Abstract
Voltage-gated proton channels are strongly inhibited by Zn2+, which binds to His residues. However, in a molecular model, the two externally accessible His are too far apart to coordinate Zn2+. We hypothesize that high-affinity Zn2+ binding occurs at the dimer interface between pairs of His residues from both monomers. Consistent with this idea, Zn2+ effects were weaker in monomeric channels. Mutation of His193 and His140 in various combinations and in tandem dimers revealed that channel opening was slowed by Zn2+ only when at least one His was present in each monomer, suggesting that in wild-type (WT) HV1, Zn2+ binding between His of both monomers inhibits channel opening. In addition, monomeric channels opened exponentially, and dimeric channels opened sigmoidally. Monomeric channel gating had weaker temperature dependence than dimeric channels. Finally, monomeric channels opened 6.6 times faster than dimeric channels. Together, these observations suggest that in the proton channel dimer, the two monomers are closely apposed and interact during a cooperative gating process. Zn2+ appears to slow opening by preventing movement of the monomers relative to each other that is prerequisite to opening. These data also suggest that the association of the monomers is tenuous and allows substantial freedom of movement. The data support the idea that native proton channels are dimeric. Finally, the idea that monomer–dimer interconversion occurs during activation of phagocytes appears to be ruled out.
Introduction
Human (HV1) and mouse (mVSOP) voltage-gated proton channels in heterologous expression systems appear to function as dimers, with a conduction pathway in each monomer (Koch et al. 2008; Lee et al. 2008; Tombola et al. 2008). Very recent studies suggest that the monomers interact during gating, although the details of this interaction remain obscure (Gonzalez et al. 2010; Tombola et al. 2010). It is not certain that the native channel is a dimer, because dimerization has been demonstrated only in heterologous expression systems, and the gating of expressed and native proton channels differs significantly (Musset et al. 2008). Here we compare certain properties of monomeric and dimeric proton channels in an attempt to gain insight into possible interactions. We also focus on Zn2+ binding sites in the channel, which provide clues about the possible dimer interface.
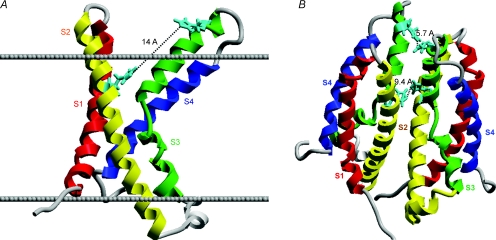
The most potent inhibitor of voltage-gated proton channels is Zn2+ (Mahaut-Smith, 1989). The effects of Zn2+ are profoundly attenuated at low pHo (Cherny & DeCoursey, 1999). This strong competition between H+ and Zn2+ for an external metal binding site in native proton channels could not be explained by simple 1:1 binding. Instead, the data were well described by models that assumed that the Zn2+ atom was coordinated between two to three titratable groups with pKa 6–7, suggestive of His residues (Cherny & DeCoursey, 1999). When the human proton channel gene was identified, the HV1 protein displayed two His residues accessible to the extracellular solution (Ramsey et al. 2006). Mutating either of these His to Ala (H140A, His140 replaced by Ala, or H193A, His193 replaced by Ala) reduced Zn2+ inhibition, and the double mutant had little Zn2+ sensitivity (Ramsey et al. 2006). One might conclude that Zn2+ inhibits proton current by binding simultaneously to His140 and His193. However, examination of a homology model of the proton channel structure (Fig. 1A) reveals a striking problem, namely that the two His residues are 14 Å apart – too far to coordinate a Zn2+ atom by overlap of their individual electron orbitals. Analysis of Zn2+ binding sites in 111 proteins revealed distances of 1.9–2.4 Å between Zn2+ and its coordinating atom (Alberts et al. 1998). Of course, Zn2+ binding to the proton channel is rapidly reversible, and a formal binding site is not expected. No plausible alternative sequence alignment, coordinate reassignment or torsioning of His sidechains resulted in a significantly shorter distance between His residues within the monomer.
Figure 1. Homology models of the HV1 proton channel as a monomer (A) or a dimer (B), showing the locations of the two His residues exposed to the extracellular solution.
In A, the extracellular surface of the membrane is indicated by the upper line, the intracellular surface by the lower line. His140 and His193 are shown in aqua. In the monomer, His140 and His193 are 14 Å apart, too far to plausibly coordinate a Zn2+ atom. In the dimer (B), the His140 from each monomer are closer together, as are the His193; either or both could comprise high-affinity Zn2+ binding sites. Thus, a high-affinity Zn2+ binding site exists only in the dimer. The recent identification of proton channel genes allows homology-based structural predictions, because the proton channel molecule bears striking homology to the voltage-sensing domain of K+ and other voltage-gated ion channels (Ramsey et al. 2006; Sasaki et al. 2006). HV1 contains four transmembrane domains resembling S1–S4 of other channels, but lacks the S5–S6 regions that comprise the ion conduction pathway in other channels.
This dilemma can be resolved by postulating that Zn2+ binds at the interface between the two monomers (Fig. 1B) to complementary His resides from each monomer. Binding between residues in different subunits can occur because the corresponding His residues approach more closely than is possible in a monomer. Thus, in the dimer model shown in Fig. 1B, the His193 of the two monomers are 5.7 Å apart and the His140 are 9.4 Å apart. In the His–Zn–His complex, the Zn–His distance would be somewhat larger than half of these distances, depending on the coordination angle (Alberts et al. 1998). Thus, both pairs of His residues form potential Zn2+ binding sites, particularly if Zn2+ binding pulls the two His towards each other. Empirically, both His residues contribute to Zn2+ effects, because both single mutants exhibited reduced, but still significant Zn2+ sensitivity (Ramsey et al. 2006). An alternate arrangement of the monomers in the dimer results in two identical Zn2+ binding sites, each comprising His140 and His193, one from each monomer (not shown). To obtain information about the Zn2+ binding site(s) on the human HV1 proton channel, we generated mutants in which His140 and His193 were modified singly or together, as well as tandem dimers with various combinations of mutations.
One way to test the novel idea that Zn2+ binds simultaneously to His residues in the two individual monomers is to express the channel in monomeric form. The Zn2+ sensitivity of the monomeric channel should be identical to that of the dimer if Zn2+ were coordinated between the His140 and His193 within each monomer. However, if Zn2+ is preferentially coordinated between two His residues, one on each monomer, then the monomeric channel should have reduced Zn2+ sensitivity. To study monomeric human proton channels, we used a construct of HV1 that terminated at Lys221 (HV1ΔC), and thus lacked the intracellular C-terminus. Coiled-coil interactions in the C-terminus stabilize the dimeric form of the channel (Lee et al. 2008; Tombola et al. 2008). To generate monomeric mouse proton channels, we truncated both C- and N-termini of mVSOP (Koch et al. 2008), to produce mVSOPΔCΔN. These truncations result in monomeric, but fully functional, voltage-gated proton channels, confirming that each monomer contains its own conduction pathway (Koch et al. 2008; Tombola et al. 2008). Since Zn2+ effects in monomeric channels were weaker, we pursued the idea that Zn2+ may prevent opening by constraining movement of the monomers relative to each other. A comparison of the kinetics and temperature dependence of gating of monomers and dimers, suggests that channel opening is more complex in the dimer.
