Abstract
Nance–Horan syndrome (NHS) is an X-linked developmental disorder, characterized by bilateral congenital cataracts, dental anomalies, facial dysmorphism and mental retardation. Null mutations in a novel gene, NHS, cause the syndrome. The NHS gene appears to have multiple isoforms as a result of alternative transcription, but a cellular function for the NHS protein has yet to be defined. We describe NHS as a founder member of a new protein family (NHS, NHSL1 and NHSL2). Here, we demonstrate that NHS is a novel regulator of actin remodelling and cell morphology. NHS localizes to sites of cell–cell contact, the leading edge of lamellipodia and focal adhesions. The N-terminus of isoforms NHS-A and NHS-1A, implicated in the pathogenesis of NHS, have a functional WAVE homology domain that interacts with the Abi protein family, haematopoietic stem/progenitor cell protein 300 (HSPC300), Nap1 and Sra1. NHS knockdown resulted in the disruption of the actin cytoskeleton. We show that NHS controls cell morphology by maintaining the integrity of the circumferential actin ring and controlling lamellipod formation. NHS knockdown led to a striking increase in cell spreading. Conversely, ectopic overexpression of NHS inhibited lamellipod formation. Remodelling of the actin cytoskeleton and localized actin polymerization into branched actin filaments at the plasma membrane are essential for mediating changes in cell shape, migration and cell contact. Our data identify NHS as a new regulator of actin remodelling. We suggest that NHS orchestrates actin regulatory protein function in response to signalling events during development.
INTRODUCTION
Nance–Horan syndrome (NHS) (MIM 302350) is an X-linked developmental disorder. Male patients exhibit severe congenital cataract, distinctive dental anomalies including supernumerary incisors and crown-shaped permanent teeth, characteristic dysmorphic features (anteverted pinnae, long face and prominent nasal bridge) and developmental delay in approximately 30–50% of cases. Heterozygous females show milder signs, typically posterior Y-sutural lens opacities (1–4). Mutations in a novel gene, NHS, have been identified as the cause of the syndrome (5,6). Interestingly, all mutations identified to date as a cause of this syndrome are null mutations (4–10).
The NHS gene is alternatively spliced and composed of 10 coding exons with at least five isoforms (Supplementary Material, Fig. S1A). Isoforms NHS-A and NHS-1A are both transcribed from exon 1 coding for a 1630 and 1651 amino acid protein, respectively. These two isoforms differ in the presence or absence of exon 3a (Supplementary Material, Fig. S1A). Null mutations in exon 1 of the NHS gene are predicted to only affect isoforms NHS-A and NHS-1A, implying that these isoforms are critical to the pathogenesis of NHS (5,6). A large insertion mutation was also identified in the first intron of the mouse Nhs gene which disrupts the expression of Nhs1, the human NHS-A equivalent, and results in bilateral total cataract (Xcat) (11). We recently identified copy number variation mutations of the NHS gene as a cause of X-linked congenital cataract in patients lacking other features of NHS (4). These non-recurrent rearrangements of the NHS gene are also predicted to result in altered transcriptional regulation.
Analysis of the mouse Nhs gene revealed expression from embryonic day 9.5 in the ventral neural tube and supports a role for NHS in the development of the lens, brain, heart and limbic system (5,11,12). NHS isoforms have been shown to be differentially expressed; isoforms containing exon 1 are expressed in epithelia and localize to the cell periphery, whereas isoforms lacking exon 1 were detected in non-epithelial tissue and localize to the cytoplasm (12,13). Interestingly, NHS-1A was recently shown to immunoprecipitate with the tight junction protein ZO-1, suggesting that NHS may have a role at tight junctions (13).
The cellular function for isoforms of the NHS protein is yet to be defined. To explore the function of the NHS protein, particularly isoforms NHS-A and NHS-1A which are critical to the pathogenesis of NHS, we investigated NHS localization and the cellular effect of NHS knockdown and identified interacting protein partners. We demonstrate that NHS is essential for maintaining cell morphology through the regulation of actin cytoskeletal dynamics and suggest that an important mechanism of remodelling of the actin cytoskeleton during development would therefore be lost in patients with NHS.
RESULTS
Localization of NHS to sites of cell–cell contact
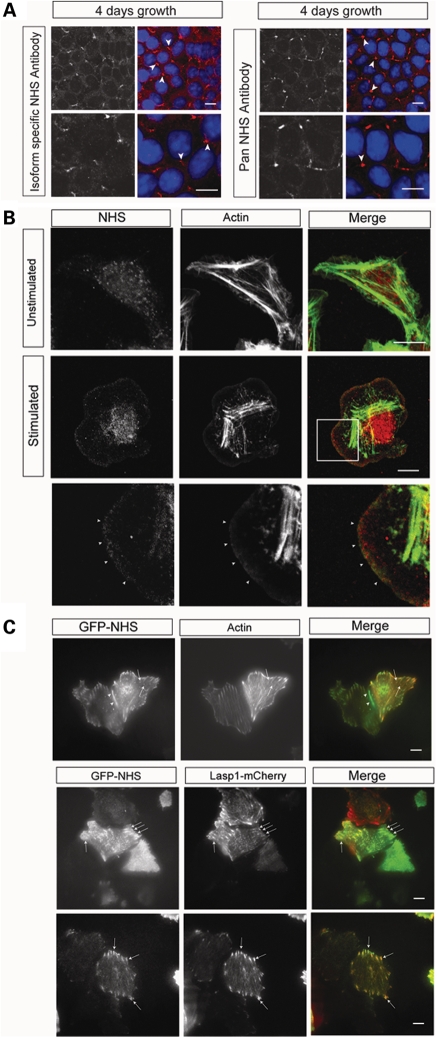
To explore the function of the NHS protein, in particular isoforms NHS-A and NHS-1A, we generated exon 1 isoform specific and pan NHS antibodies (Supplementary Material, Fig. S1A). Human epithelial colorectal adenocarcinoma (Caco-2) cells express isoform NHS-1A, determined by RT–PCR (data not shown). Both antibodies detected NHS at sites of cell–cell contact, in Caco-2 cells (Fig. 1A). NHS localization was most prominent at multicellular (tricellular) contacts (Fig. 1A) (14) and the expression level reduced as the cells differentiated (Supplementary Material, Fig. S2A). Further investigation in subconfluent cultures also revealed NHS localization at initial points of cell–cell contact (data not shown).
Figure 1.
NHS localizes to sites of cell contact, the leading edge of lamellipodia and focal adhesions. (A) Endogenous NHS (red) localized to sites of cell–cell contact in Caco-2 cells, detected by an N-terminal isoform-specific antibody (left panel) and a C-terminal pan NHS antibody (right panel). Lower panel for each antibody staining is a higher magnification. Nuclei were counterstained with DAPI. Staining for NHS (red) was prominent at tricellular contacts (arrowheads). Scale bar 10 µm. (B) Endogenous NHS (detected with pan NHS antibody; red) localized at the leading edge of lamellipodia in stimulated MTLn3 cells at the late 3 min transient (arrowheads). Cells were counterstained for F-actin (green). Top panel, unstimulated; middle panel, 3 min stimulation; bottom panel, enlargement of boxed area showing NHS at the leading edge of the lamellipod. Scale bar 10 µm. (C) Live TIRF imaging of GFP-NHS-1A (green) in MCF7 cells revealed NHS localization at cell junctions (arrowhead), the leading edge and co-localization with actin puncta (arrows) and actin fibres (red, top panel). GFP-NHS-1A co-localized with the focal adhesion complex protein Lasp1 (red, bottom two panels). Scale bar 10 µm.
