Abstract
Disrupted-in-schizophrenia 1 (DISC1) has been genetically associated with schizophrenia, and with brain phenotypes including grey matter volume and working memory performance. However, the molecular and cellular basis for these associations remains to be elucidated. One potential mechanism may be via an altered interaction of DISC1 with its binding partners. In this context, we previously demonstrated that one DISC1 variant, Leu607Phe, influenced the extent of centrosomal localization of pericentriolar material 1 (PCM1) in SH-SY5Y cells. The current study extends this work to human brain, and includes another DISC1 coding variant, Ser704Cys. Using immunohistochemistry, we first characterized the distribution of PCM1 in human superior temporal gyrus (STG). PCM1 immunoreactivity was localized to the centrosome in glia, but not in neurons, which showed widespread immunoreactivity. We quantified centrosomal PCM1 immunoreactivity in STG glia of 81 controls and 67 subjects with schizophrenia, genotyped for the two polymorphisms. Centrosomal PCM1 immunoreactive area was smaller in Cys704 carriers than in Ser704 homozygotes, with a similar trend in Phe607 homozygotes compared with Leu607 carriers, replicating the finding in SH-SY5Y cells. No differences were seen between controls and subjects with schizophrenia. These findings confirm in vivo that DISC1 coding variants modulate centrosomal PCM1 localization, highlight a role for DISC1 in glial function and provide a possible cellular mechanism contributing to the association of these DISC1 variants with psychiatric phenotypes. Whether this influence of DISC1 genotype extends to other centrosomal proteins and DISC1 binding partners remains to be determined.
INTRODUCTION
With the recent identification of potential susceptibility genes for psychiatric illnesses, the goal of many current studies has been to elucidate how genetic variation may contribute to disease susceptibility and pathophysiology. One leading candidate gene for schizophrenia and related phenotypes is disrupted-in-schizophrenia 1 (DISC1), initially identified as the gene disrupted by a balanced (1,11)(q42.1;q14.3) translocation that segregates with major mental illness in a large Scottish family (1). Although no other families with similar chromosomal abnormalities have been reported, coding and non-coding single nucleotide polymorphisms (SNPs) within DISC1 have since been associated with several mental illnesses, including schizophrenia, bipolar disorder, major depression and autism [reviewed in (2,3)]. In addition, DISC1 genetic variation has been found to influence phenotypes including hippocampal and frontal grey matter volume, hippocampal activation during memory encoding and cognition (2–5). Whilst these associations suggest that DISC1 polymorphisms are functional variants, the molecular mechanisms by which they exert their influence remain to be elucidated.
DISC1 is a pleiotropic protein with many purported functions, subcellular locations and binding partners, and has been conceptualized to act as a scaffold for its multiple protein interactions (6–12). The centrosome is one organelle where DISC1, along with several of its interacting proteins, is localized (13–16). One of these binding partners is pericentriolar material 1 [PCM1, itself a putative schizophrenia susceptibility gene (17,18)], and DISC1 regulates the recruitment of PCM1 to the centrosome (16). Microtubule assembly is PCM1 dependent, and the presence of PCM1 at the centrosome contributes to its role as the main microtubule-organizing centre in animal cells (19).
Based on the regulatory role of DISC1 upon PCM1 (16), together with the previous finding that DISC1 schizophrenia-associated SNPs influence the expression of several DISC1 binding partners (20), we hypothesized that one potential mechanism by which DISC1 SNPs may exert their pathogenic influence could be via altering the regulation of and/or affinity for its binding partners. As one test of this proposal, and given the links between DISC1, PCM1 and the centrosome, we previously examined whether DISC1 genetic variation affected centrosomal PCM1 localization. In particular, we focussed on one of the few DISC1 risk polymorphisms that is coding, Leu607Phe. Utilizing undifferentiated SH-SY5Y cells (a human neuroblastoma cell line), we reported that cells transfected with Phe607 DISC1, a putative risk variant for schizoaffective disorder (21), had reduced centrosomal PCM1 localization compared with cells transfected with Leu607 DISC1 (22). The risk variant also altered neurotransmitter release, consistent with links between the centrosome, microtubules and synaptic function (6). These findings provided in vitro evidence that DISC1 Leu607Phe may be a functional variant.
The aim of the current study was to extend these findings to human brain. Both our original variant of interest, Leu607Phe (rs6675281), and another more widely examined coding variant, Ser704Cys (rs821616), occur in the PCM1 binding region of DISC1 [amino acid 601 through to the C-terminal (16)], and we hypothesized that both substitutions may influence centrosomal PCM1 localization in vivo. In addition, we asked whether altered centrosomal localization of PCM1 occurs in schizophrenia. To investigate these questions, PCM1 immunohistochemistry was performed on post mortem tissue sections from the superior temporal gyrus (STG) of a large cohort of adults with schizophrenia and controls. We found centrosomal localization of PCM1 in glia but not in neurons, and evidence that genetic variation in DISC1 does influence the extent of centrosomal localization of PCM1 in glia in human brain, as it did in vitro. The data support this effect as being a potential cellular mechanism underlying the functionality and pathogenicity of these DISC1 coding substitutions. Finally, to investigate the finding that PCM1 was not centrosomally localized in STG neurons, we examined SH-SY5Y cells further, and found that the centrosomal localization of PCM1 is lost in these cells after differentiation.
