Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression by binding to complementary target mRNAs and either promoting their decay or inhibiting their translation. Most eukaryotic genomes studied encode miRNAs, which are processed from longer, non-coding transcripts through pathways conserved from fungi to plants to animals. MiRNAs are now understood to be key mediators of developmental transitions in a number of model organisms. With respect to the immune system, miRNAs affect all facets of immune system development, from hematopoiesis to activation in response to infection, both during the innate and the adaptive immune response. At the same time, miRNA dysregulation is a central event in the development and pathophysiology of a number of cancers of the immune system. Here we will discuss our current understanding of this general regulatory mechanism focusing on its involvement in inflammation and in oncogenesis.
Keywords: miRNA, immune system, inflammation, cancer
Historically, the small RNA revolution began with the observation of an unexplained silencing phenomenon in floral pigmentation. Specifically, overexpression of a pigment biosynthesis gene in petunia plants, which was expected to produce more vividly colored flowers, resulted instead in the production of flowers with variegated pigmentation or even complete lack of colour.[1] At the time, this phenomenon was termed “co-suppression” and it is the first example of what is now known as RNA interference. Far from being confined to plants, this gene silencing phenomenon was also observed in fungi[2] and nematodes[3] but the molecular mechanism behind it remained unclear until the landmark studies of Fire and Mello, who demonstrated that it was specifically triggered by a double-stranded RNA.[4] Within the same decade, the field of miRNA began when lin 4, a small RNA hitherto unknown as an miRNA, was found in C. elegans and noted to down-regulate expression of lin-14 by antisense complementarity to the 3’UTR of lin-14.[5, 6] This kind of RNA was then shown not to be an isolated pecularity limited to nematodes, but rather a member of a large family of small regulatory RNA particles that is widely expressed in many species.[7] Since the initial discovery, miRNAs have been continuously uncovered and implicated in many cellular processes, including but not limited to development,[8, 9] cellular proliferation,[10] apoptosis and cancer.[11] Many hundreds of miRNAs have been identified in humans,[12] and they are predicted to regulate approximately 30% of all mRNAs coding for proteins.[13, 14]
MiRNA biogenesis
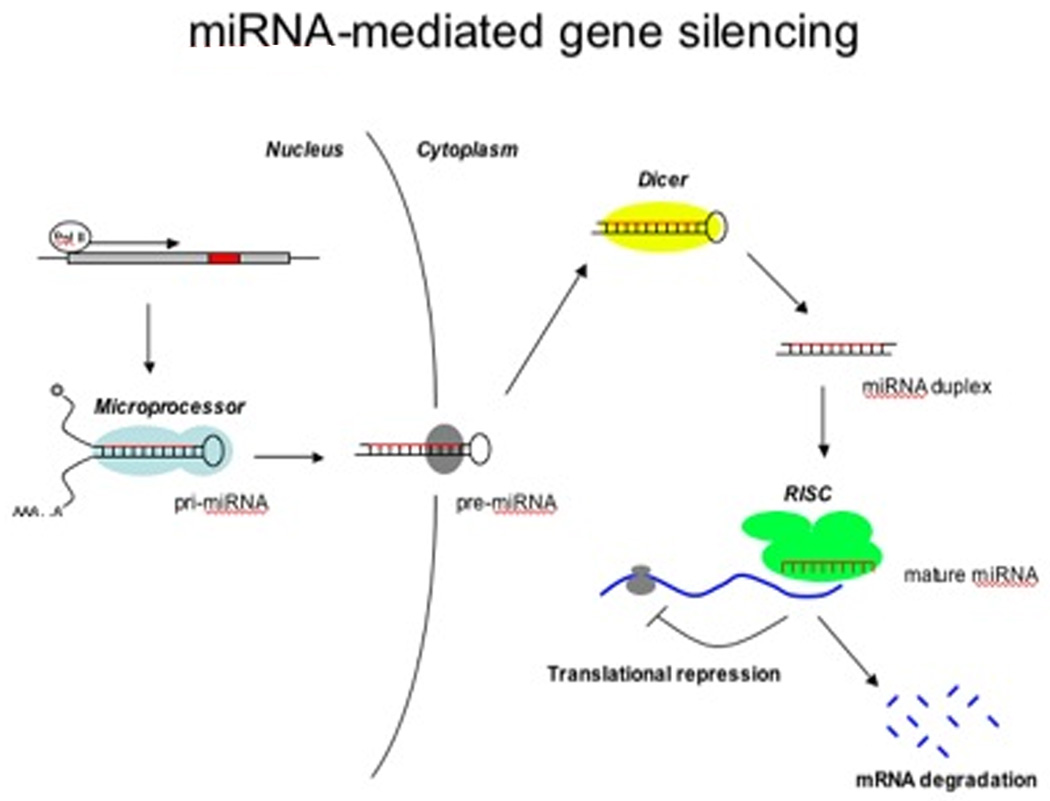
It is estimated that miRNAs comprise approximately 1% of the human genome.[15] 40–70% are encoded in introns of protein coding genes or introns and exons of non-coding RNAs, implying two or more possible mechanisms for their transcription.[16] Intronic miRNA genes are generally thought to be transcribed by RNA polymerase II;[17] while exonic miRNA genes are thought to utilize RNA polymerase III for transcription[18]. Regardless of the transcribing polymerase, miRNAs are initially synthesized as primary-miRNA (pri-miRNA) transcripts, which can extend over 1kb in length. These transcripts are processed by Drosha (a ribonuclease [RNase] III) and DGCR8, an RNA binding protein, which cleave the pri-miRNA to a ~70nt precursor-miRNA (pre-miRNA) within the nucleus.[17, 19, 20] The pre-miRNA is then exported out of the nucleus by exportin 5 in the presence of its cofactor Ran GTP, to be further processed in the cytoplasm; concordantly, depletion of Exportin 5 by RNA interference leads to a decrease in mature miRNAs and a decrease in pri- or pre-miRNA in the cytoplasm.[21, 22]