Methods
Electrophysiology
In some studies, green fluorescent protein (GFP)-tagged proton channels were studied. More often, GFP was co-transfected with the proton channel construct. Fluorescent cells were identified in the field using Nikon inverted microscopes with fluorescence capability. Micropipettes were pulled using a Flaming Brown automatic pipette puller (Sutter Instruments, San Rafael, CA, USA) from 7052 glass (Garner Glass Co., Claremont, CA, USA), coated with Sylgard 184 (Dow Corning Corp., Midland, MI, USA), and heat polished to a tip resistance ranging typically from 5 to 15 MΩ with the pipette solutions used. Electrical contact with the pipette solution was achieved by a thin sintered Ag–AgCl pellet (In Vivo Metric Systems, Healdsburg, CA, USA) attached to a Teflon-encased silver wire, or simply a chlorided silver wire. A reference electrode made from a Ag–AgCl pellet was connected to the bath through an agar bridge made with Ringer solution. The current signal from the patch clamp (EPC-9 from HEKA Elektronik, Lambrecht/Pfalz, Germany, or Axopatch 200B from Axon Instruments, Foster City, CA, USA) was recorded and analysed using Lab View, SCB-68 (National Instruments, Austin, TX, USA), Pulse and PulseFit (HEKA), or pCLAMP (Molecular Devices, Sunnyvale, CA, USA) software supplemented by Microsoft Excel, Origin 7, and Sigmaplot (SPSS Inc., Chicago, IL, USA). Seals were formed with Ringer solution (in mm: 160 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 5 Hepes, pH 7.4) in the bath, and the potential zeroed after the pipette was in contact with the cell. No liquid junction potential correction was applied.
Inside-out patches were made by forming a seal and then lifting the pipette into the air briefly. For whole-cell recording, bath and pipette solutions contained 100–200 mm buffer, 1–2 mm CaCl2 or MgCl2 (pipette solutions were Ca2+ free), 1–2 mm EGTA, and tetramethylammonium methanesulfonate or N-methyl-d-glucamine to adjust the osmolality to roughly 300 mOsm, titrated with tetramethylammonium hydroxide or methanesulfonic acid. Buffers used at various pH values were Mes at pH 5.5–6.0, BisTris at pH 6.5, Pipes at pH 7.0 and Hepes at pH 7.5–8.0. We omitted EGTA from solutions for Zn2+ measurements. No leak correction has been applied to any current records. Except where noted, measurements were done at 21°C or at room temperature (20–25°C). The bath temperature was controlled by a system from Brooks Industries (Lake Villa, IL, USA), or by an in-house built system using Peltier devices in a feedback arrangement, monitored by a resistance temperature detector element (Omega Scientific, Stamford, CT, USA) immersed in the bath. Temperature changes were transmitted to the glass recording chamber through a supporting copper plate. The temperature probe was positioned as near the pipette tip as possible.
Currents were fitted to a rising exponential to obtain the activation time constant (τact) and the proton conductance (gH), which was calculated from the steady-state current (the fitted current extrapolated to infinite time) using reversal potentials (Vrev) measured in each solution in each cell. In these fits, we ignored the initial delay in WT activation; the remaining current usually fitted a single exponential well (Fig. 3). The reversal potential (Vrev) was measured by two methods. When Vrev was negative to the threshold voltage at which the proton conductance (gH) was first activated, Vthreshold, Vrev was determined by the tail current method (Hodgkin & Huxley, 1952). If Vrev was within the range of active proton conductance, it was determined by interpolation between time-dependent inward or outward currents during test pulses (after scaling according to the tail current amplitude). The magnitude of the shift of the gH–V relationship was determined by plotting gH values semilogarithmically against voltage (e.g. Fig. 8), and then shifting the gH–V relationships along the voltage axis until they superimposed upon the control curve.
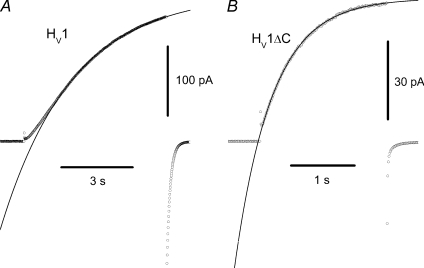
Figure 3. WT dimeric proton channels open slowly with a sigmoidal time course (A), but monomeric HV1ΔC channels open rapidly and exponentially (B).
Individual current records are shown at +50 mV from a holding potential of −40 mV in inside-out patches at pHo 7.5, pHi 7.5 at 23°C. Single exponential fits are superimposed and extrapolated on the data points shown as circles. For the records shown here, τact was 3.0 s and 0.59 s for WT and HV1ΔC, respectively.
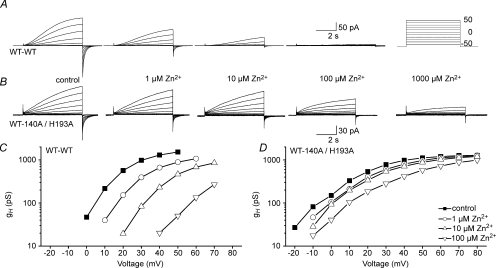
Figure 8. Greatly reduced Zn2+ sensitivity of the WT–H140A/H193A tandem dimer, compared with the WT–WT tandem dimer.
All current families in the WT–WT tandem dimer (A) or the WT–H140A/H193A tandem dimer (B) were recorded during pulses to the voltages shown in the inset at pHo 7.0, pHi 6.5. C and D, the gH–V relationships were determined from the cells in A and B using reversal potentials measured in each cell.
With overexpression of channels in small cells, large proton currents remove enough protons from the cell to increase pHi substantially (DeCoursey, 1991; Kapus et al. 1993; Demaurex et al. 1993; DeCoursey & Cherny, 1994, 1995, 1998; Musset et al. 2008; Kuno et al. 2009). As proton channel gating kinetics depends strongly on pH, proton depletion is a significant source of error. To minimize this problem, we applied pulses of different length to encompass different voltage ranges (Fig. 2). Longer pulses gave information about small depolarizations, and shorter pulses were used at large depolarizations, where activation was faster. Values for τact obtained in one cell for pulses of different lengths are plotted in the inset of Fig. 2. It is evident that despite a long interval between pulses, depletion progresses during families of pulses. Since cells survive a finite time and it is desirable to acquire data at multiple Zn2+ concentrations, some compromise is required between full pHi recovery between pulses and ideal data. We applied infrequently repeated small test pulses after each family to determine when pHi recovery was complete. Experiments were conducted with the intent of minimizing depletion, but also with the expectation that any systematic errors would apply equally to all constructs studied.
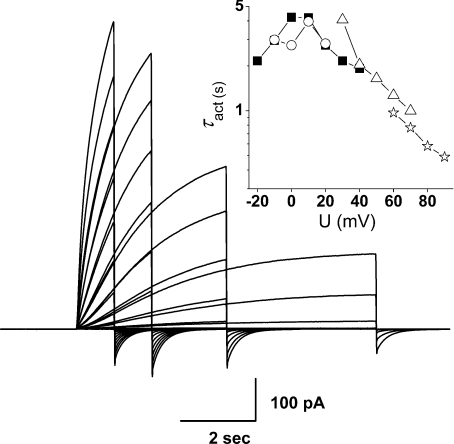
Figure 2. Use of different pulse lengths to evaluate τact at different voltage ranges, to minimize distortion of kinetics by proton depletion.
Superimposed are four pulse families of different durations in the same cell, transfected with the H193A–H140A tandem dimer at pHo 7.0, pHi 6.5 with 10 μm Zn2+. Pulses were applied in increasingly positive steps in 10 mV increments from a holding potential of −60 mV up to +20 mV (8 s pulses, ○), +40 mV (4 s, ▪), +70 mV (2 s, ▵) or +90 mV 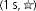 . Pulses were applied at 22 s intervals except for a 38 s interval for the 8 s family. The order of families was 4 s, 8 s, 2 s and 1 s. The inset shows τact estimated from these records.
. Pulses were applied at 22 s intervals except for a 38 s interval for the 8 s family. The order of families was 4 s, 8 s, 2 s and 1 s. The inset shows τact estimated from these records.
The slowing of τact by Zn2+ was evaluated over a wide voltage range, and each value was an average of the ratio at several voltages. The slowing by Zn2+ is roughly independent of voltage (Cherny & DeCoursey, 1999). We gave preference to τact from longer pulses, when data with different length pulses were available. As the τactvs. voltage relationship is often non-monotonic, with smaller values near Vthreshold that first increase and then decrease with depolarization (Musset et al. 2008), as evident in Fig. 2, we compared values positive to the maximum τact value.
Except where noted, statistical comparisons used Student's unpaired t test.