NHS localizes to the leading edge of lamellipodia
As the intensity of NHS staining at cell–cell contacts did not correlate with junctional maturation, we examined the subcellular localization of endogenous NHS in motile cells to test whether there was a role for NHS in cell motility. We determined that rat mammary adenocarcinoma cells (MTLn3) cells express isoform NHS-1A by RT–PCR (data not shown). These cells can be stimulated with epidermal growth factor (EGF) to generate lamellipodia, with well-studied actin dynamics (15–18). Upon EGF stimulation, MTLn3 cells exhibit early (1 min) and late (3 min) barbed end transients (17,18). Endogenous NHS localized at the leading edge of lamellipodia at both the early PLC/cofilin-dependent transient (Supplementary Material, Fig. S2B) and the late PI3-kinase/WASP family/Arp2/3 complex-dependent transient (Fig. 1B). NHS localization at the early transient implies that it may be involved in the generation of barbed ends or is associated with newly formed actin filaments. Binding to the actin network early may also result in NHS localization at the leading edge at later time points, as seen at the 3 min transient (Fig. 1B).
NHS co-localizes with focal adhesion complexes
We noted that NHS appeared to co-localize with F-actin puncta behind the lamellipodium (Fig. 1B, Supplementary Material Fig. S2B), suggestive of possible co-localization with nascent adhesions (19). Therefore, we explored further the NHS association with these actin-rich structures. A GFP-NHS-1A (GFP-NHS) construct was used for live imaging in MCF7 cells. Total internal reflection fluorescence microscopy (TIRF), which penetrates to a depth of ∼100 nm enabling selective visualization of the membrane and cytoplasmic zone close to the membrane, revealed GFP-NHS-1A localization at cell–cell contacts and at the leading edge (Fig. 1C), further confirming the localization observed for endogenous NHS in Caco-2 and MTLn3 cells (Fig. 1A and B). In addition, GFP-NHS-1A co-localized with actin fibres (Lifeact-TagRed) and at specific actin puncta (Fig. 1C). Lasp1 is a dynamic actin-binding nascent focal adhesion protein required for cell migration (20). To establish whether these structures were focal adhesions, we examined NHS and Lasp1 (Lasp1-mCherry) localization in live cells. NHS and Lasp1 were found to co-localize at focal adhesion complexes (Fig. 1C).
Collectively, the localization of NHS at cell–cell contacts, the leading edge of lamellipodia and focal adhesions suggested that NHS has an important role in dynamic actin structures. We therefore addressed the question, what is the functional link between NHS and the actin cytoskeleton?
NHS is a founder member of a new protein family with homology to the WAVE proteins
To understand whether NHS has a direct role in actin cytoskeletal dynamics, we searched for potential homologies with other proteins and specifically with actin-binding and actin-associated proteins. We previously identified an NHS paralogue, NHS-like 1 (NHSL1, Supplementary Material, Fig. S3A) (6), which prompted a wider search for NHS paralogues resulting in the identification of NHSL2 (Supplementary Material, Fig. S3A). NHS, NHSL1 and NHSL2 have conserved genomic structure and appear to be present only in vertebrate genomes (Supplementary Material, Fig. S3A). The analysis of NHSL2 transcripts in human fetal cDNA detected expression in all tissues tested and suggests that NHSL2 is also expressed during development (Supplementary Material, Fig. S3B). NHSL2 is more widely or abundantly expressed than NHS, since in a similar analysis using the same human fetal cDNA panel NHS expression was only detected in the fetal brain, thymus, lung and kidney (6). We identified nine regions of homology indicating common domains for this new protein family (Supplementary Material, Fig. S3C).
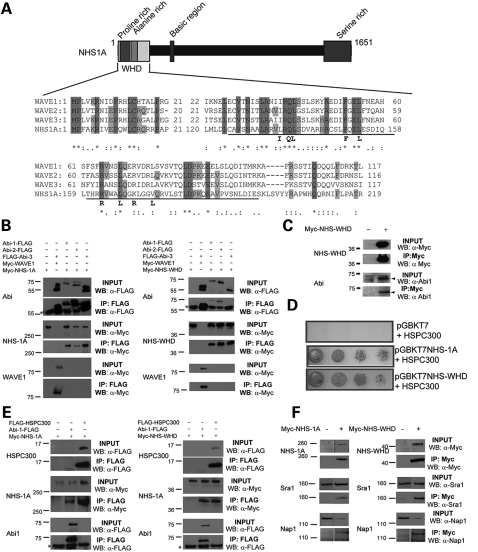
Three human WAVE (WASP family Verprolin-homologous) proteins have been identified, which are members of the Wiskott–Aldrich syndrome protein (WASP) family (21). Importantly, a sequence similarity was identified between the N-terminus of NHS and the WAVE homology domain (WHD) of the three human WAVE proteins (Fig. 2A). This domain of the NHS protein (homology domain 1) is conserved across the NHS protein family (Supplementary Material, Fig. S3C and D), supporting the hypothesis that homology to the WHD of the WAVE proteins may be functionally significant. Interestingly, this region of the NHS protein is only present in isoforms encoded by exon 1 (NHS-A and NHS-1A) which is conserved across species and critical to the pathogenesis of NHS (Supplementary Material, Fig. S1B). In mammals, the WASP protein family consists of four subclasses: WASP/N-WASP, WAVEs, WASH and WHAMM/JMY (Supplementary Material, Fig. S1C) (21–25). The WASP protein family are regulators of actin assembly, connecting signalling molecules (Rho GTPases) to the activation of the Arp2/3 complex (21–30). NHS also encodes proline-rich and basic regions, which are common domains in WASP proteins (Supplementary Material, Fig. S1C and D). WAVE proteins are functional components of a large protein complex (WAVE heteropentameric complex) that regulates actin dynamics at the leading edge of motile cells initiating lamellipodia formation downstream of the small GTPase Rac. The WHD of the WAVE proteins is required for a direct interaction with the Abl interactor (Abi) family of proteins and HSPC300 (30–34). Abi proteins are important regulators of the WASP protein family, facilitating the formation of a heteropentameric complex containing the WAVE proteins (WAVE/Abi/HSPC300/Nap1/Sra1) and influencing WASP and N-WASP-mediated processes (21–34). It is thought that the heteropentameric complex either trans-inhibits the ability of the WAVEs to activate the Arp2/3 complex (30) or is important for correct localization of the WAVEs for actin polymerization (32). Similar to WAVEs, Abi proteins localize to sites of actin polymerization in protrusive membrane structures (35).
Figure 2.
NHS contains a functional WHD which interacts with the Abi family of proteins, HSPC300, Nap1 and Sra1. (A) Schematic representation of the NHS protein and identified domains with an alignment of the N-termini of human NHS, WAVE1, WAVE2 and WAVE3. Letters in bold beneath the alignment denote conserved residues in mammalian WAVEs, Drosophila SCAR and Dictyostelium SCAR. The Abi binding domain is underlined. (B) Western blot analysis of immunoprecipitated FLAG-tagged Abi proteins to detect binding to Myc-tagged full-length NHS (NHS-1A) and NHS-WHD. IP, immunoprecipitation; WB, western blot. (C) Western blots of co-immunoprecipitated endogenous Abi1 with Myc-tagged NHS-WHD. IP, immunoprecipitation; WB, western blot. The expression of NHS-WHD resulted in increased levels of non-phosphorylated Abi1 (denoted by an arrowhead in the input panel). NHS-WHD preferentially co-immunoprecipitated non-phosphorylated Abi1. (D) Y2H library screening identified HSPC300 as a putative interactor. Both NHS-1A and NHS-WHD facilitated yeast growth on selective media with transcription of the reporter gene. The HSPC300-binding domain in NHS was therefore defined as the WHD (amino acids 1–262). Control (empty vector) did not grow on selective media. (E) Western blots of immunoprecipitated FLAG-tagged HSPC300 with Myc-tagged NHS-1A and NHS-WHD. IP, immunoprecipitation; WB, western blot. Asterisk in western blots denotes the antibody heavy chain. (F) Western blots of immunoprecipitated endogenous Nap1and Sra1 with Myc-tagged NHS-1A and Myc-tagged NHS-WHD. IP, immunoprecipitation; WB, western blot.