Once in the cytoplasm pre-miRNAs are cleaved by Dicer (an RNA endonuclease III) along with its partner protein TRBP (trans-activator RNA binding protein) to a ~21-22nt double stranded RNA molecule;[23] inhibition of Dicer leads to the accumulation of pre-miRNA in the cytoplasm.[24] This mature miRNA molecule is loaded onto the RNA-induced silencing complex (RISC), which contains Dicer, TRBP and an Argonaute protein that is present in multiple isoforms in mammalian cells.[25, 26] Within the RISC complex, the single stranded mature miRNA will guide RISC to the target mRNA.[27] At least in vertebrate systems, it is thought that targeting of miRNAs results in either target RNA destabilization,[28] or in repression of translation.[5, 29, 30] (Figure 1) However, recent data would also suggest that under certain conditions, miRNA binding leads to a boost in translation.[31, 32]
Figure 1. Biosynthesis of microRNA.
Primary-microRNA (pri-miRNA) transcription takes place in the nucleus by RNA polymerase II. Pri-miRNA is then processed in the nucleus by microprocessor complex composed of Drosha and DCGR8, producing a precursor-miRNA which is then exported out of the nucleus by Exportin-5 and Ran-GTP. In the cytoplasm, further processing by Dicer and TRPB releases a miRNA duplex. One strand is degraded while the mature miRNA is incorporated into the RNA-induced silencing complex (RISC), which leads to targeting to an mRNA leading to its destabilization or repression of translation.
MiRNA function: mRNA destabilization or translational inhibition?
Mammalian miRNAs pair to the 3’UTR of their target mRNA. It is thought that perfect or near-perfect matching of the 5’end of a miRNA (the “seed” region) to the target site within the 3’UTR was the sole requirement for miRNAs function and led to translational inhibition. Though the idea that seed matching leads to interference with translation is still current, the features within the 3’UTR of a particular mRNA that would accurately predict miRNA targeting have been expanded to include proximity of miRNA binding sites, AU-rich environments flanking the seed region and preferential sites within the UTR at both ends. Based on these parameters Grimson and colleagues have developed a database (www.targetscan.org) to better predict experimentally focused miRNA:mRNA pairing.[33]
It should be noted however that an alternate pathway of miRNA:target binding exists, which appears to rely on matching of a different stretch of sequence within the miRNA to a 3’UTR. This second pathway is thought to lead not to translational inhibition but rather, to mRNA destabilization through ARE-mediated decay.[34] These non-canonical types of miRNA:target interactions have not been extensively studied, and thus cannot be bioinformatically predicted as yet, however they could be equally consequential to the better-studied seed-matching mechanism leading to translational inhibition.
miRNA in the Immune System
The immune system provides a complex but well orchestrated defense program poised against the myriad of pathogenic infections to which an organism is exposed. For example, an infection may trigger an inflammatory response, the initiation, propagation and resolution of which must be carefully co-ordinated and balanced. Lack of proper initiation or propagation hampers the innate immune response; conversely, lack of proper resolution can lead to chronic disease states.