Culture, mutation and transfection
HEK-293 cells were maintained in 5% CO2 and 95% air in a humidified incubator at 37°C in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum, 2 mm l-glutamine, 100 U ml−1 penicillin, 250 ng ml−1 Fungizone and 100 μg ml−1 streptomycin. The coding sequence of human HV1 (Hvcn1) was cloned into either pcDNA3.1(–) or pQBI25-fC3 (to make GFP–HV1) vectors as described previously (Ramsey et al. 2006). The mouse orthologue (mVSOP) was derived from RIKEN cDNA 0610039P13 as described previously (Sasaki et al. 2006). HEK-293 cells were grown to ∼80% confluency in 35 mm cultures dishes, usually by seeding cells 1 day ahead of transfection. Cells were transfected in media without fetal bovine serum with ∼0.7 μg of the appropriate cDNA using Lipofectamine 2000 (Invitrogen). After 6 h incubation at 37°C in 5% CO2, the medium was replaced with medium containing fetal bovine serum. The next day, the cells were trypsinized and re-plated onto glass coverslips at low density for patch clamp recording. We selected green cells under fluorescence for recording.
The C-terminus of mouse mVSOP was truncated by introducing a stop codon at K217 (Quikchange site directed mutagenesis kit, Stratagene, San Diego, CA, USA). For N- and C-terminal truncations, mVSOP was amplified with primers containing a start codon at T77 and a stop codon at K217, and the PCR product was cloned in pCDNA 3.1(+). Single mutations, H140A and H193A, were generated by site-directed mutagenesis. The WT–WT, H193A–H140A and WT–H140A/H193A tandem constructs were made by removing the stop codon in the respective constructs in pCDNA and introducing a Nhe1 site and three alanines forming a six amino acid linker, ASGAAA, between the two proteins.
Homology model
A homology model of the voltage-sensing domain (VSD) of human HV1 (NCBI NP_115745.2) was created using Insight II (Accelrys, San Diego, CA, USA). The basis set consisted of the isolated voltage sensor of KvAP (1ORS) and the voltage sensor of Kv1.2 (2R9R) (Jiang et al. 2003a; Long et al. 2007). Basis set crystal structures were structurally aligned, and the sequence of the human HV1 VSD was aligned to a structure-based sequence alignment in BioEdit (Ibis Biosciences). Direct coordinate assignment was performed on structurally conserved regions. Loop assignments were performed via loop searches using a local subset of the Protein Data Bank, and using the Archpred server (Fernandez-Fuentes et al. 2006). Energy minimization routines were run to convergence on the splice points, the loops and the side-chains of the structurally conserved regions. In the final model, Procheck (Laskowski et al. 1993) revealed very few unrealistic bond lengths and bond angles, all of which resulted from crystal-based assignments. The general features of this model, including the distance between His140 and His193, were supported by an independent model generated using the automated I-TASSER method (Zhang, 2008).
Monomer models were visualized in Insight and in MolSoft ICM Pro (MolSoft), and the two monomers were moved manually (‘visual docking’), to verify the plausibility of Zn2+ binding between monomers. Further support for plausible Zn2+-binding dimer configurations was sought by submitting the model to the GrammX protein–protein docking server (Tovchigrechko & Vakser, 2006), using the homodimer option and with varying numbers of residues suggested as participating in the interface. Results of docking runs were initially visualized and inspected in VMD (visual molecular dynamics) (Humphrey et al. 1996). Dimer models retained from initial inspections were superimposed on a model of the 2R9R crystal structure in the plasma membrane obtained from the Orientations of Proteins in Membranes database (Lomize et al. 2006) to assess their ability to span the membrane appropriately. Approximately 1% of the dimers returned from the docking runs were in realistic orientations. Figure 1B is a representative example from the realistic dimers that were identified, all of which had His–His distances of 4–10 Å.
Results
Activation kinetics in monomeric and dimeric proton channels
The activation kinetics of monomeric and dimeric proton channel currents differs in two respects. First, WT dimeric proton channels activate with a sigmoidal time course (Fig. 3A), like all native proton currents (DeCoursey, 2003). In contrast, monomeric HV1ΔC proton currents activate with an exponential time course (Fig. 3B). We fitted all currents to a single rising exponential to obtain the time constant of activation, τact. For dimeric channel currents, we ignored the initial delay. Exponential opening is consistent with a simple first order opening transition in the monomer. Sigmoidal kinetics can be explained by a cooperative gating process in which multiple subunits must undergo an opening transition before conduction can occur (Hodgkin & Huxley, 1952). As the proton channel is a dimer in which each monomer contains its own conduction pathway (Koch et al. 2008; Tombola et al. 2008; Lee et al. 2008), such cooperativity is surprising.
Consistent with previous reports (Koch et al. 2008; Tombola et al. 2008), channel opening in HV1ΔC or mVSOPΔNΔC monomers was much faster than in WT dimers (Fig. 3, and not shown). Inside-out patches were used for this measurement, because proton depletion notoriously can distort gating kinetics in whole-cell studies (DeCoursey, 1991; Kapus et al. 1993; Demaurex et al. 1993; DeCoursey & Cherny, 1994, 1995, 1998; Musset et al. 2008; Kuno et al. 2009). The average ratio of τact in HV1/HV1ΔC measured (at each voltage) in inside-out patches at pHo 7.5, pHi 7.5 at +30 to +80 mV in 10 mV increments was 6.58 ± 0.44 (mean ±s.e.m. for 6 voltages, each with 4–7 cells for HV1 and 5–7 cells for HV1ΔC). A 5-fold slower time-to-half-peak was reported for mVSOP than mVSOPΔNΔC (Koch et al. 2008). Faster activation of monomeric channels suggests that interaction between monomers in the dimer slows opening or that the presence of cytoplasmic domains slows gating (Koch et al. 2008). Proton channels in monomeric form obligatorily gate independently; apparently a qualitatively different gating process occurs in the dimer.
Temperature dependence is weaker in the monomer
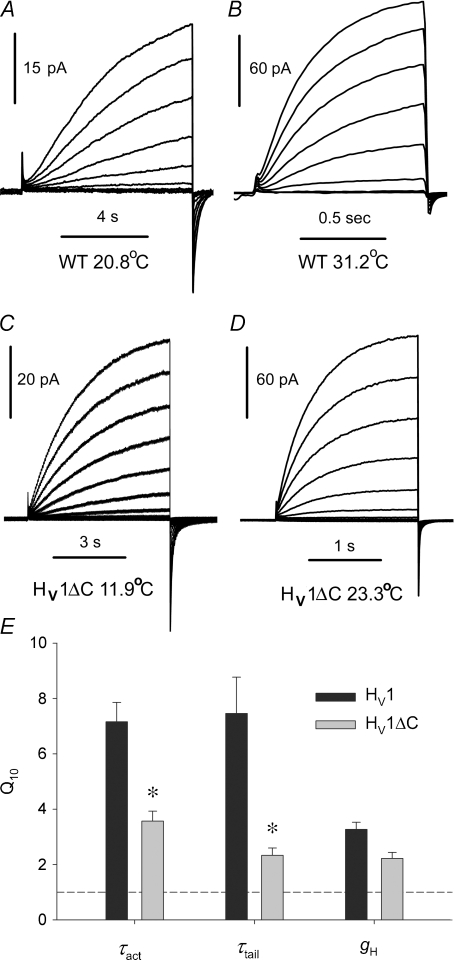
A complex gating mechanism for WT dimeric proton channels is consistent with the extreme temperature dependence of gating of native proton channels (Q10= 6–9, or 30–38 kcal mol−1 for both opening and closing kinetics) (DeCoursey & Cherny, 1998). If part of the energetic barrier arises from inter-subunit interaction, the temperature dependence of gating in the monomeric construct might be weaker. Figure 4 illustrates that this was the case for the human proton channel: gating kinetics in the HV1ΔC monomer had temperature dependence only about half that of wild type (WT) HV1. Families of WT (A and B) and HV1ΔC (C and D) proton currents are shown at roughly 10°C different temperatures. The time scales differ by a factor of 8 for WT and 3 for HV1ΔC, illustrating the much stronger temperature dependence of channel opening kinetics in the dimer. We used excised, inside-out patches of membrane for these measurements to avoid complications due to proton depletion during large currents (DeCoursey & Cherny, 1998; Kuno et al. 2009). Estimates were based on several currents from families of pulses recorded at two or more temperatures, with the assumptions that within the error of the estimates, Q10 for gating kinetics is independent of voltage and pH (DeCoursey & Cherny, 1998; Kuno et al. 2009). On average, the Q10 of channel opening (τact) was 7.16 for WT and 3.57 for HV1ΔC (Fig. 4E). The Q10 for channel closing (τtail, tail current time constant) was 7.46 for WT and 2.33 for HV1ΔC. Measurements were made over a pH range of 5.5–7.5 and within the temperature range 9–32°C. Thus, channel opening and closing are both substantially more energetically demanding in WT dimeric channels, suggesting a different gating mechanism.
