Abstract
A new coarse-grained (CG) intermolecular force field is presented for a series of zwitterionic lipids. The model is an extension of our previous work on nonionic surfactants and is designed to reproduce experimental surface/interfacial properties as well as distribution functions from all-atom molecular dynamics (MD) simulations. Using simple functional forms, the force field parameters are optimized for multiple lipid molecules, simultaneously. The resulting CG lipid bilayers have reasonable molecular areas, chain order parameters, and elastic properties. The computed surface pressure vs. area (π-A) curve for a DPPC monolayer demonstrates a significant improvement over the previous CG models. The DPPC monolayer has a longer persistence length than a PEG lipid monolayer, exhibiting a long-lived curved monolayer surface under negative tension. The bud ejected from an oversaturated DPPC monolayer has a large bicelle-like structure, which is different from the micellar bud formed from an oversaturated PEG lipid monolayer. We have successfully observed vesicle formation during CG-MD simulations, starting from an aggregate of DMPC molecules. Depending on the aggregate size, the lipid assembly spontaneously transforms into a closed vesicle or a bicelle. None of the various intermediate structures between these extremes seem to be stable. An attempt to observe fusion of two vesicles through the application of an external adhesion force was not successful. The present CG force field also supports stable multi-lamellar DMPC vesicles.
Keywords: Coarse grain force field, lipid, vesicle, surface tension, monolayer, surface pressure
1. Introduction
The recent widespread interest and development of coarse-grained (CG) molecular models of biological and polymeric systems have allowed the frontier of molecular simulations to access molecular phenomena such as the self-assembly of macromolecules into an array of complex structures.[1,2] Lipid assembly is one of the most widely studied phenomenon with CG modeling, not only because of the interesting behavior exhibiting a variety of morphologies with over 10 nm length scale (such as bicelles, vesicles, and even cubic structures), but also because of the importance in biological and nanotechnology applications. The great success of the pioneering lipid CG model proposed by Smit et al.[3,4] encouraged the subsequent CG modeling of lipids including dissipative particle dynamics from the phenomenological approaches.[5–14] There are several all-atom molecular dynamics (MD) based CG modeling approaches, which aim at establishing a link between the CG description and the underlying atomic nature of the molecule.[15–21] Among these, roughly speaking, there are three major approaches: 1) reverse Monte Carlo methods[15,16,22], 2), force matching schemes[17,18], and 3) the elaboration of CG force fields.[19–21] The reverse Monte Carlo approach, a popular method for CG modeling of polymers,[23,24] was extended to lipids by Shelley et al.[15,16] Force matching, the most systematic approach to build a CG model, is based on the underlying atomic interactions between the chosen CG segments. This method has been developed and extensively used for many molecular systems by the Voth group.[17,18] The force field approach is an extension of Smit’s approach, where the parameterization of each CG segment is done empirically using selected functional forms.[19–21] One of the most successful examples of the latter is the MARTINI force field.[19,20] Also, interested readers may refer to a recent book,[25] where more detailed accounts of a variety of CG approaches can be found.
Specifically, herein we present a new CG molecular force field for zwitterionic lipids, which was built by a force field approach. The present model is an extension of the previously reported PEG force field, where interfacial properties and the structure at the CG level are taken into account.[26,27] The previous lipid model from this group was built by the inverse Boltzmann method, where the CG model should reproduce the detailed structure of the all-atom lipid bilayer system. Here we have changed the strategy used to fix the CG force field for several reasons; 1) selected target thermodynamic quantities e.g., interfacial properties and solvation free energy, are more easily tuned in the parameterization, 2) the transferability of the force field for different lipid structures e.g., micelle, bilayer, inverse hexagonal can be expected, and 3) degradation of structural properties is minimized by choosing an appropriate functional form.
Like all-atom force fields,[28] the choice of water model is quite important for this approach. For the sake of computational efficiency, the present CG water is assumed to contain multiple water molecules. This enables application of the CG model to simulations of dilute systems such as lipid bicelles and vesicles. Having a liquid state across the temperature range of 0 to 100 °C and desired interfacial properties (experimental surface tension and density as well as a reasonable interfacial width and transfer free energy across water/oil interface), the number of water molecules to be packed into a single CG site is limited to three.[26] The functional form is also quite important to prevent an unrealistic freezing of water under ambient conditions,[26] which can be an issue for CG modeling.[20,29] The size of the water CG particle determines the CG resolution level, which in this case enhances the sampling efficiency by about three orders of magnitude, compared with all-atom MD simulations.[30]
In the next section, we describe the all-atom MD simulations used to obtain reference data for the CG modeling as well as the details of CG-MD simulations. In the section 3, the CG mapping procedure is described along with the selection of the functional forms. Then, the CG-MD results are presented in the following section. The quality of CG model is assessed by considering the structure and elastic properties of the lipid membranes. Section 5 is devoted to presenting applications using the new CG force field along with a comparison to previous models. The applications presented for monolayer collapse and vesicle formation demonstrate the potential ability of the present CG model to capture realistic macromolecular organization beyond the nanoscale.
2. Methods and Simulations
2.1. All-atom molecular dynamics
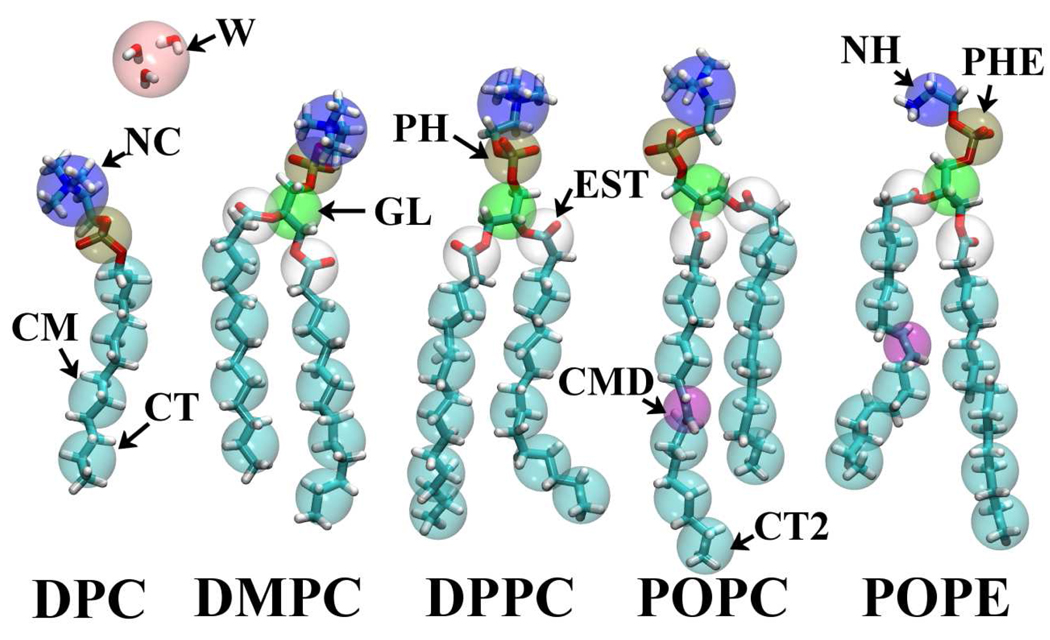
We have carried out all-atom (AA) MD runs for five systems: a dodecylphosphatidylcholine (DPC) micelle solution, and four different bilayers: dimyristoyl phosphatidylcholine (DMPC), dipalmitoyl phosphatidylcholine (DPPC), palmitoyl oleyl phosphatidylcholine (POPC), and palmitoyl oleyl phosphatidylethanolamine (POPE). (Fig.1) Temperatures are set to have a liquid crystal state for each system. The CHARMM PARAM27R force field [28,31] is employed for the lipids along with the TIP3P water potential.[32] We used 60 DPC molecules for the micellar solution. For the bilayers, we set up each system with 128 lipid molecules along with a sufficient quantity of water to mimic a saturated lipid bilayer; the number of water molecules being determined according to experiments.[33,34] The area per lipid molecule was fixed during the MD simulations using the NPnAT ensemble. The temperature was controlled by Langevin dynamics, which was coupled to all atoms except hydrogens with a 5 ps−1 damping coefficient. Normal pressure was held at 1 atm by a Nosé-Hoover Langevin piston [35] with a decay period of 5 ps and a damping time of 2.5 ps. The detailed system setup is listed in Table 1. AA-MD simulations were carried out with periodic boundary conditions using the efficient parallel molecular dynamics program NAMD 2.6.[36] Long-range electrostatics were calculated with the Particle-Mesh Ewald (PME) scheme [37] using an order-4 interpolation and a grid spacing smaller than 0.1 nm. Lennard-Jones (LJ) pair potentials were evaluated within a cutoff distance of 1.2 nm, with a smooth switching function above 1 nm. A multiple time-step scheme was employed, with a base timestep of 2 fs and an extended timestep of 4 fs for longer-range Coulomb forces. All bonds involving hydrogen atoms were constrained to their equilibrium length by SHAKE/RATTLE. [38,39]
Fig. 1.
Schematic definition of coarse-grained sites.
TABLE 1.
System and conditions for all-atomic molecular dynamics simulations
| Lipid | Lipid no. | Water | Time [ns] | Area [nm2] | T [K] | |
|---|---|---|---|---|---|---|
| Micelle | DPC | 60 | 14802 | 10 | – | 298 |
| Bilayer | DMPC | 128 | 3405 | 80 | 0.606 | 310 |
| DPPC | 128 | 4017 | 40 | 0.64 | 323 | |
| POPC* | 128 | 3963 | 100 | 0.64 (0.68) | 303 | |
| POPE | 128 | 3838 | 40 | 0.60 | 308 |
Simulation data is taken from Ref [34].
2.2 Coarse-grained molecular dynamics
To develop and evaluate the CG model, we used our in-house MD code, MPDyn.[40] NVT, NPT, NPnAT dynamics were generated using a Nosé-Hoover chain thermostat and an Andersen or Parrinello-Rahman barostat, with a response time of 0.5 and 10 ps, respectively.[41–43] The target pressure and temperature were 0.1 MPa and 303 K, respectively, though the temperature is slightly changed depending on the lipid component to simulate the liquid crystal phase. The nonbonded LJ interaction was truncated at 1.5 nm, though the Coulomb interaction was calculated by Ewald summation when charged particles were in the system.[39] The rRESPA algorithm was employed to integrate the equations of motion.[44] Bonded and short-ranged nonbonded interactions were updated every 5 fs, while long-ranged interactions were updated every 40 fs. We used XO-RESPA for NPT dynamics, [40,43] which increases the numerical stability of integration.
The LAMMPS code [45] was adopted for the large scale computations (N > 100,000) found in Sec. 5. In this case, there are some additional restrictions on the available methods: The NPT ensemble was generated by Nosé-Hoover dynamics, [46] the Coulomb interactions were evaluated by the PPPM method, [37] and a single time step of 10fs was used. These minor changes did not affect the simulation results of interfacial properties or structure, which are the target properties of the CG model.
3. Coarse-graining
The CG lipid model was developed here as an extension of our PEG (polyethyleneglycol) surfactant model. We use the same water and alkane models as in the earlier work.[26] The CG particles are defined in Fig. 1 and Table 2. The CG water particle has a different nature from the other CG particles because it represents multiple molecules. This choice warrants discussion, as some more recent CG models use a single water molecule CG representation to improve accuracy.[17,21] Packing several independent molecules into one site does impose a limitations on the present CG model. For example, we cannot expect to have a reasonable structure factor for the bulk water. Also, it is impractical to use the force-matching procedure [17,18] to construct the CG model, as shown previously.[26] It should be noted here that, in many situations (relatively dilute systems such as micelles and liposomes), the MD simulation is dominated by the calculation of water-water interactions, even though these interactions are not of primary interest. In this sense, a coarser (multi-molecule) CG water particle is preferred, even though this makes the details of the water-water structure beyond the scope of the CG model. Our interest is mainly on the lipid or surfactant self-assembled structure, while we still require the CG water to reproduce several properties: First, we selected an explicit water model in order to have hydrodynamic interactions between surfactant aggregates. Thus, the CG water should work essentially as a momentum carrier. Second, the water model should have a liquid phase in the temperature range of 0 to 100°C. Third, the CG solvent should have the correct density and surface tension. Finally, the CG model should reproduce a reasonable interfacial width with alkanes and the correct transfer free energy of solutes. To meet these conditions, we have evaluated a series of functional forms and eventually selected the Lennard-Jones (LJ) 12-4 function. Readers may refer to the previous paper for more details.[26]
TABLE 2.