Propagation of the innate response activates the adaptive immune system, which relies upon a pre-determined program of DNA rearrangements in lymphocytes, to generate specific antibodies toward the pathogen that initiated the immune response, as well as memory. This process of DNA rearrangements in lymphocytes is stringently regulated; a failure in co-ordination of cutting and repairing the DNA leads to chromosomal translocations and the seeds of oncogenesis.
Clearly, the immune response (both innate and adaptive) is extremely highly regulated; recent work from a number of laboratories has revealed that micro-RNAs play a very important role in this intricate system (Figure 2). The first step toward this understanding was the identification of multiple miRNAs in hematopoietic cells, many of which were expressed specifically in cells and tissues of immune relevance. In fact, cells of the hematopoietic system can be selectively identified from other tissues by their miRNA expression profile: they all express five highly specific miRNAs, miR-142, miR-144, miR-150, miR-155 and miR-223.[35] Further, different lineages of immune cells can also be distinguished by their unique miRNA expression profiles. For example erythrocytes show higher expression of miR-451, whereas, B- and T- Lymphocytes express miR-223.[36, 37] Additionally, although it may seem that similar expression patterns of miRNAs exist in two very different cell types, expression levels, and spatio-temporal control can be very different. For example, in comparing B and T lymphocytes, Merekova et al. report similar expression patterns of miRNAs in peripheral blood samples of both these cell types (particularly the expression of miR-16, miR-142-3 and miR-150), however, miR-342 expression was 10-fold higher in T-cells than in B-cells.[36] Others have shown differential temporal and spatial expression of miRNA in these cells, which have a distinct role on their development and function. Therefore miRNA profiles can help in delineating the different cells of the immune system, yet within each cell type, varying expression of selected miRNA can help them navigate through their developmental stages.
Figure 2. Involvement of miRNAs in B-cell Differentiation.
MicroRNAs are involved in B-cell differentiation and maturation. MicroRNAs near the arrows have been shown to be involved in that developmental stage, by affecting (downregulation-red arrows or indirectly stimulating-green arrows) the labeled factors (black) involved in that developmental transition.
We will focus on the B-cell lineage to further illustrate the contribution of miRNAs on normal development and maturation of cells in the immune system.
B-cells and miRNA
MiRNA profiling reveals differences in miRNA expression between the various maturation states of B-cells (Figure 2). Malumbres et al. show that the differentiation of naïve B-cells to centroblast, and further, of centroblast to memory B-cell is marked by radical changes in the miRNA profile while naïve and memory B-cells exhibit marked similarities.[38] For example, MiR-125b is highly expressed in centroblasts, possibly preventing these from further differentiation (miR-125b has been shown to repress PRDM1 and IRF4, two key factors in post germinal center (GC) reaction,[39] while allowing for expression of BCL6, which is important in GC reaction[38]). Similarly, a correlation between miR-223 and B-cell development has been argued. LMO2, a target of miR-223, is required for hematopoiesis during mouse embryogenesis,[40] and is also expressed in GC B-cells.[41] In addition, Zhang et al, report high expression of miR-223 in both naïve and memory cells compared to GC cells, which they postulate control the expression of LMO2, allowing for repression of this transcription factor once the cell has exited the germinal center.[42] Similar correlations are made between members of the miR-30 family and miR-9 on PRDM1, which is essential for post GC B-cell development and function.[43] Thus aside from marking distinct developmental transitions, miRNAs are also vital for maintaining a cell at its proper developmental stage.
Though a correlation can be made between miRNAs expressed in certain cells and the potential target genes they may regulate, direct demonstration that these genes are in fact targets of miRNA regulation has relied upon genetic approaches. These have come in three flavors: (a) ablation of miRNAs (gene knock-outs); (b) enforced expression of miRNAs (usually by targeted knock-in approaches) and (c) ablation of miRNA target sites from 3’UTRs of putative target genes. Though bioinformatics algorithms would suggest that miRNAs target many hundreds of genes within a single cell, and therefore genetic gain- and loss-of function studies (a and b above) may be predicted to have pleiotropic effects, that is not necessarily the case: studies with some miRNAs have shown that only a few of the target mRNAs may be critical for a particular biological process. Alternatively, there are hints that a particular miRNA may target multiple components but which belong to a common regulatory pathway, therefore the impact of miRNA control of a process may be additive and much more important that the moderate effects on individual target protein concentrations may suggest. These two considerations have eased fears that such studies will be hard to interpret.
