Tob1, p21Cip1, and p27Kip1. Little or no expression of c-Myc and cyclin D1 protein can be detected in ERK1/ERK2-deficient cells, and p27 accumulates to high levels that are comparable to those observed in serum-deprived cells. Cyclin D1, Tob1, c-Myc, and p27Kip1 are known transcriptional targets of the ERK1/2 pathway (26, 41, 63, 68, 69, 71), which also impacts on the stability of c-Myc protein (54) and on the activity of Tob1 (57). c-Myc, in turn, is required for efficient transcriptional activation of a large number of genes involved in G1-to-S-phase progression, including the E2f1, E2f2, and E2f3 genes (29, 73) and Mcm genes (25, 56, 73). Notably, gene-targeting studies have shown that E2f1-E2f3 transcription factors are absolutely essential for the ability of MEFs to enter S phase and proliferate (67). The suppression of E2f1/E2f3 transcription coupled to the functional inactivation of the Rb/E2F pathway, as a result of the inhibition of G1 cyclin expression and the accumulation of p27, explains in large part the G1 arrest observed in ERK1/ERK2-deficient fibroblasts.
Replicative senescence of late-passage fibroblasts is accompanied by a reduction in nuclear ERK1/2 activity (23, 24, 36, 60). The observation that ectopic expression of the nuclear MAP kinase phosphatase MKP2 induces premature senescence has led to the hypothesis that impaired nuclear ERK1/2 signaling may contribute to certain phenotypic aspects of the senescence process (61). Here, we provide genetic evidence that persistent inhibition of ERK1/2 signaling induces premature senescence of MEFs concomitant to G1 arrest of the cell cycle. Although the molecular mechanisms underlying the establishment of the senescent state remain to be investigated, we speculate that the stable inhibition of E2F activity and the resulting silencing of E2F target genes contribute to this process (8, 44). The Rb and p53 tumor suppressor pathways play an important role in the triggering and maintenance of cellular senescence (8). To bypass senescence and try to immortalize ERK1/ERK2-deficient MEFs, we have infected the cells with SV40 large T antigen, which binds both Rb and p53 and is able to overcome replicative senescence (22). However, we failed to established cell lines with deletions of the Erk1 and Erk2 genes. This suggests that the role of ERK1/2 signaling in cell cycle progression may be more complex than generally depicted and extend beyond inactivation of Rb.
Acknowledgments
We thank Kim Lévesque for animal care and maintenance, Sébastien Harton for expert help with tetraploid aggregation experiments, Danièle Gagné for help with FACS analysis, and Pierre Chagnon and Raphaële Lambert for quantitative PCR analysis. We thank Bernard Thorens and Trang Hoang for reagents. We are grateful to Gilles Pagès and Jacques Pouysségur for originally providing the Erk1−/− mice. We thank Philippe Lenormand for communicating results prior to publication and for discussions.
This work was supported by a grant from the National Cancer Institute of Canada. C. Frémin is recipient of a fellowship from the Cole Foundation. S. Meloche holds the Canada Research Chair in Cellular Signaling.
Footnotes
Published ahead of print on 5 April 2010.
