Abstract
Mutations in DJ-1 cause recessively transmitted early-onset Parkinson disease (PD), and oxidative damage to DJ-1 has been associated with the pathogenesis of late-onset sporadic PD. The precise biochemical function of DJ-1 remains elusive. Here, we report that DJ-1 is synthesized as a latent protease zymogen with low-intrinsic proteolytic activity. DJ-1 protease zymogen is activated by the removal of a 15-amino acid peptide at its C terminus. The activated DJ-1 functions as a cysteine protease with Cys-106 and His-126 as the catalytic diad. We show that endogenous DJ-1 in dopaminergic cells undergoes C-terminal cleavage in response to mild oxidative stress, suggesting that DJ-1 protease activation occurs in a redox-dependent manner. Moreover, we find that the C-terminally cleaved form of DJ-1 with activated protease function exhibits enhanced cytoprotective action against oxidative stress-induced apoptosis. The cytoprotective action of DJ-1 is abolished by the C106A and H126A mutations. Our findings support a role for DJ-1 protease in cellular defense against oxidative stress and have important implications for understanding and treating PD.
INTRODUCTION
Parkinson disease (PD) is the most common neurodegenerative movement disorder, characterized by the loss of dopaminergic neurons in the substantia nigra pars compacta (1). Mutations in DJ-1 have been identified as the cause for an autosomal recessive, early-onset familial form of PD (2–4). Accumulating evidence indicates that DJ-1 mutations trigger neurodegeneration via a loss-of-function pathogenic mechanism (5). Our previous studies reveal that DJ-1 is widely expressed in many tissues and cell types, including dopaminergic neurons (6,7) and that oxidative damage to DJ-1 is linked to the pathogenesis of late-onset sporadic form of PD (8). Increasing evidence suggests that DJ-1 may also be involved in other age-related neurodegenerative disorders, such as Alzheimer disease, Lewy body dementia and parkinsonism–dementia–amyotrophic lateral sclerosis complex (8–11).
DJ-1 is a 189-amino acid protein that was originally identified as an oncogene (12) and was also independently described as a regulatory subunit of a RNA-binding complex (13) and a protein involved in male infertility (14,15). Abnormal expression of DJ-1 has been associated with cancer (16,17), supporting the emerging view that there may be potential overlap in the underlying biochemical dysfunction between PD and cancer (18,19). DJ-1 has been implicated in multiple cellular processes, including oxidative stress response, protein quality control, anti-apoptotic signaling, transcriptional regulation and translational control (5). Despite recent progress in the characterization of DJ-1, the precise biochemical function of DJ-1 remains unknown.
DJ-1 is an evolutionarily conserved protein that shares significant sequence homology with the PfpI family of bacterial intracellular proteases and with heat shock protein 31 (Hsp31), an Escherichia coli chaperone which also possesses protease activity (20–25). We and other groups have solved the crystal structure of human DJ-1, which shows that DJ-1 adopts a helix-strand-helix sandwich structure similar to that of bacterial protease PH1704 and Hsp31 (26–30). However, unlike PH1704 and Hsp31, DJ-1 does not have a Cys-His-Glu/Asp catalytic triad. Instead, DJ-1 contains the putative catalytic nucleophile Cys-106 which has the potential to form a Cys-His diad with His-126 (27,28,30). Although the lack of a catalytic triad has been used to argue against a protease function of DJ-1 (26,29), it has been proposed that DJ-1 may use the Cys-His diad to carry out its protease function (28,30). A fully functional Cys-His catalytic diad has been demonstrated in several cysteine proteases, such as caspases (31–33).
Proteases perform essential functions in regulating diverse cellular processes by catalyzing the cleavage of peptide bonds (34–37). To avoid potentially hazardous consequences of unregulated protease activity, proteases generally reside in cells as latent precursors called zymogens and require an activation event for conversion into the catalytically active form (38,39). We have previously shown that full-length recombinant DJ-1 protein exhibits weak intrinsic proteolytic activity (7), suggesting that full-length DJ-1 is a protease zymogen. In this study, we investigated the mechanism for activating DJ-1 protease zymogen and characterized the role of DJ-1 protease in cytoprotection. Our results reveal that DJ-1 protease zymogen is activated by cleavage of a 15-amino acid peptide at its C terminus and support a role for DJ-1 protease in cellular defense against oxidative stress-induced apoptosis.
RESULTS
Cleavage of the C-terminal 15-amino acid peptide activates DJ-1 protease function without affecting its dimerization
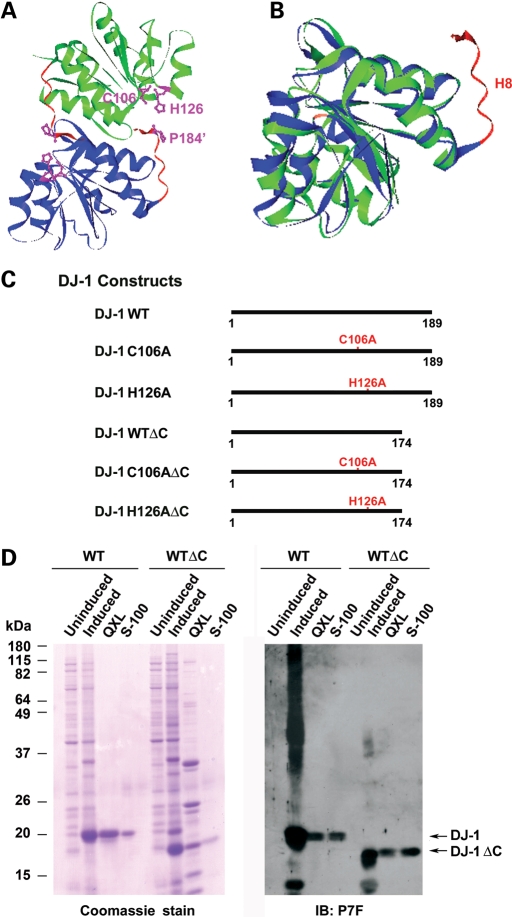
We and other groups have previously reported that DJ-1 exists as a homodimer in the crystal structure (Fig. 1A) as well as in solution (7,26–30). The monomeric structure of DJ-1 is highly homologous to that of bacterial protease PH1704 except that DJ-1 has an additional α-helix, helix H8, at the C terminus (Fig. 1B). This extra helix is generated because DJ-1 is 15 residues longer at the C terminus than PH1704 and other proteases of the PfpI family (27,30). The H8-containing, 15-amino acid C-terminal tail is evolutionally conserved in DJ-1 orthologs across different species (25). In the crystal structure of DJ-1, the 15-amino acid C-terminal tail appears to block the access of substrates to the putative catalytic site containing the highly conserved, predicted catalytic nucleophile Cys-106 (Fig. 1A) (27,28,30).
Figure 1.
Structural comparison of DJ-1 with intracellular protease PH1704 and generation of DJ-1 mutant proteins. (A) Ribbon diagram of the crystal structure of DJ-1 dimer. One monomer is colored in green and the other in blue. The α-helix H8 and its C-terminal region are highlighted in red. The side chains of Cys-106, His-126 and Pro-184′ (indicates from the other subunit of DJ-1 dimer) are in purple. The figure was produced based on the published DJ-1 structure using the VectorNTI program (Invitrogen). (B) Superposition of DJ-1 monomer structure (blue) over that of PH1704 (green). The putative catalytic cysteines in PH1704 (Cys-100) and in DJ-1 (Cys-106) are highlighted in pink and the α-helix H8-containing C-terminal region of DJ-1 in red. (C) Schematic representation of wild-type and mutant DJ-1 proteins used in this study. (D) Purification of untagged wild-type and mutant DJ-1 proteins. Lysates from un-induced or induced E. coli BL21 cells expressing indicated DJ-1 proteins and the fractions containing DJ-1 proteins purified by Sepharose QXL ion exchange chromatography and Sephacryl S-100 gel filtration chromatography were resolved on SDS–PAGE, followed by Coomassie blue stain (left panel) or immunoblotting with the anti-DJ-1 antibody P7F (right panel).
The most commonly known mechanism for the activation of protease zymogen is via specific proteolytic cleavage to remove an N-terminal peptide, often referred to as the pro-region (38). For some cysteine proteases, such as caspases, the activation involves the cleavage of an internal linker peptide (31,39). On the basis of the DJ-1 structural data (27,28,30) and sequence homology analysis (25), we hypothesized that the activation of DJ-1 protease zymogen may involve the cleavage of its H8-containing C-terminal 15-amino acid peptide (Fig. 1A and B). To test this hypothesis, we generated DJ-1 WTΔC, a truncated form of wild-type DJ-1 lacking the C-terminal 15 amino acids (residues 175–189) (Fig. 1C). WT and WTΔC DJ-1 proteins were expressed in E. coli as untagged proteins and purified as we described previously (7,27) to homogeneity, as shown by a single protein band on the Coomassie blue stain (Fig. 1D, left panel). The purified recombinant proteins were confirmed to be DJ-1 by immunoblot analysis using the anti-DJ-1 antibody (Fig. 1D, right panel).