Below, we will concentrate on a handful of miRNAs of specific relevance to B cells.
MiR-150
MiR-150, which is expressed selectively in mature B and T cells,[44] prevents pro-B to pre-B cell transition through the downregulation of c-Myb;[45] targeted deletion of c-Myb was shown to block B-cell development at the pro-B to pre-B cell transition. Furthermore, miR-150 plays a vital role in B cell survival in the spleen, and targeted deletion of c-Myb mirrors that phenotype.[46]
Overexpression of miR-150 in B-cells resulted in decreased levels of c-Myb both at the mRNA and protein levels. Transgenic mice overexpressing miR-150 are phenotypically similar to mice with a haploinsuffiency for c-Myb. Xiao et al., further showed that ectopic miR-150 expression in B-cell progenitors blocked B-cell development at the pro-B cell stage and these mice also had decreased levels of B1 cells, a subset of mature B-cells present in spleen and the peritoneal cavity, due to increased apoptosis. Upon miR-150 deletion there was an expansion of splenic and peritoneal B1 cells plus an increase in IgA levels, the preferential isotype related to this B-cell subset.[47]
As opposed to most other miRNAs it has been reported that expression of miR-150 is higher in normal B-cells over lymphoma cells.[48]
MiR-181
MiR-181 comprises a family of four non-coding RNAs (miR-181 a through d) each of which is processed from a distinct transcript but all of which contain an identical seed sequence, therefore potentially targeting the same set of mRNAs for translational repression. MiR-181 is preferentially expressed in the thymus, lung and brain with detectable levels in bone marrow and spleen. Within the bone marrow miR-181 is detectable in undifferentiated progenitor cells but upregulated in differentiated B-lymphocytes in mice. Ectopic expression of miR-181 in these progenitor cells leads to an increase in the production of B-lymphoid cells over T-cells as measured by increased expression of CD19. This was also seen in vivo, upon infection of mouse bone marrow B cells with a miR-181 expressing retrovirus and transfer into irradiated recipients,[49] therefore implicating this miRNA as an early determinant in B-cell hematopoietic lineage differentiation.
One of the members of miR-181 family, miR-181b, has also been implicated in regulation of B-cell function by affecting class-switch recombination (CSR). CSR is an intrachromosomal deletional recombination event that leads to a switch in the effector portion of the immunoglobulin gene and thus to the expression of a new Ig isotype. CSR is catalyzed by activation-induced cytidine deaminase (AID)-dependent mutation,[50] and AID may contain what has been described as an alternative target site for miR-181b on its 3’UTR (i.e. a target site that does not depend on seed match and therefore is not predicted by robust algorithms such as TargetScan). Specifically, De Yebenes et al. have shown that infection of mouse primary B-cells with retrovirus expressing miR-181b impaired class switching to IgG1 upon LPS and IL-4 stimulation. AID mRNA and protein levels were both diminished in transfected B-cells and the 3’UTR of AID was fingered as a specific target site for miR-181b by in vitro luciferase reporter assays. Although these in vitro data do not exclude a more pleiotropic contribution of miR-181b to the CSR reaction in vivo, the interpretation preferred by the authors of this study is that miR-181b helps in fine-regulating CSR by directly regulating AID.[51]
MiR-17~92
The miR-17~92 cluster consists of six miRNAs that are processed from a common precursor transcript, and which are grouped together based on their seed sequence as well as function.[52] MiR17~92 cluster, has been implicated in the pathogenesis of B-cell lymphomas, in particular diffuse large B-cell lymphoma; this will be discussed in further detail below.
However, what is miR 17~92 cluster’s contribution to normal B-cell physiology? Targeted deletion of the miR 17~92 cluster in embryonic stem cells shows normal levels of progenitor B-cells in the fetal liver, however results in a decrease in Pre-B-cells due to increased apoptosis specifically in the B-cell compartment. Adult B-cell development is also compromised. Ventura et al. treated lethally irradiated mice with fetal liver cells of wild-type or miR 17~92 deleted mice in order to reconstitute their hematopoietic system. They found that, as compared to WT, reconstitution with cells from miR 17~92-deficient mice lead to a contraction of the lymphoid compartment and this was due to a specific reduction in circulating B-cells, splenic B-cells and peritoneal B1 cells. They attribute this to increased expression of the pro-apopotic gene Bim, which has been shown to keep cell proliferation in check in Myc-induced lymphoma models.[53] Interestingly, the 3’UTR of Bim contains four predicted sites for the members of the miR 17~92 cluster and increased levels of Bim are found in pro- and pre-B-cells of transgenic mice fetal liver, and in acute deletion of miR 17~92 in adult hematopoietic cells.[54]
miR-155
miR-155 is probably the best-characterized miRNA involved in B-cell maturation and function. Original studies showed that co-expression of B-cell integration cluster or bic, a common site of insertion for avian leukosis virus, which induces lymphomas, and c-Myc, cooperate to induce cellular proliferation involving mostly cells of the hematopoietic system. Lymphomas were found to be from an early-B-cell progenitor, which lacked rearrangement of the λ-light chain locus,[55] pre-B cells. Bic has been shown to encode miR-155, which is then processed by the Drosha/Dicer pathway.