Figure 4. Temperature dependence of WT (A and B) and HV1ΔC proton currents (C and D) recorded in inside-out patches of membrane.
For each patch, families of pulses to the same voltages were applied, with the pulse duration adjusted as shown. A, WT proton currents during pulses from −60 mV to +20 mV in 10 mV increments at 20.8°C (A) and 31.2°C (B), at pHo 7.5, pHi 6.5. C, HV1ΔC proton currents in a patch elicited by families of pulses from −40 mV to +80 mV in 10 mV increments, at 11.9°C. D, pulses to the same voltages in the same patch at 23.3°C. Both at pHo 7.5, pHi 7.5. E, summary of the temperature dependence of WT and HV1ΔC proton channel gating kinetics and conductance. Measurements were made in excised inside-out patches to avoid depletion effects. Means ±s.e.m. are shown for 6–8 WT (HV1) patches and 6–9 HV1ΔC patches. *P < 0.01 by Student's t test.
Zn2+ sensitivity of the monomer is weaker than the dimer
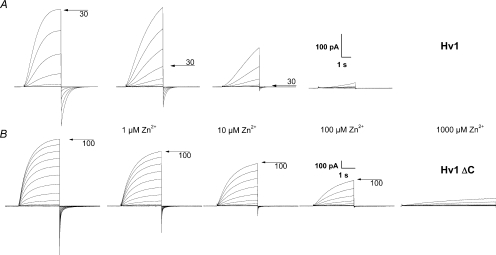
Figure 5 shows that the Zn2+ sensitivity of the monomeric HV1ΔC channel (Fig. 5B) was distinctly weaker than that of the dimeric HV1 (Fig. 5A). The main effects of Zn2+ on proton currents are to slow channel opening (larger activation time constant, τact) and to shift the proton conductance–voltage relationship, gH–V, positively. The slowing of τact was significantly greater in the WT channels than in the HV1ΔC (P < 0.01 at 1, 10 and 100 μm Zn2+, data not shown).
Figure 5. The Zn2+ sensitivity of the human monomeric proton channel is weaker than that of the dimer.
A, WT HV1 proton current families in the same cell at pHo 7.0, pHi 6.5 during pulses in 10 mV increments to +30 mV (control) or to +60 mV (Zn2+). B, weaker Zn2+ sensitivity of HV1ΔC monomeric channels at pHo 7.0, pHi 6.5. Pulses were applied in 10 mV increments up to +100 mV.
Measurements were also performed on mouse proton channels and the monomeric mVSOPΔNΔC construct, in order to determine whether the phenomena described here apply to proton currents in other species. WT mouse proton currents (mVSOP) activated sigmoidally, and the monomeric mVSOPΔNΔC currents activated exponentially (not shown). Slowing of τact by Zn2+ was evaluated as described for HV1. The Zn2+ sensitivity of the monomeric construct was significantly weaker than WT at 1, 10 and 100 μm Zn2+ (P < 0.05 for each, data not shown).
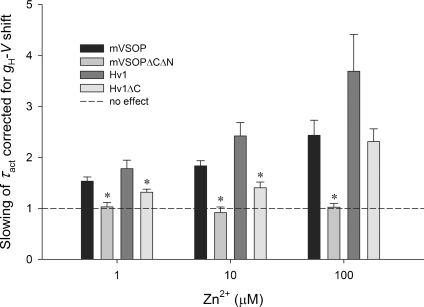
Some slowing of activation is expected to result from the shift by Zn2+ of the gating of most voltage-dependent channels, although depending on the mechanism, different gating parameters may be shifted by different amounts (Frankenhaeuser & Hodgkin, 1957; Hille, 2001; Elinder & Åhrem, 2004). To evaluate this electrostatic effect, we determined the shift of the gH–V relationship produced by each [Zn2+] in each cell and shifted the τact–V relationship negatively by that amount. Figure 6 shows the residual slowing of τact after this correction. WT murine and human proton channels are slowed by Zn2+ beyond what is expected from a simple voltage shift, but for the mVSOPΔNΔC and HV1ΔC constructs, most of the observed slowing was attributable to the voltage shift.
Figure 6. Monomeric proton channels (mVSOPΔNΔC or HV1ΔC) are less sensitive to the slowing effect of Zn2+ than WT dimeric channels (mVSOP or HV1).
Plotted is the mean ±s.e.m. residual slowing after correction for the observed shift of the gH–V relationship in each cell at each [Zn2+]. Slowing is defined as the ratio τact(Zn2+)/τact, which was determined at moderate depolarizations in the voltage range where τact is approximately exponentially dependent on voltage. For mVSOPΔNΔC and mVSOP, n= 3–4; for HV1 and HV1ΔC, n= 8–11. *P < 0.05 for truncated vs. WT channels.
Zn2+ sensitivity of histidine mutants
In order to further test the hypothesis that externally applied Zn2+ exerts its characteristic effects at one or more high affinity binding sites at which Zn2+ is coordinated between two His residues on the proton channel, we studied a series of His mutants and constructs, illustrated in the diagram in Fig. 7. The various constructs in Fig. 7 were expressed and studied at pHo 7.0 in the presence of Zn2+. Of the main effects of Zn2+ (Cherny & DeCoursey, 1999), we focused on the shift of the gH–V relationship (Figs 8 and 9A) and the slowing of τact (Fig. 9B).
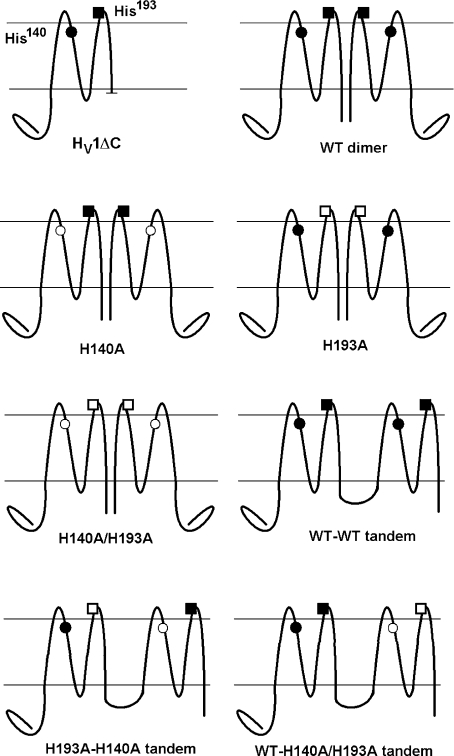
Figure 7. Diagram of the histidine mutants and constructs used and their nomenclature.
His140 is on S2 near the external surface of the membrane; His193 is on the S3–S4 linker. Filled symbols indicate His, open symbols indicate Ala substitution. The C-terminal truncation HV1ΔC expresses as a monomer (Tombola et al. 2008; Koch et al. 2008). The dimer assembles mainly by C-terminal interactions (Tombola et al. 2008; Koch et al. 2008; Lee et al. 2008). Tandem dimers were linked by insertion of –ASGAAA– between C- and N-termini of two monomers.