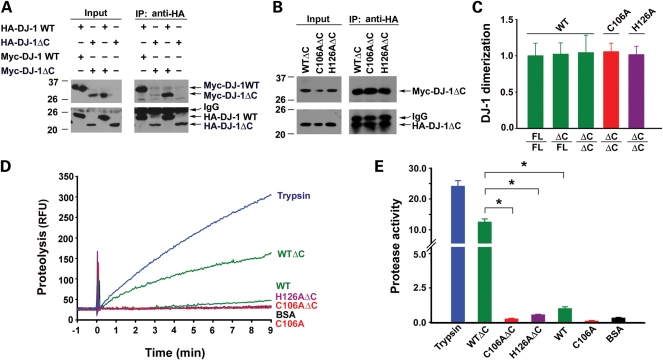
Co-immunoprecipitation analysis showed that hemaglutinin (HA)-tagged DJ-1 WTΔC protein was able to dimerize with Myc-tagged DJ-1 WTΔC protein as well as with DJ-1 WT, the full-length form of wild-type DJ-1 protein (Fig. 2A–C), confirming that the deletion of the C-terminal 15-residues has no effect on DJ-1 dimerization. We then assessed the protease activities of purified recombinant DJ-1 WTΔC and DJ-1 WT proteins (Fig. 2D) by using a continuous, real-time fluorescence-based protease assay with BODIPY FL-labeled casein as the substrate (7,40). We found that DJ-1 WTΔC protein exhibited much higher protease activity than DJ-1 WT protein (Fig. 2D and E), indicating that removal of the C-terminal 15-amino acid peptide activates the protease function of DJ-1.
Figure 2.
Deletion of the C-terminal tail enhances DJ-1 protease activity without affecting its dimerization. (A and B) Lysates from HeLa cells expressing the indicated HA- and/or Myc-tagged DJ-1 proteins were immunoprecipitated with anti-HA antibody, followed by immunoblotting with anti-Myc and anti-HA antibodies. (C) DJ-1 dimerization was determined by quantification of the intensity of co-immunoprecipitated Myc-tagged DJ-1 band and normalized to the amount of HA-tagged DJ-1 band in the same immunoprecipitate. Data represents mean ± SEM from three independent experiments. (D) Protease activity of the indicated proteins was measured by a continuous, real-time fluorescence-based protease assay using BODIPY FL-casein (10 µg/ml) as the substrate. Fluorimetric recordings of 0.6 µm trypsin (blue), 50 µm DJ-1 WT or DJ-1 WTΔC (green), DJ-1 C106A or DJ-1 C106AΔC (red), DJ-1 H126AΔC (purple) and BSA (black). RFU, relative fluorescence units. (E) Protease activity of each protein was quantified by measuring the initial velocity of the proteolytic reaction from the fluorimetric trace shown in Fig. 2D. Protease activity on the Y-axis indicates the measured protease activity of each protein after normalization to the protease activity of DJ-1 WT. Data represents mean ± SEM from three independent experiments. *P < 0.05.
The C-terminally cleaved form of DJ-1 is an active cysteine protease with a functional Cys-His catalytic diad
Although DJ-1 does not contain a canonical Cys-His-Glu catalytic triad found in the PfpI family of cysteine proteases such as PH1704, structural comparison reveals that Cys-106 of DJ-1, like the catalytic nucleophile Cys-100 of PH1704, resides in a ‘nucleophile elbow’ with a strained backbone conformation (27,28,30). Moreover, Cys-106 is located in close proximity to His-126 in the crystal structure (Fig. 1A) and has the potential to form a Cys-His catalytic diad with His-126 (28,30). A functional role of Cys-106 and His-126 is further suggested by the fact that both of these residues are evolutionally conserved in all DJ-1 orthologs across various species (25). However, whether Cys-106 and His-126 can form a catalytic diad is debatable (26–30) because in the crystal structure, the His-126 imidazole ring is in an unfavorable orientation for diad formation as it forms a hydrogen bond with the main-chain carbonyl group of Pro-184 in the helix H8 (Fig. 1A).
Our finding that removal of the helix H8-containing C-terminal 15-amino acid peptide activates DJ-1 protease function (Fig. 2) raises the possibility that elimination of the hydrogen bond formation between His-126 and Pro-184 by the C-terminal cleavage may enable His-126 to form a catalytic diad with Cys-106 for carrying out the proteolytic function of activated DJ-1 protease. To test whether Cys-106 and His-126 can form a catalytic diad in the activated DJ-1, we used site-directed mutagenesis to generate DJ-1 C106AΔC and H126AΔC mutants, which replaced Cys-106 and His-126 with an Ala residue in the C-terminally cleaved form of DJ-1, respectively (Fig. 1C). Co-immunoprecipitation analysis confirmed that neither C106AΔC nor H126AΔC mutation affected DJ-1 dimerization (Fig. 2B and C). Protease activity assays revealed that the protease activity of DJ-1 WTΔC protein was abolished by the C106A mutation of the predicted catalytic nucleophile Cys-106 (Fig. 2D and E). Similarly, the protease activity of DJ-1 WTΔC protein was also abrogated by the H126A mutation of the putative Cys-His catalytic diad component His-126 (Fig. 2D and E). Together, these data support that the C-terminally cleaved form of DJ-1 is an active cysteine protease which uses the diad formed by Cys-106 and His-126 for catalysis.
DJ-1 cysteine protease is susceptible to inactivation by oxidation
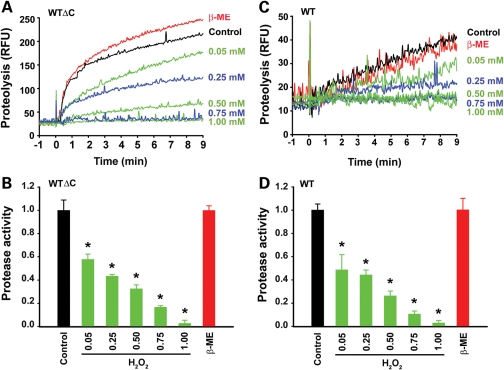
Cysteine protease usually requires a reducing environment for achieving full activity because oxidation of the catalytic thiol group on the active site cysteine abolishes its ability to mediate nucleophilic attack for breaking the peptide bond (31). Given our result suggesting that DJ-1 functions as a cysteine protease with Cys-106 as the catalytic nucleophile (Fig. 2D and E), we next assessed whether DJ-1, like other cysteine proteases, can be inactivated by oxidation. Our protease assays revealed that pre-incubation of DJ-1 WTΔC protein with increasing concentrations of the oxidizing agent H2O2 (0.05–1 mm) resulted in a dose-dependent loss of DJ-1 WTΔC protease activity (Fig. 3A and B). In contrast, pre-incubation with the reducing agent β-mercaptoethanol (β-ME; 1 mm) did not significantly alter protease activity of DJ-1 WTΔC protein (Fig. 3A and B). Similarly, protease activity of DJ-1 WT was also inhibited by pre-incubation with increasing concentrations of H2O2 but not by pre-incubation with β-ME (Fig. 3C and D). These results, together with the previous report that Cys-106 is the most sensitive cysteine residue of DJ-1 to H2O2-mediated oxidation (41), further support the function of DJ-1 as a cysteine protease and indicate that DJ-1 protease, in its active form or precursor form, is susceptible to inhibition by oxidation.
Figure 3.
Inhibition of DJ-1 cysteine protease function by oxidation. (A and C) Protease activity of 50 µm DJ-1 WTΔC (A) or 50 µm DJ-1 WT (C) was measured after pre-incubation with the indicated concentrations of H2O2 or 1 mm β-ME or mock-treated (control) at 37°C for 30 min by a real-time fluorescence-based protease assay using the fluorogenic substrate BODIPY FL-casein (10 µg/ml). RFU, relative fluorescence units. (B and D) Protease activity was quantified by measuring the initial velocity of the proteolytic reaction under the indicated conditions and normalized to the protease activity of mock-treated DJ-1 WTΔC (B) or DJ-1 WT (D). Data represents mean ± SEM from three independent experiments. *P < 0.05.