NHS encodes a functional WHD which interacts with all three members of the Abi protein family, HSPC300, Nap1 and Sra1
To address whether the NHS-WHD was functional, we first used a targeted approach to test the ability of NHS to bind Abi proteins. Full-length NHS (NHS-1A) co-immunoprecipitated with all three members of the Abi family (Fig. 2B). We then tested whether the NHS-WHD alone (amino acids 1–262) was necessary and sufficient to bind the Abi family. Co-immunopreciptation assays demonstrated the binding of the first 262 amino acids of the NHS protein with all Abi family members (Fig. 2B). The two Abi species detected on these blots represent phosphorylated (higher MW) and unphosphorylated (lower MW) forms of the Abi proteins (36). Endogenous Abi1 was also shown to co-immunoprecipitate with NHS-WHD (Fig. 2C).
To identify additional novel interactors of NHS, we also performed a yeast two-hybrid (Y2H) screening using an E17 mouse embryo library as the prey and the WHD of NHS as the bait (amino acids 1–262). An E17 embryo library was selected as the prey, since NHS has been shown to be expressed as early as E9.5 and clearly has a role in normal tissue development. Importantly, this non-targeted approach resulted in the identification of two cDNAs encoding HSPC300 from the Y2H library screening. HSPC300 is a member of the WAVE heteropentameric complex and directly interacts with the WHD of all three WAVE proteins (31). Both NHS-WHD and NHS-1A interacted with HSPC300 in yeast (Fig. 2D). This demonstrates that the interaction of NHS with HSPC300 is a direct interaction (i.e. not mediated by WAVE or Abi) since the WAVE complex is not present in yeast. The interaction of HSPC300 with NHS was further confirmed by co-immunoprecipitation (Fig. 2E).
In addition to direct binding of HSPC300 and Abi proteins to the WHD of WAVEs, the Abi proteins also bind Nap1 (Nck associated protein-1), facilitating recruitment of Nap1 to the heteropentameric WAVE complex. Sra1 (specifically Rac1-associated protein 1) is recruited to the complex through binding to Nap1 (30–32). Binding of the RhoGTPase Rac1 to Sra1 links Rho GTPase signalling to the Arp2/3 complex via the WAVE heteropentameric complex (32). Since HSPC300 interacts directly with the NHS-WHD, independent of Abi binding, and all Abi proteins also bind to the NHS-WHD, we tested the ability of NHS to co-immunoprecipitate the two remaining known WAVE heteropentameric complex members, Nap1 and Sra1. Endogenous Nap1 and Sra1 both co-immunoprecipitated with NHS-1A and NHS-WHD (Fig. 2F).
Importantly, these data demonstrate that the WHD in NHS is functionally similar to the WHD described in WAVE proteins. The identification of these novel protein partners of NHS, which are known to form a pentameric complex with the WAVE proteins, combined with our data localizing NHS to dynamic actin-rich structures, suggests that NHS has a direct role in actin dynamics. Therefore, we tested the hypothesis that NHS is required for dynamic actin cytoskeletal remodelling.
Depletion of NHS disrupts the circumferential actin ring and cell morphology
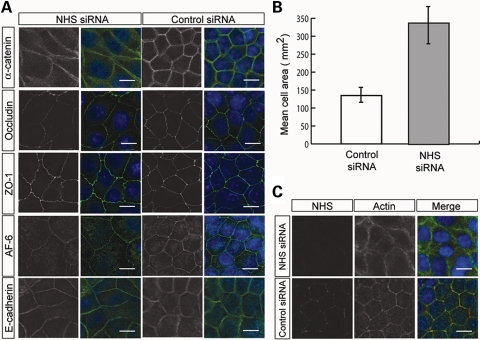
In epithelia, lamellipodial structures facilitate cell–cell adhesion, giving rise to cell–cell contacts. Clustering of cadherin junctions at points of contact between cells leads to the formation of actin bundles, which associate with cadherin clusters. New actin filaments are then recruited to the sites of cell–cell contact as they lengthen, eventually forming a circumferential actin ring (37). Polarization then occurs leading to junction maturation and a cuboidal morphology. To test whether NHS has a role in actin cytoskeletal assembly, Caco-2 cells were treated with short interfering RNA (siRNA) targeting all known NHS isoforms.
Cells treated with NHS siRNA (targeted to the 3′-UTR) appeared larger and did not exhibit a typical cuboidal morphology (Fig. 3A). Quantification revealed a 2.5-fold increase in the cell surface area of cells treated with NHS siRNA (Fig. 3B). The localization of most junctional proteins (including ZO-1) was unaffected following NHS knockdown. In contrast, α-catenin appeared more diffuse at points of cell–cell contact (Fig. 3A). Staining for F-actin in NHS-depleted cells revealed the disruption of the circumferential actin ring (Fig. 3C). In comparison to cells treated with control siRNA, NHS-depleted cells exhibited a diffuse, non-linear F-actin structure at the periphery (Fig. 3C). These data show that NHS is essential for maintaining the integrity of the circumferential actin ring and cell morphology.
Figure 3.
NHS is essential for organization of the circumferential actin ring. (A) Knockdown of NHS in Caco-2 cells resulted in larger cells, loss of a cuboidal cell shape and a diffuse localization of α-catenin (green) at the cell periphery. Scale bar 10 µm. Nuclei stained with DAPI (blue). (B) NHS siRNA treatment resulted in a 2.5-fold increase in cell area. For each condition, 100 cells from two independent experiments were measured. Error bars are ±standard deviation. (C) The thick circumferential ring of actin bundles (counterstained for F-actin, green) aligned with the cell borders was disrupted in NHS-depleted cells. Nuclei stained with DAPI (blue). Scale bar 10 µm.
NHS knockdown causes rearrangement of the actin cytoskeleton and excessive cell spreading and lamellipod extension
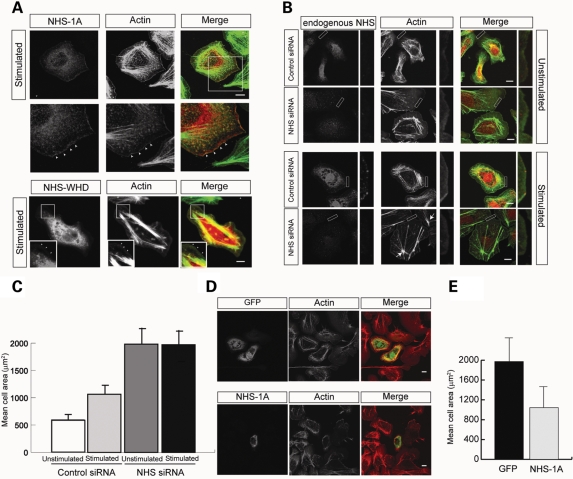
The WAVE heteropentameric complex proteins co-localize at the leading edge of lamellipodia. Localization of the WAVE proteins to the leading edge is dependent on their WHD and subsequent incorporation into the heteropentameric complex (38,39). To determine whether the WHD of NHS facilitates localization to the leading edge of lamellipodia, we expressed NHS-1A and NHS-WHD in MTLn3 cells. Both NHS-1A and NHS-WHD localized to the leading edge of lamellipodia in stimulated cells (Fig. 4A). NHS was not detected at the periphery of unstimulated MTLn3 cells (Supplementary Material, Fig. S2C). Co-expression of NHS-WHD, with either FLAG-Abi3 or FLAG-HSPC300, revealed co-localization throughout the cell and at the leading edge of lamellipodia in stimulated cells (Supplementary Material, Fig. S2D). These data show that NHS localization at the leading edge of lamellipodia is mediated, at least in part, by the NHS-WHD.