MiR-155 was also shown to be essential for normal immune function. Bic-deficient mice showed reduced humoral and cellular response to infection, succumbed more readily to Salmonella infection, had a reduced number of germinal center B-cells, an overall reduction of immunoglobin levels, deficient antigen presentation by dendritic cells, failure to activate T-cells and an overall failure to mount a memory immune response and build protection against re-infection.[56, 57]
Though humoral immunity was clearly impaired in miR-155 deficient animals, there was no deficiency in the antibody diversification processes of somatic hypermutation or CSR, both of which are catalyzed by AID. However, the levels of Ig in the blood were clearly decreased and the number of memory B-cells 42 days post immunization was decreased as well.[58]
The mechanism behind this impairment is complex: it partly involves a defect in cell differentiation and maturation by failure of B-cells to differentiate into plasmablasts, as the miR-155-/- phenotype was partly recapitulated by overexpression of Pu.1 (an important protein in B-cell development,[59] which is highly expressed in GC B-cells and down-regulated in post GC cells,[39] and which has been identified as a possible target for miR-155 by transcriptome profiling).[58] It can also be partly attributed to a lack of down-regulation of AID, itself a direct target of miR-155; directed mutation of the miR-155 target site within the 3’UTR of AICDA leads to a failure of translational repression of AID by miR-155. Consequently, mice that contain these mutations have higher levels of CSR and aberrant affinity maturation, recapitulating the miR155-/- phenotype.[60, 61]
As is clear from these recent studies, we have only begun to scratch the surface of what promises to be a very fertile field of investigation of miRNA-mediated gene expression control mechanisms during the immune response. (figure 2)
MiRNAs and lymphoid malignancies
Alterations of miRNAs have been observed in hematologic malignancies, including lymphomas, by global miRNA expression profiling as well as expression analysis of specific miRNAs. Apart from identifying abnormal expressions of specific miRNAs, the former method also allows a global view of the miRNomes. It revealed in the miRNA profiles of B-cell lymphomas genetic fingerprints of normal B cell counterparts and aberrations of a small number of miRNAs presumably acquired during transformation.[38, 42, 62] In addition, miRNA profiling identified miRNA signatures that can distinguish distinct types of lymphomas,[62, 63] or subgroups of a lymphoma type, for example, the germinal center B-cell vs. the activated B-cell type of diffuse large B-cell lymphomas (DLBCL).[38, 48] These miRNA signatures may have ancillary diagnostic roles to the current methodology based on morphology and immunophenotype. Moreover, these signatures may provide mechanistic insights to the biologic differences between lymphoma subgroups and subtypes.
Among the miRNAs that are shown to be abnormally expressed in lymphoid malignancies, two -- miR-155 and miR-17-92 polycistron deserve more detailed discussions. As briefly highlighted above, miR-155 and miR-17-92 have important functions in the normal immune system and are also implicated in the pathogenesis of lymphomas.
