SUMMARY
Mitochondria are highly mobile and dynamic organelles that continually fuse and divide. These processes allow mitochondria to exchange contents, including mitochondrial DNA (mtDNA). Here we examine the functions of mitochondrial fusion in differentiated skeletal muscle through conditional deletion of the mitofusins Mfn1 and Mfn2, mitochondrial GTPases essential for fusion. Loss of the mitofusins causes severe mitochondrial dysfunction, compensatory mitochondrial proliferation, and muscle atrophy. Mutant mice have severe mtDNA depletion in muscle that precedes physiological abnormalities. Moreover, the mitochondrial genomes of the mutant muscle rapidly accumulate point mutations and deletions. In a related experiment, we find that disruption of mitochondrial fusion strongly increases mitochondrial dysfunction and lethality in a mouse model with high levels of mtDNA mutations. With its dual function in safeguarding mtDNA integrity and preserving mtDNA function in the face of mutations, mitochondrial fusion is likely to be a protective factor in human disorders associated with mtDNA mutations.
INTRODUCTION
Mitochondrial fusion and fission have emerged as important processes that govern mitochondrial function from yeast to mammals (Detmer and Chan, 2007a; Suen et al., 2008; Okamoto and Shaw, 2005; Hoppins et al., 2007). In mammalian cells, three large GTPases are important for mitochondrial fusion, which requires the coordinated fusion of the outer and inner membranes. The mitofusins, Mfn1 and Mfn2, are located on the mitochondrial outer membrane and are involved in early steps in membrane fusion (Koshiba et al., 2004; Meeusen et al., 2004; Song et al., 2009). The dynamin-related protein OPA1 is associated with the inner membrane and is essential for inner membrane fusion (Song et al., 2009; Meeusen et al., 2006).
In addition to its well-recognized function of controlling mitochondrial morphology (Bleazard et al., 1999; Chen et al., 2003; Sesaki and Jensen, 1999), mitochondrial fusion clearly protects mitochondrial function (Detmer and Chan, 2007a). Mutations in Mfn2 and OPA1 (Alexander et al., 2000; Delettre et al., 2000; Zuchner et al., 2004) cause two neurodegenerative diseases--Charcot-Marie-Tooth type 2A and dominant optic atrophy, respectively. Neurodegeneration is also prominent in mice with a targeted mutation in Mfn2 (Chen et al., 2007). Cells that lack mitochondrial fusion have a severe defect in respiratory capacity (Chen et al., 2005). Their fragmented mitochondria become functionally divergent, resulting in heterogeneity in membrane potential and mitochondrial DNA (mtDNA) nucleoids, which encode components of the electron transport chain. These results have led to a model that mitochondrial fusion protects mitochondrial function by enabling content mixing (Detmer and Chan, 2007a; Chen et al., 2007).
Because of this content mixing, it has been hypothesized that mitochondrial fusion may be involved in the ability of human cells to tolerate high levels of pathogenic mtDNA (Nakada et al., 2009; Nakada et al., 2001). A distinguishing feature of mitochondrial genetics is that most mtDNA mutations are recessive and must accumulate to high proportions before an effect on oxidative phosphorylation is observed (DiMauro and Schon, 2003; Taylor and Turnbull, 2005). Depending on the mutation, heteroplasmic cells (containing both mutant and wildtype mtDNA) can accumulate up to 60–90% pathogenic mtDNA molecules without a noticeable decline in respiratory activity (Rossignol et al., 2003; Chomyn, 1998). In both cultured cells and mouse models, cells engineered to contain a mixture of wildtype and pathogenic mtDNA are protected from dysfunction until a high threshold of pathogenic mtDNA is breached (Nakada et al., 2009; Ono et al., 2001). These observations have led to the proposal that inter-mitochondrial mixing in heteroplasmic cells might underlie the functional complementation of pathogenic mtDNA genomes (Nakada et al., 2009; Nakada et al., 2001). However, the proposed role of mitochondrial fusion in functional complementation remains to be experimentally tested.
Other fundamental issues concerning mitochondrial fusion remain to be explored. Most studies on mammalian mitochondrial dynamics have been limited to cultured cells and neurons (Detmer and Chan, 2007a; Suen et al., 2008). These studies show that mitochondrial fusion is highly associated with mitochondrial transport along the cytoskeleton, because mitochondria fuse when they collide end-to-end or end-to-side. However, little is known about the role of mitochondrial dynamics in most tissues in vivo, including skeletal muscle fibers, which are highly metabolically active and are often affected in diseases of mitochondrial dysfunction. Elegant ultrastructural studies have documented that mitochondria in skeletal muscle are tightly packed and arranged in a stereotypic manner in relation to the sarcomeric unit (Ogata and Yamasaki, 1997). This unique organization and apparently limited mobility raises the issue of whether mitochondrial dynamics is important in this cell type.
To address these issues, we examined mice lacking mitofusin function in skeletal muscle and found that these mice develop a lethal mitochondrial myopathy. In addition, we found that mitochondrial fusion protects mtDNA function through several distinct mechanisms. It maintains mtDNA levels, preserves mtDNA fidelity, and enables cells to tolerate high levels of mtDNA mutations.
RESULTS
A role of mitofusins Mfn1 and Mfn2 in skeletal muscle
Given the unique arrangement of mitochondria in skeletal muscle, we examined whether mitochondrial fusion plays an important role in this cell type. We specifically disrupted the mitochondrial fusion genes Mfn1 and Mfn2 in skeletal muscle, using MLC-Cre transgenic mice (Bothe et al., 2000). The MLC1f promoter that drives Cre recombinase is well-characterized to express in differentiated fast-twitch muscle cells and not in myocytes (Lyons et al., 1990). Surprisingly, such double mutant mice (MLC-Cre/dm) are severely runted, reaching only 30–50% the weight of control littermates (Table 1) before dying by 6–8 weeks of age. In contrast, mice of other genotypic combinations (including Mfn1−/− Mfn2+/− and Mfn1+/− Mfn2−/−) survive and grow to normal size.
Table 1.
Physiological measurements on 7–8 week old MLC/dm mice
| Measurement | Double mutant | Control | P value |
|---|---|---|---|
| Weight (females) | 11±2.6 g, n=2 | 23±2.1 g, n=6 | <0.0001 |
| Weight (males) | 10±2.4 g, n=11 | 30±3.6 g, n=11 | <0.0001 |
| Body temperature | 35±1.1 °C, n=10 | 37±0.3 °C, n=20 | <0.0001 |
| Non-fasting blood glucose | 50±14 mg/dL, n=2 | 143±13 mg/dL, n=7 | <0.0001 |
| Fasting blood glucose | 26±2.8 mg/dL, n=2 | 78±12 mg/dL, n=10 | =0.0002 |
| Resting serum lactate | 5.7±1.7 mmol/L, n=7 | 3.9±1.2 mmol/L, n=9 | =0.02 |
| Post-exercise lactate | 9.5±2.3 mmol/L, n=5 | 4.7±0.65 mmol/L, n=5 | =0.002 |
Measurements are given as value±standard deviation. P values were calculated using the unpaired-t test. Controls are mixed genotype littermates of double mutant animals. The number of animals (n) for each measurement is indicated.