Figure 4.
NHS knockdown results in excessive cell spreading and lamellipod formation. (A) Ectopic expression of full-length (Myc-NHS-1A) and WHD (Myc-NHS-WHD) NHS constructs in MTLn3 cells after 3 min EGF stimulation. NHS constructs are in red and F-actin in green. NHS-1A and NHS-WHD localized at the leading edge after EGF stimulation (arrowheads). Scale bar 10 µm. (B) Knockdown of NHS expression results in increased cell spreading and lamellipod formation in stimulated and unstimulated MTLn3 cells. Cells were stained with the pan NHS antibody (red) and counterstained for F-actin (green). NHS siRNA-treated stimulated cells also displayed the re-arrangement of actin stress fibres (arrows) radiating from ‘spoke wheels’. Scale bar 10 µm. Lower panel for each is a higher magnification. (C) NHS knockdown cells did not spread in response to EGF stimulation. Cell area measurements for MTLn3 cells treated with control or NHS siRNAs in the presence (stimulated) and absence (unstimulated) of EGF. A total of 200 cells from two independent experiments were measured for each condition. Addition of EGF for 3 min increased cell area in control siRNA treated cells. No significant difference in cell area was observed in the presence/absence of EGF for NHS siRNA treated cells. Error bars are ±standard deviation. (D) Following siRNA treatment, the ectopic expression of siRNA-resistant NHS-1A (green) rescued the excessive cell spreading phenotype in unstimulated MTLn3 cells. Top panel: empty GFP vector control; bottom panel: NHS-1A rescue. Cells were counterstained for F-actin (red) Scale bar 10 µm. (E) Cell area measurements for unstimulated MTLn3 cells transected with either control (GFP) or NHS-1A following NHS siRNA treatment. The mean area of 26 GFP expressing cells and 21 NHS-1A expressing cells are shown ±standard deviation.
To explore the role of NHS at the leading edge, we then treated MTLn3 cells with NHS siRNA, which resulted in a striking excessive cell spreading phenotype (Fig. 4B) consistent with our previous observations with Caco-2 cells (Fig. 3). Cells with reduced NHS expression also displayed rearrangement of the actin cytoskeleton. Filamentous actin appeared to coalesce forming focal points from which actin filaments radiated, reminiscent of a spoke wheel (Fig. 4B). We then addressed whether NHS knockdown functionally altered lamellipod extension by quantification of the cell surface area in NHS siRNA-treated resting and EGF-stimulated MTLn3 cells, as cell area increase in response to EGF is a reliable marker of lamellipod extension in these cells (15–18,40). NHS knockdown led to a 3-fold increase in cell area compared with the unstimulated control siRNA-treated cells and a 2-fold increase compared with the stimulated siRNA-treated cells. The NHS siRNA-treated cells no longer increased in size on EGF stimulation, suggesting that this pathway has been dysregulated (Fig. 4C). The excessive cell spreading and lamellipod extension phenotype of NHS siRNA-treated cells was rescued by the ectopic expression of an siRNA-resistant NHS-1A construct (Fig. 4D and E).
Collectively, our data demonstrate that NHS regulates actin assembly and cell spreading. Our data also suggest that there is interplay between the WAVE heteropentameric complex and NHS at specific sites of dynamic remodelling. NHS may regulate turnover, displace or compete for the complex and negatively regulate WAVE function, which would be consistent with the observation that the absence of NHS leads to excessive cell spreading due to overactive WAVE complex activity. NHS may also have a role in co-ordinating a balance between actin-based protrusion and adhesion in lamellipodia.
NHS and WAVE complex interplay
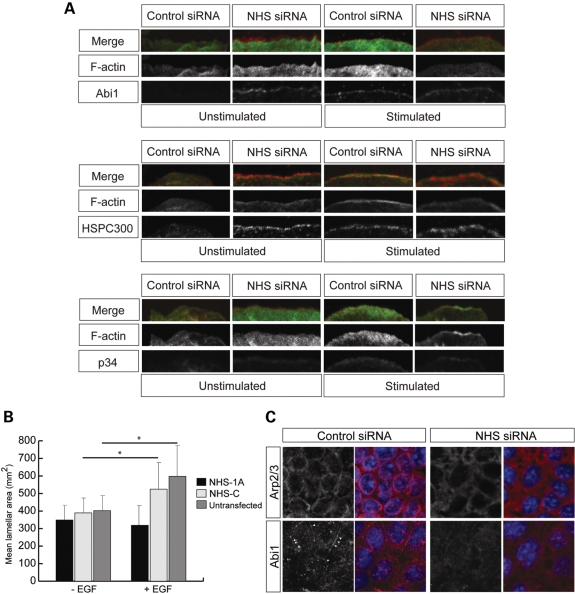
The activated WAVE complex at the leading edge of lamellipodia promotes actin polymerization at the plasma membrane by activating the Arp2/3 complex (40,41). We therefore tested whether an altered localization of complex members in the absence of NHS could be responsible for this dysregulation. WAVE and NHS protein partners Abi1 and HSPC300, and the p34 subunit of the Arp2/3 complex, consistently localized at the edges of cells treated with NHS siRNA, independent of EGF stimulation and WAVE complex activation (Fig. 5A; Supplementary Material, Fig. S4A). These data suggest that in the absence of NHS, the translocation of Abi1 is dysregulated. Furthermore, ectopic expression of NHS-1A inhibited the ability of cells to respond to EGF stimulation, as mean lamella area was significantly reduced compared with controls (Fig. 5B). These data indicate that cells overexpressing NHS lose the capacity to fully activate the WAVE complex.
Figure 5.
NHS is essential for co-ordinated actin regulatory protein dynamics. (A) Knockdown of NHS results in lamellipod formation and consistent localization of Abi1, HSPC300 and Arp2/3 at the cell membrane in the absence of EGF. Endogenous Abi1, HSPC300 and Arp2/3 were localized in unstimulated and EGF-stimulated (3 min) MTLn3 cells after treatment with control or NHS siRNAs. Abi1, HSPC300, and the p34 subunit of Arp2/3 are in red, F-actin in green. Scale bar 10 µm. (B) Ectopic expression of NHS-1A inhibited the ability of cells to respond to EGF stimulation. Cell lamella area measurements for stimulated or unstimulated MTLn3 cells transfected with either control (NHS-C) or NHS-1A ±standard deviation. Cells transfected with NHS-1A containing the WHD fail to extend lamellipods compared with the NHS-C construct lacking the WHD (*P < 0.001). (C) NHS depletion in Caco-2 cells caused mislocalization of Arp2/3 (red) and Abi1 (red). Scale bar 10 µm.
Since we already noted disruption of the circumferential actin ring in Caco-2 NHS knockdown cells (Fig. 3C), we examined endogenous Abi1 and Arp2/3 localization in these cells. Localization of Abi1 at sites of cell–cell contact was disrupted in NHS siRNA cells compared with controls (Fig. 5C). Furthermore, Arp2/3 localization at cell borders was disrupted in the absence of NHS, resulting in a diffuse localization at the cell periphery (Fig. 5C), consistent with the disrupted F-actin distribution (Fig. 3C).
These data show that localization of Abi1 and HSPC300 was altered in the absence of NHS. Importantly, in the absence of NHS, Arp2/3 localization and F-actin distribution are disrupted, suggesting that Arp2/3 actin-nucleation activity is mediated, in part, by NHS providing an additional functional link between NHS and actin. Interestingly, our data also suggest that the cellular response to EGF is bypassed in the absence of NHS or suppressed in the presence of NHS, implying that NHS may modulate responses to extracellular signalling.
