Abstract
This paper reports studies of two novel, allelic missense mutations found in the S-adenosylhomocysteine hydrolase (AHCY) gene from a new case of AHCY deficiency in an infant girl who died at age four months. The mutations lead to replacement of arginine with cysteine (p.Arg49Cys) and aspartic acid with glycine (p.Asp86Gly). Functional analysis of recombinant proteins containing the mutations detected showed that both dramatically reduce AHCY activity. The p.Arg49Cys mutant protein forms intermolecular disulphide bonds, leading to macromolecular structures that can be prevented by reducing agent DTT. The p.Asp86Gly protein tends to form enzymatically inactive aggregates and the loss of a single negative charge as a result of the mutation is involved in enzyme inactivation. We show that replacing Gly86 with negatively charged Glu86 in mutant protein restores enzymatic activity to 70% of wild-type, whereas changing Gly86 to positively charged Lys86 or uncharged Leu86 does not improve enzyme activity, indicating that the negative charge is important for maintenance of such activity. These studies significantly extend knowledge about the importance of residue 86 for AHCY activity. Residue 86 has not been implicated before in this way and the results suggest that the present model of S- adenosylhomocysteine (AdoHcy) hydrolysis may need refinement. Our functional studies provide novel insight into the molecular defect underlying AHCY deficiency and reveal that both low enzyme activity and protein stability of AHCY contribute to the clinical phenotype.
Keywords: AHCY, intermolecular disulphide bond, NADH, genotype-phenotype
Introduction
S-adenosylhomocysteine hydrolase (EC 3.3.1.1; MIM# 180960) catalyzes the hydrolysis of S-adenosylhomocysteine (AdoHcy) to adenosine (Ado) and homocysteine (Hcy) [Dela Haba and Cantoni, 1959]. The 3-dimensional structures of rat [Hu et al. 1999] and human [Turner et al., 1998; Yang et al., 2003] AHCY have been resolved and mutational studies have allowed detailed insights into the processes important for catalytic activity [Gomi et al., 1989; Gomi et al., 1990; Ault Riche et al., 1994; Komoto et al., 2000; Takata et al., 2002; Yamada et al., 2005]. Amino acids His54, Asp130, Glu155, Lys185, Asp189, and Asn190 have been pinpointed as crucial for this activity (in rat AHCY), and the detailed role of each in the reaction mechanism has been elucidated [Yamada et al., 2005].
Genetic deficiency of AHCY activity has been reported to date in only four humans [Barić et al., 2004; Barić et al., 2005; Buist et al., 2006; Ćuk et al., 2007]. These patients have been characterized clinically chiefly by myopathy and delayed development and, metabolically, by striking elevations of plasma AdoHcy, S-adenosylmethionine (AdoMet), methionine, and creatine kinase, with less marked abnormalities of liver function tests. Inhibition by AdoHcy of AdoMet-dependent methyltransferasees is considered to play a major role in the pathophysiology of this condition [Barić et al., 2005]. In this paper we report the molecular and functional characterization of two novel missense mutations from a fifth case of this deficiency, namely p.Arg49Cys and p.Asp86Gly. The clinical history and metabolic findings for this patient will be reported separately. Each of the mutations results in significant loss of enzyme activity. Each affects holoenzyme formation. p.Arg49Cys alters the oxidation state of bound co-factor NAD and gel filtration and native PAGE demonstrated the possibility of irregular disulphide bond formation due to this p.Arg49Cys exchange. Site-directed mutagenesis targeting Asp86 was carried out, replacing mutant Gly86 with either negatively charged glutamic acid (Glu86), positively charged lysine (Lys86) or uncharged leucine (Leu86) to find clues whether the negative charge located at residue 86 in wild-type protein is essential for catalytic activity of AHCY. We suggest that Asp86 be added to the list of residues important for the maintenance of the catalytic capacity of AHCY, and that missense mutations p.Arg49Cys and p.Asp86Gly show medical relevance for example to prenatal diagnosis or overall disease diagnosis. We conclude that the molecular and biochemical data correlate with the clinical severity of patients with AHCY deficiency.
Material and Methods
Cloning of Recombinant Wild-type and Mutant AHCY for Expression in E. coli
The expression vector harboring the wild-type AHCY gene (p32AHHwt) [Belužić et al., 2006] was used as template for site-directed mutagenesis in order to generate vectors pR49C and pD86G using the GeneTailor™ system (Invitrogen, Carlsbad, CA, U.S.A). Accordingly, exchanges p.Asp86Glu, p.Asp86Lys, and p.Asp86Leu were introduced by using the newly constructed pD86G vector as template (oligonucleotides listed in Table 1). The exchanges were confirmed by dideoxy sequencing using the BigDyeR® chemistry (Applied-Biosystems, Foster City, CA, USA).
Table 1.