MLC-Cre/dm mice show severe physiological aberrations. They display low blood glucose levels under fasting and non-fasting conditions and reduced body temperatures (Table 1). Moreover, we found high levels of lactate in the serum of MLC-Cre/dm mice, most pronounced after an exercise regiment but also under resting conditions. These profound metabolic aberrations suggest a rapid turnover of glucose without adequate energy production, as would result from the predominant use of glycolysis instead of oxidative phosphorylation.
Mitochondrial defects in mitofusin-deficient muscle
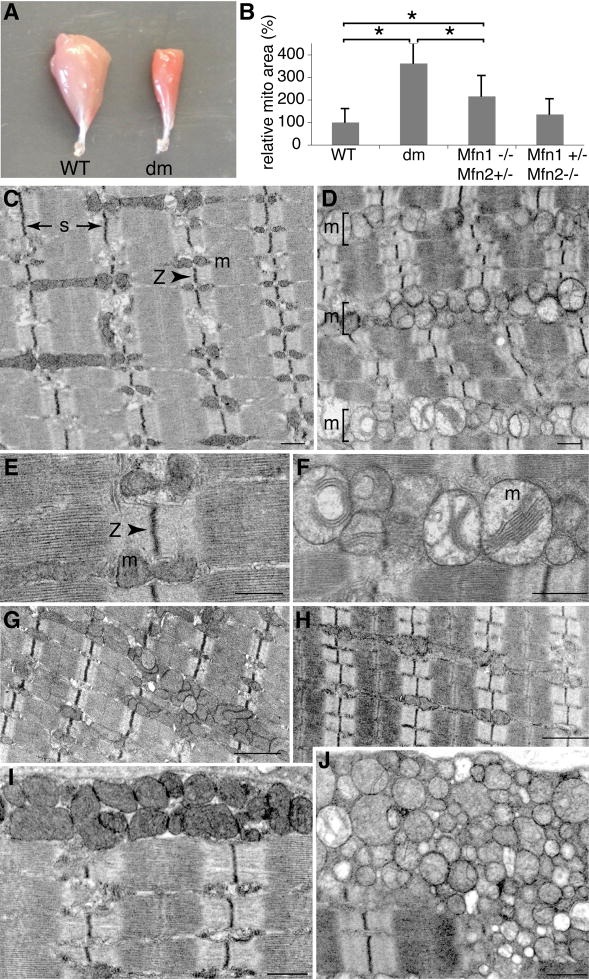
On gross examination of muscle, it is readily apparent that the MLC-Cre/dm mice have smaller muscles that are deeper red than control muscles (Figure 1A), suggesting an increase in mitochondrial mass. This interpretation is confirmed by electron microscopy of tibialis anterior (TA) muscle, which is composed of predominantly fast-twitch fibers. On longitudinal sections, wildtype 7 week old skeletal muscle displays tightly juxtaposed arrays of myofibrils that are extremely uniform and precisely aligned (Figure 1C). Consistent with previous ultrastructural studies of muscle fiber architecture (Ogata and Yamasaki, 1997), the mitochondria typically reside in pairs, positioned on either side of the Z-disc in each I band. In contrast, the mitochondria in 7 week MLC-Cre/dm muscle are fragmented into round spheres, as has been found in other cells lacking mitochondrial fusion (Chen et al., 2003; Chen et al., 2007; Chen and Chan, 2005). In addition, there is massive accumulation of mitochondria that are tightly packed throughout the space between myofibrils (Figure 1D). These aggregates of mitochondria fill the interfibrillar space and disrupt the alignment of myofibril arrays. We also observed proliferation of mitochondria in double mutant 4 week old muscle, although the increase was less extensive, and regions with normal mitochondrial content are preserved (Figure S1). Highly pronounced accumulations of mitochondria are likewise present in the subsarcolemmal space (Figure 1I, J), similar to that observed in ragged-red fibers of mitochondrial myopathies caused by mtDNA mutations (DiMauro and Schon, 2003). Furthermore, both interfibrillar and subsarcolemmal mitochondria in mutant muscle show classic ultrastructural signs of dysfunction: they are swollen and contain few of the densely packed cristae membranes characteristic of wildtype mitochondria (Figure 1E, F). Quantification of the micrographs indicates that in MLC-Cre/dm muscle, the mitochondria occupy several fold more area than in wildtype muscle (Figure 1B). In contrast, muscles carrying just one Mfn allele show much milder histological defects (Figure 1B, G, H), with occasional patches of abnormal mitochondria particularly evident in Mfn1−/− Mfn2+/− muscle (Figure 1G).
Figure 1. Mitochondrial defects in 7 week old MLC-Cre/dm muscle.
(A) Whole-mount gastrocnemius muscles. The double mutant muscle (right) is smaller and deeper red. (B) Quantification of mitochondrial area in muscle of the indicated genotypes. The area occupied by mitochondria was measured from at least 10 representative EM fields and normalized to the area occupied by mitochondria in wildtype muscle. Asterisks (P<0.0003, unpaired t-test) indicate significant changes. (C–J) Electron microscopy of tibialis anterior (TA) longitudinal sections. Labels: s, length of sarcomere; Z, Z-disc; m, mitochondria. (C, E, and I) Wildtype. (D, F, and J) Mfn double mutant. (G) Mfn1−/−, Mfn2+/−. (H) Mfn1+/−, Mfn2−/−. (I and J) Subsarcolemmal mitochondria. Scale bars: 0.5 μm in C–F, I and J; 1μm in G and H. See also Figure S1.