DISCUSSION
NHS associates with actin-rich structures and controls cell morphology
Our data show that the NHS protein, which is essential for normal embryonic development, has a key role in maintaining cell morphology. NHS stabilizes the circumferential contractile actin ring which is essential for providing strength for cell adhesion and generating cell shape. The observation that α-catenin and F-actin localization was disrupted in NHS knockdown Caco-2 cells, but other junctional protein localization was unaffected, implies that NHS is required for the formation of a stable link between the cadherin/β-catenin complex and the actin cytoskeleton, but not essential for junction formation. This hypothesis is supported by the enlarged and non-cuboidal morphology observed in NHS knockdown cells. Recent evidence suggests that different conformational states of α-catenin confer its ability to form a stable link between the cadherin/β-catenin complex and the actin cytoskeleton (42). The disruption of circumferential F-actin and α-catenin as a result of NHS knockdown, but not E-cadherin localization, supports the hypothesis that there is a dynamic interaction between α-catenin and the actin cytoskeleton. In Caco-2 cells, we also observed the downregulation of NHS expression upon differentiation. This is consistent with the observation that NHS transcript expression in the anterior lens vesicle and fibre cells becomes progressively restricted to the anterior lens epithelium during lens development (12) and supports a role for NHS early in cell differentiation as opposed to junctional maturation.
In addition to their role in facilitating cell–cell adhesion, through cell–cell contacts, lamellipodial structures are essential for cell motility. We show that NHS also controls lamellipod protrusions in motile cells. NHS localized at the leading edge of lamellipodia of stimulated MTLn3 cells. The rearranged actin cytoskeleton and excessive cell spreading phenotype observed following NHS knockdown in these cells, resulting from excessive lamellipod extension, is consistent with our observations in Caco-2 cells. These data demonstrate that NHS has a key role in maintaining the robust cytoskeletal architecture required for cell stability and in controlling cytoskeletal plasticity to allow remodelling. Our data show that NHS maintains cell shape by stabilizing cortical actin structures and suggest that lack of this control leads to overactive dynamic lamellipod formation. The cortical actin stabilization function of NHS is facilitated, at least in part, through a functional WHD.
Nascent adhesions are myosin II-independent and serve as precursors for focal adhesions. They assemble in the lamellipodium and their rate of assembly is linked to protrusion rate and Arp2/3 actin polymerization (19). Even though the lamellipodial actin structure has been well described, the precise organization of the actin filaments that emerge from focal adhesions is poorly understood. Interestingly, NHS also localized to specific actin puncta behind the leading edge of the lamellipodium and co-localized with focal adhesion complexes implicating that NHS may have a role at the site of nascent adhesion assembly.
NHS regulates actin cytoskeletal dynamics
We describe NHS as the first human protein to contain a functional WHD since the discovery of the WAVE proteins. Enormous progress has been made to understand WAVE heteropentameric complex assembly and WAVE complex activation and function in the formation of the Rac-dependent membrane structures. However, the mechanism by which Rac regulates the WAVE complex is the subject of some debate and the mechanisms controlling WAVE heteropentameric complex assembly and disassembly have yet to be defined. We show that the WHD of NHS interacts with the Abi family of proteins, HSPC300, Nap1 and Sra1, and is important for the localization of NHS to the leading edge. We hypothesize that NHS affects the WAVE function, either by controlling assembly of the WAVE complex or by regulating the rate of WAVE complex assembly. In support of this hypothesis is the similarity of the phenotype of NHS knockdown in Caco-2 and MTLn3 cells to increased cell spreading of pirA (the Sra1 homologue) null Dictyostelium discoideum cells (43). The pirA null cells continually extended new pseudopods and the phenotype is thought to be due to overactive Scar (WAVE) protein function, as pirA negatively regulates the WAVE protein (43). Furthermore, Scar (WAVE) knockout Dictyostelium cells show reduced adhesion to the substratum, indicating a conserved association between the role of WAVE complex in adhesion and in protrusion of lamellipods and pseudopods (44). Therefore, excessive cell spreading and lamellipod extension as a result of NHS knockdown is likely to be a result of loss of WAVE complex regulation, leading to overactive WAVE activity or a dysregulation of nascent focal adhesions in lamellipodia.
NHS knockdown also resulted in the mislocalization of the Arp2/3 complex and disruption of the actin cytoskeleton. This implies that NHS function in controlling actin dynamics is mediated by regulating the Arp2/3 complex, which may be indirect as a result of negative regulation of the WAVEs, or by a direct effect of NHS function. The precise mechanism by which NHS controls actin dynamics is yet to be determined, and in this study, we have focused on the N-terminal WHD of NHS, so it is likely that other domains in this large protein confer additional functions affecting the interplay between NHS, the WAVE complex and actin. Since actin assembly factors work in concert, NHS may function as a key scaffold protein which controls the balance between cell adhesion and motility by coordinating the function of multiple regulatory pathways important for actin stability and plasticity.
The NHS protein family
Our study has focused on understanding the function of the N-terminal domain, encoded by exon 1 of the NHS gene. Importantly, NHS, NHSL1 and NHSL2 are much larger proteins and differ markedly from the WAVEs, with multiple shared protein domains of unknown function. The expression of different NHS protein family members is likely to be co-ordinated in a spatiotemporal manner during development. We predict that the WHDs we identified in NHSL1 and NHSL2 will also be functional and that they will have a role in regulating actin dynamics. However, these proteins appear to have no functional redundancy in the developing lens, brain and craniofacial tissues as highlighted by the lack of NHS in NHS patients. Investigation of the cellular and subcellular localization of NHSL1 and NHSL2, and their association with actin-rich structures, will be an important next step in understanding the function of this protein family.
Loss of NHS isoforms containing a functional WHD causes NHS
The NHS gene was first identified as the gene mutated in patients with a developmental disorder, NHS. Since null mutations have been identified in exon 1, which encodes only isoforms NHS-A and NHS-1A, these isoforms are known to be critical to the pathogenesis of NHS (5,6). In this study, we show that isoforms NHS-A and NHS-1A differ from other NHS isoforms in that they contain a WHD. Interestingly, NHS could co-immunoprecipitate all three Abi proteins. Which Abi proteins are the physiological partners of NHS is not clear. For example, NHS may preferentially bind one or more Abi's in vivo, and it is also likely that specificity is governed by spatiotemporal expression of both proteins. Abi2 is highly expressed in the lens and plays a pivotal role in the development of the anterior and posterior sutures (45). This suggests that an NHS–Abi2 interaction may be physiologically important for lens development and that null mutations in the NHS gene could cause congenital cataract by disruption of this interaction and the actin cytoskeleton. In support of the potential importance of an NHS–Abi2 interaction in the pathogenesis of NHS, Abi2 null mice also exhibit deficits in learning and memory, and similarly, NHS patients often have behavioural problems and developmental delay (3,4,45).
In summary, our data demonstrate that NHS has an important role in regulating cell morphology and functions as a novel actin regulatory protein. We suggest that NHS orchestrates the activity of other actin regulatory proteins to build specific cytoskeletal structures in response to cellular signals during development. These new findings and further research on the function of NHS and other family members will impact on our understanding of how tissues are shaped during development.