C-terminal cleavage activates DJ-1 protease by enhancing substrate binding as well as catalysis
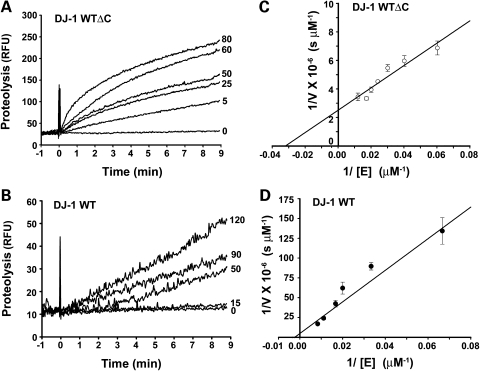
To further characterize the activation of DJ-1 protease function by the C-terminal cleavage, we performed kinetic analyses of casein proteolysis catalyzed by DJ-1 WT and DJ-1 WTΔC proteins (Fig. 4). The kinetic parameters Km and kcat were determined from the double reciprocal plots (Fig. 4C and D) as described (42) and summarized in Table 1. As a positive control, we analyzed trypsin-mediated casein proteolysis in parallel and found that trypsin has a Km of 6.5 µm and kcat of 1.4 s−1, which are similar to the values reported previously (43,44), validating our assay system. Our analyses showed that the Km value of DJ-1 WTΔC is more than 5-fold lower than that of DJ-1 WT (Table 1), suggesting that the cleavage of the C-terminal 15-amino acid peptide enhances substrate binding. Moreover, DJ-1 WTΔC has a 4.6-fold greater kcat-value than DJ-1 WT (Table 1), suggesting that the C-terminal cleavage also enhances the catalysis. The combination of these two effects leads to a 26-fold greater catalytic efficiency (kcat/Km) for DJ-1 WTΔC compared with DJ-1 WT (Table 1). The increase in catalytic efficiency after zymogen activation is often referred to as zymogenicity (31,45). Our kinetic studies thus indicate that DJ-1 protease has a zymogenicity of 26, when activated by the removal of the C-terminal 15-amino acid peptide.
Figure 4.
Kinetic analyses of the full-length and C-terminally cleaved forms of DJ-1 protease. (A and B) Protease activities of the indicated concentrations (in μm) of DJ-1 WT (A) and DJ-1 WTΔC (B) measured in a real-time protease assay using the fluorogenic substrate BODIPY FL-casein (10 µg/ml). RFU, relative fluorescence units. (C and D) Double reciprocal plot of 1/V versus 1/[E] for DJ-1 WT (C) and DJ-1 WTΔC (D). Each data point represents mean ± SEM from three independent experiments.
Table 1.
Summary of kinetic parameters
| kcat (s−1) | Km (µm) | kcat/Km (m−1 s−1) | |
|---|---|---|---|
| DJ-1 WT | 0.023 ± 0.018 | 173.4 ± 23.7 | 1.3 × 102 |
| DJ-1 WTΔC | 0.105 ± 0.011 | 31.3 ± 3.0 | 3.4 × 103 |
| Trypsin | 1.436 ± 0.234 | 6.5 ± 0.3 | 2.2 × 105 |
Endogenous DJ-1 in dopaminergic cells undergoes C-terminal cleavage in response to mild oxidative stress
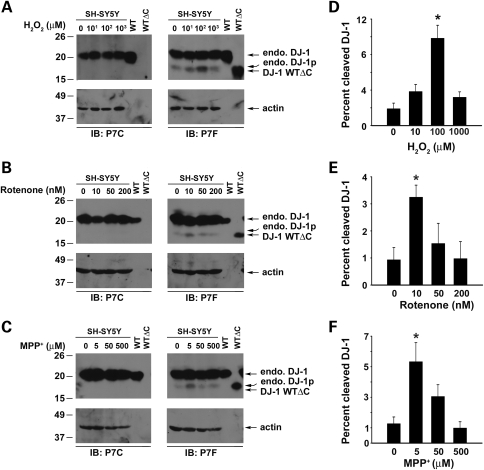
The observed activation of DJ-1 protease zymogen upon removal of its C-terminal 15 residues (Figs 2 and 4) prompted us to investigate whether endogenous DJ-1 can undergo C-terminal proteolytic processing in human SH-SY5Y dopaminergic cells. For the detection of C-terminally cleaved form of endogenous DJ-1, we took advantage of two anti-DJ-1 antibodies, P7C and P7F, which we generated and characterized previously (6,7). P7C was generated against the C-terminal 20 residues of DJ-1, and this antibody selectively recognizes full-length DJ-1 but not C-terminally cleaved form of DJ-1, such as DJ-1 WTΔC (Fig. 5A–C). In contrast, P7F was raised against purified full-length DJ-1 recombinant protein, and this antibody is capable of detecting both full-length and C-terminally cleaved forms of DJ-1 (Fig. 5A–C). Using these antibodies, we found that, under normal cell culture conditions, endogenous DJ-1 existed primarily in the full-length form in cells (Fig. 5A–C), indicating that, in common with other proteases, DJ-1 is synthesized as a latent protease zymogen. In response to oxidative stress induced by H2O2 (Fig. 5A), rotenone (Fig. 5B) or 1-methyl-4-phenylpyridinium (MPP+) (Fig. 5C), endogenous DJ-1 underwent C-terminal cleavage, resulting in a processed form of DJ-1 protein (DJ-1p) that was recognized specifically by the P7F antibody but not by the P7C antibody (Fig. 5A–C). The endogenous DJ-1p protein had an apparent molecular weight of ∼17 kDa, which was very close to that of recombinant DJ-1 WTΔC (Fig. 5A–C), supporting that the proteolytic processing of endogenous DJ-1 occurs at its C terminus near residue 174. Interestingly, the level of endogenous DJ-1p was significantly higher in cells treated with low concentrations of rotenone, MPP+, or H2O2 (Fig. 5), namely at a dosage equivalent to the LD10 of rotenone, LD25 of H2O2 or LD10 of MPP+ where there was no overt cell death (Fig. 5 compared with Supplementary Material, Fig. S1). Together, these data suggest that endogenous DJ-1 protease zymogen in dopaminergic cells is activated in response to mild oxidative stress.
Figure 5.
Analysis of C-terminal cleavage of endogenous DJ-1 under normal and oxidative stress conditions. (A–C) SH-SY5Y cells were treated with indicated concentrations of H2O2 (A), rotenone (B) or MPP+ (C) for 24 h, and the levels of full-length endogenous DJ-1 (endo. DJ-1) and C-terminally processed form of endogenous DJ-1 (endo. DJ-1p) in the cell lysates were analyzed by immunoblotting with anti-DJ-1 antibody P7C or P7F as indicated. Purified recombinant full-length DJ-1 WT and DJ-1 WTΔC proteins were included as positive controls. Actin was used as a loading control. (D–F) The levels of the cleaved DJ-1 and full-length DJ-1 proteins were quantified by measuring the intensity of the DJ-1p and the full-length DJ-1 protein band, respectively. Percent cleaved DJ-1 on the Y-axis indicates the level of cleaved DJ-1 protein as a percentage of the total level of DJ-1 proteins (full-length DJ-1 plus DJ-1p) in each cell lysate. Data represent mean ± SEM from three independent experiments. *P < 0.05.
Previous studies have shown that auto-proteolysis is involved in activation of a number of protease zymogens (46,47), therefore we sought to determine whether DJ-1 is capable of auto-proteolysis in response to oxidative stress. Purified recombinant DJ-1 WT, DJ-1 C106A or DJ-1 WTΔC proteins were incubated with various concentrations of H2O2 and the potential proteolysis of these proteins was assessed by immunoblot analysis with anti-DJ-1 antibody P7F. The result showed no detectable self-cleavage of DJ-1 proteins (data not shown), arguing against the possibility that DJ-1 is activated by auto-proteolysis.