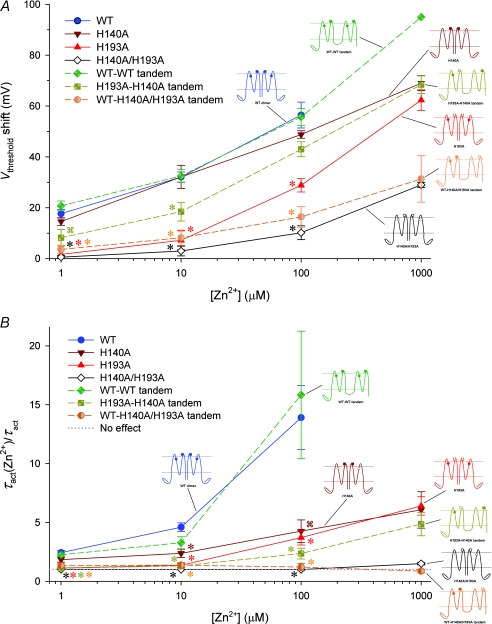
Figure 9. Effects of Zn2+ on the voltage dependence and gating kinetics of several proton channel constructs with His mutations.
A, shifts of the gH–V relationship produced by Zn2+ in various human HV1 constructs. Inset diagrams illustrate the positions of His140 and His193 residues (•, ▪) or their substitution by Ala (○, □), respectively, as also shown in Fig. 7. Tandem dimer data are connected by dashed lines. Mean ±s.e.m. are shown for numbers of cells studied at up to 100 μm Zn2+: 9–12 WT; 5–7 H140A; 5–6 H193A; 5–6 H140A/H193A; 3–12 WT–WT tandem; 5–7 H193A–H140A tandem; 5–6 WT–H140A/H193A tandem. 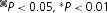 by Student's t test compared with WT. B, slowing of activation kinetics produced by Zn2+ in various HV1 constructs. Mean ±s.e.m. slowing of τact by Zn2+ (defined as the ratio τact(Zn2+)/τact) in the indicated proton channel constructs, for numbers of cells studied at up to 100 μm Zn2+: 8–11 WT; 5–7 H140A; 5–8 H193A; 4 H140A/H193A; 4–11 WT–WT tandem; 5–7 H193A–H140A tandem; 5–6 WT–H140A/H193A tandem.
by Student's t test compared with WT. B, slowing of activation kinetics produced by Zn2+ in various HV1 constructs. Mean ±s.e.m. slowing of τact by Zn2+ (defined as the ratio τact(Zn2+)/τact) in the indicated proton channel constructs, for numbers of cells studied at up to 100 μm Zn2+: 8–11 WT; 5–7 H140A; 5–8 H193A; 4 H140A/H193A; 4–11 WT–WT tandem; 5–7 H193A–H140A tandem; 5–6 WT–H140A/H193A tandem. 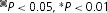 by Student's t test compared with WT.
by Student's t test compared with WT.
With regard to the shift of the gH–V relationship by Zn2+, H140A was similar to WT, but H193A was distinctly less sensitive (Fig. 9A; P < 0.001 at all [Zn2+]). This result suggests that Zn2+ bound to His193 exerts a stronger effect on the voltage sensor of the proton channel. Surprisingly, both single His mutants H140A and H193A exhibited more similar slowing effects (Fig. 9B), although H193A was again more sensitive (P < 0.05 vs. H140A at 1 and 10 μm Zn2+). The slowing of τact by Zn2+ was significantly weaker than WT in either single mutant, indicating that both His residues contribute to the slowing of channel opening by Zn2+. The double His mutant H140A/H193A exhibited essentially no slowing by Zn2+, but a weak shift of the gH–V relationship remained. The shift may reflect non-specific divalent cation effects or Zn2+ binding to sites on the channel other than His. Nevertheless, that no slowing was seen in H140A/H193A suggests that virtually the entire slowing effect can be attributed to Zn2+ binding to His140 and His193.
Zn2+ sensitivity of histidine mutants and constructs: tandem dimers
A control WT–WT tandem dimer was generated by linking two WT HV1 channels. The gating and Zn2+ sensitivity of this WT–WT tandem dimer (Fig. 8A) were indistinguishable from WT channels. This result supports the validity of the use of tandem dimers to evaluate the characteristics of Zn2+ binding to HV1. In addition, this result further supports the conclusion that the Hvcn1 gene product assembles as a dimer in several expression systems (Koch et al. 2008; Lee et al. 2008; Tombola et al. 2008).
If a high-affinity binding site for Zn2+ can exist within a single monomer, then a high-affinity site should exist on the WT–H140A/H193A tandem dimer, which retains both His residues in one monomer but neither in the other. However, Zn2+ effects were drastically attenuated in this construct (Fig. 8B). For example, in the WT–WT tandem dimer, 1 μm Zn2+ already shifts the gH–V relationship positively by ∼10 mV (Fig. 8C). In contrast, in the WT–H140A/H193A tandem dimer, little shift occurs until 100 μm Zn2+ is introduced (Fig. 8D).
The properties of the WT–H140A/H193A tandem dimer were unique in one respect. Most of the channel constructs tested activated exponentially or exponentially after a delay, with or without Zn2+. An exception is the H193A–H140A tandem dimer, whose currents were sometimes better fitted with two components in the absence of Zn2+. Proton currents in the WT–H140A/H193A tandem dimer activated with two components, but only in the presence of ≥100 μm Zn2+ (Fig. 8B). When fitted with two exponentials, the faster component had time constants <1 s, whereas the time constant of the slower component was close to τact measured in the absence of Zn2+. Thus, Zn2+ not only failed to slow activation, it appeared to introduce a novel rapid component of activation. Assuming that cooperative gating normally occurs, it might be speculated that in this construct, Zn2+ may uncouple the two monomers. In any case, the absence of slowing by Zn2+ of any component of opening of the WT–H140A/H193A tandem dimer suggests that slowing occurs when Zn2+ binds between His residues in the two monomers at the dimer interface.
The shifts of the gH–V relationship and the slowing of τact produced by Zn2+ are summarized for this and other constructs in Fig. 9. Figure 9A reveals that the shift in the gH–V relationship produced by Zn2+ was almost as weak in the WT–H140A/H193A construct which has one pair of dissimilar His residues as in the double mutant H140A/H193A that lacks His altogether.
Upon examination of the slowing of channel opening by Zn2+ (Fig. 9B), the constructs fall into three categories. Slowing is most profound for constructs in which both pairs of His193 and His140 exist (WT and WT–WT tandem). Weaker but distinct slowing also occurs when a single symmetrical pair of His140 or His193 are present (H193A or H140A) or the asymmetrical pair (H193A–H140A tandem dimer). Remarkably, there was practically no slowing of activation in the WT–H140A/H193A tandem dimer or the double mutant H140A/H193A. The simplest interpretation is that slowing occurs when Zn2+ can interact with both monomers simultaneously.
Discussion
Monomer interactions during channel opening
An apparent contradiction between structural predictions based on electrophysiological measurements of competition between Zn2+ and H+ (Cherny & DeCoursey, 1999), and a molecular homology model of the proton channel VSD (Fig. 1) led to the hypothesis that Zn2+ binds between monomers in the proton channel dimer. The obvious prediction that monomeric channels should have lower Zn2+ affinity was borne out. That Zn2+ could prevent channel opening at such a location further suggested that the two monomers are closely apposed and may interact during channel opening. Several types of evidence are consistent with this hypothesis. Monomeric channels open with an exponential time course, rather than the sigmoidal kinetics typical of all native proton channels (DeCoursey, 2003) and of cooperative gating mechanisms in general (Hodgkin & Huxley, 1952; Horrigan et al. 1999). Channel opening was 6.6 times faster in monomeric channels, consistent with a more facile opening process than in the dimer. The activation energy of WT channel gating was double that of monomeric channels, indicating a more complex opening process in the dimer. The native voltage-gated proton channel has much larger activation energy for opening than most other voltage-gated ion channels. That the Q10 was found to be similar for the delay, τact and τtail, led to the suggestion that the channel comprises multiple subunits that each undergoes a single rate-determining conformational change (DeCoursey & Cherny, 1998). Finally, Zn2+ retards proton channel opening beyond the slowing expected from a simple voltage shift (Cherny & DeCoursey, 1999), as was observed for Cd2+ effects on snail neuron proton channels (Byerly et al. 1984). Together, these observations strongly suggest pronounced interaction between monomers during proton channel opening. Two recent studies support this conclusion by providing evidence for cooperativity (Gonzalez et al. 2010; Tombola et al. 2010). In one proposal, both monomers must activate before either can conduct current (Gonzalez et al. 2010). In the other proposal, the subunits can open individually, but positive cooperativity results in both usually being in the same open or closed state (Tombola et al. 2010).