Oligonucleotide primers used for site-directed mutagenesis (purchased from Invitrogen, Carlsbad, CA, U.S.A).
| Construct | Oligonucleotide (5′-3′) forward |
Oligonucleotide (5′-3′) reverse |
Codon change |
|---|---|---|---|
| p.R49C | GGGCGCATGCATCGCTGGCT | CCAGCGATGCATGCGCCCTTCAGTGGCTT | CGC ->TGC |
| p.D86G | GCAATTGCAGCCGCCGCATGGCCCTGGGTGGAGAA | GAC ->GGC |
|
| p.G86E | TTCTCCACCCAGGAGCATGCGGCGGCTGCCATT | GCCTTGGCAATGGCAGCCGCCGCATGCTCCTGGGT | GGC ->GAG |
| p.G86K | TTCTCCACCCAGAAGCATGCGGCGGCTGCAATT | GCCTTGGCAATTGCAGCCGCCGCATGCTTCTGGGT | GGC ->AAG |
| p.G86L | TTCTCCACCCAGCTGCATGCGGCGGCTGCAATT | GCCTTGGCAATTGCAGCCGCCGCATGCAGCTGGGT | GGC ->CTG |
To construct expression plasmids based on the T7 promotor system but without thioredoxin tag, vectors p32AHHwt, pR49C and pD86G were restricted with endonuclease NdeI to remove a 345 bp fragment containing the thioredoxin tag and religated accordingly. Successful thioredoxin tag removal was confirmed via restriction (NdeI) and subsequent gel electrophoresis.
Additionally, for all constructs the whole coding and flanking regions were sequenced using specific primers for AHCY (Ahhg1: 5′-GCCGACATCGGCCTGGCTGCCT; Ahhg3: 5′-cggaatgccagccttggcaat; Ahhg5: 5′-ggcagaagctgcgggtactt; Ahhg6: 5′-AGGCATCCGAGGCATCTCTGA; Ahhg7: 5′-gcggcttgatgttcaccttct; Ahhg8: 5′-AGGTGGACCGGTATCGGTTGA; Ahhg10: 5′-AGCTGGATGAGGCAGTGGCT) and pET32b vector (T7: 5′-TAATACGACTCACTATAGGG; T7 term: 5′-CTAGTTATTGCTCAGCGGT). The reference sequence for the AHCY gene was retrieved from GenBank (NM_000687.1), and the sequence for pET32b from the manufacturer (Novagen-Merck KGaA, Darmstadt, Germany).
Plasmid pQE30 (Qiagen, Hilden, Germany) was used as host vector to construct expression plasmids pQE_R49C and pQE_D86G as described previously [Belužić et al., 2008] in order to produce mutated AHCYs with a fusion part of only 18 residues including a Histidine tag at the N-terminus.
Over-expression and affinity purification of recombinant protein
A detailed protocol for expression and purification of recombinant AHCYs in E. coli BL21 (DE3) RIL is given in the paper by Belužić and co-workers [2006].
Procedure for over-expression of p.Asp86Gly protein was adapted according to de Marco and co-workers [2005]. In brief, bacteria were grown to OD>0.7 at 37 C, cooled to 28 C, and supplemented with benzyl-alcohol to a final concentration of 0.1% (9.65mM) to induce endogenous chaperones. After 20 min of continuous agitation protein expression was induced with IPTG (0.5mM final concentration).
Additionally, to allow expression of recombinant p.Arg49Cys protein the growth temperature was shifted from 37°C to 25°C after IPTG induction.
Inclusion bodies preparation and protein refolding
Retrieval and refolding of recombinant p.Asp86Gly protein from inclusion bodies was carried out following the instructions of the iFold Protein Refolding System 1 (Novagen, Madison, USA).
Polyacrylamide Gel Electrophoresis (PAGE) and Gel Filtration Chromatography
SDS-PAGE and determination of protein concentrations followed standard laboratory procedures. Additionally, purity and electrophoretic behavior of the recombinant AHCY protein was analyzed using native polyacrylamide gel electrophoresis (native PAGE) as described previously [Belužić et al., 2006] and according to Kim and Robinson [2006] with reducing agent DTT. The molecular weights of recombinant mutant and wild-type forms of AHCY were analyzed by gel filtration chromatography according to Belužić and coworkers [2006].
Enzymatic Assays
S-Adenosylhomocysteine hydrolase activity in purified enzyme preparations was assayed according to Takata et al. [2002] by using 5 μg recombinant protein. Data were the average of three determinations, and the Km and enzyme activity values were obtained by directly fitting the data into the Michaelis-Menten equation using a Lineweaver-Burke linearization.
Quantitation of enzyme bound NAD+ and NADH was performed using a fluorescence technique described by Hohman et al. [1985] using 200 μg of recombinant protein.
Bioinformatics
Software applications used for bioinformatical analysis are described previously [Belužić et al., 2006].
RESULTS
Identification of two novel mutations in the patient’s AHCY gene
Sequencing of the complete AHCY genes of patient and parents revealed that the patient had two point mutations: a paternally derived mutation of codon 49 in exon 2 that changes arginine to cysteine (c.145C>T; p.Arg49Cys) and a maternally derived mutation of codon 86 in exon 3 that changes aspartic acid to glycine (c.257A>G; p.Asp86Gly). Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1.
Values retrieved from http://prowl.rockefeller.edu/aainfo/contents.htm [Zamyatnin, 1971; Chothia, 1976] show that arginine with a surface area (square Ångstrom; Å2) of 225 and an average residue volume (Vr) of 173.4 and aspartic acid (Å2 = 150; Vr = 111.1) possess bigger surface areas and residue volumes than do residues cysteine (Å2 = 135; Vr = 108.5) and glycine (Å2 = 75; Vr = 60.1). The latter also differ from arginine or aspartic acid, respectively, in that they contain uncharged, but polar, side-chains and are significantly more hydrophobic.