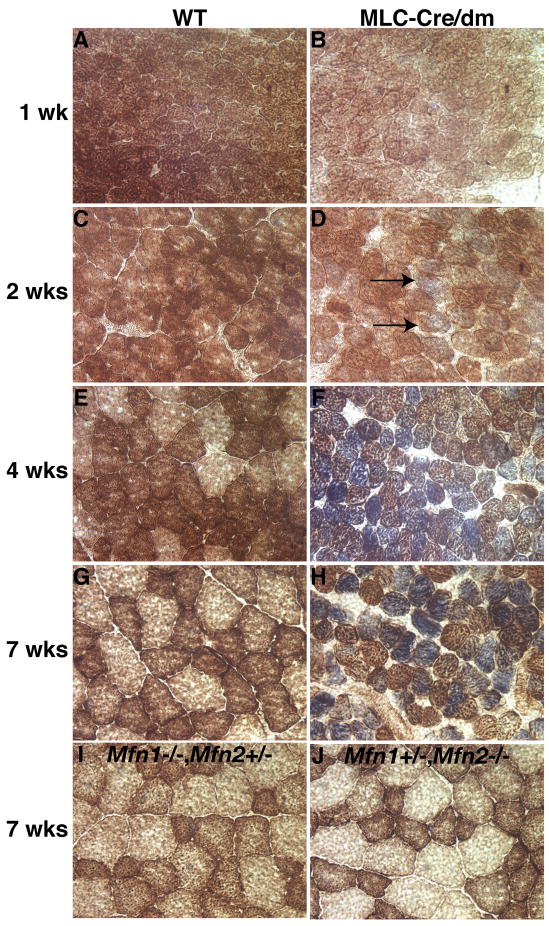
Given the physiological and ultrastructural evidence for mitochondrial dysfunction, we used histochemical staining for cytochrome c oxidase (COX, complex IV, brown stain) and succinate dehydrogenase (SDH, complex II, blue stain) activity to directly assess the function of respiratory complexes. In 1 and 2 week old wildtype mice, the muscle sections show uniform COX staining of all the muscle fibers (Figure 2A, C). Over the next several weeks, this gradually matures into the brown checkerboard appearance of adult muscle, due to the differentiation of specific muscle fiber types with varying mitochondrial activity (Figure 2E, G). In MLC-Cre/dm muscle, the staining pattern is normal at week 1, but a mildly abnormal pattern emerges at week 2 with the appearance of some fibers that stain faintly blue, indicating increased SDH and low COX activity (Figure 2D, arrows). This abnormal histological pattern becomes highly pronounced at 4 and 7 weeks, at which time many deeply blue fibers are apparent (Figure 2F, H). In human mitochondrial encephalomyopathies, this characteristic blue histological pattern is often found in cases of respiratory dysfunction due to mtDNA defects (DiMauro and Schon, 2003; Taylor and Turnbull, 2005), because SDH activity is encoded solely by the nuclear genome, whereas COX activity is dependent on the mitochondrial genome. The increased SDH staining is consistent with the increased mitochondrial accumulation observed by EM (Figure 1B, D, J). In addition, double mutant muscle fibers have notably smaller diameters at 4 and 7 weeks. This small fiber size accounts for the overall decreased muscle size, because no reduction in fiber number was found (data not shown). In contrast, Mfn1−/− Mfn2+/− and Mfn1+/− Mfn2−/− muscles demonstrate normal COX/SDH staining patterns, normal fiber size, and no systemic indicators of respiratory deficiency (Figure 2I, J). Therefore, the severe defects found in MLC-Cre/dm mice are not due to a specific function of Mfn2 versus Mfn1.
Figure 2. Temporal analysis of respiratory deficiency in MLC-Cre/dm muscle.
Transverse sections of TA muscle were stained histochemically for COX (brown) and SDH (blue) activity. (A, C, E, and G) Wildtype muscle. (B, D, F, and H) Double mutant muscle. Blue staining is indicative of mitochondrial dysfunction. Note the initial appearance of faintly blue fibers in D (arrows) and deeply blue fibers in F and H. The ages of the samples are indicated. (I and J) Genotypes and age of the muscles are indicated. 400× magnification. See also Figure S2.
Loss of mitofusins also resulted in a marked shift in the composition of skeletal muscle fiber types. MLC-Cre/dm tibialis anterior muscles show an increase in the proportion of fibers positive for myosin heavy chain 2A, a marker for oxidative type IIA fibers, and a corresponding decrease in the proportion of fast-twitch type IIB fibers (Figure S2). These results are consistent with reports that mitochondrial biogenesis can drive muscle fiber type switching (Arany et al., 2007; Lin et al., 2002).
Mitofusins are important for mtDNA stability and fidelity
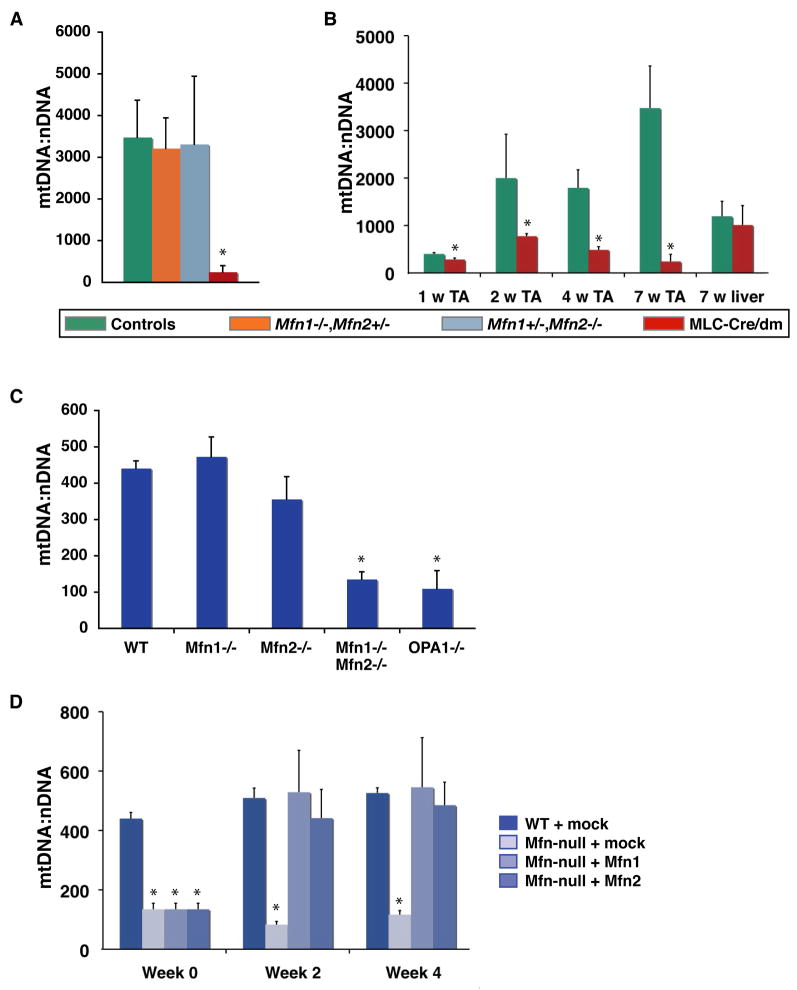
Given the histological and physiological features of mitochondrial myopathy in MLC-Cre/dm mice, we investigated the relationship between mitochondrial fusion and mtDNA copy number. Remarkably, we found that 7–8 week old muscle from double mutant mice contains only ~250 copies of mtDNA per nuclear genome, whereas muscle from age-matched controls contains ~3500 copies of mtDNA per nuclear genome (Figure 3A). Mice with single alleles of either Mfn1 or Mfn2 show normal levels of mtDNA at 7–8 weeks (Figure 3A) and later (Figure S3A).
Figure 3. Quantitative analysis of mtDNA from muscle and MEFs.
(A) Analysis of mtDNA copy number per nuclear genome in 7–8 week TA muscle. Controls include wildtype, Mfn1+/−, and Mfn2+/− littermates of the double mutants. P=0.0002. (B) Temporal analysis of mtDNA depletion in double mutant TA muscle. Ages of the samples are indicated. P<0.008. Legend indicates genotypes for (A) and (B). (C) Mitochondrial copy number in MEFs. Genotypes are indicated. P<0.0001. (D) Restoration of mtDNA levels in Mfn double null cells. MtDNA levels were measured in mutant cells infected with retrovirus expressing mito-DsRed (mock), Mfn1 or Mfn2. Mitochondria were analyzed 2 and 4 weeks after infection. P<0.0001. In all panels, asterisks denote statistically significant changes from control, and error bars indicate standard deviations from 3–5 animals or experiments. To obtain the mtDNA level for a single DNA sample, quantitative PCR was performed in quadruplicate. P-values were obtained from unpaired t-tests. See also Figure S3.