miR-155 and lymphomas
Given the role of miR-155 in normal B cell differentiation, the involvement of miR-155 in B lymphomagenesis is not surprising. Elevated levels of miR-155 compared to normal B cells have been observed in many lymphomas, including DLBCL, primary mediastinal B-cell lymphoma, Hodgkin lymphoma, and chronic lymphocytic leukemia/small lymphocytic lymphoma.[63–66] miR-155 expression in these lymphomas can be heterogeneous and associated with specific subgroups that have prognostic significance. For example, in DLBCL, miR-155 expression is highest in the activated B-cell subgroup, which have worse clinical outcome.[48, 65, 67] In chronic lymphocytic leukemia, miR-155 expression tends to be higher in cases with unmutated IgVH, a poor prognostic marker, although statistical significance has not been reached.[66] In addition, miR-155 expression itself may have prognostic significance that is independent of its association with other prognostic markers. A miRNA signature that includes high miR-155 expression correlates with shorter interval to need for treatment intervention in CLL.[68] In DLBCL, although miR-155 expression itself does not appear to correlate with overall survival in DLBCL as a group, high miR-155 in the activated B-cell subgroup is associated with better survival.[69]
The exact mechanisms that mediate miR-155 over-expression in these lymphomas are not known. MiR-155/BIC induction has been linked to the B-cell receptor and the transforming growth factor-beta pathways.[70, 71] The former is believed to play an important role in lymphomagenesis[72] and is thought to induce miR-155 through the AP-1 and NF-kB binding sites.[71, 73] A causal role of abnormal NF-kB activity in driving miR-155 over-expression is supported by the association of higher miR-155 expression with higher NF-kB activity in DLBCL cell lines and a higher miR-155/BIC levels in ABC-type DLBCLs,[67] in which NF-kB is often constitutively activated.[74] miR-155 is also induced by the latent membrane protein of EBV.[75, 76] Increased miR-155 may contribute to the pathogenesis of EBV-associated lymphoid malignancies by modulating endogenous transcription regulatory factors and by maintaining viral latency via attenuation of NF-kB activities.[76, 77]
Besides the aforementioned lists of circumstantial evidence that support a role of miR-155 in lymphoma development, miR-155 has been demonstrated directly in animal models to cause lymphomas. As previously mentioned, BIC over-expressed by retroviral vectors cooperated with c-Myc to promote lymphomagenesis in chickens;[55] as well, mice over-expressing miR-155 in B cells developed polyclonal pre-B-proliferation followed by full-blown high-grade B-cell lymphoma at 6 months of age.[78]
Obviously, further investigations are necessary to better understand how miR-155 contributes to lymphoma development. Based on the animal studies, miR-155 could promote B-cell proliferation. In normal B cells, miR-155 deficiency seems to disrupt the production of high-affinity B-cell clones that give rise to plasma cells and memory cells. [79] Thus, an over-expressed miR-155 may “arrest” the lymphoma cells in an activated proliferative state characteristic of these B-cells prior to entry into terminally differentiated B cells. In addition, mouse models that specifically disrupt the interaction of miR-155 with AICDA 3’ untranslated region demonstrated that miR-155 can target AICDA in normal B cells in vivo.[60, 61] Considering the role of AICDA in lymphomagenesis,[80] it will also be important to determine the relevance of these interactions in lymphoid malignancies.
Overall, identification of the full complement of miR-155 target genes will be instrumental to mechanistically delineate the role of miR-155 in B cell lymphomas. Several potential targets for miR-155 with relevance to lymphomagenesis have been identified through the use of gene expression arrays generated by manipulating miR-155 levels in cell lines,[77] by subgrouping primary lymphoma samples based on miR-155 levels (i.e. high vs. low),[67] or by comparing cell lines with differential miR-155 expression.[63] Further studies should aim at dissecting the relative contribution of each of those targets in miR-155-mediated lymphomagenesis for specific lymphoma types.
miR-17-92 and lymphomas
MiR-17-92 (also called oncomir-1) is a polycistronic miRNA gene located in 13q31-32 that encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92a-1). miR-17-92 and its paralogs are known to act as oncogenes in a variety of tumors, including lymphomas.[81] MiR-17-92 was first implicated in lymphomagenesis when a minimal amplicon containing the primary transcript of this polycistron (at that time called C13orf25) was identified in B cell lymphoma cell lines and primary lymphoma samples, which was associated with increased expression levels of this transcript.[52, 82] In a subsequent study, up-regulation of miR-17-92 was observed in 65% of the B-cell lymphoma patients, including those without detectable amplifications.[83]
A direct causal relationship between increased miR-17-92 and lymphomagenesis was demonstrated in a couple of mouse models. In the first one, enforced miR-17-92 expression in Em-Myc transgenic mice resulted in acceleration of lymphoma formation.[83] The tumors in these double-transgenic mice did not exhibit the high degree of apoptosis normally associated with lymphoid tumors in the Em-Myc mice, suggesting that miR-17-92 helps to suppress c-Myc-induced apoptosis. In the second model, transgenic mice that have elevated miR-17-92 levels developed a lymphoproliferative disease and autoimmunity and died prematurely. Both the peripheral B and T cell compartments were expanded.[84] These studies indicate that miR-17-92 per se can lead to abnormal expansion of the lymphoid compartment, but may require co-operating genetic lesions for full-blown malignancy.