REFERENCES
- 1.Anjum, R., and J. Blenis. 2008. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 9:747-758. [DOI] [PubMed] [Google Scholar]
- 2.Balmanno, K., S. D. Chell, A. S. Gillings, S. Hayat, and S. J. Cook. 2009. Intrinsic resistance to the MEK1/2 inhibitor AZD6244 (ARRY-142886) is associated with weak ERK1/2 signalling and/or strong PI3K signalling in colorectal cancer cell lines. Int. J. Cancer. 125:2332-2341. [DOI] [PubMed] [Google Scholar]
- 3.Bost, F., M. Aouadi, L. Caron, P. Even, N. Belmonte, M. Prot, C. Dani, P. Hofman, G. Pages, J. Pouyssegur, Y. Le Marchand-Brustel, and B. Binetruy. 2005. The extracellular signal-regulated kinase isoform ERK1 is specifically required for in vitro and in vivo adipogenesis. Diabetes 54:402-411. [DOI] [PubMed] [Google Scholar]
- 4.Boulton, T. G., and M. H. Cobb. 1991. Identification of multiple extracellular signal-regulated kinases (ERKs) with antipeptide antibodies. Cell Regul. 2:357-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton, T. G., S. H. Nye, D. J. Robbins, N. Y. Ip, E. Radziejewska, S. D. Morgenbesser, R. A. DePinho, N. Panayotatos, M. H. Cobb, and G. D. Yancopoulos. 1991. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65:663-675. [DOI] [PubMed] [Google Scholar]
- 6.Boulton, T. G., G. D. Yancopoulos, J. S. Gregory, C. Slaughter, C. Moomaw, J. Hsu, and M. H. Cobb. 1990. An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science 249:64-67. [DOI] [PubMed] [Google Scholar]
- 7.Bourcier, C., A. Jacquel, J. Hess, I. Peyrottes, P. Angel, P. Hofman, P. Auberger, J. Pouyssegur, and G. Pages. 2006. p44 mitogen-activated protein kinase (extracellular signal-regulated kinase 1)-dependent signaling contributes to epithelial skin carcinogenesis. Cancer Res. 66:2700-2707. [DOI] [PubMed] [Google Scholar]
- 8.Campisi, J., and F. d'Adda di Fagagna. 2007. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8:729-740. [DOI] [PubMed] [Google Scholar]
- 9.Chang, F., L. S. Steelman, J. G. Shelton, J. T. Lee, P. M. Navolanic, W. L. Blalock, R. Franklin, and J. A. McCubrey. 2003. Regulation of cell cycle progression and apoptosis by the Ras/Raf/MEK/ERK pathway (review). Int. J. Oncol. 22:469-480. [PubMed] [Google Scholar]
- 10.Denizot, F., and R. Lang. 1986. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 89:271-277. [DOI] [PubMed] [Google Scholar]
- 11.Di Benedetto, B., C. Hitz, S. M. Holter, R. Kuhn, D. M. Vogt Weisenhorn, and W. Wurst. 2007. Differential mRNA distribution of components of the ERK/MAPK signalling cascade in the adult mouse brain. J. Comp. Neurol. 500:542-556. [DOI] [PubMed] [Google Scholar]
- 12.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebisuya, M., K. Kondoh, and E. Nishida. 2005. The duration, magnitude and compartmentalization of ERK MAP kinase activity: mechanisms for providing signaling specificity. J. Cell Sci. 118:2997-3002. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, A. M., C. D. Katayama, G. Pages, J. Pouyssegur, and S. M. Hedrick. 2005. The role of Erk1 and Erk2 in multiple stages of T cell development. Immunity 23:431-443. [DOI] [PubMed] [Google Scholar]
- 15.Fremin, C., A. Bessard, F. Ezan, L. Gailhouste, M. Regeard, J. Le Seyec, D. Gilot, G. Pages, J. Pouyssegur, S. Langouet, and G. Baffet. 2009. Multiple division cycles and long-term survival of hepatocytes are distinctly regulated by extracellular signal-regulated kinases ERK1 and ERK2. Hepatology 49:930-939. [DOI] [PubMed] [Google Scholar]
- 16.Fremin, C., F. Ezan, P. Boisselier, A. Bessard, G. Pages, J. Pouyssegur, and G. Baffet. 2007. ERK2 but not ERK1 plays a key role in hepatocyte replication: an RNAi-mediated ERK2 knockdown approach in wild-type and ERK1 null hepatocytes. Hepatology 45:1035-1045. [DOI] [PubMed] [Google Scholar]