To determine the temporal relationship between the mtDNA depletion and the muscle fiber defects, we analyzed mtDNA levels in younger MLC-Cre/dm muscle (Figure 3B). Even at the earliest time point, 1 week of age, we measured a significantly lower level of mtDNA in MLC-Cre/dm muscle compared to control littermates. The discrepancy in mtDNA levels between MLC-Cre/dm and wildtype muscle enlarges over the next several weeks, due to the rapid increase in mtDNA content in wildtype muscle. In contrast, mtDNA in mutant muscle fails to expand during postnatal development. Because this mtDNA depletion becomes evident prior to histological evidence for gross mitochondrial dysfunction (Figure 2), it is likely that reduced mtDNA levels in the mutants underlie, at least in part, the muscle pathology observed later.
We made similar observations of mtDNA loss in mouse embryonic fibroblasts (MEFs) that lack both Mfn1 and Mfn2 (Figure 3C), consistent with the impaired respiratory capacity of these cells (Chen et al., 2005). In contrast, MEFs with only a partial defect in fusion (Mfn1−/− or Mfn2−/− cells) harbor normal levels of mtDNA (Figure 3C) and respire normally (Chen et al., 2005). Importantly, overexpression of either Mfn1 or Mfn2 in Mfn-double null MEFs rapidly restores mtDNA to wildtype levels (Figure 3D), again emphasizing that the shared function of mitochondrial fusion by Mfn1 and Mfn2 is responsible for the defects. Moreover, we found that OPA1-null cells, which are deficient for fusion of the inner mitochondrial membrane, but proficient for outer membrane fusion (Song et al., 2009), also display mtDNA depletion (Figure 3C), suggesting that lack of mixing of the matrix compartment is ultimately responsible for loss of mtDNA in fusion-deficient cells.
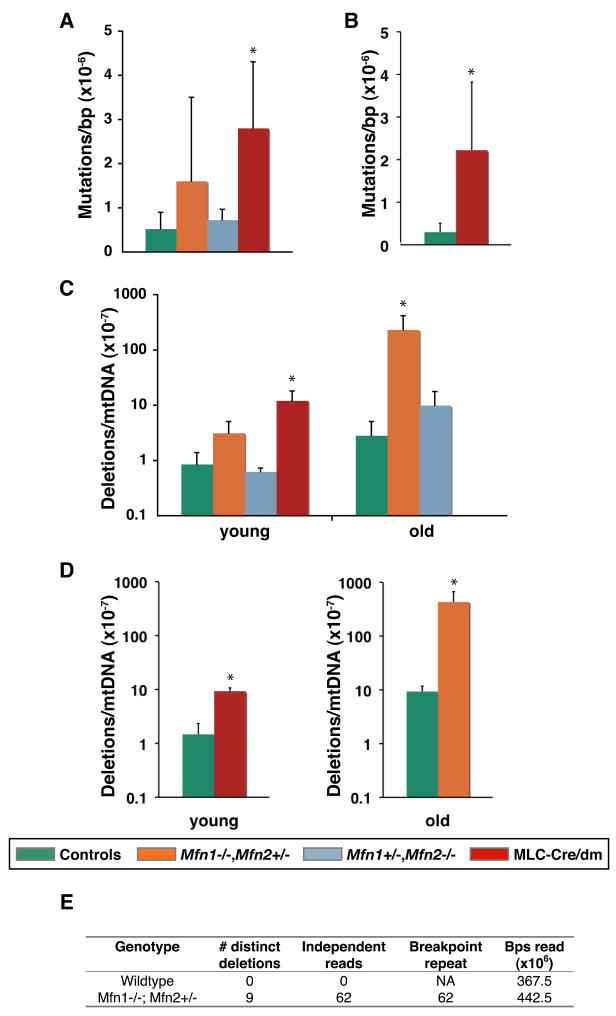
To further explore the connection between mitochondrial fusion and mtDNA, we interrogated the fidelity of the mitochondrial genome by screening mtDNA for point mutations and deletions. We found that muscle from 7–8 week old MLC-Cre/dm mice harbors a 5-fold increase in mtDNA point mutations (Figure 4A) and a 14-fold increase in deletions (Figure 4C) compared to wildtype animals. We confirmed both of these measurements by interrogating additional loci in the mitochondrial genome (Figure 4B,D). Most deletions occurred between short homologous repeats in both double mutant and wildtype tissue, whereas there was a shift in the spectrum of single base pair substitutions in double mutant mice (Figure S4B, C).
Figure 4. Quantitative analysis of mtDNA mutations.
(A–D) Quantification of mtDNA mutations in TA muscle by the random mutation capture assay. Legend for genotypes applies to panels A–D. Error bars indicate standard deviations from 3–5 animals. Asterisks denote statistically significant changes from control. P-values were obtained from unpaired t-tests. (A, B) Quantification of point mutation frequency per basepair at two independent sites. P=0.03 in (A); P=0.05 in (B). (C, D) Quantification of deletion frequency per mtDNA genome at two independent sites. In (C), P=0.01 for young muscle (7–8 weeks), and P=0.02 for old muscle (8–13 months). In (D), P=0.0003 for young muscle, and P=0.04 for old muscle. In total, approximately 250 million basepairs were screened for point mutations, and 700 million genomes for deletion. (E) Tabulation of mtDNA deletion events from Solexa sequencing of mtDNA from 10-month skeletal muscle of the indicated genotypes. “Breakpoint repeat” indicates the number of deletions involving direct repeats of 6–14 basepairs. See also Figure S4 and Table S1.
Because mitochondrial mutations accumulate with age (Vermulst et al., 2007; Vermulst et al., 2008a), we also examined mtDNA from older animals carrying single alleles of Mfn1 or Mfn2. Remarkably, we found that muscle tissue from 8–13 month old Mfn1−/− Mfn2+/− mice exhibits an ~80-fold increase in mtDNA deletions (2.3×10−5/genome) compared to control muscle (2.8×10−7/genome) (Figure 4C). This increase was confirmed by analysis at an independent site (Figure 4D). However, we found no significant change in the frequency of point mutations or mtDNA copy number of Mfn1−/− Mfn2+/− or Mfn1+/− Mfn2−/− muscle (Figure S3A, S4A), suggesting that the mechanism underlying the generation of mtDNA deletions is more sensitive to a reduction in mitochondrial fusion.