The physical interpretation of the present results is not clear-cut. Ascribing the delay in H+ current activation to a gating step (between closed states) preceding opening in each (independent) monomer would not account for the absence of delay in monomeric channels. One possibility is that in the dimer, the gating of each monomer is slowed by the presence of the other monomer. Alternatively, perhaps both channel subunits must enter the ‘open’ configuration before either can conduct current. This interpretation is suggested by the observation that the monomeric channel activates with an exponential time course, whereas dimeric channel currents exhibit a sigmoidal time course (Fig. 3), reminiscent of classical Hodgkin–Huxley kinetics (Hodgkin & Huxley, 1952). Similarly, for the Ciona proton channel (CiVSOP), monomeric constructs activated exponentially, whereas the dimer opened with sigmoidal kinetics (Gonzalez et al. 2010). The relatively brief delay and slower subsequent activation suggest that a slow concerted gating process occurs after both monomers have activated (Horrigan et al. 1999). In either case, the slower activation of dimeric channels suggests a complex gating process involving interaction between the two monomers. Intriguingly, direct measurements of the single-channel current of voltage-gated proton channels provided estimates that were roughly twice the value determined from current fluctuation analysis (Cherny et al. 2003). This observation is compatible with a gating process in which both channels in the dimer open approximately synchronously often enough to be interpreted as a single opening event (with twice the unitary conductance), but in which most gating transitions (that contribute to the fluctuations) involve individual conduction pathways. This phenomenology appears to be more consistent with the proposal of Tombola et al. (2010) than that of Gonzalez et al. (2010).
The sigmoidal activation kinetics (DeCoursey, 2003) and strong temperature dependence of gating of native proton currents (DeCoursey & Cherny, 1998; Kuno et al. 2009) closely resemble the corresponding behaviour of expressed WT (dimeric) proton channels, but not monomeric constructs. These properties therefore support the idea that the native proton channel, like expressed proton channels, assembles as a dimer, despite the difference in voltage dependence of expressed and native proton channels (Musset et al. 2008).
Teleological advantages of cooperative gating
Why would cooperative gating at the expense of slower activation be advantageous for proton channels? Cooperativity increases the voltage sensitivity of channel opening (Sigworth, 1993). Proton channels exist in many non-excitable cells. In many situations that result in channel opening, speed typically is not an important consideration. For example, the phagocyte respiratory burst continues for roughly 1 min to 1 h depending on the stimulus (DeCoursey & Ligeti, 2005). It may be more important that proton channels open with steep voltage dependence than rapidly. Proton efflux limits membrane depolarization, which is desirable during the respiratory burst, because NADPH oxidase enzyme activity is voltage dependent, and is inhibited at large positive voltages (DeCoursey et al. 2003; Petheő & Demaurex, 2005).
Deducing channel architecture from Zn2+ effects
Simple models of competition between H+ and Zn2+ indicated that Zn2+ binds to multiple titratable groups with pKa 6.2–7.0 (Cherny & DeCoursey, 1999). The human proton channel has two His residues predicted to be exposed to the extracellular solution, His140 and His193, and mutation of either one to Ala reduced Zn2+ sensitivity of the channel; the double mutant H140A/H193A was even less sensitive (Ramsey et al. 2006; see also Fig. 9). These observations point to Zn2+ coordination between two His residues. The two His in each monomer were too far apart to coordinate Zn2+ in the homology model (Fig. 1A), and the weaker Zn2+ effects in monomeric HV1ΔC are consistent with the distance between His140 and His193 being too great for Zn2+ to bind to both His within a single monomer. However, potential high-affinity Zn2+ binding sites occur at the interface between monomers in the dimer model. Plausible dimer models were identified in which potential binding sites were comprised of His140–His140 or His193–His193 pairs (Fig. 1B). An alternative site formed by His140–His193 from complementary subunits can be obtained by small, opposite rotations of each monomer in the dimer shown in Fig. 1B, around an axis parallel to the membrane. The resulting His140–His193 distance is approximately 7 Å (not shown).
An alternate interpretation is that in the WT channel, strong Zn2+ effects are the result of Zn2+ binding independently to each channel subunit. Two arguments oppose this interpretation. First, the competition between H+ and Zn2+ required that at least two titratable groups coordinate Zn2+ at its primary site of action (Cherny & DeCoursey, 1999). Second, assuming that neither channel can conduct until both undergo an opening transition (Gonzalez et al. 2010), if Zn2+ could bind independently within each monomer, then the Zn2+ effects observed in the WT–H140A/H193A tandem dimer (Fig. 9) would be expected to be more profound. In contrast, this construct was nearly as insensitive to Zn2+ as the double mutant that lacks both His (H140A/H193A).
Since Zn2+ effects do not saturate at achievable concentrations, distinguishing differential binding affinity from differential efficacy at distinct sites is not straightforward. Nevertheless, some conclusions appear warranted. When Zn2+ binds either to His140 (in H193A) or His193 (in H140A) the gH–V relationship shifts, although the shift is greater at any given [Zn2+] when Zn2+ binds at His193 (in H140A or H193A–H140A tandem). This result suggests that Zn2+ exerts a stronger effect on the voltage sensor of the proton channel when bound to His193 than to His140. However, it is also possible that the affinity of Zn2+ is greater for His193 than for His140.
Interpreting Zn2+ effects on the shift of the gH–V relationship and slowing of τact is further complicated by the interrelatedness of these parameters. If the channel opening rate is decreased, the gH–V relationship will shift positively, because the open probability will decrease at all potentials. Electrostatic effects generally shift voltage-dependent parameters along the voltage axis (Frankenhaeuser & Hodgkin, 1957; Hille, 2001; Elinder & Åhrem, 2004); in the simplest form of this type of mechanism, all parameters shift equally. However, the slowing of τact in proton channels by Zn2+ not only exceeds that predicted from the shift of the gH–V relationship, but results in τact values at high Zn2+ that are slower than at any voltage without Zn2+ (Cherny & DeCoursey, 1999). It appears that Zn2+ slows channel opening by a distinct mechanism, and the data presented here indicate that this slowing effect occurs when Zn2+ binds to His residues. That no slowing was seen in H140A/H193A suggests that virtually the entire slowing effect can be attributed to Zn2+ binding to His140 and His193. In addition, because the rank order of sensitivity to Zn2+ effects is different among the constructs tested for slowing and shift of the gH–V relationship, these effects are, at least to some extent, separable.
The slowing of channel opening in the constructs tested (Fig. 9B) supports the hypothesis that the dimer interface allows His140 and His193 from each monomer to pair with the corresponding His from the other monomer to form a Zn2+ binding site. Profound slowing by Zn2+ occurs when all four His residues are present in the dimer (WT and WT–WT tandem). Moderate slowing also occurs when a single His residue is present in each monomer (H140A, H193A or H193A–H140A tandem dimer). The slowing of τact by Zn2+ was significantly weaker than WT in either single mutant, indicating that both His residues contribute to the slowing of channel opening. The weak but distinct slowing in the H193A–H140A tandem dimer might reflect coordination of Zn2+ between dissimilar His or between His193 and Glu192 or Glu196 in the opposite monomer. Glu192 and Glu196 are 9 Å and 11 Å, respectively, from His193 in the model, but they are in the flexible S3–S4 linker and thus might approach more closely. Remarkably, there was no slowing of τact in the WT–H140A/H193A tandem dimer. Taken together, these data suggest that when Zn2+ binds to His140 or His193 residues at the interface between monomers, neither monomeric channel can undergo the physical movement required for opening. Zn2+ may retard the movement of one monomer relative to the other during gating. Alternatively, Zn2+ may constrain the dimer in a conformation from which it cannot open.