CG segment name and the corresponding all-atomic functional group
| CG segment | All-atom |
|---|---|
| NC* | -CH2CH2-N- (CH3)3 (+1) |
| NH* | -CH2CH2-NH3 (+1) |
| PH* | -PO4- (—1) |
| PHE* | -PO4- (—1) (for PE headgroup) |
| GL | -CH2CH-CH2- |
| EST1 | -CH2CO2- (in the sn-2 chain) |
| EST2 | -CH2CO2- (in the sn-1 chain) |
| CMD2 | -HC=CH- (cis) |
| CM | -CH2CH2CH2- |
| CT | CH3CH2CH2- |
| CT2 | CH3CH2- |
| W | (H2O)3 |
These are the charged CG particles.
In our parameterization, we use simple functional forms for the inter- and intra-particle potentials. For intramolecular interactions, we assume harmonic bond stretching and angle bending potentials for 1–2 and 1-2-3 bonded pairs, while the pairs separated by more than two bonds interact via the nonbonded force. We have also a correction term for 1–3 interaction, which is the same as the nonbonded interaction but being truncated-shifted to get zero at r = rs.
| (1) |
| (2) |
| (3) |
The last term is optional but sometimes needed as a correction to prevent an implausibly bent structure in CG-MD. In the CG model, the bending force constant can be comparable to the nonbonded potential depth. This sometimes causes a complete angle collapse where a strong 1–4 nonbonded interaction overwhelms the 1-2-3 bending interaction. The last term should contribute only when the artificial collapse of an angle happens. A choice of rs = rmin, which is the distance of the LJ potential minimum used for the nonbonded interactions, meets this condition and does not affect the angle distribution in a normal situation. Therefore, the parameters for previous CG models [26,27] are useful without any modifications. Actually, it is quite a rare event to have 1–3 pair CG sites closer than rmin. Our target quantity for the parameterization of the force constants is bond and angular distributions obtained from AA-MD simulations. In this approach, the detailed structural correlation at the resolution finer than the CG particle size is beyond the scope of the model. For example, although we sometimes have a bimodal distribution of bond distance from AA simulations, we have only unimodal distributions with the simple harmonic potential. In this case, the parameters are chosen to reproduce the average and dispersion of the corresponding AA distribution function.
After a series of investigations of structural and interfacial properties using different functional forms,[26] we selected two types of LJ functions, (12-4) and (9-6), for the nonbonded interaction potential;
| (4) |
As shown in the previous work, [26,27] for pure solvents, we fixed the LJ parameters, ε and σ, by fitting to surface tension and density data. This procedure was applied to alkenes and esters. The alkene-water interaction was determined using the interfacial tension data, while the ester-water interaction parameters were fixed to give the experimental hydration free energy.
The above mentioned strategy was systematically applied to fix the parameters using the segmental molecules (chemical functional groups) composing the lipids, when the bulk molecules are in the liquid state at the target temperature of 303 K. Unfortunately for the lipid head group this is not the case. To optimize the head group parameters, we used structural data of lipid assemblies, e.g., experimental molecular area and distribution functions obtained from AA-MD simulations. In this case, we had several parameters to be determined simultaneously. By using multiple lipid molecules as shown in Fig.1, we optimized the model to perform well for a series of lipids using the same transferable parameters.
The main difference between the lipid model and the previous PEG model is that each of the lipid headgroup CG sites (PH, NC, and NH) has a partial charge. We calculated the interaction using the Ewald sum with the standard Coulomb functional form. [39]
| (5) |
where qi is the partial charge of i-th particle and ε0 is the permittivity in vacuum. The relative permittivity was selected to take into account the screening effects of water implicitly. It should be emphasized that we evaluated the Coulomb interaction with no truncation using an Ewald summation. For the sake of computational efficiency, attempts were made to introduce a cutoff based Coulomb interaction with a damping function. When we attempted to apply the latter approach to systems having heterogeneity in the charge distribution, e.g., an ionic surfactant monolayer at the air/water interface, the cutoff could not reproduce the correct asymptotic behavior of the counter ion distribution away from the interface. Specifically, despite the choice of damping function and parameters, the counter ion distribution does not decay as an exponential function, though theoretically it should do so. These details will appear in a forthcoming paper. The correct ionic distribution is a necessary condition, even though not a sufficient condition, to have a reasonable interaction between two charged surfaces at large separations. For CG-MD, the lack of a valid long-ranged interaction is a significant flaw because the computations should give insight into the mesoscopic organization of the macromolecules. Therefore, even though a non-cutoff scheme requires extra computation, we evaluate the Coulomb interaction without a truncation.
All CG parameters determined are tabulated in Tables 3–5.
TABLE 3.
Bond stretching parameters
| Bond pair | kb [kcal/mol/Å2] | r0 [Å] | |
|---|---|---|---|
| NC | PH | 4.80 | 4.25 |
| PH | CM | 12.00 | 3.69 |
| PH | GL | 8.90 | 3.52 |
| GL | EST1 | 30.00 | 2.88 |
| GL | EST2 | 8.40 | 3.48 |
| EST1 | CM | 4.70 | 3.55 |
| EST2 | CM | 5.10 | 3.61 |
| NH | PHE | 9.40 | 3.60 |
| PHE | GL | 8.90 | 3.52 |
| CM | CM | 6.16 | 3.64 |
| CM | CT | 6.16 | 3.65 |
| CT2 | CM | 9.00 | 3.13 |
| CMD2 | CM | 8.00 | 3.03 |
| CT | CMD2 | 8.00 | 3.09 |
TABLE 5.
Nonbonded interaction parameters
| Pair | LJ-Type | ε [kcal/mol] | σ [Å] | |
|---|---|---|---|---|
| PH | CM | LJ9-6 | 0.300 | 4.9530 |
| PH | CT | LJ9-6 | 0.320 | 4.9925 |
| NC | CM | LJ9-6 | 0.400 | 5.1280 |
| NC | CT | LJ9-6 | 0.420 | 5.1675 |
| PH | W | LJ12-4 | 1.000 | 4.0300 |
| NC | W | LJ12-4 | 0.900 | 4.6100 |
| PH | PH | LJ9-6 | 1.400 | 5.4000 |
| NC | NC | LJ9-6 | 0.700 | 5.7500 |
| NC | PH | LJ9-6 | 1.150 | 4.2000 |
| GL | GL | LJ9-6 | 0.420 | 4.5060 |
| GL | EST1 | LJ9-6 | 0.470 | 4.4030 |
| GL | EST2 | LJ9-6 | 0.470 | 4.4030 |
| GL | CM | LJ9-6 | 0.420 | 4.5060 |
| GL | CT | LJ9-6 | 0.444 | 4.5455 |
| GL | W | LJ12-4 | 0.640 | 4.4385 |
| GL | CT2 | LJ9-6 | 0.362 | 4.3635 |
| PH | CT2 | LJ9-6 | 0.280 | 4.8105 |
| NC | CT2 | LJ9-6 | 0.320 | 4.9855 |
| CMD2 | GL | LJ9-6 | 0.312 | 4.2555 |
| CMD2 | EST1 | LJ9-6 | 0.440 | 4.0050 |
| CMD2 | EST2 | LJ9-6 | 0.440 | 4.0050 |
| PH | CMD2 | LJ9-6 | 0.300 | 4.7025 |
| NC | CMD2 | LJ9-6 | 0.350 | 4.8775 |
| GL | NC | LJ9-6 | 0.650 | 4.6200 |
| GL | PH | LJ9-6 | 0.300 | 4.7500 |
| EST1 | NC | LJ9-6 | 0.750 | 4.4750 |
| EST2 | NC | LJ9-6 | 0.750 | 4.4750 |
| EST1 | PH | LJ9-6 | 0.500 | 4.5500 |
| EST2 | PH | LJ9-6 | 0.500 | 4.5500 |
| PHE | PHE | LJ9-6 | 1.400 | 4.6000 |
| PHE | CM | LJ9-6 | 0.300 | 4.9530 |
| PHE | CMD2 | LJ9-6 | 0.300 | 4.7025 |
| PHE | CT | LJ9-6 | 0.320 | 4.9925 |
| PHE | CT2 | LJ9-6 | 0.280 | 4.8105 |
| PHE | GL | LJ9-6 | 0.300 | 4.7500 |
| PHE | EST1 | LJ9-6 | 0.500 | 4.5500 |
| PHE | EST2 | LJ9-6 | 0.500 | 4.5500 |
| PHE | W | LJ12-4 | 1.000 | 4.0300 |
| NH | CM | LJ9-6 | 0.330 | 4.5530 |
| NH | CMD2 | LJ9-6 | 0.300 | 4.3025 |
| NH | CT2 | LJ9-6 | 0.320 | 4.4105 |
| NH | CT | LJ9-6 | 0.340 | 4.5925 |
| NH | NH | LJ9-6 | 1.100 | 4.6000 |
| NH | PHE | LJ9-6 | 1.200 | 3.8000 |
| NH | GL | LJ9-6 | 0.750 | 4.1900 |
| NH | EST1 | LJ9-6 | 0.850 | 4.1100 |
| NH | EST2 | LJ9-6 | 0.850 | 4.1100 |
| NH | W | LJ12-4 | 0.800 | 3.9500 |
| PH | PHE | LJ9-6 | 1.400 | 5.0000 |
| NH | PH | LJ9-6 | 1.200 | 3.8000 |
| NC | PHE | LJ9-6 | 1.150 | 4.2000 |
| NC | NH | LJ9-6 | 0.880 | 5.1750 |
| CT | CT | LJ9-6 | 0.469 | 4.5850 |
| CT | CM | LJ9-6 | 0.444 | 4.5455 |
| CM | CM | LJ9-6 | 0.420 | 4.5060 |
| CMD2 | CMD2 | LJ9-6 | 0.232 | 4.0050 |
| CMD2 | CT | LJ9-6 | 0.330 | 4.2950 |
| CMD2 | CT2 | LJ9-6 | 0.269 | 4.1130 |
| CMD2 | CM | LJ9-6 | 0.312 | 4.2555 |
| CMD2 | W | LJ12-4 | 0.270 | 4.1880 |
| EST1 | CM | LJ9-6 | 0.470 | 4.4030 |
| EST2 | CM | LJ9-6 | 0.470 | 4.4030 |
| EST1 | CT | LJ9-6 | 0.470 | 4.4425 |
| EST2 | CT | LJ9-6 | 0.470 | 4.4425 |
| EST1 | CT2 | LJ9-6 | 0.390 | 4.2605 |
| EST2 | CT2 | LJ9-6 | 0.390 | 4.2605 |
| EST1 | EST1 | LJ9-6 | 0.495 | 4.3000 |
| EST1 | EST2 | LJ9-6 | 0.495 | 4.3000 |
| EST2 | EST2 | LJ9-6 | 0.495 | 4.3000 |
| EST1 | W | LJ12-4 | 0.820 | 4.2900 |
| EST2 | W | LJ12-4 | 0.820 | 4.2900 |
| W | CT | LJ12-4 | 0.360 | 4.4780 |
| W | CM | LJ12-4 | 0.340 | 4.4385 |
| CT2 | CT2 | LJ9-6 | 0.312 | 4.2210 |
| CT2 | CM | LJ9-6 | 0.362 | 4.3635 |
| W | CT2 | LJ12-4 | 0.290 | 4.2960 |
| CT2 | CT | LJ9-6 | 0.383 | 4.4030 |
| W | W | LJ12-4 | 0.895 | 4.3710 |
4. Results
In the first step of coarse-graining, the bond and angle parameters were adjusted to reproduce the bond and angle distributions obtained from AA-MD simulations. Because we have just two parameters to fit, i.e., the force constant and equilibrium length or angle, the average and dispersion of the distribution are enough to fix the parameters uniquely. Although we sometimes find bimodal distributions for the target bond or angle, we still use the simple harmonic potential model, ignoring the detailed bimodality in this CG model. This approximation limits the effective resolution of the CG model, since the missing resolution due to this approximation is smaller than the size of the CG particle. In the second step, we fixed the nonbonded parameters. Due to the (re)parameterization of nonbonded interactions, the bond and angle distributions are also changed in general and have to be modified. However, the required correction for the bond and angle parameters is usually quite minor. Therefore, we can efficiently fix all parameters in this manner. As mentioned above, the 1–3 correction van der Waals-term seldom affects the angular distribution, but works to prevent a non-realistic bent structure.