Studies of p.Arg49Cys
Approx. 3–5 mg of homogeneous p.Arg49Cys protein was obtained from a 500 ml culture when growth was maintained at 25°C after IPTG induction. Expression at higher temperature did not yield significant amounts of recombinant protein. T7-promotor-driven expression of pET32 constructs yielded higher amounts of recombinant protein than the T5 lac-promoter-driven expression provided by the pQE30 vector system.
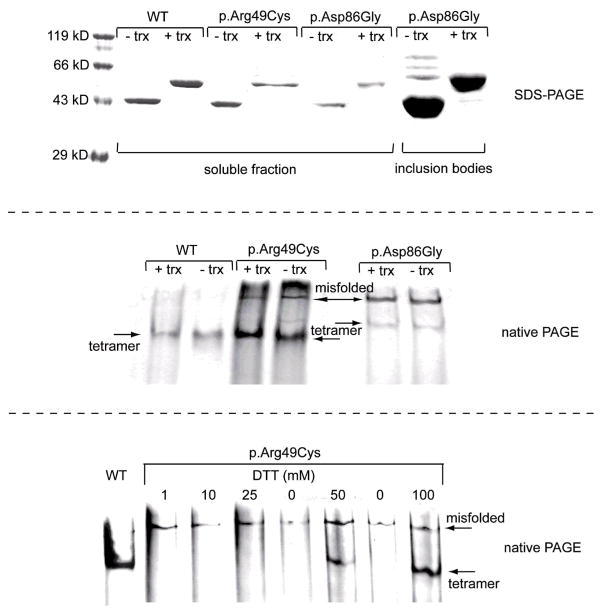
Surprisingly, during standard native-PAGE analysis, p.Arg49Cys, unlike recombinant wild-type protein does not form a tetrameric complex (Figure 1, lower panel). However, in the presence of reducing agent DTT (100 mM), a single band appears on the gel, migrating at an almost identical position as the wild-type tetramer. Similar results for the effect of reducing agent DTT on the dissociation of intermolecular disulfide bonds have been shown by Kim and Robinson during their studies of P22 tailspike protein [Kim and Robinson, 2006]. We suppose that during native-PAGE the shift in temperature caused by electrode heat leads to changes in the dissociation constants of the buffer with a decrease in [H+]. In these circumstances, and without DTT, the formation of intermolecular disulphides occurs, leading to tetramer disassembly. Sensitivity to temperature is in agreement with our expression studies showing that E. coli produces soluble recombinant protein only at growth temperatures of 25°C or below. Furthermore, incubation of p.Arg49Cys at 37°C for 10 min resulted in almost complete precipitation of the protein.
Figure 1.
Upper panel: SDS PAGE of freshly prepared Ni-NTA affinity-purified recombinant p.Arg49Cys, p.Asp86Gly, and wild-type (WT) AHCY retrieved from the soluble protein fraction; p.Asp86Gly protein retrieved from inclusion bodies indicates over-expression of p.Asp86Gly as insoluble protein. The ratio of insoluble and soluble p.Asp86Gly protein obtained from over-expressing E. coli is calculated to > 200:1. Shown is protein with or without thioredoxin tag (trx). Middle panel: Native PAGE of Ni-NTA affinity-purified p.Arg49Cys, p.Asp86Gly and wild-type AHCYs with and without thioredoxin tag. Both tagged and untagged recombinant protein is forming AHCY tetramers. Lower Panel: Native PAGE under reducing or non-reducing conditions of Ni-NTA affinity-purified recombinant p.Arg49Cys and wild-type AHCY; 100 mM DTT aids in formation of p.Arg49Cys tetramer. p.Arg49Cys without addition of 100 mM DTT is unable to form tetramers. Protein was resolved on 7.5% native PAGE or 12% SDS-PAGE, respectively.
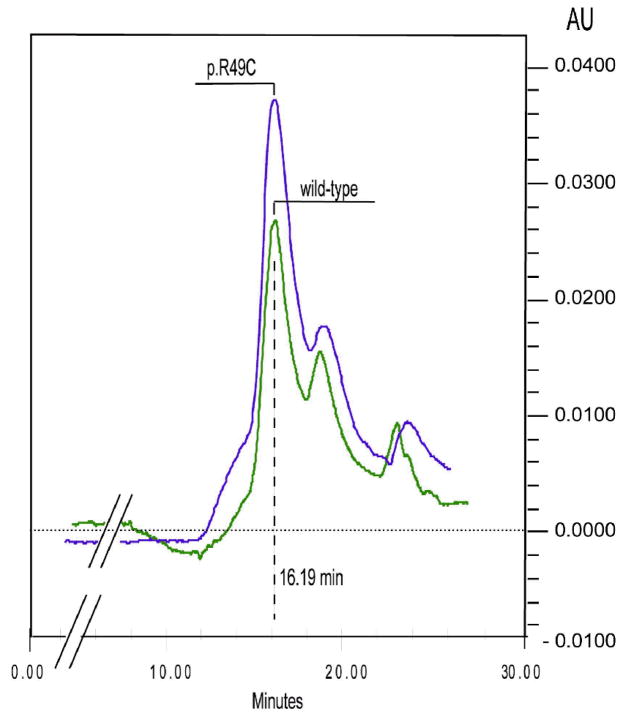
Gel filtration chromatography was carried out at room temperature (20°C) and conditions maintaining pH 7.2. Under these circumstances p.Arg49Cys does not form intermolecular disulphide bonds and the p.Arg49Cys protein eluted with the same retention time as the wild-type protein, as a single symmetrical peak at a position corresponding to Mr of 260,000 (Figure 2), indicating that both p.Arg49Cys and wild-type recombinant enzyme can form tetramers, as does the native enzyme found in human tissues.