miR-17-92 is likely to contribute to lymphoma development through promotion of cell cycle entry and suppression of apoptosis. B and T lymphocytes isolated from miR-17-92 transgenic mice have enhanced proliferation and reduced apoptosis, which correlates with down-regulation of two miR-17-92 target genes, the tumor suppressor PTEN and the pro-apopotic Bim.[84] miR-17-92 may also regulate cell cycle and apoptosis of lymphocytes through regulation of other target genes. miR-17-92 targets E2F and is an integral component of the E2F/c-Myc regulatory circuitry controlling cell cycle and apoptosis. Complex positive and negative feedback loops exist among E2F, c-Myc and miR-17-92, which ultimately determine the levels of E2F and whether the cells would progress from G1 into S phase, arrest at G1 or undergo apoptosis.[85–88] The critical role of miR-17-92 in this circuitry may be particularly relevant in the pathogenesis of c-Myc-mediated lymphoid malignancies. In the presence of a deregulated c-Myc, which can up-regulate E2F, particularly E2F1, over-expression of miR-17-92 may modulate its level to facilitate cell cycle progression rather than apoptosis. Mir-17-92 may also promote cell proliferation by targeting CDKN1A/p21, a negative regulator of the G1-S checkpoint,[89, 90] or by down-regulating transforming growth factor beta-signaling.[91] Interestingly, miR-17-92 may contribute to lymphomagenesis by down-regulating distinct targets in different lymphoma subtypes, depending on the genetic constitution of the tumor cells.[90] In lymphomas with co-existing genetic events that down-regulate Bim (e.g. a deletion) or up-regulate BCL2 (e.g. a translocation), miR-17-92 may promote lymphoma development by down-regulating p21 and promoting cell cycle progression. On the other hand, in lymphomas where there is already high proliferative potential (e.g. c-Myc driven), miR-17-92 may facilitate lymphoma formation by down-regulating Bim.
MiRNAs and inflammation
MiRNAs provide a new layer in the multi-tiered regulation of cellular physiology. Thus far, we have tried to illuminate how miRNAs partake in cellular development and differentiation by highlighting their involvement in normal B-cell maturation. We have also touched upon the role of two miRNAs important for B cell oncogenesis. Although we did not discuss the roles of miRNAs in development and maturation of other immune cell types (T cells, dendritic cells, macrophages) some of the miRNAs we did discuss are important in the inflammatory response. For example, Baltimore and colleagues have shown that miR-155 is induced early in macrophages as a consequence of exposure to a broad range of inflammatory mediators.[92] However, all the miRNAs we highlighted thus far are thought to interact with their cognate target mRNAs in a canonical fashion, that is to say through their seed region through base complementarity with the target 3’UTR. Therefore, in all these cases miRNA regulation is through translational repression.
But what of miRNAs with base complementarity to a 3’UTR that does not require interaction with the canonical seed region? Currently, one such example exists in the literature, and centers on the interaction of miR-16 with mRNAs whose 3’UTRs contain AU-rich elements (AREs).[93] MiR-16 contains an 8-nt long stretch of sequence (UAAAUAUU) that is complementary to the ARE sequence; within the 3’UTR, this sequence targets mRNAs for ARE-mediated degradation (requiring a number of mRNA binding proteins such as tristetraprolin or TTP, BRF1 etc).[94] Indeed, Jin et al have reported that miR-16 is required for targeting specific ARE-containing mRNAs (the very labile mRNAs of TNF-alpha and cox-2) for degradation by delivering it to a RISC complex, in a process that requires both Dicer and TTP. Complementing this pioneering work is a recent study by Calin and colleagues, where exogenous expression of miR-16 followed by array expression profiling revealed that mRNAs with ARE elements are more frequently downregulated upon miR-16 overexpression. Based on these two studies, we hypothesize that a miR-16-like, non-canonical mRNA targeting mechanism is not uncommon, though at present bioinformatics tools do not exist to directly predict such occurrences.
Should such a non-canonical mechanism be widely operative, the best system to study it would appear to be the immune inflammatory response. There is clear evidence in the literature that ARE-mediated degradation directly influences the initiation, propagation and resolution of the inflammatory response. Most recently, an elegant study by Hao and Baltimore has clearly shown that differences in mRNA stability exert a strong influence on the temporal order of gene expression after stimulation of fibroblasts or macrophages with pro-inflammatory stimuli (TNF and LPS). Of direct relevance was the observation that mRNA expressed early in the response had abundant AREs in their 3’UTRs, whereas those expressed later had fewer or none.[95] Although this study focused on the notion that mRNA instability is an intrinsic property of gene structure, in light of the miR-16 study detailed above the implication is that ARE-mediated, mRNA instability during the inflammatory response could well be either miR-16 mediated, or mediated by an as yet uncharacterized miR with ARE-base complementation properties. We are looking forward to future studies aiming to find such miRNAs; perhaps the inflammatory response will offer a paradigm in this regard.