The nonbonded parameters are fixed to reproduce experimental surface tension and density data as long as the experimental data are available. In the previous study, we fixed parameters for a series of alkane and PEG chains in this fashion,[26] and here, we applied the same procedures to alkenes and esters. Alkenes are especially needed for unsaturated lipids, e.g., the oleyl chain, which is found in an important lipid class. Table 6 summarizes the surface tension and density data for small molecules including unsaturated bonds and ester groups, which are used for the parameter fitting. We used interfacial tension to fix the nonbonded parameters for the alkene – water (CMD (=-CH=CH-) - W) interaction. Our model yields 49±1 mN/m for 4-cis-octane-water interfacial tension, which experimentally is ~50 mN/m. For the ester-water interaction, we used experimental hydration free energy values for the parameter fitting. For example, the hydration free energy of propylbutyrate, (CT2-EST-CTE), is calculated as 2.4±0.1 kcal/mol, which is in good agreement with experimental value of 2.5 kcal/mol.
TABLE 6.
Surface tension and density for esters and alkenes.
| System | Molecular Structure | Interaction | Surface Tension [mN/m] | Density [103kg/m3] | ||
|---|---|---|---|---|---|---|
| exp | MD | Exp | MD | |||
| polypentanoate | CT-EST-CTE | CT-EST | 24.768 | 24.8 | 0.862 | 0.8629 |
| EST-EST | ||||||
| polybutyrate | CT2-EST-CTE | CT2-EST | 24.04 | 24.4 | 0.8633 | 0.8610 |
| Propyloctanoate | CT-CM-EST-CTE | EST-CM | 28.7±3 | 27.0 | 0.872 | 0.8576 |
| Hexyloctanoate | CT-CM-EST-CME-CT | 29.3±3 | 28.0 | 0.867 | 0.8545 | |
| 4-cis-octene | CT-CMD-CT | CT-CMD | 21.06 | 20.9 | 0.713 | 0.7120 |
| CMD-CMD | ||||||
| 3-cis-hexene | CT2-CMD-CT2 | CT2-CMD | 17.39 | 17.8 | 0.6704 | 0.6694 |
CTE represents (CH3CH2CH2-), which is identical with CT, though CTE is CT segments connecting to oxygen atom in ester group. To distinguish between the bonds CTE-EST and CT-EST, we conventionally use CTE, which has the same non bonded parameters as CT.
The above mentioned method is the same as used in the CG modeling of PEG surfactants,[26] where fitting surface/interfacial tension, density, and hydration free energy of small molecules was used to estimate the net interaction between the CG segments. This approach enabled a step-by-step parameterization in a systematic way. However, this is applicable only when the selected small molecules are in a liquid state at the target temperature. This is not the case for the lipid head groups. For example, phosphatidylcholine (PC) should be in a solid state at 303 K. We did not use thermodynamic data of the solid material because the effective interaction between headgroups of liquid-crystalline lipid (bilayers) is likely to be different from that of a PC solid. Therefore, as an alternative we used structural properties of the lipid micelle and bilayers, such as pair-distribution functions from AA-MD simulations.
As mentioned above, since the present CG model is designed to simulate the liquid (or liquid-crystalline) state of lipids, a minimal temperature change is required to prevent the freezing of the alkane chains of the lipid bilayer. For example, because the melting point of a DPPC bilayer is 315 K, we set the simulation temperature at 323 K to examine the CG model bilayer, as we have done for the AA-MD. The model is not designed to reproduce the melting point indeed, but is designed to produce thermodynamic, interfacial and structural properties in the liquid crystal phase.
Figure 2 gives the distribution functions for the DPC micelle system. In Fig. 2a, radial distributions from the micelle center for each segmental unit of DPC are plotted for both CG and AA models, respectively. The micelle structure obtained from the CG model is quite similar to that of the AA model. This is confirmed by examination of the pair-distribution functions among headgroup segments as shown in Fig. 2b. Comparing to the previous model made by the inverse Boltzmann method and using a fully tabulated potential,[15] the structural accuracy is slightly degraded. However, using simple functions for pair-interaction, the quality of the CG model is still surprisingly good. In addition, unlike the earlier tabulated potential model, our new model shows versatility in that it can be applied to several different lipid molecules as shown later.
Fig. 2.
(a) Probability density of each CG segments as a function of distance from the DPC micelle center. (b) Radial distribution functions between DPC headgroup segments. Solid lines: CG-MD, dashed lines: AA-MD.
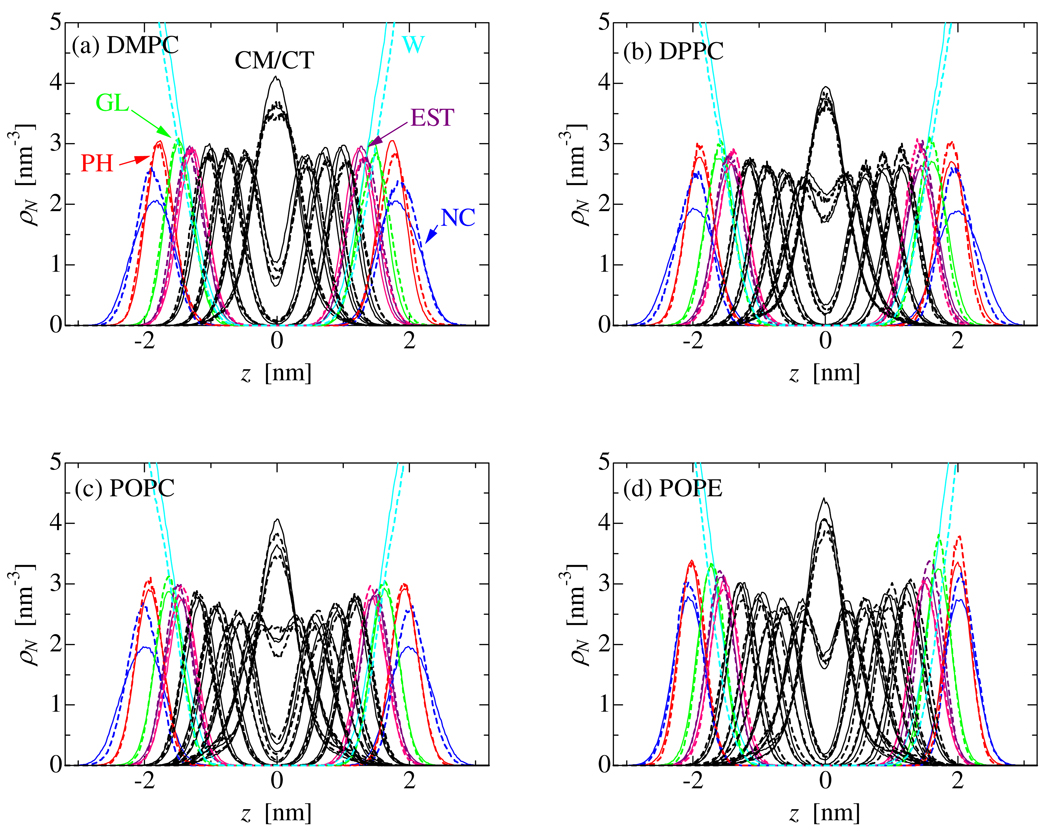
A series of CG-MD simulations was carried out to parameterize full sets of interaction parameters for PC and PE lipid molecules (DMPC, DPPC, POPC, and POPE). Literally hundreds of trial MD simulations were needed to fix all the parameters for head group-head group, head group-lipid tails and head group-water interactions. For the CG-MD, the NPT ensemble was used to simulate tension-free membranes. For membrane simulations, interfacial tension and density were also selected as target properties for the parameter fitting. Therefore, as shown in table 7, the CG model accurately reproduces the experimental molecular area and lamellar repeat spacing of these lipid bilayers. Note that our target area of 0.64 – 0.66 nm2 for POPC has a slightly smaller value than the experimental one, namely 0.68 nm2. This is because AA-MD runs of a POPC bilayer in the NPnAT yields better agreement with the NMR order parameter profile with the molecular area fixed at 0.64 nm2 rather than 0.68 nm2.[34] Figure 3 plots the probability distributions of each CG segment along the normal to the bilayer membrane for each of these lipids. Again, the agreement between the CG and AA-MD calculations is excellent, though the probability distribution of the choline group (NC) is slightly broader in the CG model than in the AA model. The water penetration into the membrane seems slightly enhanced, though the shift of the water distribution is less than 0.09 nm. In order to evaluate the alkane-water interaction, we have checked the partitioning of water into hexadecane. For this examination, we have carried out an NPnAT-MD of the binary system of 566 hexadecane molecules and 3200 water CG particles, each of these two molecules make a slab in the simulation box because of their immiscibility. The interfacial area in the x-y plane is fixed at 6 × 6 nm, though the box length along the interface normal fluctuates around 1.6 nm to adjust the system pressure at 0.1MPa. Temperature was kept at 318 K to compare to the experimental data. Water migration into the hexadecane region rarely happened, though 100ns MD is enough to see leakages of water and to obtain reasonably converged solubility data. The water solubility in hexadecane was calculated as 370±10 µg/cm3, which is larger than the experimental one of 95 µg/cm3 [47] by a factor of about 4. WThus, we observe a bit too much leakage of water into the hydrocarbon region. As shown in the previous paper, the interfacial tension is selected as a primary target property in our parameterization, though by selecting the functional forms for the nonbonded interaction the experimental transfer free energy of hexane from its bulk into water is also reproduced. Since our CG modeling is designed to predict lipid self-assembly in solution and an alkane chain is a key part of a lipid, the solubility of alkane to water is more important than that of water into alkane. A slight degradation of calculated water solubility into alkane cannot alone explain the slight shift of the water distribution in lipid membranes, because there is no such shift in the DPC micelle system. Thus, there is still some room to improve the total performance of the CG model for the target properties.
TABLE 7.
Structural parameters of lipid bilayers obtained from coarse-grained MD simulations
Fig. 3.
Probability distribution of the CG segments along the bilayer normal. Solid lines: CG-MD, dashed-lines: AA-MD.