Figure 2.
Gelfiltration chromatography of recombinant wild-type and p.Arg49Cys protein. 20 g of protein was applied to a BIO-SIL SEC 250-5 column (Biorad, Munich), equilibrated with a buffer containing 50 mM potassium phosphate (pH 7.2) and 150 mM NaCl. Elution was performed in the same buffer at a flow rate of 1 ml/min. The protein was monitored at 280 nm.
The total NAD+ plus NADH content of recombinant p.Arg49Cys enzyme was 0.7–0.9 molecules NAD per monomer, in agreement with previously published data for the wild type protein that indicate 4 molecules of coenzyme bound per tetramer [Belužić et al., 2006]. However, in contrast to wild-type protein in which more than 80% of the bound coenzyme is NAD+ [Belužić et al., 2006], for p.Arg49Cys approximately 95.7% of the total bound coenzyme was in the form of NADH.
As shown previously, the enzymatic activities of recombinant wild-type protein expressed with either the pET32 or the pQE30 vector system are identical [Belužić et al., 2008]. Using the human placental AHCY as reference, the pET32-based recombinant AHCY has an increased molecular weight (63.9 kD) due to the additional 152 amino acid residues contributed by the plasmid expression vector tag region [Belužić et al., 2006], whereas the pQE30 expressed AHCY is a fusion protein with only 18 additional residues. However, due to yield and solubility considerations we preferred using the pET32-based system for AHCY over-expression. Therefore, we constructed pET32-based expression vectors without thioredoxin tag for wild-type, and mutant proteins p.Arg49Cys and p.Asp86Gly, respectively. Such recombinant AHCY protein has 38 amino acids fused to its N-terminus, and migrates on native PAGE at almost identical position as its thioredoxin-tagged counterpart. This indicates that the tag region does not interfere with holoenzyme formation, nor does it affect enzymatic activity. A summary for all investigated constructs is given in Table 2.
Table 2.
Overview of AHCY constructs and amino acid sizes of tagged and untagged protein.
| AHCY Expression Vectors | T7 based: with Trx tag |
T7 based: without Trx tag |
pQE30 based |
|---|---|---|---|
| Fusion protein amino acid count | 584 aa | 469 aa | 450 aa |
| Fusion part | 152 aa | 38 aa | 18 aa |
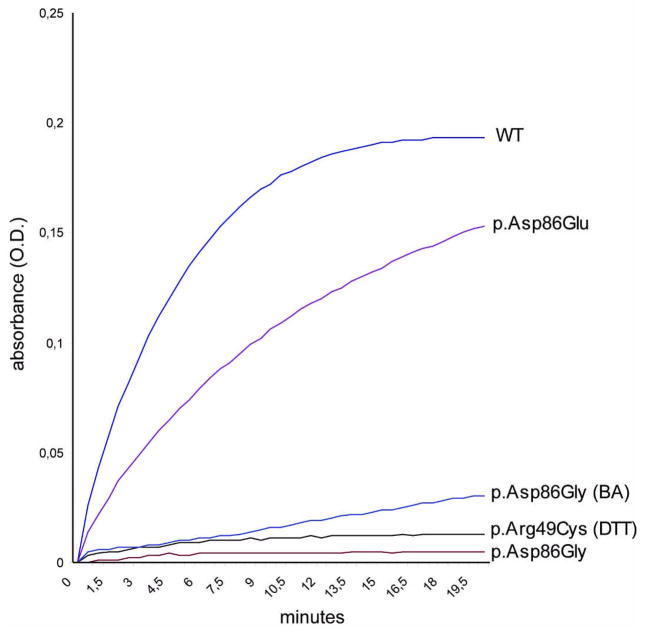
The catalytic activity of thioredoxin-tagged p.Arg49Cys, determined in the direction of AdoHcy hydrolysis, was 6.7% that of the wild-type protein (Figure 3 and Table 3). Similar values have been determined for p.Arg49Cys protein without thioredoxin-tag (6.9% that of wild-type protein).
Figure 3.
Graphical representation of enzymatic activities in the hydrolytic direction of untagged recombinant wild-type and mutant enzymes p.Arg49Cys (supplemented with DTT), p.Asp86Gly (expressed without benzyl-alcohol), p.Asp86Gly (BA) (expressed with benzyl-alcohol), and p.Gly86Glu. Thioredoxin –tagged proteins show almost identical AHCY activity and are omitted from the graph for clarity. The reaction for specific determination of AHCY activity was performed in buffer containing 50 mM potassium phosphate (pH 7.2) and 1 mM EDTA, and started by the addition of various amounts of a 1 mM stock solution of SAH. Hydrolysis of SAH generates Ado, which is converted to inosine by Ado Deaminase. Disappearance of Ado is measured at a wavelength of 265 nm. To graphically show an increase in enzymatic activity the O.D. values (absorbance) for disappearing Ado have been inverted and plotted against elapsed time. 5 μg of freshly prepared Ni-NTA affinity-purified recombinant wild-type, or mutant proteins were used (mutant p.Arg49Cys without DTT supplementation, p.Asp86Lys and p.Asp86Leu are omitted from the graph in view of lack of activity).