DJ-1 protease activation by C-terminal cleavage enhances its cytoprotective function
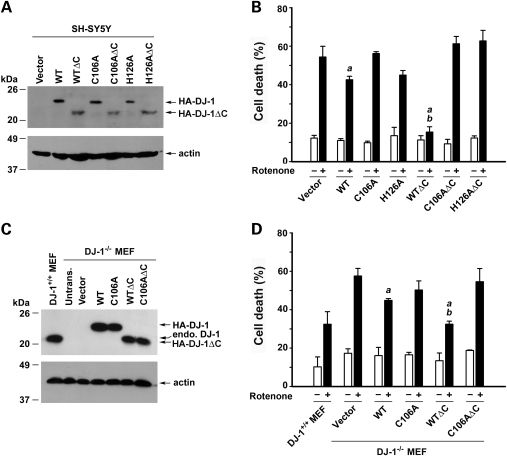
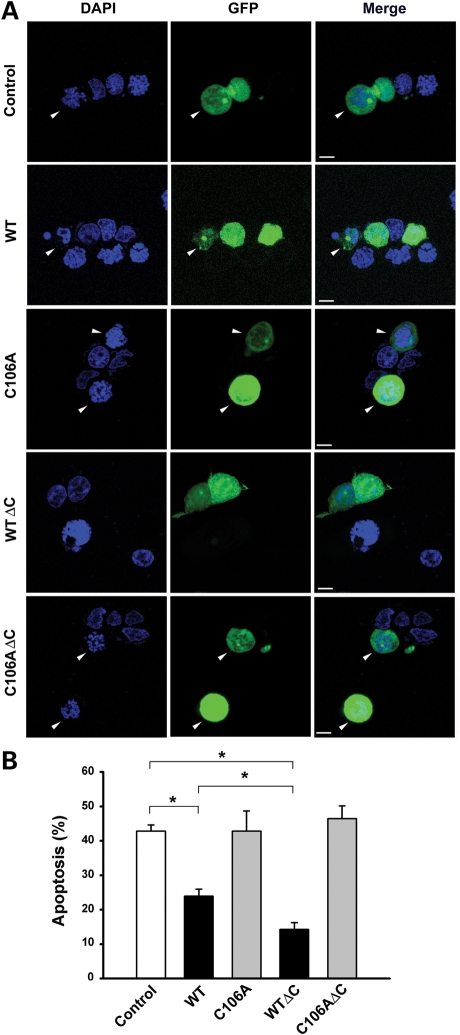
To determine the functional consequence of DJ-1 protease activation in cells, we first examined the effect of DJ-1 C-terminal cleavage on the vulnerability of SH-SY5Y cells to oxidative stress induced by PD-relevant toxicant rotenone. Consistent with previous reports (48–50), we found that over-expression of full-length DJ-1 WT protein in SH-SY5Y cells significantly reduced rotenone-induced apoptotic cell death, as measured by the lactate dehydrogenase (LDH) release assay (Fig. 6A and B) and apoptotic nuclear morphology analysis (Fig. 7). The cytoprotective action of DJ-1 was further enhanced by the C-terminal cleavage, as transfected SH-SY5Y cells expressing DJ-1 WTΔC were significantly more resistant to rotenone-induced apoptosis than cells expressing similar level of DJ-1 WT (Figs 6A, B, and 7). The ability of DJ-1 WT and DJ-1 WTΔC to protect SH-SY5Y cells against rotenone-induced apoptosis were abrogated by the catalytic site mutations C106A and H126A (Fig. 6A and B), indicating that the protease activity of DJ-1 is required for its cytoprotective action.
Figure 6.
C-terminal cleavage enhances DJ-1 cytoprotective action and this effect is dependent on DJ-1 protease activity. (A and C) Lysates from SH-SY5Y cells (A) or DJ-1−/− MEFs (C) transfected with the pCHA vector or indicated HA-tagged wild-type or mutant DJ-1 were analyzed by immunoblotting with anti-HA antibody (A) or anti-DJ-1 antibody P7F (C). The amount of cell lysates loaded in each lane was verified using anti-actin antibody. (B and D) SH-SY5Y cells (B) or DJ-1−/− MEFs (D) transfected with indicated plasmids were treated with 100 nm rotenone for 24 h. The extent of cell death was assessed by measuring LDH released to the culture medium and normalized to the total level of LDH release upon cell lysis. Data represent mean ± SEM from four independent experiments. aSignificantly different from the corresponding vector-transfected control (P < 0.05). bSignificantly different from the corresponding DJ-1 WT-transfected control (P < 0.05).
Figure 7.
The ability of DJ-1 to protect against rotenone-induced apoptosis is augmented by its C-terminal cleavage. (A) SH-SY5Y cells co-transfected with an expression vector encoding green fluorescent protein (GFP) and vehicle (control) or indicated DJ-1 plasmids were treated with 100 nm rotenone for 24 h. Transfected cells were shown by GFP fluorescence (green) and nuclear integrity was assessed by DAPI staining (blue). Arrowheads indicate transfected cells with apoptotic nuclei. Scale bar, 10 µm. (B) Apoptosis is expressed as the percentage of transfected cells with apoptotic nuclear morphology. Data represent mean ± SEM from four independent experiments. *P < 0.05.
To provide further evidence for the involvement of DJ-1 protease activation in cytoprotection, we assessed the role of DJ-1 C-terminal cleavage in cellular defense against oxidative stress using the mouse embryonic fibroblasts (MEFs) cultured from the DJ-1 knockout (DJ-1−/−) mice (6,51). The DJ-1−/− MEF cells lack DJ-1 expression, as confirmed by immunoblot analysis with anti-DJ-1 antibody P7F (Fig. 6C), and thus allow experiments to directly compare the cytoprotective actions of the full-length and C-terminally cleaved forms of DJ-1 without interference from endogenous DJ-1. We found that DJ-1−/− MEFs were significantly more susceptible than DJ-1+/+ MEFs to rotenone-induced cell death (Fig. 6C and D). The pro-apoptotic phenotype of DJ-1−/− MEFs were suppressed more effectively by expressing DJ-1 WTΔC than by expressing DJ-1 WT in these cells (Fig. 6C and D), indicating that the C-terminally cleaved form of DJ-1 has a stronger cytoprotective capability than the full-length DJ-1. The capability of DJ-1 WTΔC and DJ-1 WT to suppress the pro-apoptotic phenotype of DJ-1−/− MEFs was abolished by the C106A mutation of the putative catalytic nucleophile (Fig. 6C and D). These data, together with the observed effects of DJ-1 truncation and mutations on its protease activity (Fig. 2), support that DJ-1 protease activation by C-terminal cleavage enhances its cytoprotective function.
Activated DJ-1 protease exerts enhanced neuroprotection against oxidative stress-induced apoptosis
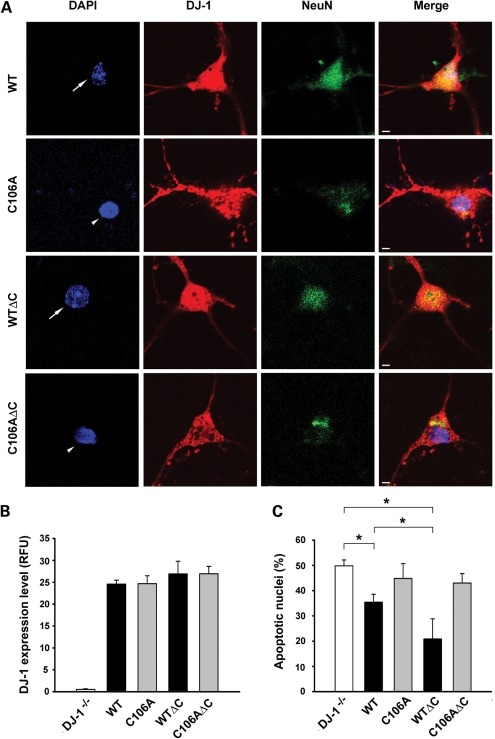
To directly determine the role of DJ-1 protease activation in neuroprotection, we expressed the full-length and C-terminally cleaved form of wild-type and mutant DJ-1 in primary cortical neurons cultured from DJ-1−/− mice. Immunostaining of transfected DJ-1−/− cortical neurons with anti-DJ-1 antibody P7F confirmed that DJ-1 WT and DJ-1 WTΔC and their C106A mutant forms were expressed at similar levels with an intracellular distribution pattern (Fig. 8A and B) similar to that of endogenous DJ-1 (7). Apoptosis assays revealed that the expression of DJ-1 WTΔC protein was significantly more effective in reducing rotenone-induced apoptosis in DJ-1−/− cortical neurons than full-length DJ-1 WT protein (Fig. 8). The ability of DJ-1 WT and DJ-1 WTΔC to rescue the pro-apoptotic phenotype of DJ-1−/− cortical neurons was abrogated by the protease-dead C106A mutation (Fig. 8), indicating that the protease activity of DJ-1 is required for its neuroprotective action. We then repeated the same experiments in primary mid-brain neurons cultured from DJ-1−/− mice and found that, compared with full-length DJ-1 WT protein at a similar expression level, the C-terminally cleaved form of DJ-1 exerted a stronger neuroprotective action in DJ-1−/− mid-brain neurons in a DJ-1 protease-dependent manner (Fig. 9). Together, these findings provide strong evidence that DJ-1 protease activation by C-terminal cleavage enhances its neuroprotective function.
Figure 8.
DJ-1 WTΔC is more effective than DJ-1 WT in suppressing the pro-apoptotic phenotype of DJ-1−/− cortical neurons. (A) DJ-1−/− cortical neurons transfected with indicated DJ-1 plasmids were treated with 5 nm rotenone for 24 h. Neurons were stained with anti-NeuN antibody (green), transfected neurons were identified by anti-DJ-1 (P7F) immunostaining (red) and nuclear integrity was assessed by DAPI staining (blue). Arrows indicate neurons with normal nuclei, whereas arrowheads indicate neurons with apoptotic nuclei. Scale bar, 10 µm. (B) The expression level of wild-type or mutant DJ-1 protein was quantified and is presented as the fluorescence intensity of DJ-1 immunofluorescence in transfected neurons. Data represent mean ± SEM from three independent experiments. RFU, relative fluorescence units. (C) Apoptosis is expressed as the percentage of transfected neurons with apoptotic nuclear morphology. Data represent mean ± SEM from four independent experiments. *P < 0.05.