To further confirm this finding of increased mtDNA deletions in a genome-wide, unbiased manner, we analyzed mtDNA from 10-month wildtype and Mfn1−/− Mfn2+/− muscle with Solexa sequencing. For each sample, we obtained 75-nucleotide sequence reads that, in aggregate, cover 367.5 and 442.5 million basepairs of mtDNA, respectively. Both ends of each sequence read were mapped onto the mtDNA genome, and sequences with ends that mapped to different locations were selected as candidates for deletions. To minimize the chances of artifactual deletions, we only considered deletions that were identified by 4 or more independent sequence reads (Figure 4E). In wildtype muscle, we found no such deletions. In contrast, the Mfn1−/− Mfn2+/− sample contained 9 distinct deletions that were identified by 62 independent sequence reads, and therefore represent 62 deletion events. In each case, the breakpoint junction occurred between repeats of 6–14 nucleotides (Table S1), in agreement with our earlier sequence analysis of deletions breakpoints (Figure S4C). These 9 deletions may comprise particularly vulnerable regions in the mtDNA genome of mitofusin-deficient mice.
Previous results indicate that fusion-defective cells have mitochondrial populations that are heterogeneous for membrane potential and the presence of mtDNA nucleoids (Chen et al., 2007; Chen et al., 2005). To extend this finding, we examined whether Mfn-double null cells have a generalized heterogeneity in protein content. An unbalanced mitochondrial proteome could lead to mtDNA instability, because precise stoichiometries of proteins involved in mtDNA metabolism are likely to be important for mtDNA stability and fidelity. Therefore, we quantitatively analyzed wildtype and Mfn-double null MEFs immunostained for cytochrome c and hsp60, two abundant mitochondrial proteins (Figure S3B, C). In the mutant MEFs, the signal intensity ratios for cytochrome c versus hsp60 showed substantially greater variance within the mitochondrial population than in wildtype cells (Figure S3D, E). This increased variance was highly significant and resulted in lower correlation coefficients for the two fluorophores in double mutant MEFs (P=0.0001, n=10). Similar heterogeneity was found upon expression of three fluorescent proteins to the mitochondrial matrix (data not shown). These observations imply that mitochondrial proteomes may become unbalanced in the absence of content mixing.
Synthetic lethality between an error-prone mtDNA polymerase and loss of Mfn1
We expanded our study to examine a separate issue concerning the relationship between mitochondrial dynamics and mtDNA mutations. It has been proposed that mitochondrial fusion might underlie the remarkable ability of mammalian cells to tolerate high loads of pathogenic mtDNA (Nakada et al., 2009; Nakada et al., 2001). To test this idea, we generated Mfn1−/− mice carrying homozygous alleles of PolgAD257A. The PolgAD257A knock-in allele encodes a mitochondrial DNA polymerase with a deficient proofreading domain, causing cells to accumulate mtDNA mutations at an accelerated rate (Kujoth et al., 2005; Trifunovic et al., 2004). Remarkably, whereas single mutants for either Mfn1 or PolgA survive well into adulthood, the combination of the two mutations leads to a synthetic neonatal lethality (Table 2). Moreover, PolgAD257A heterozygous mice, which also have an increased mutation burden (Vermulst et al., 2007), show reduced viability in the absence of Mfn1 as well. The synthetic lethality of PolgAD257A/D257A Mfn1−/− mice is not due to an increased mutation rate, because we detected no significant difference in mutation frequency between PolgAD257A/D257A and PolgAD257A/D257A Mfn1−/− embryos (Figure S5). It would be interesting to test for a similar synthetic interaction between PolgAD257A and deletion of Mfn2. However, this interaction is more difficult to assess because loss of Mfn2 causes a severe neurodegenerative disorder that leads to early lethality (Chen et al., 2007).
Table 2.
Survival of mice carrying the PolgAD257A allele in an Mfn1 deficient background.
| PolgA Genotype | Mfn1 Genotype | Actual Number of Mice | Actual Frequency (%) | Expected Frequency (%) |
|---|---|---|---|---|
| +/+ | +/+ | 48 | 7 | 6.25 |
| +/+ | +/− | 81 | 12 | 12.5 |
| +/+ | −/− | 31 | 5 | 6.25 |
| D257A/+ | +/+ | 99 | 15 | 12.5 |
| D257A/+ | +/− | 207 | 32 | 25 |
| D257A/+* | −/− | 51 | 8 | 12.5 |
| D257A/D257A | +/+ | 34 | 5 | 6.25 |
| D257A/D257A | +/− | 102 | 16 | 12.5 |
| D257A/D257A** | −/− | 1 | 0 | 6.25 |
Mice of the indicated genotypes are tabulated. A Chi-squared test was used to compare observed and expected populations.
P=0.0003,
P<0.0001.
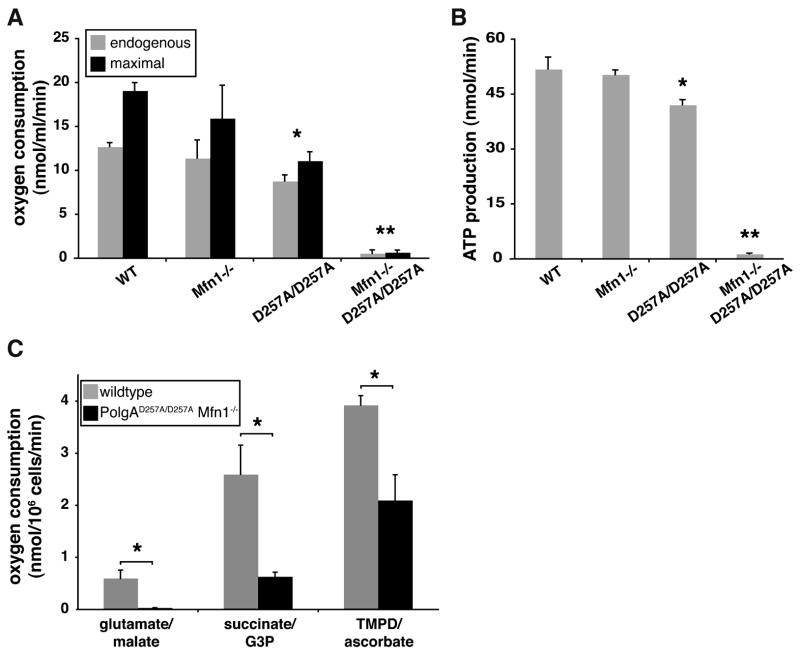
To examine this synergism at a cellular level, we established MEFs and measured their respiratory capacity and ATP production. Oxygen polarography demonstrated a profound decrease in the endogenous (4.5% of wildtype) and maximum rates (3.7% of wildtype) of oxygen consumption in PolgAD257A/D257A Mfn1−/− cells compared to controls (Figure 5A). These oxygen consumption rates were tightly correlated with kinetic measurements of ATP production (Figure 5B), with PolgAD257A/D257A Mfn1−/− cells producing only 2.7% the amount of ATP present in wildtype cells. To determine whether specific segments of the respiratory chain are affected, we measured substrate-driven oxygen consumption rates. The most profound defect was found in respiratory complex I (20-fold reduction), with less severe reductions in Complexes III (4-fold) and IV (2-fold) (Figure 5C). The severe reduction in Complex I is likely to be largely responsible for the overall respiratory decline, because the majority of the respiration in these cells occurs via Complex I and is sensitive to the Complex I inhibitor rotenone. The severity of the Complex I defect in PolgAD257A/D257A Mfn1−/− cells may reflect the fact that 7 mtDNA-encoded subunits are present in Complex I, far more than for any of the other respiratory complexes. Complex I is therefore more likely than the other complexes to be inactivated by an mtDNA mutation due to error-prone PolgAD257A function. These results underscore the importance of an intermixing mitochondrial network to ameliorate the damaging effects of mtDNA mutations.