- 17.Fukunaga, R., and T. Hunter. 1997. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16:1921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giasson, E., and S. Meloche. 1995. Role of p70 S6 protein kinase in angiotensin II-induced protein synthesis in vascular smooth muscle cells. J. Biol. Chem. 270:5225-5231. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, F. A., D. L. Raden, and R. J. Davis. 1991. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem. 266:22159-22163. [PubMed] [Google Scholar]
- 20.Hatano, N., Y. Mori, M. Oh-hora, A. Kosugi, T. Fujikawa, N. Nakai, H. Niwa, J. Miyazaki, T. Hamaoka, and M. Ogata. 2003. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells 8:847-856. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Jha, K. K., S. Banga, V. Palejwala, and H. L. Ozer. 1998. SV40-mediated immortalization. Exp. Cell Res. 245:1-7. [DOI] [PubMed] [Google Scholar]
- 23.Kim, H. S., M. C. Song, I. H. Kwak, T. J. Park, and I. K. Lim. 2003. Constitutive induction of p-Erk1/2 accompanied by reduced activities of protein phosphatases 1 and 2A and MKP3 due to reactive oxygen species during cellular senescence. J. Biol. Chem. 278:37497-37510. [DOI] [PubMed] [Google Scholar]
- 24.Kim-Kaneyama, J., K. Nose, and M. Shibanuma. 2000. Significance of nuclear relocalization of ERK1/2 in reactivation of c-fos transcription and DNA synthesis in senescent fibroblasts. J. Biol. Chem. 275:20685-20692. [DOI] [PubMed] [Google Scholar]
- 25.Koppen, A., R. Ait-Aissa, J. Koster, P. G. van Sluis, I. Ora, H. N. Caron, R. Volckmann, R. Versteeg, and L. J. Valentijn. 2007. Direct regulation of the minichromosome maintenance complex by MYCN in neuroblastoma. Eur. J. Cancer 43:2413-2422. [DOI] [PubMed] [Google Scholar]
- 26.Lavoie, J. N., G. L'Allemain, A. Brunet, R. Muller, and J. Pouyssegur. 1996. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 271:20608-20616. [DOI] [PubMed] [Google Scholar]
- 27.Lefloch, R., J. Pouyssegur, and P. Lenormand. 2008. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol. Cell. Biol. 28:511-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefloch, R., J. Pouyssegur, and P. Lenormand. 2009. Total ERK1/2 activity regulates cell proliferation. Cell Cycle 8:705-711. [DOI] [PubMed] [Google Scholar]
- 29.Leung, J. Y., G. L. Ehmann, P. H. Giangrande, and J. R. Nevins. 2008. A role for Myc in facilitating transcription activation by E2F1. Oncogene 27:4172-4179. [DOI] [PubMed] [Google Scholar]
- 30.Lewis, T. S., P. S. Shapiro, and N. G. Ahn. 1998. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74:49-139. [DOI] [PubMed] [Google Scholar]
- 31.Li, J., and S. E. Johnson. 2006. ERK2 is required for efficient terminal differentiation of skeletal myoblasts. Biochem. Biophys. Res. Commun. 345:1425-1433. [DOI] [PubMed] [Google Scholar]
- 32.Lips, D. J., O. F. Bueno, B. J. Wilkins, N. H. Purcell, R. A. Kaiser, J. N. Lorenz, L. Voisin, M. K. Saba-El-Leil, S. Meloche, J. Pouyssegur, G. Pages, L. J. De Windt, P. A. Doevendans, and J. D. Molkentin. 2004. MEK1-ERK2 signaling pathway protects myocardium from ischemic injury in vivo. Circulation 109:1938-1941. [DOI] [PubMed] [Google Scholar]
- 33.Liu, X., S. Yan, T. Zhou, Y. Terada, and R. L. Erikson. 2004. The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene 23:763-776. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd, A. C. 2006. Distinct functions for ERKs? J. Biol. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loonstra, A., M. Vooijs, H. B. Beverloo, B. A. Allak, E. van Drunen, R. Kanaar, A. Berns, and J. Jonkers. 2001. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 98:9209-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenzini, A., M. Tresini, M. Mawal-Dewan, L. Frisoni, H. Zhang, R. G. Allen, C. Sell, and V. J. Cristofalo. 2002. Role of the Raf/MEK/ERK and the PI3K/Akt(PKB) pathways in fibroblast senescence. Exp. Gerontol. 37:1149-1156. [DOI] [PubMed] [Google Scholar]