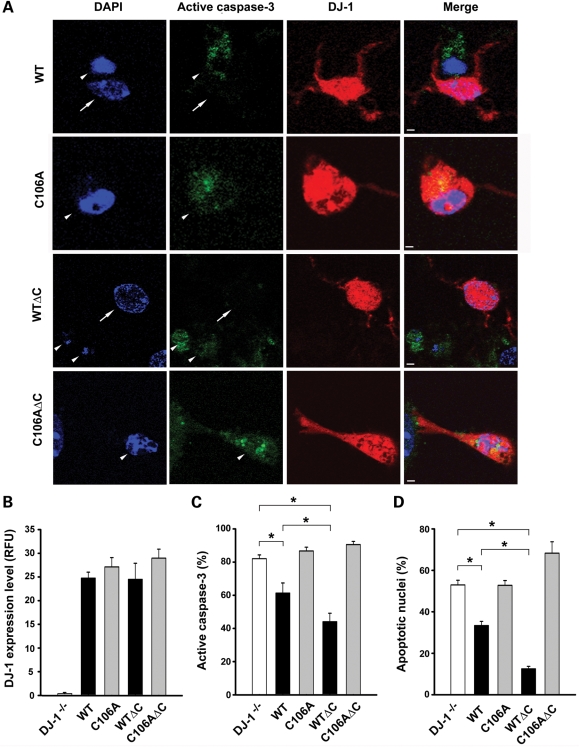
Figure 9.
C-terminal cleavage enhances the ability of DJ-1 to rescue the pro-apoptotic phenotype of DJ-1−/− mid-brain neurons. (A) DJ-1−/− mid-brain neurons transfected with indicated DJ-1 plasmids were treated with 5 nm rotenone for 24 h. Transfected neurons were identified by anti-DJ-1 (P7F) immunostaining (red), caspase-3 activation was assessed with anti-active-caspase-3 antibody (green) and nuclear morphology visualized by DAPI staining (blue). Arrows indicate neurons with normal nuclei and no active caspase-3 staining, whereas arrowheads indicate neurons with apoptotic nuclei and active caspase-3 staining. Scale bar, 20 µm. (B) The expression level of wild-type or mutant DJ-1 protein was quantified and is presented as the fluorescence intensity of DJ-1 immunofluorescence in transfected neurons. Data represent mean ± SEM from three independent experiments. RFU, relative fluorescence units. (C and D) The percentage of transfected neurons with active caspase-3 staining (C) or with apoptotic nuclei (D) was quantified. Data represent mean ± SEM from four independent experiments. *P < 0.05.
DISCUSSION
The identification of homozygous mutations in DJ-1 as a cause for autosomal recessive PD (2–4) and the association of abnormal DJ-1 expression with cancer (16,17) highlight the importance of understanding the biochemical function of this ubiquitously expressed protein. Although DJ-1 has been implicated in several cellular processes (5), the precise biochemical function of DJ-1 remains elusive. The present study reveals that DJ-1 is synthesized as a latent protease zymogen and its C-terminal cleavage activates DJ-1 cysteine protease function. Our data indicate that DJ-1 protease plays a role in cellular defense against oxidative stress-induced apoptosis and provide new insight into the mechanism of action for DJ-1 in normal physiology and PD pathogenesis.
We and other groups have previously shown that DJ-1 exhibits conspicuous structural and sequence similarity to the PfpI family of bacterial intracellular proteases (26–30). However, whether DJ-1 can function as a protease remains unclear because unlike PfpI proteases, DJ-1 lacks a canonical Cys-His-Glu catalytic triad and in the crystal structure, access to its predicted catalytic nucleophile Cys-106 appears to be blocked by the extra helix (helix H8) at its C terminus (Fig. 1A). Our biochemical analyses reveal that full-length DJ-1 protein is a cysteine protease zymogen which can be activated by cleavage of its helix H8-containing C-terminal 15-amino acid peptide (residues 175–189). We find that the activated DJ-1 protease, in common with several known cysteine proteases such as caspases (31–33), uses a Cys-His diad formed by its Cys-106 and His-126 for catalysis. Our kinetic analyses show that the removal of C-terminal 15-amino acid peptide results in a 5-fold decrease in the Km and a 4.6-fold increase in the kcat-value, indicating that the C-terminal cleavage activates DJ-1 protease by facilitating substrate binding as well as catalysis. These results are consistent with predictions from the crystal structure of full-length DJ-1 (26–30) and support a model in which the C-terminal cleavage converts DJ-1 into an active conformation by removing the helix H8 that blocks substrate entry and eliminating the interactions (e.g. the hydrogen bond between His-126 and Pro-184) that stabilize the inactive conformation of the zymogen.
Our finding that the protease activity of the C-terminally cleaved form of DJ-1 as well as the full-length DJ-1 is abolished by the C106A mutation supports the role of Cys-106 as the catalytic nucleophile of DJ-1 protease. We have shown that, DJ-1, in common with other cysteine proteases (31), requires a reducing environment for the catalytic thiol group to function and its protease activity is inhibited by oxidation. The oxidation-induced inactivation of DJ-1 protease could explain the failure to detect the protease activity of full-length DJ-1 in previous studies using non-reducing conditions during purification of DJ-1 protein and/or in the protease assays (29,52). The active site cysteine, Cys-106, was found to be oxidized during purification under non-reducing conditions (26), and similar oxidation of the active site cysteine during purification has also been reported for other cysteine proteases such as caspases (31).
Results from our group and many others have clearly established that DJ-1 exists as a homodimer in cells (7,53,54). Our finding that C-terminal cleavage activates DJ-1 protease function without affecting its dimerization suggests that DJ-1 dimerization per se is not involved in DJ-1 activation or inactivation. Our previous study has shown that PD-associated DJ-1 L166P mutation disrupts DJ-1 dimerization and abolishes DJ-1 protease activity and suggests that dimerization is required for DJ-1 protein stability and function (7). Indeed, monomeric form of DJ-1 protein was found to be unstable in cells and undergoes rapid degradation via the ubiquitin–proteasome pathway (7,53,54).
Our cellular studies indicate that endogenous DJ-1, in common with other proteases, resides in cells as a latent protease zymogen and requires limited proteolysis at its C terminus for activation. Our finding that DJ-1 zymogen is activated by C-terminal proteolytic processing in response to mild oxidative stress points to a role for DJ-1 protease in cellular defense against oxidative stress. The observation that the C-terminal cleavage of endogenous DJ-1 is reduced under strong oxidative stress conditions suggests that the protease responsible for cleaving endogenous DJ-1 might be a cysteine protease whose activity is inhibited by oxidation. Our results argue against the possibility that DJ-1 is activated via auto-proteolysis, although we cannot exclude this possibility completely because auto-proteolysis of DJ-1 may require a cofactor(s), as in the case of caspase-9 activation by apoptotic peptidase activating factor-1 apoptosome (39). Alternatively, DJ-1 zymogen may be activated by a yet unidentified protease, similar to the activation of executor caspases by the initiator caspases or the serine protease granzyme B (39,55). Future studies are needed to further elucidate the mechanism of DJ-1 protease activation.
The crystal structure of full-length DJ-1 also shows a high degree of structural similarity to that of E. coli chaperone protein Hsp31 (26–30), and full-length DJ-1 has been reported to have chaperone activity (29,52). It has been proposed that DJ-1 chaperone interacts with its client proteins via a hydrophobic patch composed of a cluster of surface-exposed hydrophobic residues primarily from helix H7 and helix H8 (29). Consistent with this idea deletion of DJ-1 helix H8-containing C-terminal 16-amino acid peptide (residues 174–189), which is predicted to disrupt the proposed hydrophobic patch, results in a significant reduction in the chaperone activity of DJ-1 (29). The opposite effects of C-terminal truncation on DJ-1 chaperone and protease activities raise the possibility that DJ-1 is a chaperone–protease dual function enzyme that switches from a chaperone to a cysteine protease upon oxidative stress-induced C-terminal cleavage. Previously, a chaperone–protease dual function has been described for E. coli heat shock protein DegP, which switches from a chaperone to a serine protease at elevated temperature (37,56,57). In addition, Hsp31 has been suggested to switch from a chaperone to a cysteine protease in a temperature-dependent manner (20).