Figure 5. Oxygen polarography and ATP production in MEFs.
(A) Respiration rates of MEFs of the indicated genotypes. Endogenous respiration is the oxygen consumption rate of untreated cells, and maximal respiration is the oxygen consumption rate after addition of the uncoupler dinitrophenol. Standard deviations from 3 experiments are shown. (B) ATP production via Complex I. ATP production was measured using a luciferase-based assay on 1×106 permeabilized cells. Standard deviations from 3 experiments are indicated. (C) Substrate-driven respiration rates of MEFs of the indicated genotypes. Oxygen consumption rates driven by the substrates glutamate/malate (Complex I), succinate/G3P (Complex III), and TMPD/ascorbate (Complex IV) were measured. Standard deviations from 2 experiments are shown. In all panels, single asterisks (P<0.05) and double asterisks (P<0.01) represent significant changes compared to wildtype. See also Figures S5.
DISCUSSION
Our studies provide insight into the in vivo functions of mitochondrial fusion in mammals. Mitochondrial fusion plays an essential role in skeletal muscle fibers, in spite of its compact subcellular architecture and precise placement of mitochondria. In the absence of mitochondrial fusion, a pathological profile emerges that bears striking similarities to human mitochondrial myopathies, particularly those associated with mtDNA depletion syndromes (Copeland, 2008).
Our studies indicate that mitochondrial fusion safeguards mtDNA function through three distinct pathways. First, mitochondrial dynamics maintains cellular health by stabilizing mtDNA copy number. In budding yeast, loss of the mitofusin Fzo1 results in rapid and complete loss of mtDNA (Hermann et al., 1998; Rapaport et al., 1998), resulting in absence of respiratory function. In contrast, deletion of mammalian mitofusins or OPA1 results in loss of mtDNA nucleoids from only a subpopulation of mitochondria (Chen et al., 2007). Here, we show that this unequal distribution of nucleoids is caused, at least in part, by long-term mtDNA depletion. The dramatic reduction of mtDNA levels to 7% of wildtype levels at 2 months is likely a key cause for the mitochondrial myopathy observed in mitofusin-deficient mice. Consistent with a causative role for mtDNA depletion, the reduction in mtDNA levels is found prior to overt histological evidence for muscle fiber dysfunction. Moreover, whereas wildtype muscle shows a rapid post-natal increase in mtDNA from weeks 1 to 7, mitofusin-deficient muscle fail to increase mtDNA levels (Figure 3), suggesting a possible defect in mtDNA replication. For comparison, MLC-Cre/Tfam mice--which lack a key DNA-binding protein involved in mtDNA packaging, transcription, and replication--retain greater than 30% mtDNA levels at 4 months of age (Wredenberg et al., 2002). The MLC-Cre/dm mice therefore provide an animal model for human mtDNA depletion syndromes associated with mitochondrial myopathy (Copeland, 2008). We do not, however, think that mtDNA depletion solely accounts for the mitochondrial dysfunction in mutant muscle; it is likely that the inability to mix mitochondrial contents further undermines mitochondrial function.
Second, we discovered that loss of mitochondrial fusion in skeletal muscle leads to an increase in mtDNA point mutations and deletions. In double mutant mice, the absolute levels of mutations are too low to account for the severe physiological defects. For comparison, double mutant mice have a several-fold increase in mtDNA point mutations, whereas PolgAD257A/D257A mutator mice have a 2–3 orders-of-magnitude increase (Vermulst et al., 2007). In old Mfn1 mutant mice, mtDNA deletion levels are comparable to that of PolgAD257A/D257A mice (Vermulst et al., 2008a). Although their relationship to the physiological phenotypes is unclear, these substantial increases in point mutations and deletions demonstrate the importance of mitochondrial fusion for mtDNA fidelity. It is interesting to note that some cases of dominant optic atrophy caused by dysfunction of the mitochondrial fusion gene OPA1 are associated with respiration-deficient muscle fibers and accumulation of mtDNA deletions (Amati-Bonneau et al., 2008; Hudson et al., 2008).
This accumulation of mtDNA mutations could be driven by multiple mechanisms. In principle, the accumulation of mtDNA mutations can be due to an increase in mtDNA damage, a failure to repair damaged mtDNA, or perhaps a failure to clear mitochondria with damaged mtDNA. Our finding that Mfn-double null cells have great protein heterogeneity, from one mitochondrion to another, provides a plausible mechanism that may contribute to each of these processes. If protein stoichiometries are improperly balanced, protein complexes critical for mtDNA replication, maintenance, repair, and clearance may operate inefficiently or with lower fidelity. We anticipate that additional mechanisms leading to mtDNA instability will be uncovered with future studies.
The demonstration of a link between mitochondrial fusion and mtDNA fidelity raises the issue of whether mitochondrial fusion plays a protective role in conditions associated with mtDNA mutations. For instance, age-related neurodegeneration and muscle atrophy are closely associated with mtDNA mutations (Chomyn and Attardi, 2003; Krishnan et al., 2007). It will be interesting to examine whether mitochondrial fusion in humans is a modifying factor that affects the rate at which mtDNA mutations occur.
Finally, our finding that loss of Mfn1 is incompatible with an error-prone mtDNA polymerase suggests that mitochondrial fusion can dramatically dampen the deleterious effects of pre-existing mtDNA mutations to preserve respiratory function. The synthetic lethality of mice containing PolgAD257A/D257A and Mfn1−/− mutations indicates that a critical cell type has been compromised. Previous experiments show that Mfn1 and Mfn2 play largely redundant roles in mitochondrial fusion, and it is the expression patterns of the two proteins that often dictate which tissues are selectively affected by the deletion of either gene (Chen et al., 2003; Chen et al., 2007; Chen et al., 2005). Therefore, the affected cells in PolgAD257A/D257A Mfn1−/− mice may express Mfn1 more highly than Mfn2. Alternatively, they may be exquisitely sensitive to changes in mitochondrial function and may be affected by deletion of either mitofusin.