Table 3.
Kinetic parameters of wild-type and mutant AHCY enzymes.
| Km (μM) hydrolysis | Enzymatic activity (μmol min−1mg−1) hydrolysis | |
|---|---|---|
| WT | 15.09 | 0.748±0.013 (100%) |
| p.Asp86Glu | 16.4 | 0.525±0.019 (70.2%) |
| p.Asp86Gly | N/A | 0.116±0.013 (15.5%) |
| p.Arg49Cys | N/A | 0.050±0.011 (6.7%) |
Km for p.Arg49Cys and p.Asp86Gly was not determined due to the extremely low average change in OD265 per minute (Δ OD265) and thus unreliable Vi values. The data represent 3 technical replicates shown as mean values ± S.D. Values in parentheses are percentage of wild-type.
Studies of p.Asp86Gly
The p.Asp86Gly protein was almost impossible to express in E. coli in soluble form, whatever conditions were tried and regardless of the vector system used. Yield was approximately 100 μg of soluble protein per 500 ml culture. On the other hand, more than 5 mg of denatured protein could be retrieved from a 500 ml culture by solubilization of bacterial inclusion bodies (Figure 1, upper panel). However, attempts to refold the recombinant protein retrieved from inclusion bodies failed, even when a commercial refolding kit was used.
During electrophoresis p.Asp86Gly protein migrated significantly more slowly than did recombinant wild-type. An additional high-molecular band was visible in native PAGE (Figure 1, middle panel) that might indicate the formation of clusters or aggregates of mutant protein, and gel filtration of p.Asp86Gly protein did not yield interpretable results most likely due to formation of aggregates during chromatography. Initially, p.Asp86Gly was found to have a small detectable AHCY activity, although it was only 1.5% that of the wild-type protein. After modification of the expression protocol by inducing endogenous bacterial chaperones with benzyl alcohol, AHCY activity could be boosted to approx. 15.5% that of the wild-type protein (Figure 3 and Table 3). Similar values have been determined for p.Asp86Gly protein without thioredoxin tag (14.9% that of wild-type protein).
To our knowledge, despite numerous mutational analyses of the mechanism of AHCY catalysis, no evidence has, until now, been found to indicate D86 as a residue involved in the catalytic process. However, the dramatic decrease in enzymatic capability of mutant p.Asp86Gly protein indicates that Asp86 must be of high importance for enzyme function.
Accordingly, our next step was to investigate whether loss of the negative charge of Asp86 as a consequence of the mutation is a major reason for enzyme malfunction. We performed site directed mutagenesis to replace G86 in mutant p.Asp86Gly protein with residues showing similar physico-chemical properties such as charge (positive or negative) or molecular weight. Gly86 (uncharged) was replaced with Glu86 (negatively charged), Lys86 (positively charged) or Leu86 (apolar and uncharged). Glutamic acid represents the only alternative for aspartic acid in terms of its negatively charged side chain. Leucine (Mw 131) was chosen for its similar molecular weight and structure comparing to aspartic acid (Mw 133). Lysine (Mw 146) was chosen due to molecular weight and charge considerations. Exchanges were then introduced into pD86G expression vector to produce p.Gly86Glu, p.Gly86Lys, and p.Gly86Leu protein. Protein with the Glu86 modification (negatively charged) regained enzymatic activity from <16% to 70.2% compared to wild-type (Figure 3 and Table 3), whereas Lys86 or Leu86 protein remained relatively inactive (not shown). Introduction of Glu86 is far from being an optimal substitution of Asp86 although enzymatic activity is dramatically enhanced, because the p.Gly86Glu tetramer is unstable and tends to disassemble rather quickly as evaluated by native PAGE (at 4°C, 2–3 days). Wild-type protein is extremely robust and maintains activity for months under the same conditions. Interestingly, untagged mutant AHCY protein tends to precipitate over night, whereas thioredoxin-tagged mutant AHCY remained in solution for the duration of this study (at 4°C).
DISCUSSION
The mechanism of oxidation of AdoHcy and Ado during the AHCY cycle involves several steps: upon substrate binding, the enzyme undergoes conformational changes and the cleft between the catalytic (AdoHcy and Ado binding domain; Figure 4A) and the NAD-binding domain is closed. Bioinformatical analysis of the 3-D model of AHCY (PDB: 1a7aA) shows that at the ends of beta-sheets 1 and 3 are amino acids H55 and D131, with their side-chains facing the catalytic cleft (Figure 4B). Both residues are crucial for substrate binding and catalytic activity, and proximity of Asp131 to the C4′-H of Ado is essential for the reaction [Yamada et al., 2005]. Further, Yamada and co-workers suggest residue His55 (His54 in rat AHCY) as the amino acid residue that acts as a general acid/base to cleave the C5′-SD bond of AdoHcy.