MATERIALS AND METHODS
Cell culture and stimulation
BE and Caco-2 cells were cultured in a Dulbecco's modified Eagle's medium (DMEM) medium (Gibco BRL), MTLn3 cells were cultured in an MEMα medium (Gibco BRL) and CHO cells in a DMEM–F12 medium (Gibco BRL). All cell culture mediums contained 10% fetal bovine serum (Gibco BRL), supplemented with 100 IU/ml penicillin and 100 IU/ml streptomycin (Invitrogen). Cells were grown at 37°C in the presence of 5% CO2. Cells were transfected with 100 ng of each DNA plasmid using Lipofectamine plus (Invitrogen) according to the manufacturer's instructions. Glass cover slips were pre-treated prior to seeding MTLn3 cells. Briefly, chamber slides were washed for 5 min in 1 M HCl, rinsed with 1× PBS, washed in 95% ethanol for 2 min, and then rinsed twice in 1× PBS. MTLn3 cells were cultured, starved and stimulated with murine natural EGF (Invitrogen) as previously described (37). MCF7 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum and 2 mm l-glutamine (Gibco BRL) and maintained at 37°C with 5% CO2. MCF7 cells were transfected using Amaxa Nucleofector Kit V (Lonza, VCA-1003). For live imaging of MCF7 cells, DMEM/F12 (1:1) medium (Gibco BRL) supplemented with 10% fetal bovine serum and 2 mm l-glutamine was used to culture cells for microscopy.
Antibodies and reagents
An N-terminal, isoform specific anti-peptide antibody (NHS-1A and NHS-A) was raised against amino acids 2–13 (PFAKRIVEPQWL) of the NHS protein and affinity purified (Eurogentec, Belgium). A second pan NHS anti-peptide antibody was raised against the C terminus of NHS, amino acids 1639–1652 (DGSPHDDRSSQSST) (Sigma Genosys). The following antibodies were used: mouse monoclonal anti-c-Myc 9E10 and M2 clone anti-FLAG (Sigma), rabbit polyclonal anti-p34, mouse monoclonal C4 anti-actin (Chemicon). Mouse monoclonal antibodies against occludin and ZO-1 were obtained from Zymed. Anti-GAPDH mouse monoclonal antibody was purchased from Sigma. Polyclonal rabbit Abi1 antibody was a kind gift from Alexis Gautreau. The polyclonal rabbit HSPC300 antibody was a generous gift from Theresia Stradal. A goat polyclonal antibody against E-cadherin, rabbit polyclonal against α-catenin and mouse monoclonal against AF-6 were a kind gift from Karl Matter. Alexa Fluor 488 phalloidin was obtained from Invitrogen. Rabbit polyclonal antibodies against Nap1 and Sra1 were obtained from Upstate Biotech.
Expression constructs
Full-length human NHS (amino acids 1–1652; NHS1A), the N terminus (amino acids 1–262; NHS-WHD) and the C terminus (amino acids 492–1651) were amplified by RT-PCR from human BE carcinoma cells and cloned into pCMV-Tag3b (Stratagene) to create constructs Myc-NHS1A, Myc-NHS-WHD and Myc-NHS-C, respectively. Full-length human NHS (amino acids 1–1652) was also cloned into pEGFP-C2 (Clontech) to create GFP-NHS1A for live imaging. Abi1, Abi2 and Abi3 were amplified from human cDNA clones: AU77-e10, AU7-b4 and AU34-b2 (MRC geneservice), respectively, and cloned into either pCMV-Tag2b (Stratagene) or p3 × FLAG-CMV™-14 (Sigma). The pCDNA5-FLAG-HA-HSPC300 construct was a gift from Alexis Gautreau. Empty pcDNA3-TagRed was a gift from Kurt Anderson and Lasp1-mChery was a gift from Bradford Ozanne. Lifeact was released from lifeact-GFP (a gift from Roland Wedlich-Soldner) and cloned into pDNA3-TagRed.
Y2H screening
Novel NHS interacting partners were identified using the Matchmaker GAL4 two-hybrid system 2 (Clontech). A 17-day mouse embryo cDNA library was obtained from Clontech in the vector pGADT7-Rec, pre-transformed into Saccharomyces cerevisiae yeast strain Y187. Full-length human NHS-1A cDNA (nt 1–4956) and the WHD domain of human NHS (nt 1–788; NHS-WHD) were cloned into the yeast expression vector pGBKT7 in frame with the GAL4 DNA-binding domain. The NHS-WHD bait construct was used to transform S. cerevisiae yeast strain AH109 using the manufacturers protocols (Clontech). The mating procedure was performed according to the manufacturer's protocols (Clontech). Approximately 2.4 × 107 clones from the 17-day mouse embryo library were screened. Positive colonies were picked and re-streaked on fresh SD/-Ade/-His/-Leu/-Trp agar plates. Extracted yeast plasmids were electroporated into Escherichia coli JM109 cells and plasmid DNA was purified using manufacturer's protocols (Sigma). Co-transformation of the bait pGBKT7-NHS plasmids (full-length and partial cDNAs) with each of the prey-positive plasmids was performed in AH109 to confirm positive interactions. Positive prey plasmids, confirmed by co-transformation, were sequenced and cDNA sequences were initially analysed using the BLAT algorithm at UCSC.
Immunoprecipitation assays
CHO cells were transfected with 100 ng of either, Myc-NHS1A (amino acids 1–1652), Myc-NHS-WHD (amino acids 1–262; WHD-NHS) or Myc-WAVE1 and 100 ng of Flag-tagged Abi1, Abi2, Abi3 or HSPC300. For co-immunoprecipitation of endogenous Abi1, CHO cells were transfected with 100 ng Myc-NHS-WHD. Twenty-four hours later, cells lysed in Kinase lysis buffer (150 mm NaCl, 10 mm sodium phosphate pH 7.4, 1% Triton X-100, phosphatase inhibitors: Cantharidin, Bromotetramisole, Microcystin LR, protease inhibitors: AEBSF, Aprotinin, Leupeptin, Bestatin, Pepstatin A, E-64) were cleared by centrifugation (20 000g, 30 min) and incubated overnight with 4 µg of anti-FLAG antibody. The following day, 50 µl of a 50% slurry of G Sepharose beads (GE Healthcare) in Kinase lysis buffer was added to the overnight lysates and incubated at 4°C for 3 h. Beads were washed four times in 40-fold bed volume of stringent Kinase lysis buffer (500 mm NaCl) pH 7.4 and then resuspended in 50 µl 2× SDS sample buffer. A volume of 10 µl of bead sample and 5 µl supernatant were analysed by SDS–PAGE and western blotting. For co-immunoprecipitation of endogenous Nap1 and Sra1, A431 cells were transfected with 100 ng of either Myc-NHS-1A or Myc-NHS-WHD. Cells were lysed 24 h post-transfection by incubating with TNE buffer (Tris 50 mm, NaCl 150 mm, 1% Triton X-100, 1 mm EDTA, 1 mm protease inhibitors) on ice for 10 min. Cell lysates were centrifuged at 20 000 g for 10 min. Supernatant was aspirated and mixed with 50 µl anti-c-Myc Agarose beads (Sigma) prewashed six times with 1× PBS and incubated overnight at 4°C. Beads were then washed three times with TNE buffer. Washed beads were boiled with NuPAGE reducing agent and 4× loading buffer. Samples were centrifuged and the supernatant analysed by SDS–PAGE and western blotting.