Our findings may be applicable to the pathogenesis of mitochondrial encephalomyopathies caused by mtDNA point mutations and deletions. Such diseases typically show a “threshold effect”, in that clinical signs manifest only when the level of pathogenic mtDNA in a particular cell lineage breaches a critical level (DiMauro and Schon, 2003; Rossignol et al., 2003). From studies of patient samples and experimental cybrids, the threshold level for mtDNA deletions is around 60%, whereas the threshold for point mutations can be as high as 90% (Rossignol et al., 2003; Chomyn, 1998; Chomyn et al., 1992). It has been proposed that protection in the face of mtDNA mutations might result from the ability of mitochondrial fusion, through content mixing, to allow mtDNA genomes with distinct mutations to complement each other (Nakada et al., 2009; Nakada et al., 2001). Such complementation would be possible even if genomes within a single mitochondrion do not physically interact (Gilkerson et al., 2008). Our results provide strong experimental support for this hypothesis and suggest that mitochondrial fusion may ameliorate the clinical severity of inherited mtDNA encephalomyopathies. In future work, it will be important to address whether mitochondrial fusion plays an analogous protective role against age-associated mtDNA deletions, which similarly cause cellular dysfunction only upon clonal expansion to high levels (Bender et al., 2006; Kraytsberg et al., 2006; Wanagat et al., 2001).
EXPERIMENTAL PROCEDURES
Mouse Breeding and Physiological Experiments
MLC-Cre/dm mice and littermate controls were bred by crossing MLC-Cre,Mfn1+/−,Mfn2+/− mice to Mfn1loxP/loxP,Mfn2loxP/loxP mice. PolgA,Mfn1 animals were generated by crossing PolgAD257A/+,Meox2-Cre,Mfn1+/− mice to PolgAD257A/+,Mfn1loxP/loxP mice. The Mfn1 mutant (Chen et al., 2003; Chen et al., 2007), Mfn2 mutant (Chen et al., 2003; Chen et al., 2007), PolgAD257A mutant (Kujoth et al., 2005), MLC-Cre (Bothe et al., 2000), and Meox2-Cre mice (Tallquist and Soriano, 2000) have been described previously.
Physiological measurements were performed on 7–8 week old mice. Body temperature was measured with a rectal probe, using the Thermalert Monitoring Thermometer, TH-5 (Braintree Scientific). Blood glucose levels were measured using one drop of blood from the tail vein with the OneTouch Ultra2 Blood Glucose Monitoring System (LifeScan). To obtain lactate measurements, eye bleeds were performed on mice anesthetized with avertin, and the blood was analyzed in CG4+ cartridges with the iSTAT system (Abbott). Mice were exercised by swimming for 20 min at ~37°C before being anesthetized. All experiments were approved by the Institutional Animal Care and Use Committee at Caltech.
Histological and EM Analysis
TA muscle was used for histological analysis, since this muscle contains a high percentage of fast-twitch fibers that express MLC-Cre. For COX/SDH staining, freshly dissected muscle was embedded in Optimal Cutting Temperature (OCT) compound (Tissue-Tek) and frozen in liquid nitrogen. Slides were stained for COX activity, washed in water, stained for SDH activity, washed, and mounted in GelMount (Biomeda). Staining protocols were obtained from the Washington University Neuromuscular Disease Center web site (http://www.neuro.wustl.edu/neuromuscular/index.html).
For EM, mice were perfused with 3% paraformaldehyde, 1.5% glutaraldehyde, 100 mM cacodylate (pH7.4), and 2.5% sucrose. The TA was immobilized in an outstretched position by tying onto a toothpick splint prior to excision. The splint-attached muscle was postfixed for 1 hour and then processed and imaged as described previously (Chen et al., 2007).
For analysis of muscle fiber types, fresh-frozen TA muscle was transversely cryosectioned and immunostained with mouse IgG1 anti-MHC2A (ATCC HB-277) or mouse IgM anti-MHC2B (ATCC HB-283). Cy3-sheep anti-mouse (Jackson ImmunoResearch) or Alexa Fluor 555-goat anti-mouse IgM (Invitrogen) were used as secondary antibodies.
Cell Culture and Respiration Measurements
Previously established MEFs were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with either 10% fetal bovine serum (Mfn-double null and OPA1-null cells) or 10% bovine calf serum (wildtype, Mfn1−/−, and Mfn2−/− cells). MEFs for polarography were derived from e12.5 embryos, infected with a retrovirus expressing SV40 large T antigen to promote immortalization, and maintained in DMEM, 10% fetal bovine serum, 1 mM pyruvate, and 50 μg/ml uridine.
Oxygen consumption was measured in intact cells using a Clark oxygen electrode (Oxygraph, Hansatech Instruments), as described previously (Chen et al., 2005; Villani and Attardi, 2001). Substrate-driven oxygen consumption rates were measured essentially as described (Duan et al., 2003) with the following modifications. The first wash in Respiration buffer I was omitted, succinate and glycerol-3-phosphate were used to drive respiration through complex III, and N,N,N′,N′-tetramethyl-p-phenyldiamine dihydrochloride (TMPD)-driven respiration was measured in Respiration buffer I instead of Respiration buffer II. The substrate and inhibitor concentrations were as follows: 5 mM glutamate, 5 mM malate, 200 nM rotenone, 5 mM succinate, 5 mM glycerol-3-phosphate, 12 nM antimycin, 400 μM TMPD, and 10 mM ascorbate.
For measurement of ATP production, cells were grown to 80% confluency, harvested, permeabilized with 50 μg/ml digitonin for 1 minute, and resuspended in reaction buffer (150 mM KCl, 25 mM Tris-HCl, 0.4 mM EDTA, 0.1% BSA, 10 mM potassium phosphate, 0.5 mM MgCl2 and 0.15 mM P1,P5-di(adenosine) pentaphosphate). After addition of 0.25 mM Tris-acetate, 40 nM luciferin and 1 μg/ml luciferase, luciferase activity was measured in a Tecan Infinite M200 luminescence meter with 1×106 cells per measurement, in the presence of 1 mM malate and 1 mM pyruvate, with and without 2 μg/ml rotenone. The ATP measurements in the presence of rotenone were subtracted to give ATP production via Complex I.
Expression of Mfn1 or Mfn2 in MEFs was done via retroviral transduction as described previously (Chen et al., 2003). Both Mfn constructs overexpress greater than 10-fold over endogeneous Mfn levels, and exclusive mitochondrial localization was found, consistent with previous studies (Chen et al., 2003; Chen et al., 2005; Detmer and Chan, 2007b).
DNA Isolation
To isolate mitochondrial DNA, cells or tissue were first homogenized and a mitochondrial fraction was isolated according to previously published protocols (Frezza et al., 2007). Mitochondria were then lysed in the presence of 0.5% SDS and 0.2 mg/ml proteinase K in 10mM Tris-HCl, 0.15M NaCl and 0.005M EDTA (Vermulst et al., 2008b). MtDNA was then purified by phenol/chloroform extraction and ethanol precipitation. Total DNA was isolated using standard protocols.
MtDNA Quantitation
To quantify the amount mtDNA present per nuclear genome, we used the following primers:
mtDNA forward primer, CCTATCACCCTTGCCATCAT;
mtDNA reverse primer, GAGGCTGTTGCTTGTGTGAC.
To quantify nuclear DNA, we used a primer set that detects the Pecam gene on chromosome 6:
nuclear DNA forward primer, ATGGAAAGCCTGCCATCATG;
nuclear DNA reverse primer, TCCTTGTTGTTCAGCATCAC.