Our data reveal that DJ-1 protease activation by C-terminal cleavage enhances its cytoprotective function against oxidative stress-induced apoptosis in both neuronal (SH-SY5Y cells, primary cortical neurons and primary mid-brain neurons) and non-neuronal (MEFs) cells. We find that the cytoprotective action of DJ-1 depends on its cysteine protease activity, as the catalytic site mutations C106A and H126A abolish the cytoprotective capability of the C-terminally cleaved DJ-1 as well as full-length DJ-1. Our findings raise the possibility that DJ-1 cysteine protease may exert cytoprotection against oxidative stress by mediating limited proteolysis of its substrate(s) or acting as a deubiquitinating or deSUMOylating enzyme for regulating ubiquitination or SUMOylation of its substrate(s). Future studies to identify substrate(s) of DJ-1 cysteine protease should advance our understanding of DJ-1 cytoprotective action and its dysregulation in PD and other neurodegenerative diseases.
MATERIALS AND METHODS
Expression constructs and antibodies
The pCHA-DJ-1, pMyc-DJ-1, pET3E-DJ-1 and the corresponding C106A mutant forms of DJ-1 have been described previously (7). Conventional molecular biological techniques were used to generate the following additional DJ-1 constructs in the pCHA, pMyc and pET3E vectors: DJ-1 H126A, DJ-1 WTΔC (residues 1–174) and its mutant forms DJ-1 C106AΔC and H126AΔC. All expression constructs were sequenced to confirm that the fusion was in the correct reading frame and there were no unwanted changes in the codons.
The rabbit polyclonal anti-DJ-1 antibodies, P7F and P7C, generated against purified full-length recombinant DJ-1 protein and a synthetic DJ-1 C-terminal peptide (residues 171–189), respectively, were described in our previous studies (6,7). Other antibodies used in this study include the following: anti-HA (12CA5), anti-Myc (9E10.3, Neomarkers), anti-actin (Chemicon), anti-NeuN (Millipore) and anti-active-caspase-3 (Cell Signaling). All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc.
DJ-1 knockout mice and primary cell cultures
DJ-1 knockout mice were generated by Dr Charles R. Gerfen at the National Institute of Mental Health and described previously (51). A breeding colony of DJ-1 knockout mice was established from breeding pairs obtained from the National Institute for Neurological Disease and Stroke/University of California Los Angeles Repository for Parkinson disease Mouse Models using standard mouse husbandry procedures (6). The genotype of the offspring was determined by PCR and the elimination of the DJ-1 protein was confirmed by western blot analysis. Primary cortical and mid-brain neuronal cultures were prepared from postnatal 0.5 day (P0.5) wild-type and DJ-1 knockout pups as described previously (58–60). Primary cortical and mid-brain neuronal cultures were maintained in NeuroBasal Media (Gibco) supplemented with cytosine arabinoside (AraC) (Sigma) for 3–7 days. MEF cultures were prepared from embryonic day 13 mice as described (59,61), and early passage cells were used for all experiments.
Protein expression and purification
Wild-type and mutant DJ-1 proteins were expressed in E. coli BL21 cells (CodonPlus) as untagged proteins and purified under reducing conditions as we described previously (7,27) by ammonium sulfate fractionation, Sepharose QXL ion exchange chromatography and Sephacryl S-100 gel filtration chromatography (Amersham). Fractions containing DJ-1 were identified by SDS–PAGE and confirmed by immunoblot analysis with anti-DJ-1 antibody P7F. Protein concentration was determined by using the bicinchoninic acid assay (Pierce).
Protease activity assays
Protease activities of DJ-1 WT and its mutants were measured by using a continuous, real-time fluorescence-based protease assay as described (7,40). Briefly, purified wild-type or mutant DJ-1 protein was incubated with BODIPY FL-labeled casein (10 µg/ml; Molecular Probes) in a protease assay buffer containing 50 mm Tris–HCl, pH 7.4, 200 mm NaCl and 5 mm CaCl2 at 37°C. An increase in fluorescence upon protease-catalyzed hydrolysis of casein was monitored continuously in a PerkinElmer Life Sciences LS55 luminescence spectrometer using an excitation wavelength of 505 nm and emission wavelength of 513 nm. Kinetic parameters (Km and kcat) were obtained as described (42) by fitting the data to the equation: V = kcat [S]0/(1 + Km [E]−1). Trypsin (Athena Environmental Sciences, Inc.) was used as a positive control for the protease activity assay. For the analysis of the effect of redox state on DJ-1 protease activity, purified recombinant DJ-1 proteins were treated with 1 mm β-ME, 0.05, 0.25, 0.50, 0.75, 1.00 mm H2O2 or mock-treated at 37°C for 30 min, dialyzed against the protease assay buffer using the D-tube dialyzer (Novagen), and then subjected to protease activity assays.
DJ-1 cleavage analysis
For the detection of endogenous DJ-1 cleavage in vivo, SH-SY5Y cells were treated with various concentrations of H2O2, rotenone or MPP+ for 24 h as indicated. The levels of full-length and processed forms of endogenous DJ-1 in the cell lysates were analyzed by immunoblotting with anti-DJ-1 antibodies P7C and P7F and quantified by using Scion Image software (NIH). For the analysis of DJ-1 self-cleavage in vitro, 7.5 µg of purified recombinant DJ-1 WT, DJ-1 C106A or DJ-1 WTΔC proteins were incubated with various concentrations of H2O2 at 37°C for 30 min in a total reaction volume of 20 µl containing 50 mm Tris–HCl, pH 7.4, 200 mm NaCl and 5 mm CaCl2. The reaction was stopped by boiling in Laemmli sample buffer, and the proteins were resolved by SDS–PAGE followed by immunoblotting with the P7F antibody.
Cell transfection, immunoprecipitation and immunoblot analysis
Cells were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cell lysates were prepared from transfected cells and immunoprecipitations were carried out as described previously (7,59–62) using anti-HA antibody followed by the recovery of immunocomplexes with protein G-Sepharose beads (Sigma). Immunocomplexes were then dissociated by boiling in the Laemmli sample buffer, resolved by SDS–PAGE, and immunoblotted with the indicated primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Results were visualized using enhanced chemiluminescence.
Immunofluorescence confocal microscopy
Cells were fixed in 4% paraformaldehyde, stained with appropriate primary and secondary antibodies and processed for indirect immunofluorescence microscopy as described previously (7,59–62). Analysis and acquisition were performed using a Zeiss LSM 510 confocal laser-scanning microscope. Images were imported in TIFF format using LSM-510 software (Carl Zeiss MicroImaging, Inc.) and processed using Adobe Photoshop (Adobe Systems, Inc.) to adjust the contrast and brightness.
Cell viability and apoptosis assays
Cell viability was assessed using either the LDH release assay as described (63) or the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described (60,64). For the analysis of apoptosis in transfected SH-SY5Y cells, transfected cells were identified by the green fluorescence emitted from co-transfected GFP expression vector and apoptotic cell death was measured by morphological assessment of nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI, Molecular Probes) as described (64). The percentage of transfected cells with nuclear fragmentation and chromatin condensation was scored for apoptosis.
For the analysis of apoptosis in transfected DJ-1−/− cortical neurons, cells were triple-stained with anti-NeuN as a neuronal cell marker, anti-DJ-1 antibody P7F for the detection of transfected wild-type or mutant DJ-1, and DAPI for the assessment of nuclear morphology. The percentage of DJ-1-transfected neurons with apoptotic nuclei was scored for apoptosis. For the analysis of apoptosis in transfected DJ-1−/− mid-brain neurons, transfected neurons were identified by immunostaining with anti-DJ-1 antibody P7F, and the extent of apoptotic cell death was determined by the measurement of caspase-3 activation with anti-active-caspase-3 antibody and assessment of nuclear integrity with DAPI staining. The percentage of DJ-1-transfected neurons with active caspase-3 staining or with apoptotic nuclei was scored for apoptosis as described (65). Quantification of DJ-1 expression level in transfected DJ-1−/− neurons was performed on unprocessed images of DJ-1 immunostained neurons using NIH Image J software as described (66). For each wild-type or mutant DJ-1 protein, the fluorescence intensity of DJ-1 immunostaining was quantified from 20 randomly selected transfected neurons and is presented as the mean ± SEM from three independent experiments.