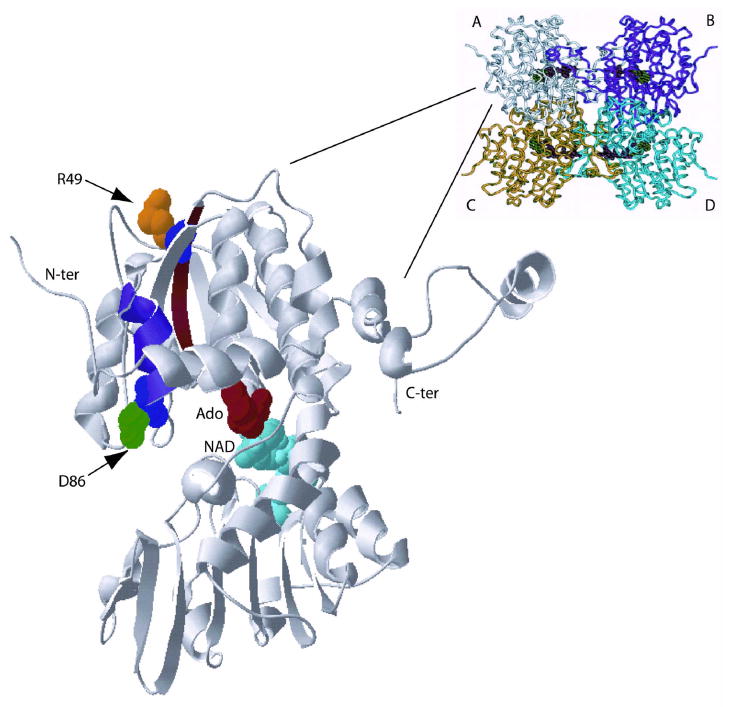
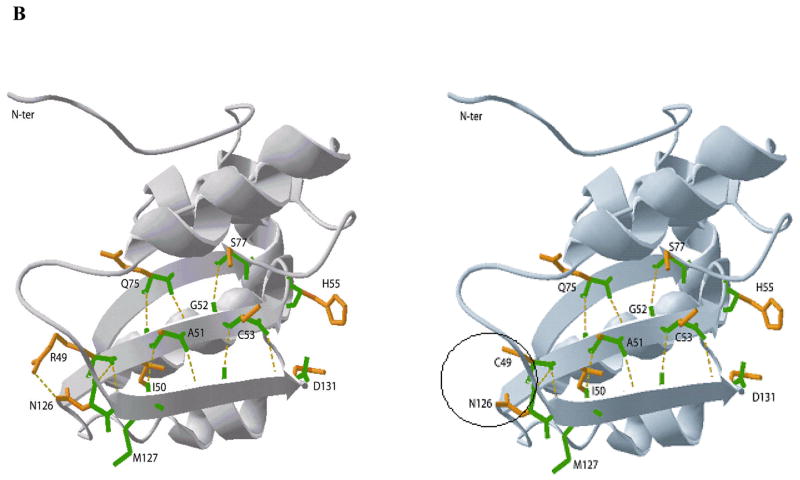
Figure 4.
A: Ribbon diagram of the AHCY tetramer showing subunits A to D. The enlargement shows subunit A including Ado and NAD. Mutated residues are shown in color and are represented with side-chains in 3-D rendering. Alpha-helix 3 is shown in blue, and beta-sheet 1 in brown color. B: (overleaf) Ribbon diagrams of a portion of the catalytic domain of human AHCY. The enlargements show the delicate hydrogen-bond network between beta-sheets 1, 2 and 3 of human AHCY.
The region containing Arg49 is located at the outer surface of the catalytic domain in beta-sheet 1 (Figure 4A). In the wild-type protein a hydrogen bond exists between Arg49 and Asn126, forming part of a sophisticated H-bond network between beta-sheets 1, 2 and 3 that plays an important role in protein substructure. Although major changes in the secondary structure are not predicted in the region containing Arg49 after conversion to cysteine, in the p.Arg49Cys mutant the Arg49-Asn126 hydrogen bond is lost (Figure 4B). Thus, it may be that loss of the H-bond connecting sheets 1 and 3 leads to conformational changes, which slightly change the geometry of the active site and move Asp131 away from His55 into an unfavorable position. As a consequence, the C5′-SD bond cleavage of 3′-keto-4′-dehydro-AdoHcy to produce 3′-keto-4′5′-dehydro-Ado and Hcy may be inefficient, and NADH accumulation would tend to occur, as does indeed happen for p.Arg49Cys protein in which 95.7% of cofactor is found as NADH. Similar accumulation of NADH has been described for mutant p.Tyr143Cys protein, found in the first identified AHCY-deficient patient [Barić et al., 2004; Belužić et al., 2006]: and by mutational analysis of human Lys426Arg, and rat Asp189Asn and His54Asn AHCY, respectively [Ault-Riche et al., 1994; Takata et al., 2002]. NADH accumulation is accompanied by loss of overall catalytic activity due to an imbalance in the redox reaction.