Quantification of relative copy number differences was carried out using both analysis of the difference in threshold amplification between mtDNA and nuclear DNA (delta delta C(t) method), and analysis with a standard curve of a reference template. Both methods provided identical results.
MtDNA Mutation Assays
The random mutation capture assays were performed as previously described (Vermulst et al., 2008b). Briefly, mtDNA was digested with TaqI for 5 hours, with the addition of 100 units of TaqI every hour. MtDNA was then diluted in a 96 well format and probed with primers flanking the TaqI restriction site in order to detect mtDNA genomes that contain a mutation in the TaqI restriction site. A second pair of primers was used to determine the amount of mtDNA genomes that was interrogated. PCR steps were as follows: Step 1, 37°C for 10 minutes; Step 2, 95°C for 10 minutes; Step 3, 95°C for 30 seconds; Step 4, 58/60°C for 1 minute; Step 5, 72°C for 1.5 minutes; Step 6, go to step three 44 times; Step 7, 72°C for 4 minutes; Step 8, melting curve from 65°C to 95°C; Step 9, hold at 4°C indefinitely. PCR reactions were carried out in 25 μl reactions using the Brilliant SYBR-green I master mix (Stratagene), containing 10 pM of forward and reverse primers and 1 unit of uracil DNA glycosylase. The following primers were used for DNA amplification:
MtDNA control primer forward, CCTATCACCCTTGCCATCAT;
MtDNA control primer reverse, GAGGCTGTTGCTTGTGTGAC;
MtDNA primer flanking Taq634 forward, ACTCAAAGGACTTGGCGGTA;
MtDNA primer flanking Taq634 reverse, AGCCCATTTCTTCCCATTTC;
MtDNA primer flanking Taq7667 forward, AATTTCATCTGAAGACGTCCT C;
MtDNA primer flanking Taq7667 reverse, AACGCTCTTAGCTTCATAGTGA;
MtDNA deletion forward site 1, CAAGGCCACCACACTCCTAT;
MtDNA deletion reverse site 1, GCTTCCGAATGCTAGGCGTT;
MtDNA deletion forward site 2, ACTGACTTCCAATTAGTAGATTCTG;
MtDNA deletion reverse site 2, GAGAGATTTTATGGGTGTAATGC.
All putative mutations were verified by sequencing and TaqI digestion.
Solexa sequencing was performed at the Caltech Genomics Facility. Libraries of mtDNA were constructed according to the manufacturer’s instructions, and paired-end sequence reads of 75 nucleotides were obtained. To identify candidate deletions, each sequence read was trimmed to 70 nucleotides, and the 15 nucleotides at each end were mapped using the program Bowtie to the mouse mtDNA genome. Sequence reads with ends mapping to separate regions of the mtDNA genome were retrieved as candidate deletions. With this algorithm, deletions occurring within the central 40 nucleotide region of a sequence read could be identified. The candidate deletions were then manually verified. To minimize the false-positive rate, only deletions identified by 4 or more independent reads were included in the analysis.
Supplementary Material
Acknowledgments
We thank Drs. S. Burden and P. Soriano for generously providing the MLC-Cre and Meox2-Cre mouse strains. We are grateful to Igor Antoshechkin, director of the Caltech Genomics Facility, for help with analysis of Solexa sequence data. This work was supported by grants to DCC (R01 grant GM062967 and an Ellison Medical Foundation Senior Scholar Award) and JMM (NCRR grant 1 S10 RR023454-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007a;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Nakada K, Sato A, Hayashi J. Mitochondrial functional complementation in mitochondrial DNA-based diseases. Int J Biochem Cell Biol. 2009;41:1907–1913. doi: 10.1016/j.biocel.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Nakada K, Inoue K, Hayashi J. Interaction theory of mammalian mitochondria. Biochem Biophys Res Commun. 2001;288:743–746. doi: 10.1006/bbrc.2001.5838. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A. The myoclonic epilepsy and ragged-red fiber mutation provides new insights into human mitochondrial function and genetics. Am J Hum Genet. 1998;62:745–751. doi: 10.1086/301813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Isobe K, Nakada K, Hayashi JI. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat Genet. 2001;28:272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- Ogata T, Yamasaki Y. Ultra-high-resolution scanning electron microscopy of mitochondria and sarcoplasmic reticulum arrangement in human red, white, and intermediate muscle fibers. Anat Rec. 1997;248:214–223. doi: 10.1002/(SICI)1097-0185(199706)248:2<214::AID-AR8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Bothe GW, Haspel JA, Smith CL, Wiener HH, Burden SJ. Selective expression of Cre recombinase in skeletal muscle fibers. Genesis. 2000;26:165–166. [PubMed] [Google Scholar]
- Lyons GE, Ontell M, Cox R, Sassoon D, Buckingham M. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J Cell Biol. 1990;111:1465–1476. doi: 10.1083/jcb.111.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14(Spec No 2):R283–289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, Chin S, Spiegelman BM. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008a;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson NG. Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci U S A. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissiere A, Campos Y, Rivera H, de la Aleja JG, Carroccia R, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain. 2008;131:338–351. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- Hudson G, Amati-Bonneau P, Blakely EL, Stewart JD, He L, Schaefer AM, Griffiths PG, Ahlqvist K, Suomalainen A, Reynier P, et al. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain. 2008;131:329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Attardi G. MtDNA mutations in aging and apoptosis. Biochem Biophys Res Commun. 2003;304:519–529. doi: 10.1016/s0006-291x(03)00625-9. [DOI] [PubMed] [Google Scholar]
- Krishnan KJ, Greaves LC, Reeve AK, Turnbull DM. Mitochondrial DNA mutations and aging. Ann N Y Acad Sci. 2007;1100:227–240. doi: 10.1196/annals.1395.024. [DOI] [PubMed] [Google Scholar]
- Chomyn A, Martinuzzi A, Yoneda M, Daga A, Hurko O, Johns D, Lai ST, Nonaka I, Angelini C, Attardi G. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc Natl Acad Sci U S A. 1992;89:4221–4225. doi: 10.1073/pnas.89.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. Faseb J. 2001;15:322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Villani G, Attardi G. In vivo measurements of respiration control by cytochrome c oxidase and in situ analysis of oxidative phosphorylation. Methods Cell Biol. 2001;65:119–131. doi: 10.1016/s0091-679x(01)65007-6. [DOI] [PubMed] [Google Scholar]
- Duan S, Hajek P, Lin C, Shin SK, Attardi G, Chomyn A. Mitochondrial outer membrane permeability change and hypersensitivity to digitonin early in staurosporine-induced apoptosis. J Biol Chem. 2003;278:1346–1353. doi: 10.1074/jbc.M209269200. [DOI] [PubMed] [Google Scholar]
- Detmer SA, Chan DC. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J Cell Biol. 2007b;176:405–414. doi: 10.1083/jcb.200611080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Bielas JH, Loeb LA. Quantification of random mutations in the mitochondrial genome. Methods. 2008b;46:263–268. doi: 10.1016/j.ymeth.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.