RNA interference
Pools of siRNAs directed against human NHS (sense strand: 5′-GCGCUAAACCCUCAGCAUA-3′, 5′-CGACAAUGGUUUAUGGACU-3′, 5′-GUGGACAUGUGGUCAAUUA -3′, 5′-GUACAGAGACUGAGCCUAU-3′) and control siRNAs (ON-TARGETplus SMARTpools) were purchased from Dharmacon (Lafayette, CO, USA). Dharmafect transfection reagents 1 and 4 were used to transfect MTLn3 and Caco-2 cells, respectively. Caco-2 cells or MTLn3 cells were attached to 0.8 cm2 eight-well glass chamber slides at a seeding density of 10 000 cells. Cells were transfected the following day with siRNA oligonucleotides (100 nM) for 72 h using Dharmafect transfection reagent in complete, antibiotic-free media. The cultured cells were then fixed and processed for immunofluorescence analysis.
siRNA deconstruction and rescue
Individual siRNAs from the NHS siRNA pool were obtained from Dharmacon and tested separately in MTLn3 cells. siRNA sequence 5′-GUGGACAUGUGGUCAAUUA-3′ targeting the 3′-UTR of the NHS gene resulted in effective NHS knockdown and the same phenotype observed for the NHS siRNA pool. To rescue the NHS knockdown phenotype obtained with the NHS 3′-UTR siRNA, cells were treated with 50 nM siRNA for 48 h, then transfected with 100 ng of an siRNA-resistant Myc-NHS1A construct and incubated for 24 h before fixation and staining. The control experiment was run in parallel in which MTLn3 cells treated with 50 nM NHS 3′-UTR siRNA were transfected with 100 ng of pEGFP-N1 (Clontech).
Immunofluorescence confocal microscopy
Caco-2 cells were fixed in 100% methanol for 10 min. MTLn3 cells were fixed for 15 min in 4% formaldehyde and permeabilized in 0.3% Triton X-100 for 15 min. Cells were then incubated with primary antibody for 2 h, followed by incubation with secondary antibody for 1 h. F-actin was detected by staining with Alexa Fluor 488 Phalloidin. DNA was counterstained with DAPI (4,6-diamidino-2-phenylindole). All cell images were captured using a Zeiss LSM 510 confocal microscope.
Total internal reflection fluorescence microscopy
After transfection, 5 × 104–1 × 105 MCF7 cells were plated on collagen G-coated 35 mm glass bottom dishes (IWAKI). Cells were then incubated, as described, overnight. Just prior to imaging, the medium was replaced with DMEM/F12 (1:1) supplemented with 10% fetal bovine serum and 2 mm l-glutamine for live cell imaging. TIRF was performed on a Nikon Eclipse TE 2000-U microscope. The Nikon Epi-fluorescence condenser was replaced with a custom condenser in which laser light was introduced into the illumination pathway directly from the output of a 3.5 µm optical fibre oriented parallel to the optical axis of the microscope. GFP and RFP excitation were performed at 473 and 561 nm, respectively, using individually coupled diode lasers (Omicron) controlled by a DAC 2000 card running MetaMorph (Molecular Devices). All cell images were captured with a Cascade 512F EMCCD camera (Photometrics UK).
Bioinformatics
NHS orthologues were identified either through protein database searches using the BLAST algorithms TBLASTP and TBLASTX or through BLASTX against species-specific databases at NCBI or through BLAT analysis using the UCSC genome browser. To identify potential homologies with other proteins/classes of protein, the human NHS protein sequence was used as a query to perform a position-specific iterated BLAST (PSI-BLAST) search.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by The Wellcome Trust (077477/Z/05/Z to A.J.H., M.E.C. and M.B.) and Fight for Sight (to A.J.H.). Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alexis Gautreau and Theresia Stradal for providing us with the FLAG-tagged HSPC300 construct and antibodies for HSPC300 and Abi1, Karl Matter for the E-cadherin, α-catenin and AF-6 antibodies, Kurt Anderson for pcDNA3-TagRed, Bradford Ozanne for Lasp1-mCherry and Roland Wedlich-Soldner for lifeact-GFP. We also thank Belen Martin-Martin for help with the MTLn3 cells and Peter Munro for assistance with confocal microscopy.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Nance W.E., Warburg M., Bixler D., Helveston E.M. Congenital X-linked cataract, dental anomalies and brachymetacarpalia. Birth Defects Orig. Artic. Ser. 1974;10:285–291. [PubMed] [Google Scholar]
- 2.Horan M.B., Billson F.A. X-linked cataract and Hutchinsonian teeth. Aust. Paediatr. 1974;10:98–102. [Google Scholar]
- 3.Toutain A., Ayrault A.D., Moraine C. Mental retardation in Nance–Horan syndrome: clinical and neuropsychological assessment in four families. Am. J. Med. Genet. 1997;71:305–314. doi: 10.1002/(sici)1096-8628(19970822)71:3<305::aid-ajmg11>3.0.co;2-o. doi:10.1002/(SICI)1096-8628(19970822)71:3<305::AID-AJMG11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 4.Coccia M., Brooks S.P., Webb T.R., Christodoulou K., Wozniak I.O., Murday V., Balicki M., Yee H.A., Wangensteen T., Riise R., et al. X-linked cataract and Nance–Horan syndrome are allelic disorders. Hum. Mol. Genet. 2009;8:2643–2655. doi: 10.1093/hmg/ddp206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burdon K.P., McKay J.D., Sale M.M., Russell-Eggitt I.M., Mackey D.A., Wirth M.G., Elder J.E., Nicoll A., Clarke M.P., FitzGerald L.M., et al. Mutations in a novel gene, NHS, cause the pleiotropic effects of Nance–Horan syndrome, including severe congenital cataract, dental anomalies, and mental retardation. Am. J. Hum. Genet. 2003;73:1120–1130. doi: 10.1086/379381. doi:10.1086/379381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks S.P., Ebenezer N.D., Poopalasundaram S., Lehmann O.J., Moore A.T., Hardcastle A.J. Identification of the gene for Nance–Horan syndrome (NHS) J. Med. Genet. 2004;41:768–771. doi: 10.1136/jmg.2004.022517. doi:10.1136/jmg.2004.022517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramprasad V.L., Thool A., Murugan S., Nancarrow D., Vyas P., Rao S.K., Vidhya A., Ravishankar K., Kumaramanickavel G. Truncating mutation in the NHS gene: phenotypic heterogeneity of Nance–Horan syndrome in an asian Indian family. Invest. Ophthalmol. Vis. Sci. 2005;46:17–23. doi: 10.1167/iovs.04-0477. doi:10.1167/iovs.04-0477. [DOI] [PubMed] [Google Scholar]
- 8.Florijn R.J., Loves W., Maillette de Buy Wenniger-Prick L.J., Mannens M.M., Tijmes N., Brooks S.P., Hardcastle A.J., Bergen A.A. New mutations in the NHS gene in Nance–Horan syndrome families from the Netherlands. Eur. J. Hum. Genet. 2006;14:986–990. doi: 10.1038/sj.ejhg.5201671. doi:10.1038/sj.ejhg.5201671. [DOI] [PubMed] [Google Scholar]
- 9.Huang K.M., Wu J., Brooks S.P., Hardcastle A.J., Lewis R.A., Stambolian D. Identification of three novel NHS mutations in families with Nance–Horan syndrome. Mol. Vis. 2007;13:470–474. [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma S., Burdon K.P., Dave A., Jamieson R.V., Yaron Y., Billson F., Van Maldergem L., Lorenz B., Gécz J., Craig J.E. Novel causative mutations in patients with Nance–Horan syndrome and altered localization of the mutant NHS-A protein isoform. Mol. Vis. 2008;14:1856–1864. [PMC free article] [PubMed] [Google Scholar]
- 11.Huang K.M., Wu J., Duncan M.K., Moy C., Dutra A., Favor J., Da T., Stambolian D. Xcat, a novel mouse model for Nance–Horan syndrome inhibits expression of the cytoplasmic-targeted Nhs1 isoform. Hum. Mol. Genet. 2006;15:319–327. doi: 10.1093/hmg/ddi449. doi:10.1093/hmg/ddi449. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S., Ang S.L., Shaw M., Mackey D.A., Gécz J., McAvoy J.W., Craig J.E. Nance–Horan syndrome protein, NHS, associates with epithelial cell junctions. Hum. Mol. Genet. 2006;15:1972–1983. doi: 10.1093/hmg/ddl120. doi:10.1093/hmg/ddl120. [DOI] [PubMed] [Google Scholar]