- 37.Marchi, M., A. D'Antoni, I. Formentini, R. Parra, R. Brambilla, G. M. Ratto, and M. Costa. 2008. The N-terminal domain of ERK1 accounts for the functional differences with ERK2. PLoS One 3:e3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massague, J. 2004. G1 cell-cycle control and cancer. Nature 432:298-306. [DOI] [PubMed] [Google Scholar]
- 39.Mazzucchelli, C., C. Vantaggiato, A. Ciamei, S. Fasano, P. Pakhotin, W. Krezel, H. Welzl, D. P. Wolfer, G. Pages, O. Valverde, A. Marowsky, A. Porrazzo, P. C. Orban, R. Maldonado, M. U. Ehrengruber, V. Cestari, H. P. Lipp, P. F. Chapman, J. Pouyssegur, and R. Brambilla. 2002. Knockout of ERK1 MAP kinase enhances synaptic plasticity in the striatum and facilitates striatal-mediated learning and memory. Neuron 34:807-820. [DOI] [PubMed] [Google Scholar]
- 40.Meloche, S. 1995. Cell cycle reentry of mammalian fibroblasts is accompanied by the sustained activation of p44mapk and p42mapk isoforms in the G1 phase and their inactivation at the G1/S transition. J. Cell. Physiol. 163:577-588. [DOI] [PubMed] [Google Scholar]
- 41.Meloche, S., and J. Pouyssegur. 2007. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 26:3227-3239. [DOI] [PubMed] [Google Scholar]
- 42.Morrison, D. K., and R. J. Davis. 2003. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19:91-118. [DOI] [PubMed] [Google Scholar]
- 43.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 45.Nekrasova, T., C. Shive, Y. Gao, K. Kawamura, R. Guardia, G. Landreth, and T. G. Forsthuber. 2005. ERK1-deficient mice show normal T cell effector function and are highly susceptible to experimental autoimmune encephalomyelitis. J. Immunol. 175:2374-2380. [DOI] [PubMed] [Google Scholar]
- 46.Pagès, G., S. Guerin, D. Grall, F. Bonino, A. Smith, F. Anjuere, P. Auberger, and J. Pouyssegur. 1999. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science 286:1374-1377. [DOI] [PubMed] [Google Scholar]
- 47.Papkoff, J., R. H. Chen, J. Blenis, and J. Forsman. 1994. p42 mitogen-activated protein kinase and p90 ribosomal S6 kinase are selectively phosphorylated and activated during thrombin-induced platelet activation and aggregation. Mol. Cell. Biol. 14:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson, G., F. Robinson, T. Beers Gibson, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22:153-183. [DOI] [PubMed] [Google Scholar]
- 49.Raman, M., W. Chen, and M. H. Cobb. 2007. Differential regulation and properties of MAPKs. Oncogene 26:3100-3112. [DOI] [PubMed] [Google Scholar]
- 50.Robbins, D. J., E. Zhen, H. Owaki, C. A. Vanderbilt, D. Ebert, T. D. Geppert, and M. H. Cobb. 1993. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J. Biol. Chem. 268:5097-5106. [PubMed] [Google Scholar]
- 51.Rodier, G., C. Makris, P. Coulombe, A. Scime, K. Nakayama, K. I. Nakayama, and S. Meloche. 2005. p107 inhibits G1 to S phase progression by down-regulating expression of the F-box protein Skp2. J. Cell Biol. 168:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saba-El-Leil, M. K., F. D. Vella, B. Vernay, L. Voisin, L. Chen, N. Labrecque, S. L. Ang, and S. Meloche. 2003. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 4:964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarbassov, D. D., L. G. Jones, and C. A. Peterson. 1997. Extracellular signal-regulated kinase-1 and -2 respond differently to mitogenic and differentiative signaling pathways in myoblasts. Mol. Endocrinol. 11:2038-2047. [DOI] [PubMed] [Google Scholar]
- 54.Sears, R., F. Nuckolls, E. Haura, Y. Taya, K. Tamai, and J. R. Nevins. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14:2501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Servant, M. J., P. Coulombe, B. Turgeon, and S. Meloche. 2000. Differential regulation of p27(Kip1) expression by mitogenic and hypertrophic factors: Involvement of transcriptional and posttranscriptional mechanisms. J. Cell Biol. 148:543-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shohet, J. M., M. J. Hicks, S. E. Plon, S. M. Burlingame, S. Stuart, S. Y. Chen, M. K. Brenner, and J. G. Nuchtern. 2002. Minichromosome maintenance protein MCM7 is a direct target of the MYCN transcription factor in neuroblastoma. Cancer Res. 62:1123-1128. [PubMed] [Google Scholar]