Acknowledgements
We apologize to the many investigators whose work we were unable to cite because of space limitations. We thank Dr. G. Teng for developing the diagram presented in Figure 1, and Rebecca Delker for help with editing. Supported by the US National Institutes of Health.
References
- 1.Napoli C, Lemieux C, Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 3.Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 4.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 9.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Letters. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 10.Murchison EP, et al. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biological reviews of the Cambridge Philosophical Society. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Lim LP, et al. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez A, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 19.Denli AM, et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 20.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund E, et al. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 22.Yi R, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ketting RF, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 25.Hammond SM, et al. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 26.Gregory RI, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 28.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 29.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 30.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 31.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya SN, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 37.Kluiver J, et al. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20:1931–1936. doi: 10.1038/sj.leu.2404387. [DOI] [PubMed] [Google Scholar]
- 38.Malumbres R, et al. Differentiation-stage-specific expression of microRNAs in B-lymphocytes and diffuse large B-cell lymphomas. Blood. 2008 doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cattoretti G, et al. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol. 2006;177:6930–6939. doi: 10.4049/jimmunol.177.10.6930. [DOI] [PubMed] [Google Scholar]
- 40.Warren AJ, et al. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 41.Natkunam Y, et al. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109:1636–1642. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, et al. Patterns of microRNA expression characterize stages of human B cell differentiation. Blood. 2009 doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calame K. Activation-dependent induction of Blimp-1. Curr Opin Immunol. 2008;20:259–264. doi: 10.1016/j.coi.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 44.Monticelli S, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin YC, et al. c-Myb is an evolutionary conserved miR-150 target and miR-150/c-Myb interaction is important for embryonic development. Mol Biol Evol. 2008;25:2189–2198. doi: 10.1093/molbev/msn165. [DOI] [PubMed] [Google Scholar]
- 46.Thomas MD, et al. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 48.Lawrie CH, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 49.Chen CZ, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 50.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Yébenes VG, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. Journal of Experimental Medicine. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 53.Egle A, et al. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tam W, et al. Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis. J Virol. 2002;76:4275–4286. doi: 10.1128/JVI.76.9.4275-4286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 58.Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKercher SR, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 60.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roehle A, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 63.Gibcus JH, et al. Hodgkin lymphoma cell lines are characterized by a specific miRNA expression profile. Neoplasia. 2009;11:167–176. doi: 10.1593/neo.08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kluiver J, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 65.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fulci V, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 67.Rai D, et al. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet Cytogenet. 2008;181:8–15. doi: 10.1016/j.cancergencyto.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 69.Jung I, Aguiar RC. MicroRNA-155 expression and outcome in diffuse large B-cell lymphoma. Br J Haematol. 2009;144:138–140. doi: 10.1111/j.1365-2141.2008.07424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kong W, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin Q, et al. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J Biol Chem. 2008;283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gururajan M, Jennings CD, Bondada S. Cutting edge: constitutive B cell receptor signaling is critical for basal growth of B lymphoma. J Immunol. 2006;176:5715–5719. doi: 10.4049/jimmunol.176.10.5715. [DOI] [PubMed] [Google Scholar]
- 73.Kluiver J, et al. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 74.Davis RE, et al. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rahadiani N, et al. Latent membrane protein-1 of Epstein-Barr virus induces the expression of B-cell integration cluster, a precursor form of microRNA-155, in B lymphoma cell lines. Biochem Biophys Res Commun. 2008;377:579–583. doi: 10.1016/j.bbrc.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 76.Lu F, et al. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J Virol. 2008;82:10436–10443. doi: 10.1128/JVI.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin Q, et al. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calame K. MicroRNA-155 function in B Cells. Immunity. 2007;27:825–827. doi: 10.1016/j.immuni.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 80.Pasqualucci L, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 81.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 83.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O'Donnell KA, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 86.Sylvestre Y, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 87.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 88.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–145. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ivanovska I, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inomata M, et al. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 91.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 92.O'Connell RM, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Calin GA, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jing Q, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 95.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]