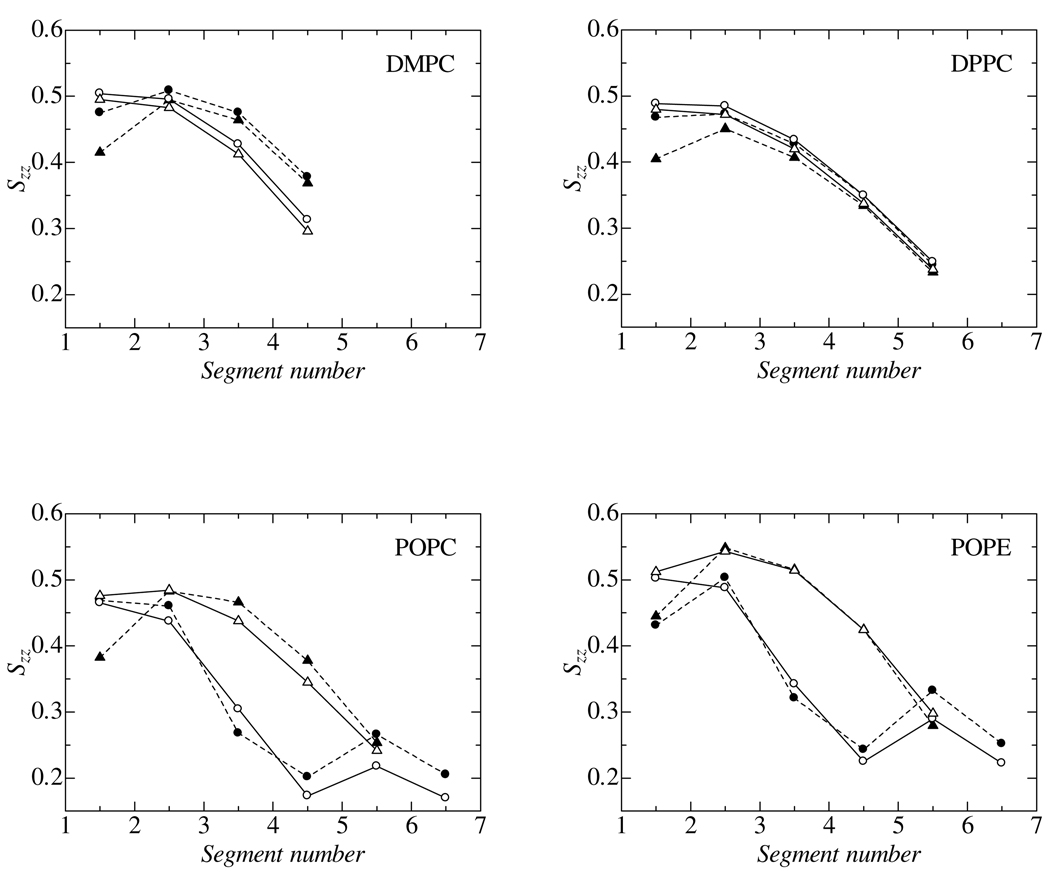
To characterize the hydrocarbon chain structure in the bilayer core, the order parameter is defined in a similar manner to the ordinal deuterium NMR order parameter:
| (6) |
where b is the unit bond vector connecting two adjacent CG beads in the hydrocarbon chain and n is the unit vector along the bilayer normal. This is an analysis for the CG configurations, though we applied the same analysis to the AA configurations using the fictitious CG sites calculated as the center of mass of the associated segments. (Fig. 1) The calculated order parameters are plotted in Fig. 4. The segment number in the abscissa was given for each hydrophobic chain segments starting from EST to the tail segment. The figure demonstrates that the CG model accurately reproduces the whole order parameter profiles and captures several important features. Details such as plateaus in the profiles and the gradual disorder along the profile for straight chains are captured by the CG model. Another detail reproduced by the model is the slightly lower order in the sn-2 than in the sn-1 chains of DMPC and DPPC molecules. Finally, the specific disorder around the double bonded segments in the unsaturated chains of POPC and POPE are present in the CG model. The agreement is excellent, though the DMPC model slightly underestimates the order parameter near the tail. It should be noted that there is room to improve even the CHARMM force field used in AA-MD, especially for the order parameters near the ester group.[34] Thus, we conclude that the quality of the current CG model meets expectations with regards to the structural properties.
Fig 4.
Segmental order parameters, Szz, in hydrophobic chains. Segment number 1 is “EST1” or “EST2”. Filled symbols with dashed lines: AA-MD, open symbols with solid lines: CG-MD. Circles and triangles denote the sn-1 and sn-2 chains, respectively.
We now turn our attention to elastic properties of CG lipid bilayers. Using a trajectory from NPT-MD, the area expansion modulus, KA, was calculated by the following relation derived from linear response theory:[48]
| (7) |
where A is the averaged cross-sectional area per lipid and Nl is the number of lipid molecules aligned in a lipid leaflet of the bilayer. The bracket denotes the ensemble average. In order to calculate the bending modulus of CG membranes, we also have done a spectral analysis of the membrane undulatory and peristaltic motions. To this end, we employed the model proposed by Brannigan and Brown,[49] which provides a consistent analysis for thermal fluctuations of membranes. MD simulations were carried out for a large lipid membrane patch consisting of 2048 lipid molecules in the periodically replicated simulation box; this system is large enough to analyze the membrane fluctuation due to the undulatory and peristaltic modes. The simulation length was over 100 ns for each system, which generated sufficiently long trajectories to obtain converged spectral data. The “GL” segment of each lipid was selected to define the membrane surface. Since no flip-flop motion was detected during the simulations, upper and lower leaflets of the membrane were clearly divided. A grid of 32 × 32 points was assigned to each monolayer giving one lipid molecule on each grid point on average. When multiple lipids were found in a grid, the height of monolayer surface was calculated as an arithmetic average of heights of “GL” segments in the grid. When no lipid occupied a grid, the height of the grid was calculated as the arithmetic average of neighboring grids. No smoothing filter to average over grids was used because it eliminates the protrusion mode though it does not affect the bending modulus.
The calculated area expansion and bending moduli are listed in Table 8. A typical area expansion modulus for common PCs and PEs is limited to the range of 200 to 300 mN/m.[50,51] The present CG model shows good agreement with the experimental value.[51] Bending moduli are more challenging quantities to be precisely evaluated by MD simulations, because the moduli are rather sensitive to the spectrum data at the lower k-values. A 100 ns-MD run was needed to obtain converged values. Although a precise estimation of the bending modulus does not seem to be straightforward even experimentally, the calculated modulus for DMPC as determined by the present method is in reasonable agreement with experiment.
TABLE 8.
Calculated elastic properties of lipid bilayer membranes.
| KA [mN/m] | KC [×10−20J] | |||
|---|---|---|---|---|
| CG-MD | Expt. | CG-MD | Expt. | |
| DMPC | 266 | 234 ± 23 a | 6.9 | 5.6 ± 0.7a, ~10 |
| DPPC | 233 | 6.4 | ~10 | |
| POPC | 222 | 5.7 | ||
| POPE | 296 | 6.5 | ||
Experimental data from Ref. [51]
5. Applications
5.1 Self-assembled bilayer
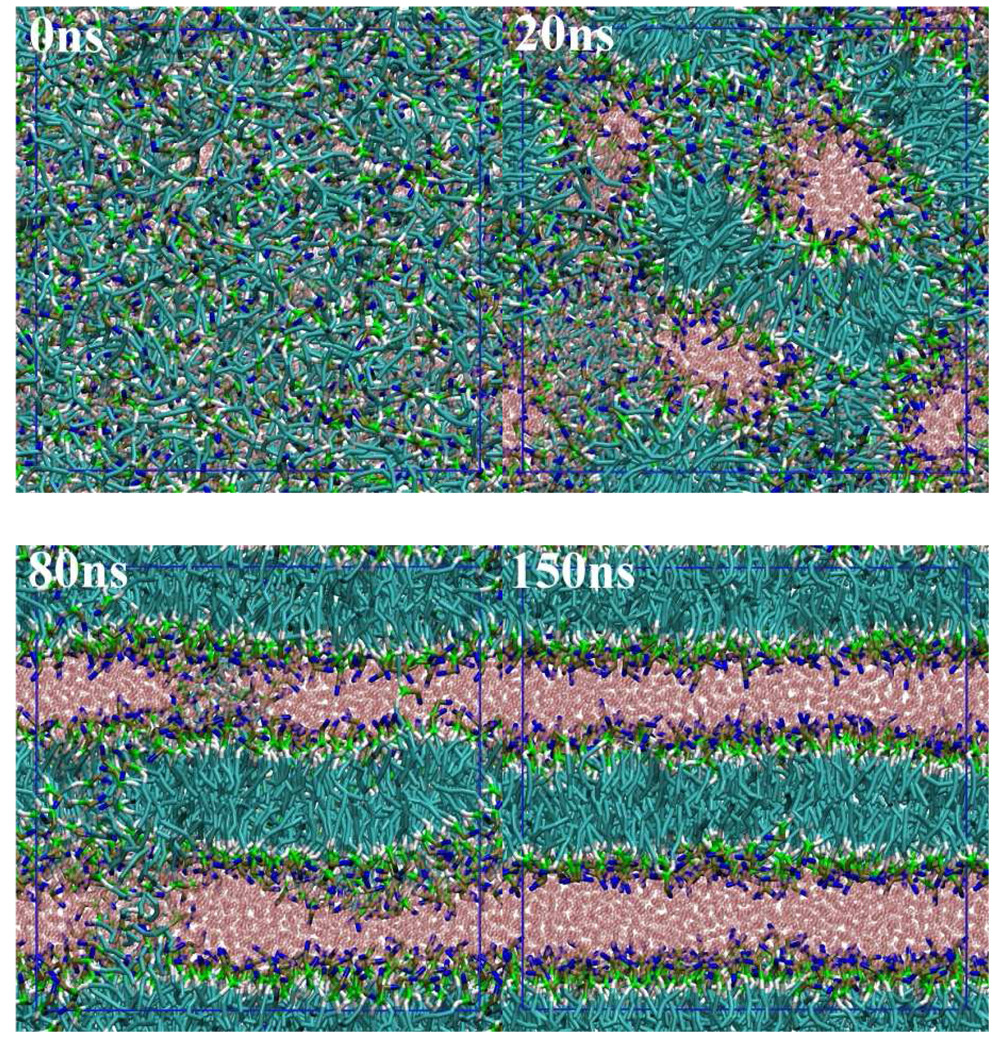
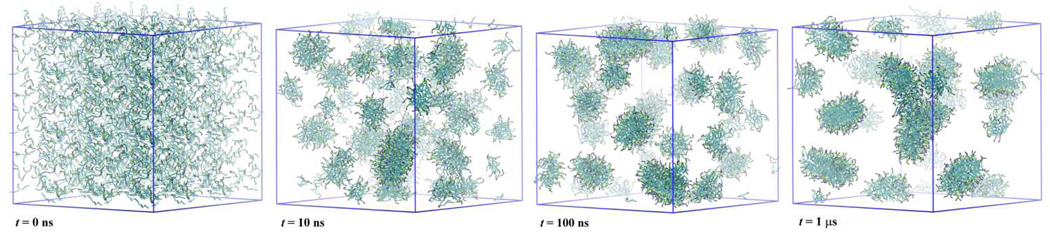
The self-assembly process of lipids is now accessible by MD simulations using the CG molecular model. Figure 5 shows a series of snapshots of a DMPC/water simulation started from a random initial configuration. The system is composed of 932 DMPC molecules and 8264 CG water particles, which corresponds to the saturated DMPC membrane condition.[33] We carried out the NPT-MD simulation for 150 ns. During the early stage of the MD simulation, we observed a rapid organization of random aggregates of lipids that orient their head group toward water. After 20 ns, a continuous aggregate having a highly branched bilayer structure had grown in the simulation box. Reorganization into a clear lamellar structure occurred by 80ns. At this stage, water channels were found in the lipid bilayers. Simultaneously, thin lipid channels bridging two bilayers still exist. The bridging lipid channel persisted for a rather long time (~ 50 ns), compared with the case of a single chained PEG surfactant, C12E2 (< 5 ns).[26] This observation cannot be explained only by the difference of diffusion rate of the lipids, but also by the different molecular structure. Namely, some of the DMPC lipids extended each of their two hydrophobic tails into separate neighboring bilayers, which stabilizes the bridging structure. A bridge is observed in the snapshot at 80 ns shown in Fig. 5. To heal these defects in the lamellar structure, another 50 ns was needed. During this period, lipid molecules go across the bilayers relatively easily, which would minimize the strain due to the unequally distributed lipids between leaflets of the bilayers. The final numbers of lipids in four leaflets of two bilayers are almost identical: 233, 231, 231, and 237. During the MD simulation, the pressure control scheme was changed at 100 ns (where the bilayer formation was nearly complete) from isotropic to anisotropic coupling of the barostat; in the latter scheme, the membrane area and lamella repeat spacing can fluctuate independently so that further relaxation of lamella structure is facilitated. The molecular area and lamellar repeat spacing averaged over the last 20 ns trajectory of the DMPC lipid bilayer are 0.621 nm2 and 5.99 nm, respectively, which are identical to those obtained by the AA-MD simulation of the prepared DMPC bilayer. (Table 7)
Fig. 5.
Formation of lamellar structure from a random initial configuration of DMPC/water. Color codes are the same as in Fig. 1. Blue lines denote the simulation box.
5.2 Langmuir monolayer
Air/water interfacial properties are altered by addition of small amounts of amphiphiles, such as surfactants and lipids, because these molecules strongly adsorb at interfaces. Understanding the nature of this adsorption on a molecular level is an important objective. One of the most appealing consequences of our approach is that the parameterization of the CG model guarantees the correct partitioning (adsorption) of the molecules due to the well-fitted transfer free energy and the correct surface/interfacial tension with/without amphiphiles.[26,27]
Since DPPC is a primary component of lung surfactant, the monolayer at the air/water interface has been extensively investigated both by experiments and computer simulations. Here we also employed DPPC to examine how the CG lipid monolayer behaves upon compression. First of all, we check the quality of our CG model using the surface pressure-area isotherm, which is typically used to characterize a lipid monolayer.