Statistical analysis
All experiments were repeated at least three times. Data were subjected to statistical analysis by ANOVA and a P-value of less than 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL
FUNDING
The confocal imaging analysis was performed in Emory Neuroscience Core Facility supported in part by National Institutes of Health Grants P30 NS055077. This work was supported by National Institutes of Health Grants NS050650 and AG034126 (L.S.C.) and ES015813 and GM082828 (L.L.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Charles R. Gerfen and the National Institute of Neurological Disorders and Stroke/University of California Los Angeles Repository for Parkinson Disease Mouse Models for providing the DJ-1 knockout mice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lang A.E., Lozano A.M. Parkinson's disease—first of two parts. N. Engl. J. Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. doi:10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- 2.Hague S., Rogaeva E., Hernandez D., Gulick C., Singleton A., Hanson M., Johnson J., Weiser R., Gallardo M., Ravina B., et al. Early-onset Parkinson's disease caused by a compound heterozygous DJ-1 mutation. Ann. Neurol. 2003;54:271–274. doi: 10.1002/ana.10663. doi:10.1002/ana.10663. [DOI] [PubMed] [Google Scholar]
- 3.Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., Dekker M.C.J., Squitieri F., Ibanez P., Joosse M., et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset Parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. doi:10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 4.Abou-Sleiman P.M., Healy D.G., Quinn N., Lees A.J., Wood N.W. The role of pathogenic DJ-1 mutations in Parkinson's disease. Ann. Neurol. 2003;54:283–286. doi: 10.1002/ana.10675. doi:10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 5.da Costa C.A. DJ-1: a new comer in Parkinson's disease pathology. Curr. Mol. Med. 2007;7:650–657. doi: 10.2174/156652407782564426. doi:10.2174/156652407782564426. [DOI] [PubMed] [Google Scholar]
- 6.Olzmann J.A., Bordelon J.R., Muly E.C., Rees H.D., Levey A.I., Li L., Chin L.-S. Selective enrichment of DJ-1 protein in primate striatal neuronal processes: implications for Parkinson's disease. J. Comp. Neurol. 2007;500:585–599. doi: 10.1002/cne.21191. doi:10.1002/cne.21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olzmann J.A., Brown K., Wilkinson K.D., Rees H.D., Huai Q., Ke H., Levey A.I., Li L., Chin L.-S. Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 2004;279:8506–8515. doi: 10.1074/jbc.M311017200. doi:10.1074/jbc.M311017200. [DOI] [PubMed] [Google Scholar]
- 8.Choi J., Sullards M.C., Olzmann J.A., Rees H.D., Weintraub S.T., Bostwick D.E., Gearing M., Levey A.I., Chin L.-S., Li L. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J. Biol. Chem. 2006;281:10816–10824. doi: 10.1074/jbc.M509079200. doi:10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grazia A., Giovanni S., Pierfrancesco P., Marco D.A., Patrizia T., Paolo R., Vincenzo La B., Tommaso P., Donatella C., Ferdinanda A., et al. DJ-1 mutations and parkinsonism–dementia–amyotrophic lateral sclerosis complex. Ann. Neurol. 2005;58:803–807. doi: 10.1002/ana.20666. doi:10.1002/ana.20666. [DOI] [PubMed] [Google Scholar]
- 10.Rizzu P., Hinkle D.A., Zhukareva V., Bonifati V., Severijnen L.-A., Martinez D., Ravid R., Kamphorst W., Eberwine J.H., Lee V.M.Y., et al. DJ-1 colocalizes with tau inclusions: a link between parkinsonism and dementia. Ann. Neurol. 2004;55:113–118. doi: 10.1002/ana.10782. doi:10.1002/ana.10782. [DOI] [PubMed] [Google Scholar]
- 11.Neumann M., Müller V., Görner K., Kretzschmar H., Haass C., Kahle P. Pathological properties of the Parkinson's disease-associated protein DJ-1 in α-synucleinopathies and tauopathies: relevance for multiple system atrophy and Pick's disease. Acta Neuropathol. 2004;107:489–496. doi: 10.1007/s00401-004-0834-2. doi:10.1007/s00401-004-0834-2. [DOI] [PubMed] [Google Scholar]
- 12.Nagakubo D., Taira T., Kitaura H., Ikeda M., Tamai K., Iguchi-Ariga S.M.M., Ariga H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with Ras. Biochem. Biophys. Res. Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. doi:10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 13.Hod Y., Pentyala S.N., Whyard T.C., El-Maghrabi M.R. Identification and characterization of a novel protein that regulates RNA-protein interaction. J. Cell. Biochem. 1999;72:435–444. doi:10.1002/(SICI)1097-4644(19990301)72:3<435::AID-JCB12>3.0.CO;2-H. [PubMed] [Google Scholar]
- 14.Klinefelter G.R., Welch J.E., Perreault S.D., Moore H.D., Zucker R.M., Suarez J.D., Roberts N.L., Bobseine K., Jeffay S. Localization of the sperm protein SP22 and inhibition of fertility in vivo and in vitro. J. Androl. 2002;23:48–63. doi: 10.1002/jand.2002.23.1.48. [DOI] [PubMed] [Google Scholar]
- 15.Klinefelter G.R., Laskey J.W., Ferrell J., Suarez J.D., Roberts N.L. Discriminant analysis indicates a single sperm protein (SP22) is predictive of fertility following exposure to epididymal toxicants. J. Androl. 1997;18:139–150. [PubMed] [Google Scholar]
- 16.Yaacov H. Differential control of apoptosis by DJ-1 in prostate benign and cancer cells. J. Cell. Biochem. 2004;92:1221–1233. doi: 10.1002/jcb.20159. doi:10.1002/jcb.20159. [DOI] [PubMed] [Google Scholar]
- 17.Yuen H.-F., Chan Y.-P., Law S., Srivastava G., El-tanani M., Mak T.-W., Chan W. DJ-1 could predict worse prognosis in esophageal squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2008;17:3593–3602. doi: 10.1158/1055-9965.EPI-08-0214. doi:10.1158/1055-9965.EPI-08-0214. [DOI] [PubMed] [Google Scholar]
- 18.West A.B., Dawson V.L., Dawson T.M. To die or grow: Parkinson's disease and cancer. Trends Neurosci. 2005;28:348–352. doi: 10.1016/j.tins.2005.05.002. doi:10.1016/j.tins.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Kim R.H., Mak T.W. Tumours and tremors: how PTEN regulation underlies both. Br. J. Cancer. 2006;94:620–624. doi: 10.1038/sj.bjc.6602994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malki A., Caldas T., Abdallah J., Kern R., Eckey V., Kim S.J., Cha S.-S., Mori H., Richarme G. Peptidase activity of the Escherichia coli Hsp31 chaperone. J. Biol. Chem. 2005;280:14420–14426. doi: 10.1074/jbc.M408296200. doi:10.1074/jbc.M408296200. [DOI] [PubMed] [Google Scholar]
- 21.Halio S.B., Bauer M.W., Mukund S., Adams M.W.W., Kelly R.M. Purification and characterization of two functional forms of intracellular protease PfpI from the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 1997;63:289–295. doi: 10.1128/aem.63.1.289-295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du X., Choi I.-G., Kim R., Wang W., Jancarik J., Yokota H., Kim H. Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution. Proc. Natl Acad. Sci. USA. 2000;97:14079–14084. doi: 10.1073/pnas.260503597. doi:10.1073/pnas.260503597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halio S.B., Blumentals I.I., Short S.A., Merrill B.M., Kelly R.M. Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1996;178:2605–2612. doi: 10.1128/jb.178.9.2605-2612.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdallah J., Kern R., Malki A., Eckey V., Richarme G. Cloning, expression, and purification of the general stress protein YhbO from Escherichia coli. Protein Expr. Purif. 2006;47:455–460. doi: 10.1016/j.pep.2005.11.011. doi:10.1016/j.pep.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 25.Lucas J.I., Marin I. A new evolutionary paradigm for the Parkinson disease gene DJ-1. Mol. Biol. Evol. 2007;24:551–561. doi: 10.1093/molbev/msl186. doi:10.1093/molbev/msl186. [DOI] [PubMed] [Google Scholar]
- 26.Wilson M.A., Collins J.L., Hod Y., Ringe D., Petsko G.A. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc. Natl Acad. Sci. USA. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. doi:10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huai Q., Sun Y., Wang H., Chin L.-S., Li L., Robinson H., Ke H. Crystal structure of DJ-1/RS and implication on familial Parkinson's disease. FEBS Lett. 2003;549:171–175. doi: 10.1016/s0014-5793(03)00764-6. doi:10.1016/S0014-5793(03)00764-6. [DOI] [PubMed] [Google Scholar]
- 28.Honbou K., Suzuki N.N., Horiuchi M., Niki T., Taira T., Ariga H., Inagaki F. The crystal structure of DJ-1, a protein related to male fertility and Parkinson's disease. J. Biol. Chem. 2003;278:31380–31384. doi: 10.1074/jbc.M305878200. doi:10.1074/jbc.M305878200. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.-J., Kim S.J., Kim I.-K., Ko J., Jeong C.-S., Kim G.-H., Park C., Kang S.-O., Suh P.-G., Lee H.-S., et al. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J. Biol. Chem. 2003;278:44552–44559. doi: 10.1074/jbc.M304517200. doi:10.1074/jbc.M304517200. [DOI] [PubMed] [Google Scholar]