The double His mutant H140A/H193A exhibited essentially no slowing by Zn2+ although a weak but distinct shift of the gH–V relationship remained. The shift may reflect non-specific divalent cation effects or binding to residues on the channel other than His. In enzymes containing structural or catalytic Zn2+, three or four amino acids (His, Cys, Glu or Asp) coordinate the Zn2+ (Vallee & Auld, 1990; Alberts et al. 1998). Of course, Zn2+ binding to the proton channel is neither structural nor catalytic and is rapidly reversible and consequently, a formal binding site is not expected.
Conformational changes during gating are unlikely to bring His140 and His193 closer together in the monomer
The homology model in Fig. 1 was based on what is presumed to be an open conformation of K+ channels (Jiang et al. 2003a; Long et al. 2007). Closing of K+ channels is thought to involve inward motion of S4, changing its orientation relative to other parts of the voltage-sensing domain (VSD; the S1–S4 complex) (Bezanilla, 2000; Jiang et al. 2003b; Starace & Bezanilla, 2004; Tombola et al. 2006; Swartz, 2008). Roughly analogous motion of S4 in HV1 is suggested by recent studies (Gonzalez et al. 2010; Tombola et al. 2010), although evidence that truncation of the C-terminus between the second and third Arg residues in S4 did not abolish proton selectivity or gating raises questions about the extent of similarity (Sakata et al. 2010). Positioned within a long helix, His140 is probably stationary. The kink in the S3 helix might allow His193 to swing toward the S2 helix and His140, but the length of the S3b helix probably precludes an approach close enough for intramonomer Zn2+ binding. A model of the closed state of Kv1.2 (Pathak et al. 2007) does not show any significant decrease in the intramonomer distance between residues in positions homologous to those of His140 and His193. This buttresses the idea that the HV1 monomer does not support tight Zn2+ binding. In the dimer, conformational rearrangements (caused by S4 motion, for example) could orient the His pairs more favourably for inter-monomer Zn2+ binding in the closed, rather than in the open conformation, which would be consistent with the preferential effect of Zn2+ on channel opening over closing (Cherny & DeCoursey, 1999; Elinder & Åhrem, 2004).
Comparison of the proposed dimer interface with previous models
The dimer interface in Fig. 1B, which is supported by the Zn2+ studies, differs from previous proposals. FRET studies in the Ciona proton channel indicated 42 Å between corresponding Ser242 residues in the dimer (Koch et al. 2008), which like His193 in the human channel, is located in the S3–S4 linker. A study of HV1 based on a series of Cys mutants led to a topological model (Lee et al. 2008) with a dimer interface different from that in Fig. 1B, in which the two His193 are far apart. However, in that study, distinct cross-links were also detected at position 194 in HV1, at the tip of the voltage-sensor paddle (Lee et al. 2008), which indicates that His193 residues from each monomer are sometimes in close proximity. All of these observations can be reconciled if the proton channel dimer is loosely organized and occasionally samples alternative associations. This idea is supported by the fact that the channel appears as a monomer in Western blots (Ramsey et al. 2006; Koch et al. 2008; Lee et al. 2008, 2009; Capasso et al. 2010). Configurations in which His residues from both monomers approach each other may not occur frequently, but when they do, the opportunity for Zn2+ to bind exists. Zn2+ binding may then constrain the dimer in a conformation that precludes channel opening. Solving the crystal structure (Lee et al. 2009; Li et al. 2009) might shed light on these possibilities, but it may simply illustrate one of many possible orientations, or perhaps HV1 will crystallize as a monomer. An alternative possibility that cannot be ruled out is that when the HV1 dimer adopts a conformation with close apposition of S1 regions as proposed by Lee et al. (2009), Zn2+ binds between His residues exposed at the edges of dimers, promoting tetramer formation. Evidence consistent with tetramer or higher order polymer formation at least under specific conditions has been observed (Lee et al. 2009).
The C-terminus of HV1 was crystallized recently, and structural changes were observed at different pH values (Li et al. 2010). Given that both the WT–WT tandem dimer (in which coiled-coil interaction along the entire C-termini may be disrupted by linking N- and C-termini together) and constructs lacking the C-terminus altogether, such as HV1ΔC, appeared to have normal pH dependence (Koch et al. 2008; Sakata et al. 2010), it seems unlikely that the pH dependence of gating resides in the C-terminus.
The enhanced gating mode does not reflect monomer–dimer interconversion
During the respiratory burst in neutrophils and eosinophils that accompanies phagocytosis and reflects NADPH oxidase activation, proton channels exhibit profoundly enhanced gating (Bánfi et al. 1999; DeCoursey et al. 2000). The enhanced gating strongly promotes proton channel opening and was predicted by modelling to improve the efficiency of NADPH oxidase by limiting the depolarization required to open enough proton channels to compensate for the electrogenic activity of the oxidase (Murphy & DeCoursey, 2006). Depolarization directly inhibits NADPH oxidase activity (DeCoursey et al. 2003; Petheő & Demaurex, 2005). Koch et al. (2008) speculated that the conversion of phagocyte proton channels to the ‘enhanced gating mode’ might reflect interconversion between monomeric and dimeric channel arrangements. As Zn2+ binds with high affinity between monomers in the dimer, but weakly to monomeric proton channels, one would predict different Zn2+ sensitivity of the two modes of gating. However, in eosinophils, the Zn2+ sensitivity of the proton channel in resting and enhanced gating modes is identical (DeCoursey et al. 2001). Since the monomeric proton channel (HV1ΔC) has distinctly weaker Zn2+ sensitivity, it appears that the phagocyte proton channel remains a dimer during enhanced gating. Enhanced gating can be explained by phosphorylation of the dimeric channel (Morgan et al. 2007; Musset et al. 2010).
Summary
Taken together, the evidence suggests profound intermonomer interaction during opening in the dimeric voltage-gated proton channel, despite each monomer having a separate conduction pathway. Interaction between monomers during gating is suggested by the slower opening rate of the dimeric than the monomeric channel, the higher activation energy of channel gating of the HV1 dimer, and sigmoidal activation kinetics of the dimer compared with exponential kinetics of the monomer. Perhaps a concerted opening process occurs in the dimer. Alternatively, both monomers in the dimer may undergo identical gating movements as a prerequisite to conduction. Zn2+ slows channel opening most effectively when symmetrical His residues are available in both monomeric channels. As Zn2+ effects are weaker in monomeric than dimeric proton channels, the phagocyte proton channel in both resting and enhanced gating modes is evidently a dimer.
Acknowledgments
We thank I. Scott Ramsey and David E. Clapham for providing HV1ΔC plasmid, and Tatiana Iastrebova for technical assistance. This work was supported by the Professor Adolf Schmidtmann Stiftung (B.M.), by NIDDK (DK-075706 to S.R.), by the Heart, Lung and Blood Institutes of Health (HL-61437 to T.E.D.) and by the National Science Foundation (T.E.D. and S.S.).
Glossary
Abbreviations
- gH
proton conductance
- gH–V
proton conductance–voltage relationship
- HV1
human proton channel protein
- HV1ΔC
C-terminal truncation of HV1
- mVSOP
mouse proton channel
- mVSOPΔNΔC
N- and C-terminal truncation of mVSOP
- τact
activation time constant
- τtail
deactivation (tail current) time constant
- VSD
voltage-sensing domain
Author contributions
S.M.E.S., B.M., V.V.C., S.R. and T.E.D. contributed to conception and design; S.M.E.S., S.S. and S.R. contributed new tools; S.M.E.S. built the molecular model; B.M., V.V.C. and D.M. collected, analysed and interpreted data. T.E.D. wrote the paper; all authors critiqued the manuscript and approved the final version. Patch clamp studies were done at Rush University, constructs were generated at the University of Chicago and Emory University, molecular modelling was done at Emory University.