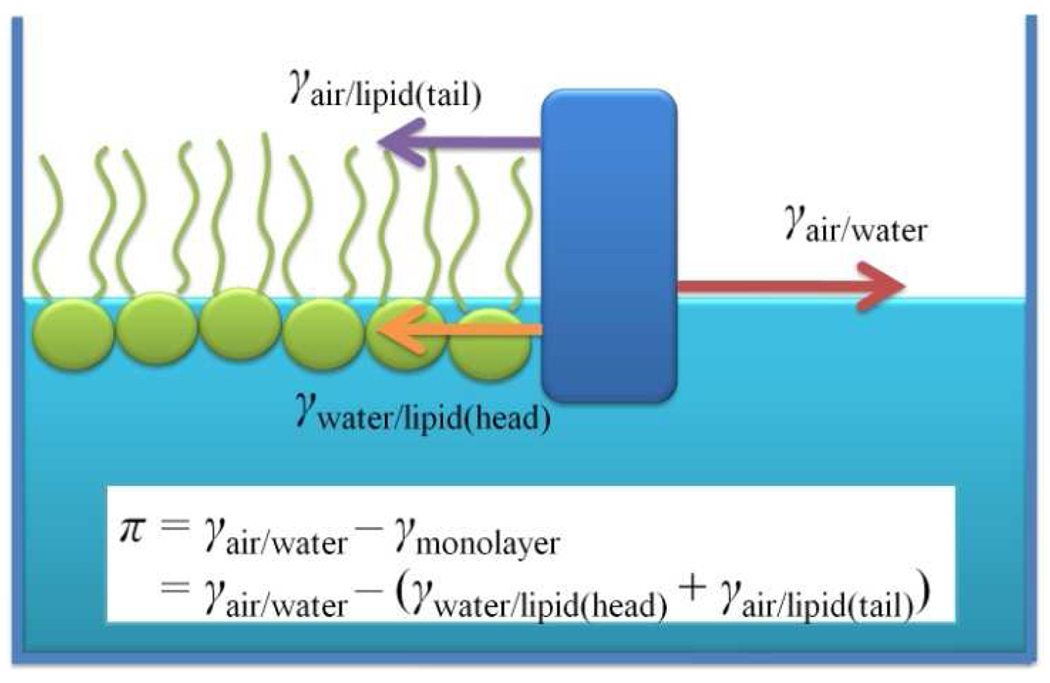
Surface pressure is defined as,
| (8) |
Here γ is the surface tension of the monolayer system and γ0 is the surface tension of pure water; γ0= 71 mN/m is attained in our model at 303 K. Moreover, our CG water exhibits transferability of the surface tension to different temperatures, which is not typically expected for a CG model. Indeed, there is an almost perfect reproduction of the surface tension in the liquid temperature range of 273 to 373K.[26] The surface tension of a monolayer, γ, can be decomposed into two contributions; one is from the water/lipid interface, γwater/lipid, and the other is from the lipid/air interface, γair/lipid. (see Fig.6) When the monolayer is condensed, the latter is similar to the surface tension at the alkane/air interface, which is also quantitatively guaranteed in the present scheme.[26] Thus, our CG model gives a fully consistent picture of experiments with respect to surface tension by yielding the correct surface pressure of the monolayer. It should be noted that almost all empirical models (including all-atom) lack this consistency for the surface tension.[52,53] For example, for the pure water case, a commonly used water model typically underestimates the surface tension by 10 to 20 mN/m.[52,53] Furthermore, the MARTINI CG force field yields a water surface tension value as low as 33 mN/m at room temperature,[20] which is less than half the experimental value of 72 mN/m. As seen from the definition, the monolayer surface pressure is a measure how much the surface tension is reduced due to the adsorbed lipids at the air/water interface. Therefore, having the correct reference for the surface tension at the air/water interface is important for having consistent discussions regarding the “surface pressure” of a lipid monolayer. This should be especially true for case of low-coverage lipid monolayers. If this condition is not met, a surface pressure that agrees with experimental data is a consequence of a compensation of errors.[56] Moreover, it is not clear how these compensating errors affect the physical properties of a high-coverage lipid monolayer or bilayer membranes.
Fig. 6.
Schematic explanation of the surface pressure of a lipid monolayer system, π.
We have carried out a series of NVT-MD simulations of a DPPC monolayer with different areas per molecule. We prepared the system typically with a water slab with a thickness of about 20 nm in a 10×10×35 nm simulation box and place the DPPC monolayers at the two air/water interfaces. The surface tension is calculated by
| (9) |
where Lz is the length of the simulation box along the interface normal, z.
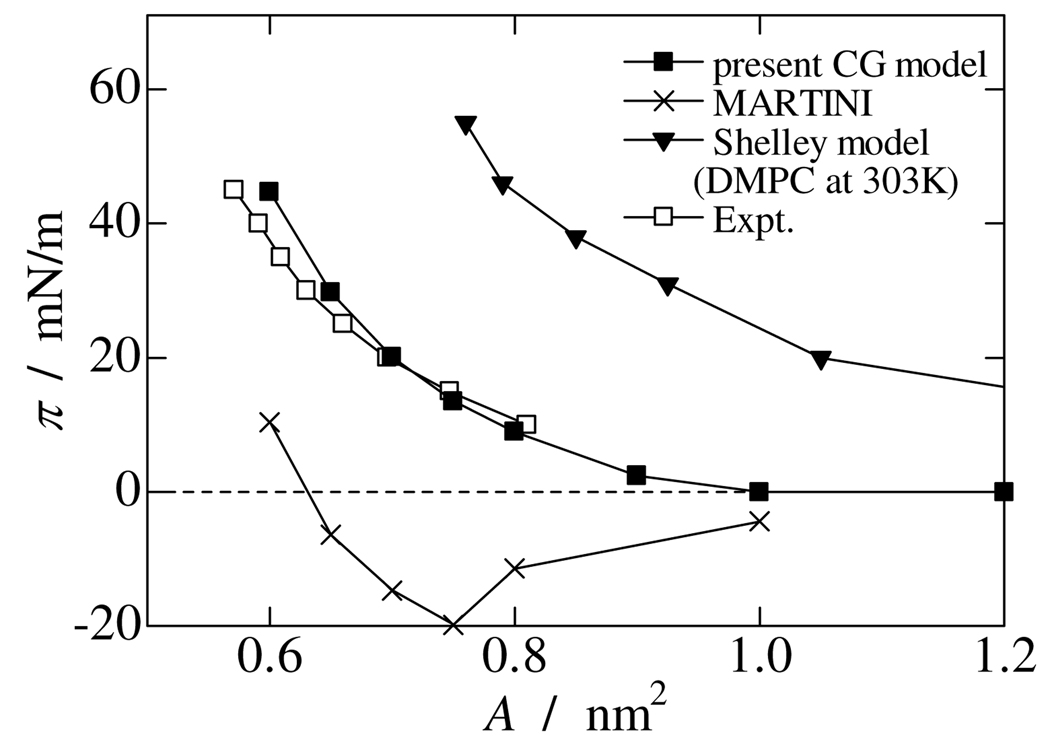
Figure 7 plots the simulated surface pressure as a function of molecular area of DPPC at the air/water interface (π – A curve) with some relevant data from the literature at 323K.[54] At the areas of 1.0 and 1.2 nm2, respectively the DPPC monolayer was not uniform, containing a hole or a low DPPC density region on the surface. This suggests coexisting liquid extended and gas phases, which are experimentally well known over a wide range of areas and were also observed in recent atomistic and CG MD studies.[55–57] As shown in Figure 7, surface pressure did not change in that area range. On decreasing the molecular area, the DPPC monolayer starts forming a defect-less uniform layer over the surface and gives a non-zero surface pressure. The present CG model demonstrates reasonable agreement over the full π – A curve with experimental data at 323 K,[54] though a slight deviation is found at small areas. Comparing the present result with our previous work (noted as the “Shelley model” in the figure),[59] which was built using the inverse Boltzmann method,[15] the present CG model significantly improves the monolayer behavior for the surface pressure over a wide range of areas. Furthermore, we also evaluate the MARTINI force field [19,20] for a DPPC monolayer at 323 K for a comparison.[59] As mentioned above, the MARTINI water significantly underestimates the surface tension (29 mN/m at 323 K). Using the latter value, we evaluated the surface pressure of the DPPC monolayer for a series of areas per lipid. For the area range 0.65 to 1.00 nm2, the MARTINI force field shows a negative surface pressure, which means the surface tension of air/water interface increases with increasing numbers of DPPC molecules on the surface. On addition of DPPC molecules, the surface tension reaches a peak at about 0.75 nm2 and then starts decreasing. This is an unphysical behavior for a lung surfactant even in a qualitative sense. A similar observation was noted previously.[56] However, apart from this issue, the MARTINI force field works reasonably for the high-coverage lipid monolayer. [56,57,60]
Fig. 7.
Surface pressure vs. area (π – A) curve of simulated DPPC monolayers. Experimental data are taken from Ref. [54]. Data for the Shelley model is taken from Ref. [58].
The correct behavior of the surface pressure – area relation encouraged us to use the new CG force field to investigate the nature of monolayer collapse. As shown in Fig. 7, the surface pressure goes up with decreasing molecular area. When the surface pressure exceeds γ0, i.e., the surface tension of monolayer becomes negative, the interface is thermodynamically unstable. The collapse can happen either to give a subphase in the solution or to yield a partial lipid overlayer on top of the monolayer.[61] The present CG model, which has a fully consistent tension at any interface, is straightforwardly used to tackle this question.
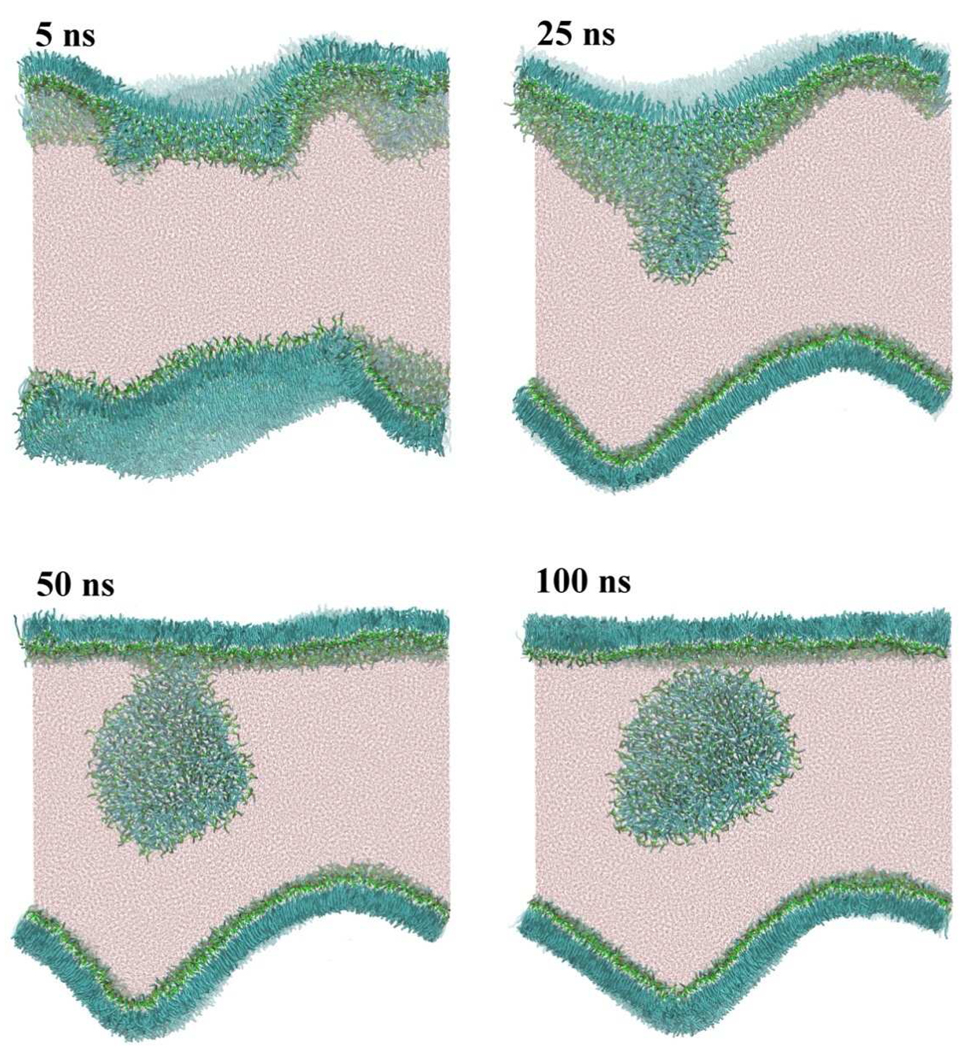
We generated an oversaturated DPPC monolayer on a water slab with a thickness of 20 nm in a large simulation box with dimensions 30×30×35 nm. The initial DPPC molecular area is set to 0.40 nm2, which is small enough to cause a negative surface tension. Figure 8 demonstrates a series of snapshots during the subsequent CG-MD trajectory. After only 5ns, a significant buckling of the monolayer was found and a few small buds were detected. One of these small buds started to grow from one monolayer and adsorbed the other small buds (25ns). Once a bud has grown to a critical size, the budding process seems to be accelerated by accumulation of monolayer stress due to the bud. This clearly shows that the correlation length of the stress propagation is quite large for a DPPC monolayer. A similar result was observed in a CG-MD simulation of compressed DPPC-POPG monolayer.[62] After 50ns, a disk-like structure evolved from the surface, but still connected to the monolayer. Another 50ns was needed to see the disk completely detach from the monolayer surface. Interestingly, the other monolayer did not give any buds during the trajectory. At 25ns, the buckling of the monolayer was minimized by adopting a sinusoidal curved surface compatible with the periodic boundary condition. Even though the curved monolayer was energetically unfavorable, this metastable structure persisted for 100 ns. Lipids in this curved monolayer were found to be in a solid state, with their lateral diffusion completely suppressed. The same is true for the early state of the other monolayer, though after birth of the bud, the lipids show lateral diffusion in the resulting flat monolayer.