- 30.Tao X., Tong L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson's disease. J. Biol. Chem. 2003;278:31372–31379. doi: 10.1074/jbc.M304221200. doi:10.1074/jbc.M304221200. [DOI] [PubMed] [Google Scholar]
- 31.Stennicke H., Salvesen G.S. Catalytic properties of the caspases. Cell Death Differ. 1999;6:1054–1059. doi: 10.1038/sj.cdd.4400599. doi:10.1038/sj.cdd.4400599. [DOI] [PubMed] [Google Scholar]
- 32.Walker R.K., Krishnaswamy S. The activation of prothrombin by the prothrombinase complex. The contribution of the substrate-membrane interaction to catalysis. J. Biol. Chem. 1994;269:27441–27450. [PubMed] [Google Scholar]
- 33.Wilson K.P., Black J.-A.F., Thomson J.A., Kim E.E., Griffith J.P., Navia M.A., Murcko M.A., Chambers S.P., Aldape R.A., Raybuck S.A., et al. Structure and mechanism of interleukin-1 beta converting enzyme. Nature. 1994;370:270–275. doi: 10.1038/370270a0. doi:10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 34.Wickner S., Maurizi M.R., Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. doi:10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 35.Tomimatsu Y., Idemoto S., Moriguchi S., Watanabe S., Nakanishi H. Proteases involved in long-term potentiation. Life Sci. 2002;72:355–361. doi: 10.1016/s0024-3205(02)02285-3. doi:10.1016/S0024-3205(02)02285-3. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Otin C., Bond J.S. Proteases: multifunctional enzymes in life and disease. J. Biol. Chem. 2008;283:30433–30437. doi: 10.1074/jbc.R800035200. doi:10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clausen T., Southan C., Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. doi:10.1016/S1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 38.Neurath H. The versatility of proteolytic enzymes. J. Cell. Biochem. 1986;32:35–49. doi: 10.1002/jcb.240320105. doi:10.1002/jcb.240320105. [DOI] [PubMed] [Google Scholar]
- 39.Donepudi M., Grutter M.G. Structure and zymogen activation of caspases. Biophys. Chem. 2002;101–102:145–153. doi: 10.1016/s0301-4622(02)00151-5. doi:10.1016/S0301-4622(02)00151-5. [DOI] [PubMed] [Google Scholar]
- 40.Jones L.J., Upson R.H., Haugland R.P., Panchuk-Voloshina N., Zhou M., Haugland R.P. Quenched BODIPY dye-labeled casein substrates for the assay of protease activity by direct fluorescence measurement. Anal. Biochem. 1997;251:144–152. doi: 10.1006/abio.1997.2259. doi:10.1006/abio.1997.2259. [DOI] [PubMed] [Google Scholar]
- 41.Kinumi T., Kimata J., Taira T., Ariga H., Niki E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2004;317:722–728. doi: 10.1016/j.bbrc.2004.03.110. doi:10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- 42.Batra R., Khayat R., Tong L. Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease. Nat. Struct. Biol. 2001;8:810–817. doi: 10.1038/nsb0901-810. doi:10.1038/nsb0901-810. [DOI] [PubMed] [Google Scholar]
- 43.Erin J.F., Jason R.C., Kelvin H.L. Kinetic characterization of sequencing grade modified trypsin. Proteomics. 2005;5:2319–2321. doi: 10.1002/pmic.200401268. doi:10.1002/pmic.200401268. [DOI] [PubMed] [Google Scholar]
- 44.Farmer W.H., Yuan Z. A continuous fluorescent assay for measuring protease activity using natural protein substrate. Anal. Biochem. 1991;197:347–352. doi: 10.1016/0003-2697(91)90403-g. doi:10.1016/0003-2697(91)90403-G. [DOI] [PubMed] [Google Scholar]
- 45.Tachias K., Madison E.L. Converting tissue-type plasminogen activator into a zymogen. J. Biol. Chem. 1996;271:28749–28752. doi: 10.1074/jbc.271.46.28749. doi:10.1074/jbc.271.46.28749. [DOI] [PubMed] [Google Scholar]
- 46.Goll D.E., Thompson V.F., Li H., Wei W.E.I., Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 47.Boatright K.M., Salvesen G.S. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. doi:10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Canet-Aviles R.M., Wilson M.A., Miller D.W., Ahmad R., Mclendon C., Bandyopadhyay S., Baptista M.J., Ringe D., Petsko G.A., Cookson M.R. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl Acad. Sci. USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. doi:10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taira T., Saito Y., Niki T., Iguchi-Ariga S., Takahashi K., Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. doi:10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W., Freed C.R. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T α-synuclein toxicity. J. Biol. Chem. 2005;280:43150–43158. doi: 10.1074/jbc.M507124200. doi:10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 51.Manning-Bog A.B., Caudle W.M., Perez X.A., Reaney S.H., Paletzki R., Isla M.Z., Chou V.P., McCormack A.L., Miller G.W., Langston J.W., et al. Increased vulnerability of nigrostriatal terminals in DJ-1-deficient mice is mediated by the dopamine transporter. Neurobiol. Dis. 2007;27:141–150. doi: 10.1016/j.nbd.2007.03.014. doi:10.1016/j.nbd.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 52.Shendelman S., Jonason A., Martinat C., Leete T., Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits α-synuclein aggregate formation. PLoS Biol. 2004;2:1764–1773. doi: 10.1371/journal.pbio.0020362. doi:10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore D.J., Zhang L., Dawson T.M., Dawson V.L. A missense mutation (L166P) in DJ-1, linked to familial Parkinson's disease, confers reduced protein stability and impairs homo-oligomerization. J. Neurochem. 2003;87:1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 54.Miller D.W., Ahmad R., Hague S., Baptista M.J., Canet-Aviles R., McLendon C., Carter D.M., Zhu P.-P., Stadler J., Chandran J., et al. L166P mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin–proteasome system. J. Biol. Chem. 2003;278:36588–36595. doi: 10.1074/jbc.M304272200. doi:10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
- 55.Shi Y. Caspase activation: revisiting the induced proximity model. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. doi:10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 56.Spiess C., Beil A., Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. doi:10.1016/S0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 57.Krojer T., Garrido-Franco M., Huber R., Ehrmann M., Clausen T. Crystal structure of DegP (HtrA) reveals a new protease–chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. doi:10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 58.Ahlemeyer B., Baumgart-Vogt E. Optimized protocols for the simultaneous preparation of primary neuronal cultures of the neocortex, hippocampus and cerebellum from individual newborn (P0.5) C57Bl/6J mice. J. Neurosci. Methods. 2005;149:110–120. doi: 10.1016/j.jneumeth.2005.05.022. doi:10.1016/j.jneumeth.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 59.Giles L.M., Chen J., Li L., Chin L.-S. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum. Mol. Genet. 2008;17:2712–2722. doi: 10.1093/hmg/ddn173. doi:10.1093/hmg/ddn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee J.T., Wheeler T.C., Li L., Chin L.-S. Ubiquitination of α-synuclein by Siah-1 promotes α-synuclein aggregation and apoptotic cell death. Hum. Mol. Genet. 2008;17:906–917. doi: 10.1093/hmg/ddm363. doi:10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- 61.Olzmann J.A., Li L., Chudaev M.V., Chen J., Perez F.A., Palmiter R.D., Chin L.-S. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell. Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. doi:10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirk E., Chin L.-S., Li L. GRIF1 binds Hrs and is a new regulator of endosomal trafficking. J. Cell. Sci. 2006;119:4689–4701. doi: 10.1242/jcs.03249. doi:10.1242/jcs.03249. [DOI] [PubMed] [Google Scholar]
- 63.Gerstner B., DeSilva T.M., Genz K., Armstrong A., Brehmer F., Neve R.L., Felderhoff-Mueser U., Volpe J.J., Rosenberg P.A. Hyperoxia causes maturation-dependent cell death in the developing white matter. J. Neurosci. 2008;28:1236–1245. doi: 10.1523/JNEUROSCI.3213-07.2008. doi:10.1523/JNEUROSCI.3213-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pridgeon J.W., Olzmann J.A., Chin L.-S., Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5:1494–1503. doi: 10.1371/journal.pbio.0050172. doi:10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young J.E., Garden G.A., Martinez R.A., Tanaka F., Miguel Sandoval C., Smith A.C., Sopher B.L., Lin A., Fischbeck K.H., Ellerby L.M., et al. Polyglutamine-expanded androgen receptor truncation fragments activate a Bax-dependent apoptotic cascade mediated by DP5/Hrk. J. Neurosci. 2009;29:1987–1997. doi: 10.1523/JNEUROSCI.4072-08.2009. doi:10.1523/JNEUROSCI.4072-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cubells L., de Muga S.V., Tebar F., Bonventre J.V., Balsinde J., Pol A., Grewal T., Enrich C. Annexin A6-induced inhibition of cytoplasmic phospholipase A2 is linked to caveolin-1 export from the Golgi. J. Biol. Chem. 2008;283:10174–10183. doi: 10.1074/jbc.M706618200. doi:10.1074/jbc.M706618200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.