In addition to the tendency of p.Arg49Cys to abnormally accumulate NADH, our evidence shows that it engages in intermolecular disulphide formation that leads to formation of irregular protein complexes. This is due to interchain disulfide shuffling that might occur between free subunits. The byproducts of these interactions might be multimers with interchain disulfide bonds. Formation of such intermolecular disulphide bonds can be prevented under reducing conditions and by maintaining environmental conditions below 25°C. Whether such intermolecular bond formation could be prevented in vivo, and whether that would be clinically beneficial are subjects for further investigation
Residue Asp86 is located opposite to Arg49 in the catalytic domain in alpha-helix 3, protruding into the cleft space with its charged side-chain (Figure 4A). Interestingly, Asp86 is separated by only two residues from the previously described p.Ala89Val mutant that is also a cause of AHCY deficiency [Belužić et al., 2008]. The reason for malfunction of the p.Asp86Gly mutant enzyme is not obvious, and, as in the p.Ala89Val mutant, bioinformatical analysis predicts neither a change in the hydrogen-bond network, nor in the secondary structure of alpha-helix 3. However, changing aspartic acid to glycine leads to increased hydrophobicity and polarity, and eliminates the negative charge in the region. Increased hydrophobicity might push away some of the six water molecules normally located in the cleft between the NAD and substrate binding domains [Yamada et al., 2005] with possible consequences to the general holoenzyme structure. Namely, during the catalytic cycle, Michael-type addition of water or Hcy to the central intermediate 3′-keto-4′,5′-dehydroadenosine, and reduction of 3′-keto by the NADH formed results in the formation of the final product, Ado or AdoHcy. In that context, Turner et al. [1998] suggested that a disordered water molecule near C4′ participates in abstracting the proton of C4′. The ‘antenna-like’ feature of Asp86 is missing in the p.Asp86Gly mutant, raising the question whether the charged side-chain of aspartic acid is needed to keep such a water molecule in an appropriate position in regard to the intermediate 3′-keto-4′,5′-dehydroadenosine. Another possibility is that Asp86 acts as a general acid/base for a yet unidentified process during the catalytic reaction.
At the moment, until explanatory crystallographic evidence becomes available, we suggest that formation of aggregates other than the regular AHCY tetramer plays a role in p.Arg49Cys’s and p.Asp86Gly’s AHCY inactivity under physiological conditions. We conclude this from our attempts to induce endogenous chaperones. A major improvement for enzymatic activity of p.Asp86Gly protein was achieved by inducing bacterial chaperones with benzyl alcohol, boosting AHCY activity to approx. 16% that of the wild-type protein. Bacterial chaperones might ameliorate formation of misfolded mutant protein to some degree, thereby enlarging the pool of p.Asp86Gly tetramers with residual AHCY activity. This is in agreement with measurements from patients’ red blood cell lysates that show residual AHCY enzymatic activity of approx. 10% when compared to values from unaffected individuals (S.H. Mudd, personal communication). In the case of p.Arg49Cys mutant, only DTT mitigates aggregate formation in vitro so far.
Nevertheless, the p.Asp86Gly mutation teaches us that the single negative charge of aspartic acid is needed for enzyme activity, and that certain aspects of the AHCY mechanism of catalysis remain elusive. Expression of p.Asp86Gly in E. coli yielded amounts of proteins far lower than those obtained with other AHCY mutant proteins studied. Circumvention of this problem might be a first step in resolving the underlying mechanism for enzyme malfunction and towards a refinement of the AHCY catalytic mechanism.
Although there are only a few cases of AHCY deficiency so far, we emphasize that the molecular and biochemical data are related to the clinical phenotype of patients with AHCY deficiency. The patients with the mildest clinical phenotype carry the mutations p.Ala89Val and/or p.Tyr143Cys leading to decreased AHCY protein stability, but considerably high residual enzymatic activity (20–30% of wild-type) [Barić et al., 2004; Barić et al., 2005; Buist et al., 2006; Belužić et al., 2006; Belužić et al., 2008]. Treatment by dietary methionine restriction and administration of creatine and phosphatidylcholine appears to have been clinically beneficial in the two patients for whom this has been tried previously [Barić et al., 2005]. In contrast, the most severely affected patient carries the mutations described in this paper leading to drastically decreased protein stability, very low residual enzymatic activity, and to death very soon after birth. Considering the available mutational data and the characteristic clinical aspects such as myopathy and delayed development and, metabolically, by striking elevations of plasma AdoHcy, S-adenosylmethionine (AdoMet), methionine, and creatine kinase, early detection of AHCY deficiency in newborn, and determination of its grade of severity might assist in preventing a lethal outcome.
Acknowledgments
This work was supported by grants 0098086 and 098-0000000-2463 (OV) of the Ministry of Science, Education and Sports from the Republic of Croatia. Special thanks go to Igor Jurak for critical reading and helpful comments.