References
- Alberts IL, Nadassy K, Wodak SJ. Analysis of zinc binding sites in protein crystal structures. Protein Sci. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánfi B, Schrenzel J, Nüsse O, Lew DP, Ligeti E, Krause K-H, Demaurex N. A novel H+ conductance in eosinophils: unique characteristics and absence in chronic granulomatous disease. J Exp Med. 1999;190:183–194. doi: 10.1084/jem.190.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- Byerly L, Meech R, Moody W., Jr Rapidly activating hydrogen ion currents in perfused neurones of the snail, Lymnaea stagnalis. J Physiol. 1984;351:199–216. doi: 10.1113/jphysiol.1984.sp015241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso M, Bhamrah MK, Henley T, Boyd RS, Langlais C, Cain K, et al. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nat Immunol. 2010;11:265–272. doi: 10.1038/ni.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny VV, DeCoursey TE. pH-dependent inhibition of voltage-gated H+ currents in rat alveolar epithelial cells by Zn2+ and other divalent cations. J Gen Physiol. 1999;114:819–838. doi: 10.1085/jgp.114.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny VV, Murphy R, Sokolov V, Levis RA, DeCoursey TE. Properties of single voltage-gated proton channels in human eosinophils estimated by noise analysis and direct measurement. J Gen Physiol. 2003;121:615–628. doi: 10.1085/jgp.200308813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey TE. Hydrogen ion currents in rat alveolar epithelial cells. Biophys J. 1991;60:1243–1253. doi: 10.1016/S0006-3495(91)82158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol Rev. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- DeCoursey TE, Cherny VV. Voltage-activated hydrogen ion currents. J Membr Biol. 1994;141:203–223. doi: 10.1007/BF00235130. [DOI] [PubMed] [Google Scholar]
- DeCoursey TE, Cherny VV. Voltage-activated proton currents in membrane patches of rat alveolar epithelial cells. J Physiol. 1995;489:299–307. doi: 10.1113/jphysiol.1995.sp021051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey TE, Cherny VV. Temperature dependence of voltage-gated H+ currents in human neutrophils, rat alveolar epithelial cells, and mammalian phagocytes. J Gen Physiol. 1998;112:503–522. doi: 10.1085/jgp.112.4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey TE, Cherny VV, DeCoursey AG, Xu W, Thomas LL. Interactions between NADPH oxidase-related proton and electron currents in human eosinophils. J Physiol. 2001;535:767–781. doi: 10.1111/j.1469-7793.2001.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey TE, Cherny VV, Zhou W, Thomas LL. Simultaneous activation of NADPH oxidase-related proton and electron currents in human neutrophils. Proc Natl Acad Sci U S A. 2000;97:6885–6889. doi: 10.1073/pnas.100047297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey TE, Ligeti E. Regulation and termination of NADPH oxidase activity. Cell Mol Life Sci. 2005;62:2173–2193. doi: 10.1007/s00018-005-5177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey TE, Morgan D, Cherny VV. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 2003;422:531–534. doi: 10.1038/nature01523. [DOI] [PubMed] [Google Scholar]
- Demaurex N, Grinstein S, Jaconi M, Schlegel W, Lew DP, Krause KH. Proton currents in human granulocytes: regulation by membrane potential and intracellular pH. J Physiol. 1993;466:329–344. [PMC free article] [PubMed] [Google Scholar]
- Elinder F, Århem P. Metal ion effects on ion channel gating. Q Rev Biophys. 2004;36:373–427. doi: 10.1017/s0033583504003932. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fuentes N, Zhai J, Fiser A. ArchPRED: a template based loop structure prediction server. Nucleic Acids Res. 2006;34:W173–W176. doi: 10.1093/nar/gkl113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenhaeuser B, Hodgkin AL. The action of calcium on the electrical properties of squid axons. J Physiol. 1957;137:218–244. doi: 10.1113/jphysiol.1957.sp005808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C, Koch HP, Drum BM, Larsson HP. Strong cooperativity between subunits in voltage-gated proton channels. Nat Struct Mol Biol. 2010;17:51–56. doi: 10.1038/nsmb.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. 3rd edn. Sunderland, MA: Sinauer Associates, Inc.; 2001. [Google Scholar]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrigan FT, Cui J, Aldrich RW. Allosteric voltage gating of potassium channels I. Mslo ionic currents in the absence of Ca2+ J Gen Physiol. 1999;114:277–304. doi: 10.1085/jgp.114.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003a;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003b;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- Kapus A, Romanek R, Qu AY, Rotstein OD, Grinstein S. A pH-sensitive and voltage-dependent proton conductance in the plasma membrane of macrophages. J Gen Physiol. 1993;102:729–760. doi: 10.1085/jgp.102.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HP, Kurokawa T, Okochi Y, Sasaki M, Okamura Y, Larsson HP. Multimeric nature of voltage-gated proton channels. Proc Natl Acad Sci U S A. 2008;105:9111–9116. doi: 10.1073/pnas.0801553105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno M, Ando H, Morihata H, Sakai H, Mori H, Sawada M, Oiki S. Temperature dependence of proton permeation through a voltage-gated proton channel. J Gen Physiol. 2009;134:191–205. doi: 10.1085/jgp.200910213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Lee S-Y, Letts JA, MacKinnon R. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc Natl Acad Sci U S A. 2008;105:7692–7695. doi: 10.1073/pnas.0803277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-Y, Letts JA, Mackinnon R. Functional reconstitution of purified human Hv1 H+ channels. J Mol Biol. 2009;387:1055–1060. doi: 10.1016/j.jmb.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Zhao Q, Zhou Q, Unno H, Zhai Y, Sun F. The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1. J Biol Chem. 2010 doi: 10.1074/jbc.M109.040360. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Zhao Q, Zhou Q, Zhai Y. Expression, purification, crystallization and preliminary crystallographic study of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:279–281. doi: 10.1107/S1744309109003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Mahaut-Smith M. The effect of zinc on calcium and hydrogen ion currents in intact snail neurones. J Exp Biol. 1989;145:455–464. doi: 10.1242/jeb.145.1.455. [DOI] [PubMed] [Google Scholar]
- Morgan D, Cherny VV, Finnegan A, Bollinger J, Gelb MH, DeCoursey TE. Sustained activation of proton channels and NADPH oxidase in human eosinophils and murine granulocytes requires PKC but not cPLA2α activity. J Physiol. 2007;579:327–344. doi: 10.1113/jphysiol.2006.124248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, DeCoursey TE. Charge compensation during the phagocyte respiratory burst. Biochim Biophys Acta. 2006;1757:996–1011. doi: 10.1016/j.bbabio.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Musset B, Capasso M, Cherny VV, Morgan D, Bhamrah M, Dyer MJS, DeCoursey TE. Identification of Thr29 as a critical phosphorylation site that activates the human proton channel Hvcn1 in leukocytes. J Biol Chem. 2010;285:5117–5121. doi: 10.1074/jbc.C109.082727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset B, Cherny VV, Morgan D, Okamura Y, Ramsey IS, Clapham DE, DeCoursey TE. Detailed comparison of expressed and native voltage-gated proton channel currents. J Physiol. 2008;586:2477–2486. doi: 10.1113/jphysiol.2007.149427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak MM, Yarov-Yarovoy V, Agarwal G, Roux B, Barth P, Kohout S, Tombola F, Isacoff EY. Closing in on the resting state of the Shaker K+ channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Petheő GL, Demaurex N. Voltage- and NADPH-dependence of electron currents generated by the phagocytic NADPH oxidase. Biochem J. 2005;388:485–491. doi: 10.1042/BJ20041889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata S, Kurokawa T, Nørholm MHH, Takagi M, Okochi Y, von Heijne G, Okamura Y. Functionality of the voltage-gated proton channel truncated in S4. Proc Natl Acad Sci U S A. 2010;107:2313–2318. doi: 10.1073/pnas.0911868107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- Sigworth FJ. Voltage gating of ion channels. Q Rev Biophys. 1993;27:1–40. doi: 10.1017/s0033583500002894. [DOI] [PubMed] [Google Scholar]
- Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- Tombola F, Ulbrich MH, Isacoff EY. The voltage-gated proton channel HV1 has two pores, each controlled by one voltage sensor. Neuron. 2008;58:546–556. doi: 10.1016/j.neuron.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F, Ulbrich MH, Kohout SC, Isacoff EY. The opening of the two pores of the Hv1 voltage-gated proton channel is tuned by cooperativity. Nat Struct Mol Biol. 2010;17:44–50. doi: 10.1038/nsmb.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee BL, Auld DS. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]