- 57.Suzuki, T., J. K-Tsuzuku, R. Ajima, T. Nakamura, Y. Yoshida, and T. Yamamoto. 2002. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes Dev. 16:1356-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tam, P. P., and J. Rossant. 2003. Mouse embryonic chimeras: tools for studying mammalian development. Development 130:6155-6163. [DOI] [PubMed] [Google Scholar]
- 59.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tresini, M., A. Lorenzini, L. Frisoni, R. G. Allen, and V. J. Cristofalo. 2001. Lack of Elk-1 phosphorylation and dysregulation of the extracellular regulated kinase signaling pathway in senescent human fibroblast. Exp. Cell Res. 269:287-300. [DOI] [PubMed] [Google Scholar]
- 61.Tresini, M., A. Lorenzini, C. Torres, and V. J. Cristofalo. 2007. Modulation of replicative senescence of diploid human cells by nuclear ERK signaling. J. Biol. Chem. 282:4136-4151. [DOI] [PubMed] [Google Scholar]
- 62.Vantaggiato, C., I. Formentini, A. Bondanza, C. Bonini, L. Naldini, and R. Brambilla. 2006. ERK1 and ERK2 mitogen-activated protein kinases affect Ras-dependent cell signaling differentially. J. Biol. 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verykokakis, M., C. Papadaki, E. Vorgia, L. Le Gallic, and G. Mavrothalassitis. 2007. The RAS-dependent ERF control of cell proliferation and differentiation is mediated by c-Myc repression. J. Biol. Chem. 282:30285-30294. [DOI] [PubMed] [Google Scholar]
- 64.Voisin, L., C. Julien, S. Duhamel, K. Gopalbhai, I. Claveau, M. K. Saba-El-Leil, I. G. Rodrigue-Gervais, L. Gaboury, D. Lamarre, M. Basik, and S. Meloche. 2008. Activation of MEK1 or MEK2 isoform is sufficient to fully transform intestinal epithelial cells and induce the formation of metastatic tumors. BMC Cancer 8:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waskiewicz, A. J., A. Flynn, C. G. Proud, and J. A. Cooper. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waskiewicz, A. J., J. C. Johnson, B. Penn, M. Mahalingam, S. R. Kimball, and J. A. Cooper. 1999. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19:1871-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto, T., M. Ebisuya, F. Ashida, K. Okamoto, S. Yonehara, and E. Nishida. 2006. Continuous ERK activation downregulates antiproliferative genes throughout G1 phase to allow cell-cycle progression. Curr. Biol. 16:1171-1182. [DOI] [PubMed] [Google Scholar]
- 69.Yang, J. Y., C. S. Zong, W. Xia, H. Yamaguchi, Q. Ding, X. Xie, J. Y. Lang, C. C. Lai, C. J. Chang, W. C. Huang, H. Huang, H. P. Kuo, D. F. Lee, L. Y. Li, H. C. Lien, X. Cheng, K. J. Chang, C. D. Hsiao, F. J. Tsai, C. H. Tsai, A. A. Sahin, W. J. Muller, G. B. Mills, D. Yu, G. N. Hortobagyi, and M. C. Hung. 2008. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 10:138-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao, Y., W. Li, J. Wu, U. A. Germann, M. S. Su, K. Kuida, and D. M. Boucher. 2003. Extracellular signal-regulated kinase 2 is necessary for mesoderm differentiation. Proc. Natl. Acad. Sci. U. S. A. 100:12759-12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yasuda, T., H. Sanjo, G. Pages, Y. Kawano, H. Karasuyama, J. Pouyssegur, M. Ogata, and T. Kurosaki. 2008. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity 28:499-508. [DOI] [PubMed] [Google Scholar]
- 72.Yoon, S., and R. Seger. 2006. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 24:21-44. [DOI] [PubMed] [Google Scholar]
- 73.Zeller, K. I., A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng, P., H. A. Wagoner, O. H. Pescovitz, and R. Steinmetz. 2005. RNA interference (RNAi) for extracellular signal-regulated kinase 1 (ERK1) alone is sufficient to suppress cell viability in ovarian cancer cells. Cancer Biol. Ther. 4:961-967. [DOI] [PubMed] [Google Scholar]