Fig. 8.
Budding and fission from an oversaturated DPPC monolayer at the air/water interface. The initial area per DPPC molecule on the flat monolayer at the air/water interface was set to 0.40 nm2. Color codes are the same as in Fig. 1.
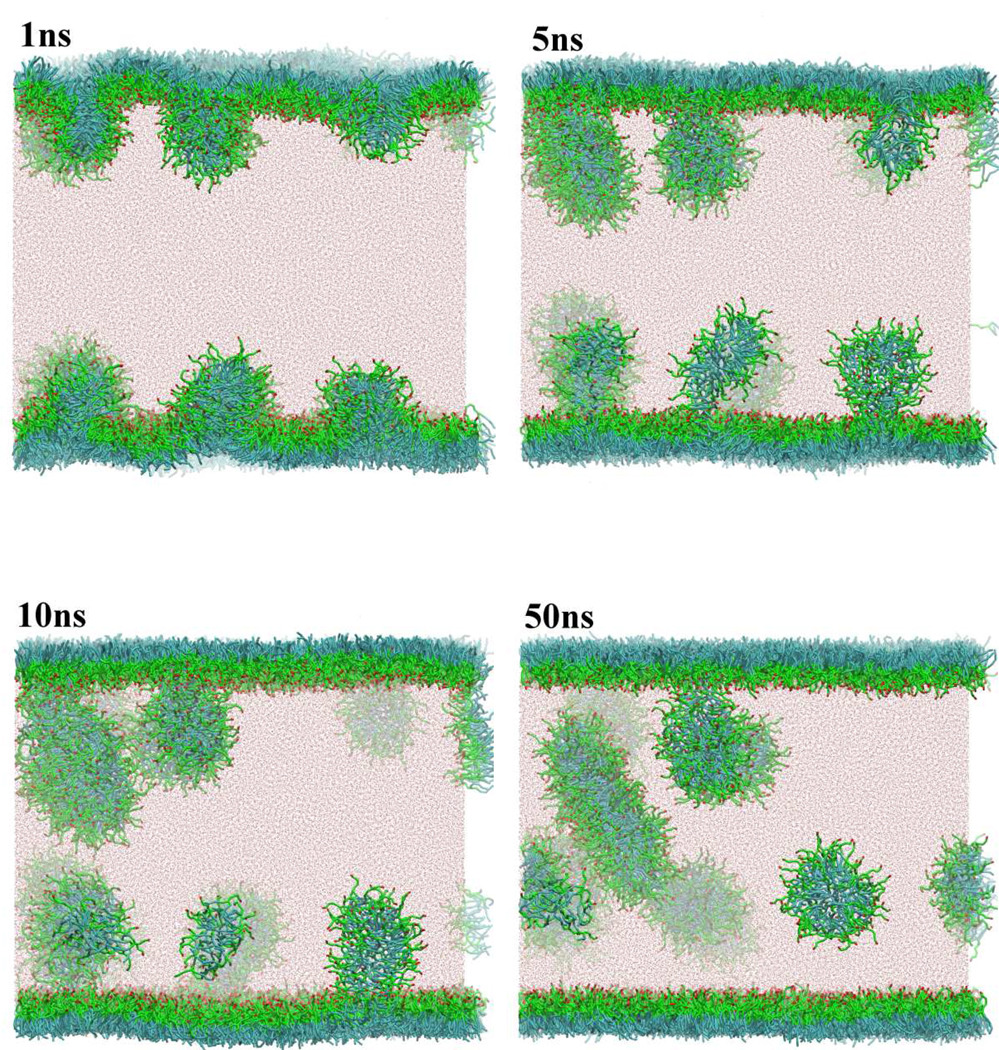
We carried out a similar examination for a PEG lipid monolayer with the simulation box of 30×30×40 nm. We used two-tailed hydrocarbon PEG surfactant, which was also used in our previous work.[27] (HO(CH2CH2O)5CH(CH2O(CH2)11CH3)2: corresponds to OA-(EO)5-(EO-(CM)3-CT2)2 in the CG notation) We confirmed that the CG model quantitatively reproduced the experimental π – A curve. Figure 9 shows a series of snapshots obtained from the CG-MD run. After only 1ns, many buds protruded from a slightly buckled monolayer. However, correlation among the buds was relatively weak so that several buds grew almost independently in 10 ns. The buds typically had an elongated micelle structure, though a disk-like shape was also found in larger aggregates. The major difference between DPPC and PEG lipid monolayers is in the persistence length, i.e., correlation among the lateral lipids. The zwitter-ionic head groups of DPPC give rise to long range interactions in a monolayer surface, while PEG head groups (ethyleneoxide; EO5) do not, which in turn causes a difference in the structure of the bud. DPPC clearly shows a disk-like bicelle, whereas PEG has typically elongated micelles. Commonly for both systems, however, the monolayer collapse happened toward the water solution. Similar observations were made by others using MD studies.[56, 62] Note that here we examined the monolayer collapse from a prepared oversaturated lipid monolayer. However, on the dynamical compression of equilibrium monolayers, we may see a different collapse mechanism depending on the compression rate.[58]
Fig 9.
Budding and fission from an oversaturated PEG lipid (HO(CH2CH2O)5CH(CH2O(CH2)11CH3)2) monolayer at the air/water interface. Color codes are the same as in Fig.1 except for PEG head group, which are shown in green (EO; -CH2-O-CH2-) and red (OA; -CH2-OH).
5.3 Vesicles
5.3.1 Vesicle formation
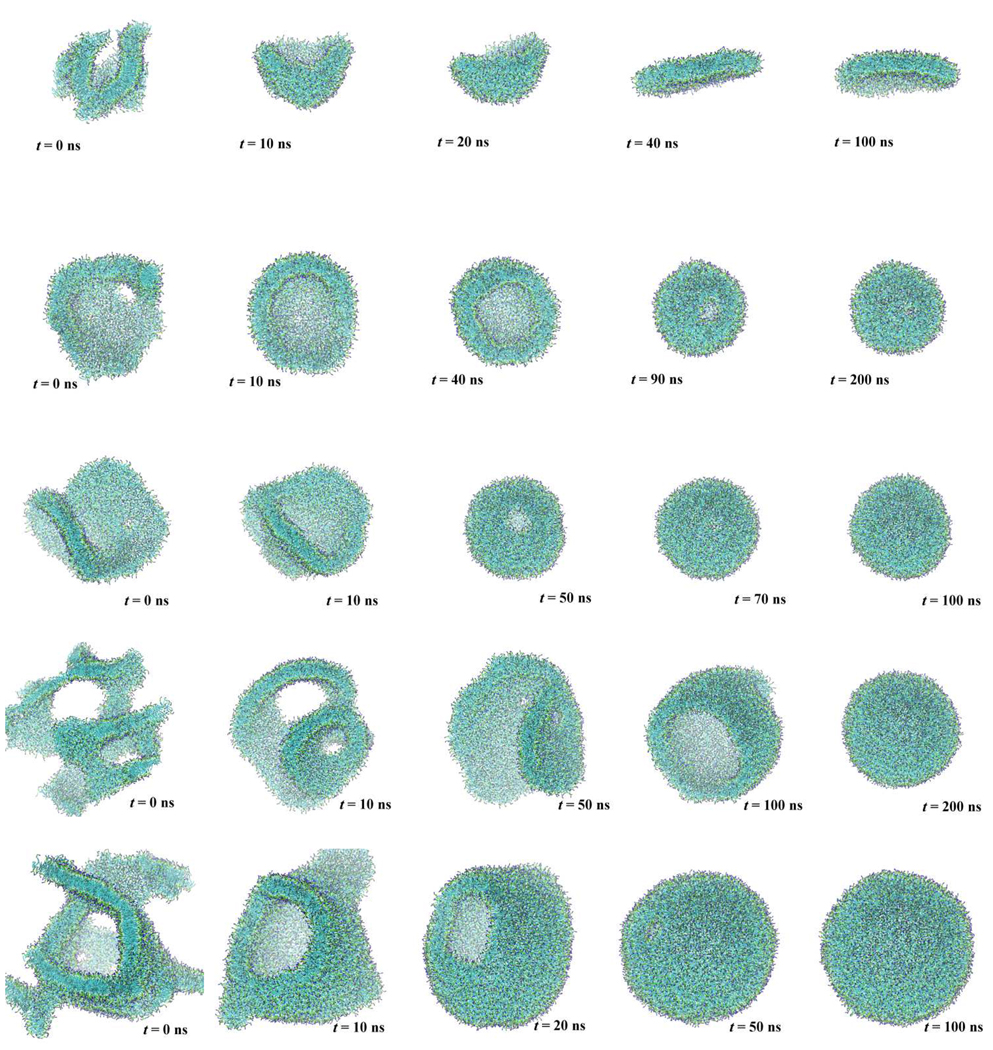
One of the most interesting and popular applications of CG molecular models is to the curved membrane like vesicle. We first tried to observe the spontaneous formation of a vesicle of 1512 DMPC lipid molecules with 100 water particles per lipid, which represents 300 water molecules per lipid. We prepared a random distribution of DMPC and water in a cubic simulation box and then performed an NPT-MD simulation over 1 µs in length. Temperature and pressure were held to 310 K and 0.1 MPa, respectively. Even after 1µs simulation time we did not obtain one large aggregate as found in a micellar growth of PEG surfactants.[27] (Fig. 10) Note this is not 1µs physical time, but longer by a factor of 4 ~ 7, in the light of the difference in lateral diffusion rate of lipids; for example, the lateral diffusion coefficient of CG DPPC molecule is calculated as 14.1×10−7 cm2/s, though the coefficient from CHARMM was 3.5 ×10−7 cm2/s [63] and the experimental coefficient of egg-yolk lipid is reported as 2.1×10−7 cm2/s.[64]. The initial stage of the vesicle formation process, which is to make small aggregates is rather rapid. On the other hand, the later stage involving cluster-cluster aggregation to increase the aggregate size, is significantly slowed down because a weak repulsive interaction between the smaller aggregates that arises due to the exposed lipid head group. A previous CG-MD simulation of vesicle formation using the MARTINI force field shows, however, a much faster organization of a vesicle from a random initial condition.[65] This may be the result of the long-range Coulomb interactions; a very short cutoff of 1.2 nm is used in the MARTINI force field, though no truncation of the Coulomb interaction is employed in the present CG model. In order to obtain a larger aggregate efficiently, we reduced the quantity of water to 33 water particles per lipid, and ran an MD simulation for several tens of nanoseconds, which was long enough to obtain a single lipid aggregate (domain) expanding through the simulation box. Then, we placed the aggregate in a larger water box having 100 CG water particles per lipid. Figure 11 (second row) demonstrates a series of snapshots from the MD simulation of a 1512 DMPC lipid system. In this figure, water is not shown for clarity. At the beginning of the MD run, the aggregate had defects in the bilayer structure, where the alkyl chains were exposed to water. These chains were rapidly covered by lipid head group shortly after continuing the MD run. A 10 ns-long MD simulation was enough to obtain a defect-less membrane, which had a bowl shaped structure. At the edge of the bowl, even though no alkyl chain is directly exposed to water, unfavorable bending of the DMPC molecule occurred and required an energetic penalty, which is thermodynamically understood in terms of a line tension. The DMPC aggregate was large enough to wrap up to eliminate the edge effect at the cost of bending free energy of the bilayer. Of course, the energy cost for this bilayer bending is significantly reduced by changing the number of DMPC molecules between inner and outer leaflets of the vesicular bilayer membrane. Thus, the bicelle-like aggregate wrapped up into a unilamellar vesicle in about 160 ns, eliminating the unfavorable bilayer edge. This transformation takes place much faster than does the cluster-cluster aggregation as seen above. The rate-limiting event in the transformation is, especially in the late stages, lipid flipping at the edge of the bilayer. Therefore, we observed a long standing small pore in the later stage of the transformation, where the edge portion is relatively small. Once the small pore closed, no exchange of lipid molecules between inner and outer leaflets was observed for the following 1 µs. This is consistent with the experimental observation that a flip-flop event takes place in hours.