- 13.Sharma S., Koh K.S., Collin C., Dave A., McMellon A., Sugiyama Y., McAvoy J.W., Voss A.K., Gécz J., Craig J.E. NHS-A isoform of the NHS gene is a novel interactor of ZO-1. Exp. Cell Res. 2009;14:2358–2372. doi: 10.1016/j.yexcr.2009.05.008. doi:10.1016/j.yexcr.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Ikenouchi J., Furuse M., Furuse K., Sasaki H., Tsukita S., Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol. 2005;6:939–945. doi: 10.1083/jcb.200510043. doi:10.1083/jcb.200510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailly M., Macaluso F., Cammer M., Chan A., Segall J.E., Condeelis J.S. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J. Cell Biol. 1999;145:331–345. doi: 10.1083/jcb.145.2.331. doi:10.1083/jcb.145.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koestler S.A., Auinger S., Vinzenz M., Rottner K., Small J. Differentially oriented populations of actin filaments generated in lamellipodia collaborate in pushing and pausing at the cell front. Nat. Cell Biol. 2008;10:306–313. doi: 10.1038/ncb1692. doi:10.1038/ncb1692. [DOI] [PubMed] [Google Scholar]
- 17.Chan A.Y., Bailly M., Zebda N., Segall J.E., Condeelis J.S. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.151.5.1119. doi:10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouneimne G., Soon L., DesMarais V., Sidani M., Song X., Yip S.C., Ghosh M., Eddy R., Backer J.M., Condeelis J. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 2004;166:697–708. doi: 10.1083/jcb.200405156. doi:10.1083/jcb.200405156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., Horwitz A.R. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. doi:10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray C.H., McGarry L.C., Spence H.J., Riboldi-Tunnicliffe A., Ozanne B.W. Novel beta-propeller of the BTB-Kelch protein Krp1 provides a binding site for Lasp-1 that is necessary for pseudopodial extension. J. Biol. Chem. 2009;30:30498–30507. doi: 10.1074/jbc.M109.023259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takenawa T., Suetsugu S. The WASP–WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. doi:10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 22.Takenawa T., Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- 23.Liu R., Abreu-Blanco M.T., Barry K.C., Linardopoulou E.V., Osborn G.E., Parkhurst S.M. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–2860. doi: 10.1242/dev.035246. doi:10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campellone K.G., Webb N.J., Znameroski E.A., Welch M.D. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. doi:10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuchero J.B., Coutts A.S., Quinlan M.E., Thangue N.B., Mullins R.D. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. doi:10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M.W. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. doi:10.1016/S0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 27.Suetsugu S., Miki H., Takenawa T. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem. Biophys. Res. Commun. 1999;260:296–302. doi: 10.1006/bbrc.1999.0894. doi:10.1006/bbrc.1999.0894. [DOI] [PubMed] [Google Scholar]
- 28.Machesky L.M., Mullins R.D., Higgs H.N., Kaiser D.A., Blanchoin L., May R.C., Hall M.E., Pollard T.D. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl Acad. Sci. USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. doi:10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. doi:10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 30.Eden S., Rohatgi R., Podtelejnikov A.V., Mann M., Kirschner M.W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. doi:10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 31.Gautreau A., Ho H.Y., Li J., Steen H., Gygi S.P., Kirschner M.W. Purification and architecture of the ubiquitous Wave complex. Proc. Natl Acad. Sci. USA. 2004;101:4379–4383. doi: 10.1073/pnas.0400628101. doi:10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L.B., Steffen A., Stradal T.E., Di Fiore P.P., Carlier M.F., Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. doi:10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 33.Echarri A., Lai M.J., Robinson M.R., Pendergast A.M. Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and WAVE-1 levels. Mol. Cell Biol. 2004;24:4979–4993. doi: 10.1128/MCB.24.11.4979-4993.2004. doi:10.1128/MCB.24.11.4979-4993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Innocenti M., Gerboth S., Rottner K., Lai F.P., Hertzog M., Stradal T.E., Frittoli E., Didry D., Polo S., Disanza A., et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 2005;7:969–976. doi: 10.1038/ncb1304. doi:10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- 35.Stradal T., Courtney K.D., Rottner K., Hahne P., Small J.V., Pendergast A.M. The Abl interactor proteins localize to sites of actin polymerization at the tips of lamellipodia and filopodia. Curr. Biol. 2001;11:891–895. doi: 10.1016/s0960-9822(01)00239-1. doi:10.1016/S0960-9822(01)00239-1. [DOI] [PubMed] [Google Scholar]
- 36.Courtney K.D., Grove M., Vandongen H., Vandongen A., LaMantia A.S., Pendergast A.M. Localization and phosphorylation of Abl-interactor proteins, Abi-1 and Abi-2, in the developing nervous system. Mol. Cell Neurosci. 2000;16:244–257. doi: 10.1006/mcne.2000.0865. doi:10.1006/mcne.2000.0865. [DOI] [PubMed] [Google Scholar]
- 37.Adams C.L., Nelson W.J., Smith S.J. Quantitative analysis of cadherin–catenin–actin reorganization during development of cell–cell adhesion. J. Cell Biol. 1996;135:1899–1911. doi: 10.1083/jcb.135.6.1899. doi:10.1083/jcb.135.6.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nozumi M., Nakagawa H., Miki H., Takenawa T., Miyamoto S. Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci. 2003;116:239–246. doi: 10.1242/jcs.00233. doi:10.1242/jcs.00233. [DOI] [PubMed] [Google Scholar]
- 39.Stovold C.F., Millard T.F., Machesky L.M. Inclusion of Scar/WAVE3 in a similar complex to Scar/WAVE1 and 2. BMC Cell Biol. 2005;6:11. doi: 10.1186/1471-2121-6-11. doi:10.1186/1471-2121-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao D., Forge A., Munro P.M.G., Bailly M. Arp2/3 complex-mediated actin polymerisation occurs on specific pre-existing networks in cells and requires spatial restriction to sustain functional lamellipod extension. Cell Motil. Cytoskeleton. 2006;63:395–414. doi: 10.1002/cm.20131. doi:10.1002/cm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DesMarais V., Macaluso F., Condeelis J., Bailly M. Synergistic interaction between the Arp2/3 complex and coflin drives stimulated lamellipod extension. J. Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. doi:10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drees F., Pokutta S., Yamada S., Nelson W.J., Weis W.I. Alpha-catenin is a molecular switch that binds E-cadherin–beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. doi:10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blagg S.L., Stewart M., Sambles C., Insall R.H. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 2003;13:1480–1487. doi: 10.1016/s0960-9822(03)00580-3. doi:10.1016/S0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- 44.Caracino D., Jones C., Compton M., Saxe C.L. The N-terminus of Dictyostelium Scar interacts with Abi and HSPC300 and is essential for proper regulation and function. Mol. Biol. Cell. 2007;18:1609–1620. doi: 10.1091/mbc.E06-06-0518. doi:10.1091/mbc.E06-06-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grove M., Demyanenko G., Echarri A., Zipfel P.A., Quiroz M.E., Rodriguiz R.M., Playford M., Martensen S.A., Robinson M.R., Wetsel W.C., et al. ABI2-deficient mice exhibit defective cell migration, aberrant dendritic spine morphogenesis, and deficits in learning and memory. Mol. Cell. Biol. 2004;24:10905–10922. doi: 10.1128/MCB.24.24.10905-10922.2004. doi:10.1128/MCB.24.24.10905-10922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.