References
- Ault-Riche DB, Yuan CS, Borchardt RT. A single mutation at lysine 426 of human placental S-adenosylhomocysteine hydrolase inactivates the enzyme. J Biol Chem. 1994;269:31472–31478. [PubMed] [Google Scholar]
- Barić I, Fumić K, Glenn B, Ćuk M, Schulze A, Finkelstein JD, Jill James S, Mejaški-Bošnjak V, Pažanin L, Pogribny IP, Radoš M, Sarnavka V, Šćukanec-Špoljar M, Allen RH, Stabler S, Uzelac L, Vugrek O, Wagner C, Zeisel S, Mudd SH. S-adenosylhomocysteine hydrolase deficiency in a human: A genetic disorder of methionine metabolism. Proc Natl Acad Sci USA. 2004;101:4234–4239. doi: 10.1073/pnas.0400658101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barić I, Ćuk M, Fumić K, Vugrek O, Allen RH, Glenn B, Maradin M, Pažanin L, Pogribny I, Radoš M, Sarnavka V, Schulze A, Stabler S, Wagner C, Zeisel S, Mudd SH. S-Adenosylhomocysteine hydrolase deficiency: A second patient, the younger brother of the index patient, and outcomes during therapy. J Inh Metab Dis. 2005;28:885–902. doi: 10.1007/s10545-005-0192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belužić R, Ćuk M, Pavkov T, Fumić K, Barić I, Mudd SH, Jurak I, Vugrek O. A single mutation at tyrosine 143 of human S-adenosylhomocysteine hydrolase renders the enzyme thermosensitive and effects the oxidation state of bound co-factor NAD. Biochem J. 2006;400:245–253. doi: 10.1042/BJ20060749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belužić R, Ćuk M, Pavkov T, Barić I, Vugrek O. S-Adenosylhomocysteine hydrolase (AdoHcyase) deficiency: Enzymatic capabilities of human AdoHcyase are highly effected by changes to codon 89 and its surrounding residues. Biochem Biophys Res Commun. 2008;368:30–36. doi: 10.1016/j.bbrc.2008.01.042. [DOI] [PubMed] [Google Scholar]
- Buist NRM, Glenn B, Vugrek O, Wagner C, Stabler S, Allen RH, Pogribny I, Schulze A, Zeisel SH, Barić I, Mudd SH. S-Adenosylhomocysteine hydrolase deficiency in a 26-year-old man. J Inh Metab Dis. 2006;29:538–545. doi: 10.1007/s10545-006-0240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia CJ. The nature of the accessible and buried surfaces in proteins. Mol Biol. 1976;105:1–14. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- Ćuk M, Lovrić M, Fumić K, Vugrek O, Mudd SH, Sarnavka V, Barić I. The fourth S-adenosylhomocysteine hydrolase deficient patient: Further evidence of congenital myopathy. Clin Chem Lab Med. 2007;45:A43. [Google Scholar]
- Dela Haba G, Cantoni GL. The enzymatic synthesis of S-Adenosyl-L-homocysteine from adenosine and homocysteine. J Biol Chem. 1959;234:603–608. [PubMed] [Google Scholar]
- de Marco A, Vigh L, Diamant S, Goloubinoff P. Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol overexpressed molecular chaperones. Cell Stress & Chaperones. 2005;10:329–339. doi: 10.1379/CSC-139R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi T, Date T, Ogawa H, Fujioka M, Aksamit RR, Backlund PS, Jr, Cantoni GL. Expression of rat liver S-adenosylhomocysteinase cDNA in Escherichia coli and mutagenesis at the putative NAD binding site. J Biol Chem. 1989;264:16138–16142. [PubMed] [Google Scholar]
- Gomi T, Takata Y, Date T, Fujioka M, Aksamit RR, Backlund PS, Jr, Cantoni GL. Site-directed mutagenesis of rat liver S-adenosylhomocysteinase: Effect of conversion of aspartic acid 244 to glutamic acid on coenzyme binding. J Biol Chem. 1990;265:16102–16107. [PubMed] [Google Scholar]
- Hu Y, Komoto J, Huang Y, Gomi T, Ogawa H, Takata Y, Fujioka M, Takusagawa F. Crystal structure of S-adenosylhomocysteine hydrolase from rat liver. Biochemistry. 1999;38:8323–8333. doi: 10.1021/bi990332k. [DOI] [PubMed] [Google Scholar]
- Kim J, Robinson AS. Dissociation of intermolecular disulfide bonds in P22 tailspike protein intermediates in the presence of SDS. Protein Sci. 2006;15:1791–1793. doi: 10.1110/ps.062197206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoto J, Huang Y, Gomi T, Ogawa H, Takata Y, Fujioka M, Takusagawa F. J Biol Chem. 2000;275:32147–32156. doi: 10.1074/jbc.M003725200. [DOI] [PubMed] [Google Scholar]
- Takata Y, Yamada T, Huang Y, Komoto J, Gomi T, Ogawa H, Fujioka M, Takusagawa F. Catalytic mechanism of S-adenosylhomocysteine hydrolase: Site-directed mutagenesis of Asp-130, Lys-185, Asp-189, and Asn-190. J Biol Chem. 2002;277:22670–22676. doi: 10.1074/jbc.M201116200. [DOI] [PubMed] [Google Scholar]
- Turner MA, Yuan CS, Borchardt RT, Hershfield MS, Smith GD, Howell PL. Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength. Nat Struct Biol. 1998;5:369–376. doi: 10.1038/nsb0598-369. [DOI] [PubMed] [Google Scholar]
- Yamada T, Takata Y, Komoto J, Huang Y, Gomi T, Ogawa H, Fujioka M, Takusagawa F. Catalytic mechanism of S-adenosylhomocysteine hydrolase: Roles of His 54, Asp130, Glu155, Lys185, and Aspl89. Int J Biochem Cell Biol. 2005;37:2417–2435. doi: 10.1016/j.biocel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Yang X, Hu Y, Yin DH, Turner MA, Wang M, Borchardt RT, Howell PL, Kuczera K, Schowen RL. Catalytic strategy of S-adenosyl-L-homocysteine hydrolase: Transition-state stabilization and the avoidance of abortive reactions. Biochemistry. 2003;42:1900–1909. doi: 10.1021/bi0262350. [DOI] [PubMed] [Google Scholar]
- Zamyatnin AA. Protein volume in solution. Prog Biophys Mol Biol. 1971;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]