Fig. 10.
Liposome formation from a DMPC bilayer fragment. Water is not shown for clarity. Color codes are the same as in Fig. 1.
Fig. 11.
Transformation of self-assembled lipid aggregates in a large water box. The five rows represent lipid aggregates with 1000, 1512, 2500, 3500, and 5000 DMPC molecules, respectively. Color codes are the same as in Fig. 1.
5.3.2 Size effect
We have carried out a series of similar MD runs while varying the number (500, 1000, 2500, 3500, and 5000) of DMPC molecules. After making a single aggregate from a random initial configuration with a smaller hydration number, we placed each of the aggregates of 500, 1000, 2500, 3500, 5000 DMPC molecules, in a larger water box to have more than 100 water particles per lipid for each system. Each aggregate with more than 2500 DMPC molecules was observed to transform into a unilamellar vesicle within 200 ns, irrespective of the size. (Fig. 11) The required time, of course, depends on the initial configuration, i.e., structure of the prepared aggregate organized from randomly distributed DMPC molecules. Although lacking enough statistics to measure a precise transformation time, it is suggested that transformation repairing an unfavorable hole or exposure of bilayer edge will happen in a relatively short time irrespective of the aggregate size, compared to the aggregation process.
On the other hand, smaller aggregates with 500 or 1000 DMPC molecules were found to transform into a disk, i.e., a bicelle structure, as shown in Fig. 11. This is simply because the bilayer leaflet was too small to form a closed vesicle, where a highly curved surface inevitably enforces a bilayer structure. More specifically, a transformation to a vesicle costs too much bending energy. Once a bicelle structure was reached, the aggregate showed rather large shape fluctuations while retaining the planar structure during another 1µs–MD run.
It should be noted that the DMPC aggregates selected a bicelle or vesicle structure while the intermediate bowl-shaped structure was not stable. This implies that bicelle and vesicle structures are in a free energy minimum separated by a free energy barrier. A similar behavior was suggested by continuum theory. [66] To confirm this, we started a MD run with 1512 DMPC molecules with the completely flat disk structure and found that the prepared bicelle structure remained for 700ns. The transformation from a completely flat disk to a vesicle should take a very long time, because it involves breaking the symmetry of the flat system.Quantification of this process would require a free energy computation, which is beyond the scope of the present paper.
5.3.3 Vesicle-vesicle interaction
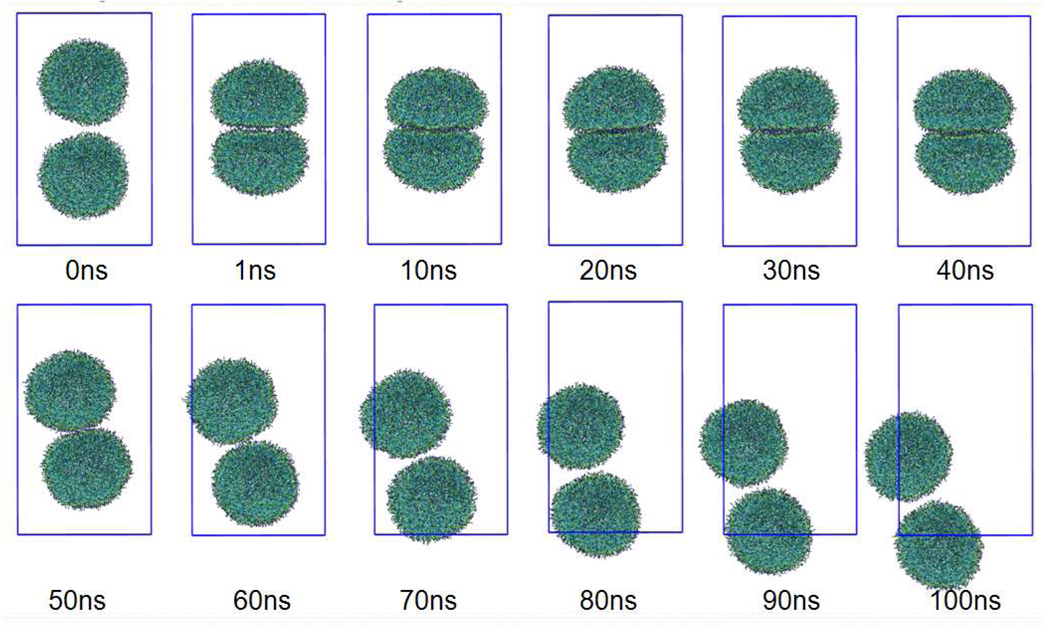
To assess the stability of the present vesicles, we carried out a MD simulation containing two vesicles. Each of the vesicles was composed of 1512 DMPC lipid molecules, which is the smallest aggregate to show vesicle formation in our CG-MD runs. The initial configuration of the vesicles was taken from the last frame of a MD run of the vesicle formation in the previous section. Without any external force, no fusion or adhesion like events took place for 150ns, with each of the two vesicles showing only random diffusion. Therefore, we applied an external force to encourage contact between the two vesicles. The external force was applied using the option “fix drag” in the LAMMPS software,[45] which applies a force to each atom in a group to drag the group toward a desired point, in this case the origin. Here we took each vesicle as a separate group and applied a constant force to each CG particle of the lipids to drag each vesicle toward the origin. This constant force is set to 0.005 kcal/mol/Å, which is much smaller than the interaction force among the neighboring CG particles but large enough to cause adhesion of the two vesicles, as shown in Fig. 12. Even though we kept this external force for 40 ns, we observed only adhesion and deformation of two small vesicles due to the external force, but could not find any sign of fusion. The small vesicles were elastic, with each vesicle flattening along the adhesion interface, but keeping an individual closed vesicle during this period. This observation is different from that found in several studies where a fusion event was easily observed without any specific external forces.[67,68] After 40ns, we removed this external force from the system. Each of the vesicles quickly reshaped to a sphere, but after this, did not repel each other. Another 60ns-MD shows that the two vesicles stay close to each other and migrate in the simulation box together. This clearly shows that two vesicles have an attractive interaction.
Fig. 12.
Two small vesicles brought in contact with each other by an external force. The external force is applied during the first 40ns and then removed. Details are given in the text. Each vesicle consists of 1512 DMPC molecules. Water is not shown for clarity. Color codes are the same as in Fig. 1.
In comparison with the previous studies on fusion processes using phenomenological CG models, where spontaneous fusion occurs without any additional forces,[67,68] the present CG lipid model forms a stable vesicle. One may question whether this finding is realistic or not. Due to the nanometer size of the simulation vesicles, it is difficult to perform a lab experiment to do the same operation as carried out here. However, we can find evidence to support our observation. For example, although the detailed mechanism is still an open question, vesicle fusion usually involves much more complex players like ions, proteins, and their complexes, e.g., SNARE complexes for neurotransmitter release,[69–71]. The fusion process is also sensitive to the lipid species; in particular, lipids that form a bilayer with non-zero spontaneous curvature would play an important role in the early stage of the fusion process. Therefore, a single component, zero-spontaneous curvature membrane like a DMPC bilayer can form a very stable vesicle like we found in our CG model that would be expected to be fusion resistant. The challenge of investigating realistic fusion processes by MD simulations will be aided by developing a systematically generated CG amino acid model.[72] The stability of DMPC vesicles is reasonably supported by the fact that a multilamellar vesicle is thermodynamically more stable than a unilamellar vesicle. We have also carried out CG-MD of a mulilamellar vesicle and confirmed it to be stable over a 1µs trajectory.
5.3.4 Mixed lipids
We have carried out an MD run for a mixture of POPC and POPE lipid molecules in a similar manner to the previous DMPC case. For these simulations, we use 1024 molecules for each lipid component. In a large water box, the prepared aggregate transformed to a unilamellar vesicle in about 40 ns. Counting the number of lipids in the inner and outer leaflets of vesicle membrane, 376 POPE and 353 POPC molecules were found in the inner leaflet while 648 POPE and 671 POPC molecules were found in the outer leaflet. Thus, POPE is more likely found in the internal leaflet of the bilayer membrane of the vesicle. Partitioning of lipids into each leaflet occurs spontaneously, which indicates that a smaller PE head group is preferential for the inner membrane leaflet of bilayer vesicle. This is consistent with the observation that a POPE membrane has a negative spontaneous curvature. A recent CG-MD study of a mixed-lipid vesicle reported a similar observation finding PE enrichment in the inner leaflet of vesicle.[73]
6. Conclusions
We have developed a new coarse-grained force field for lipids based on fitting thermodynamic and structural properties. The CG model guarantees reasonable surface/interfacial properties, bulk fluid densities as well as molecular distribution functions obtained by all-atom MD simulations. These details were preserved while keeping the interaction potential functions as simple as possible. The bilayer membrane simulated by the present model shows reasonable molecular areas and elastic moduli, which should be of key importance in characterizing the membrane structure at meso-scale resolution. We emphasize that our CG model is designed to be useful for interfacial systems, which allows us to use the CG model to discuss the surface pressure of a lipid monolayer at the air/water interface. The monolayer collapse presented in sec. 5 is a good example demonstrating the power of this CG modeling approach. Vesicle formation was observed as a transformation from a random aggregate with more than 1512 DMPC molecules. Aggregates with less than 1000 DMPC molecules preferred a disk (bicelle) structure instead of a closed vesicle. An intermediate structure between a vesicle and disk, such as a bowl, was not stable, suggesting a free energy barrier between the vesicle and disk structures. In mesoscopic continuum theory, this is understood as a competition between the energy costs of bending and bilayer-edge formation. However, vesicle formation from a bilayer raft is not simply a bending process, because lipids from the inner leaflet of the curved bilayer migrate across the membrane during the vesicle formation to reduce the bending energy. More detailed computations of the free energy profile along the formation pathway will be needed to address this question from a molecular perspective. High resistance of small DMPC vesicles to fusion was found even with the application of an external force; an observation seemingly at odds with earlier studies using CG models where spontaneous fusion occurs with or without a perturbation.[67,68] The present results suggests the necessity of fusion enhancers, such as the SNARE complex, to facilitate the lipid vesicle fusion event for small vesicles. Further development of an appropriate CG protein [74] force field will no doubt contribute to the investigation of the molecular mechanism of the lipid vesicle fusion process [75]
TABLE 4.
Angle bending parameters
| Angle pair | kθ [kcal/mol/rad2] | θ0 [degree] | ||
|---|---|---|---|---|
| NC | PH | CM | 3.300 | 112.0 |
| PH | CM | CM | 1.100 | 178.0 |
| NC | PH | GL | 3.100 | 112.0 |
| PH | GL | EST1 | 1.400 | 124.0 |
| PH | GL | EST2 | 2.000 | 138.0 |
| GL | EST1 | CM | 0.800 | 168.0 |
| GL | EST2 | CM | 0.800 | 172.0 |
| EST1 | GL | EST2 | 1.000 | 95.0 |
| EST1 | CM | CM | 1.000 | 178.0 |
| EST2 | CM | CM | 1.000 | 178.0 |
| NH | PHE | GL | 4.000 | 102.0 |
| PHE | GL | EST1 | 1.400 | 124.0 |
| PHE | GL | EST2 | 2.000 | 138.0 |
| CM | CM | CM | 1.190 | 173.0 |
| CM | CM | CT | 1.190 | 175.0 |
| CT2 | CM | CM | 1.600 | 172.0 |
| CT2 | CM | CT | 1.600 | 172.0 |
| CT2 | CM | CT2 | 1.700 | 173.0 |
| CT | CM | CT | 1.093 | 175.5 |
| CT | CMD2 | CT | 7.700 | 116.0 |
| CT2 | CMD2 | CT2 | 12.000 | 110.0 |
| CMD2 | CM | CM | 1.900 | 161.0 |
| CM | CMD2 | CM | 6.000 | 110.0 |
Acknowledgements
We thank Kelly Andersen, Preston Moore, Takenobu Nakamura, and Steve Nielsen, for many helpful comments, Jérôme Hénin for discussion and access to his AA-MD simulations, Grace Brannigan for insights on calculating elastic properties of bilayers, and Axel Kohlmeyer for implementing our new force field into LAMMPS. This research was supported in part by the NIH under GM40712 and by CREST-JST, and the Next Generation Super Computing Project, Nanoscience Program, MEXT, Japan.
REFERENCES
- 1.Marrink SJ, de Vries AH, Tieleman DP. Biochim. Biophys. Acta. 2009;1778:2308. doi: 10.1016/j.bbamem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Klein ML, Shinoda W. Science. 2008;321:798. doi: 10.1126/science.1157834. [DOI] [PubMed] [Google Scholar]
- 3.Smit B, Hilbers PAJ, Esselink K, Rupert LAM, van Os NM, Schlijper AG. Nature. 1990;348:624. [Google Scholar]
- 4.Smit B, Schlijper AG, Rupert LAM, van Os NM. J. Phys. Chem. 1990;94:6933. [Google Scholar]
- 5.Noguchi H, Takasu M. Phys. Rev. E. 2001;64:041913. doi: 10.1103/PhysRevE.64.041913. [DOI] [PubMed] [Google Scholar]
- 6.Goetz R, Lipowsky R. J. Chem. Phys. 1998;108:7387. [Google Scholar]
- 7.Groot RD, Rabone KL. Biophys. J. 2001;81:725. doi: 10.1016/S0006-3495(01)75737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shillcock JC, Lipowsky R. J. Chem. Phys. 2002;117:5048. [Google Scholar]
- 9.Kranenburg M, Smit B. J. Phys. Chem. B. 2005;109:6553. doi: 10.1021/jp0457646. [DOI] [PubMed] [Google Scholar]
- 10.Brannigan G, Lin LCL, Brown FLH. Eur. Biophys. J. Biophys. Lett. 2006;35:104. doi: 10.1007/s00249-005-0013-y. [DOI] [PubMed] [Google Scholar]
- 11.Cooke IR, Deserno M. J. Chem. Phys. 2005;123:224710. doi: 10.1063/1.2135785. [DOI] [PubMed] [Google Scholar]
- 12.Lyubartsev AP. Eur. Biophys. J. Biophys. Lett. 2005;35:53. doi: 10.1007/s00249-005-0005-y. [DOI] [PubMed] [Google Scholar]
- 13.Stevens MJ. J. Chem. Phys. 2004;121:11942. doi: 10.1063/1.1814058. [DOI] [PubMed] [Google Scholar]
- 14.Farago O. J. Chem. Phys. 2003;119:596. [Google Scholar]
- 15.Shelley JC, Shelly MY, Reeder RC, Bandyopadhyay S, Klein ML. J. Phys. Chem. B. 2001;105:4464. [Google Scholar]
- 16.Shelley JC, Shelly MY, Reeder RC, Bandyopadhyay S, Moore PB, Klein ML. J. Phys. Chem. B. 2001;105:9785. [Google Scholar]
- 17.Izvekov S, Voth GA. J. Phys. Chem. B. 2005;109:2469. doi: 10.1021/jp044629q. [DOI] [PubMed] [Google Scholar]
- 18.Izvekov S, Voth GA. J. Chem. Phys. 2005;123:134105. doi: 10.1063/1.2038787. [DOI] [PubMed] [Google Scholar]
- 19.Marrink SJ, de Vries AH, Mark AE. J. Phys. Chem. B. 2004;108:750. [Google Scholar]
- 20.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. J. Phys. Chem. B. 2007;111:7812. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 21.Orsi M, Haubertin DY, Sanderson WE, Essex JW. J. Phys. Chem. B. 2008;112:802. doi: 10.1021/jp076139e. [DOI] [PubMed] [Google Scholar]
- 22.Lyubartsev AP, Laaksonen A. Phys. Rev. E. 1995;52:3730. doi: 10.1103/physreve.52.3730. [DOI] [PubMed] [Google Scholar]
- 23.Tschop W, Kremer K, Batoulis J, Burger T, Hahn O. Acta Polym. 1998;49:61. [Google Scholar]
- 24.Sun Q, Faller R. J. Chem. Theor. Comput. 2006;2:607. doi: 10.1021/ct600065v. [DOI] [PubMed] [Google Scholar]
- 25.Voth GA, editor. Coarse-Graining of Condensed Phase and Biomolecular Systems. Francis: CRC / Taylor; 2008. [Google Scholar]
- 26.Shinoda W, Devane R, Klein ML. Mol. Simul. 2007;33:27. [Google Scholar]
- 27.Shinoda W, DeVane R, Klein ML. Soft Matter. 2008;4:2454. [Google Scholar]
- 28.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 29.Winger M, Trzesniak D, Baron R, van Gunsteren WF. Phys. Chem. Chem. Phys. 2009;11:1934. doi: 10.1039/b818713d. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen SO, Lopez CF, Srinivas G, Klein ML. J. Phys.-Condes. Matter. 2004;16:R481. [Google Scholar]
- 31.Klauda JB, Brooks BR, MacKerell AD, Venable RM, Pastor RW. J. Phys. Chem. B. 2005;109:5300. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen WL, Chandrashekar J, Madura JD, Impey R, Klein ML. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 33.Nagle JF, Tristram-Nagle S. Biochim. Biophys. Acta. 2000;1469:159. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hénin J, Shinoda W, Klein ML. J. Phys. Chem. B. 2008;112:7008. doi: 10.1021/jp800687p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feller SE, Zhang YH, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103:4613. [Google Scholar]
- 36.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel LRDKale´, Schulten K. J. Comput. Chem. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frenkel D, Smit B. Understanding of Molecular Simulation. 2nd. Ed. San Diego: Academic Press; 2002. [Google Scholar]
- 38.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327. [Google Scholar]
- 39.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford, U.K: Clarendon Press; 1987. [Google Scholar]
- 40.Shinoda W, Mikami M. J. Comp. Chem. 2003;24:920. doi: 10.1002/jcc.10249. http://staff.aist.go.jp/w.shinoda/MPDyn. [DOI] [PubMed]
- 41.Martyna GJ, Tobias DJ, Klein ML. J. Chem. Phys. 1994;101:4177. [Google Scholar]
- 42.Martyna GJ, Tuckerman ME, Klein ML. J. Chem. Phys. 1992;97:2638. [Google Scholar]
- 43.Martyna GJ, Tuckerman ME, Tobias DJ, Klein ML. Mol. Phys. 1996;87:1117. [Google Scholar]
- 44.Tuckerman M, Berne BJ, Martyna GJ. J. Chem. Phys. 1992;97:1990. [Google Scholar]
- 45.Plimpton SJ. J. Comp. Phys. 1995;117:1. [Google Scholar]
- 46.Hoover WG. Phys. Rev. A. 1986;34:2499. doi: 10.1103/physreva.34.2499. [DOI] [PubMed] [Google Scholar]
- 47.Schatzberg P. J. Polym Sci. C. 1965;10:87. [Google Scholar]
- 48.Shinoda W, Okazaki S. J. Chem. Phys. 1998;109:1517. [Google Scholar]
- 49.Brannigan G, Brown FL. Biophys. J. 2006;90:1501. doi: 10.1529/biophysj.105.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans E, Rawicz W. Phys. Rev. Lett. 1990;64:2094. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- 51.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Biophys. J. 2000;79:328. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen F, Smith PE. J. Chem. Phys. 2007;126:221101. doi: 10.1063/1.2745718. [DOI] [PubMed] [Google Scholar]
- 53.Vega C, de Miguel E. J. Chem. Phys. 2007;126:154707. doi: 10.1063/1.2715577. [DOI] [PubMed] [Google Scholar]
- 54.Crane JM, Putz G, Hall SB. Biophys. J. 1999;77:3134. doi: 10.1016/S0006-3495(99)77143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knecht V, Muller M, Bonn M, Marrink SJ, Mark AE. J. Chem. Phys. 2005;122:024704. doi: 10.1063/1.1825992. [DOI] [PubMed] [Google Scholar]
- 56.Baoukina S, Monticelli L, Marrink SJ, Tieleman DP. Langmuir. 2007;23:12617. doi: 10.1021/la702286h. [DOI] [PubMed] [Google Scholar]
- 57.Duncan SL, Larson RG. Biophys. J. 2008;94:2465. doi: 10.1529/biophysj.107.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nielsen SO, Lopez CF, Moore PB, Shelley JC, Klein ML. J. Phys. Chem. B. 2003;107:13911. [Google Scholar]
- 59.For a comparison, simulations using MARTINI potential were performed for air/water interface using the Gromacs simulation software. Surface tension calculations were performed using the Gromacs g_energy tool. All simulations were carried out at 323 K in an NVT ensemble with temperature controlled by coupling to Berendsen thermostats. For pure water surface tension calculations, the box dimensions were 6.5 nm in x and y with a total of 5128 CG water sites. For the DPPC monolayer simulations, each system consisted of 64 monomers in each monolayer leaflet with roughly 5000 CG waters. Each system was equilibrated for 40 ns followed by 100 ns production runs with a 20 fs timestep
- 60.Adahangale PS, Gaver III DP. Mol. Phys. 2006;104:3011. doi: 10.1080/00268970600935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ybert C, Lu WX, Moller G, Knobler CM. J. Phys. Chem. B. 2002;106:2004. [Google Scholar]
- 62.Baoukina S, Monticelli L, Risselada HJ, Marrink SJ, Tieleman DP. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10803. doi: 10.1073/pnas.0711563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shinoda W, Mikami M, Baba T, Hato M. Chem. Phys. Lett. 2004;390:35. doi: 10.1063/1.1806791. [DOI] [PubMed] [Google Scholar]
- 64.Baba T, Minamikawa H, Hato M, Handa T. Biophys. J. 2001;81:3377. doi: 10.1016/S0006-3495(01)75970-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marrink SJ, Mark AE. J. Am. Chem. Soc. 2003;125:15233. doi: 10.1021/ja0352092. [DOI] [PubMed] [Google Scholar]
- 66.Fromherz P. Chem. Phys. Lett. 1983;94:259. [Google Scholar]
- 67.Noguchi H, Takasu M. J. Chem. Phys. 2001;115:9547. [Google Scholar]
- 68.Smeijers AF, Markvoort AJ, Pieterse K, Hilbers PAJ. J. Phys. Chem. B. 2006;110:13212. doi: 10.1021/jp060824o. [DOI] [PubMed] [Google Scholar]
- 69.Skehel JJ, Wiley DC. Cell. 1998;95:871. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 70.Bonifacino JS, Glick BS. Cell. 2004;116:153. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 71.Rizo J, Rosenmund C. Nature Struct. Mol. Biol. 2008;15:665. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeVane R, Shinoda W, Moore PB, Klein ML. J. Chem. Theory Comput. 2009;5:2115. doi: 10.1021/ct800441u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Risselada HJ, Marrink SJ. Phys. Chem. Chem. Phys. 2009;11:2056. doi: 10.1039/b818782g. [DOI] [PubMed] [Google Scholar]
- 74.Fuhrmans M, Knecht V, Marrink SJ. J. Am. Chem. Soc. 2009;131:9166. doi: 10.1021/ja903224q. [DOI] [PubMed] [Google Scholar]
- 75.Wu S, Guo HX. J. Phys. Chem. B. 2009;113(3):589. doi: 10.1021/jp808776z. [DOI] [PubMed] [Google Scholar]