RESULTS
Western blotting for PCM1
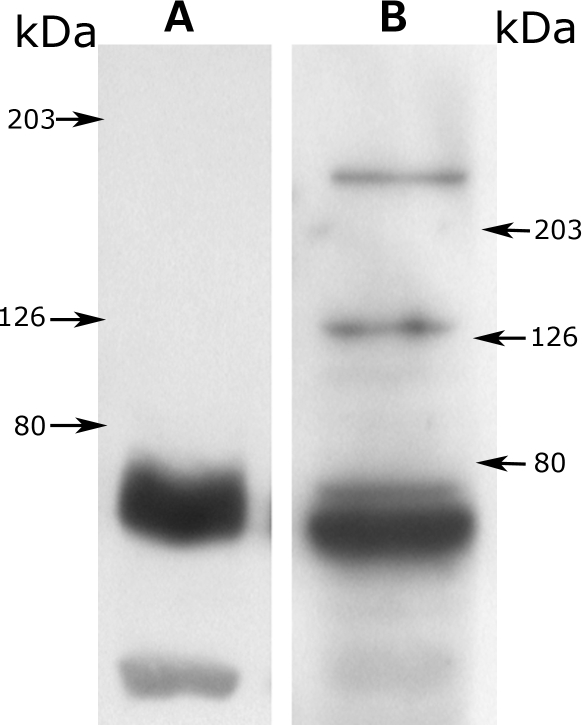
Western blotting of human STG protein (Fig. 1A) with the PCM1 antibody detected a major band of 60–65 kDa and a fainter band at 40 kDa. For HEK293 cells, in addition to these two bands, two fainter bands were detected: one at 130 kDa and another, corresponding to full-length PCM1, at 228 kDa (Fig. 1B). The PCM1 antibody also detected a band of the predicted size (90 kDa) for blots performed utilizing PCM1 fusion protein (containing amino acids 1–620), but not in cell lysates from bacteria not induced with isopropyl-β-d-thiogalactopyranoside (IPTG; data not shown). Antibody specificity was also demonstrated by the absence of immunoreactive bands for westerns performed without the primary antibody, and from the finding that a second antibody revealed the same, albeit weaker, pattern of PCM1 immunoreactivity in human STG (data not shown).
Figure 1.
Western blots using anti-PCM1 antibody. (A) Human superior temporal gyrus. (B) HEK293 cells. Arrows indicate the positions of the molecular weight markers.
Subcellular distribution of PCM1 in neurons and glia of human STG
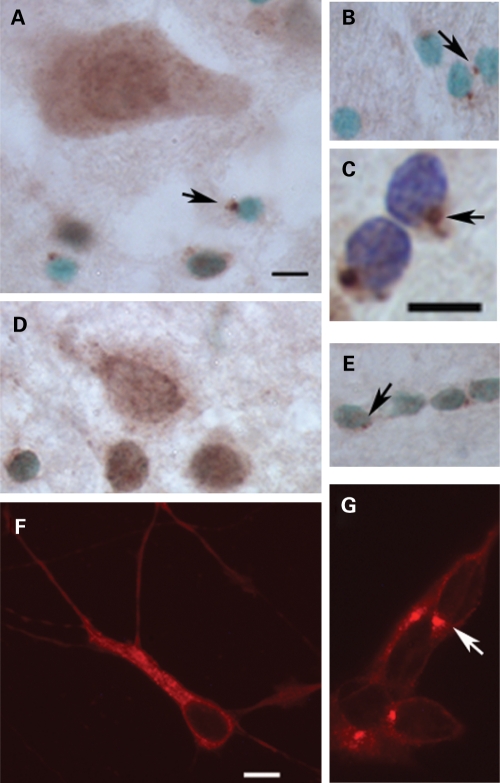
Based on our in vitro study of PCM1 in a neuronal cell line (22), we predicted that PCM1 would be likewise localized to the centrosome in neurons of the STG. In fact, centrosomal localization was restricted to cells with small round nuclei (visualized with methyl green) located in the grey matter (Fig. 2A) and predominantly in the white matter (Fig. 2B). We conclude that these cells are glia, especially since cresyl violet revealed a dense nuclear staining, a lack of a visible nucleolus and an absence of cytoplasmic staining (Fig. 2C). In contrast, in neurons, PCM1 was widely distributed throughout the soma and proximal dendrites (Fig. 2A). Against this background, no centrosomal PCM1 staining could be distinguished. To exclude the possibility that these findings were related to perimortem variables or some aspect of how the human brain tissue was processed, we repeated our immunohistochemical protocol in rat brain. We found the same pattern of PCM1 immunostaining, with PCM1 being observed throughout the neuronal cell body (Fig. 2D), while glial cells (Fig. 2E) exhibited discrete centrosomal staining. We conclude that in the brain PCM1 is concentrated at the centrosome in glia, but has a more widespread distribution in neurons.
Figure 2.
Cellular profile of PCM1 immunoreactivity. (A)–(C) Adult human superior temporal gyrus. (D) and (E) Adult rat brain. (F) Differentiated SH-SY5Y cells. (G) Undifferentiated SH-SY5Y cells. (A) PCM1 immunoreactive neuron in lamina III. PCM1 immunoreactivity was also seen at the centrosome in cells with small nuclei (arrowed) which are putative glia. (B) Glial cells in STG white matter also exhibited centrosomal staining (arrows). (C) Enlargement of glial staining observed in the white matter, showing easily identifiable centrosomal staining (arrow). (D) Neuronal PCM1 staining in layer 3 of adult rat cortex. (E) PCM1 staining of glial cells (arrowed) in adult rat brain white matter. (F) PCM1 immunofluorescence in a representative example of a differentiated SH-SY5Y cell. Note that centrosomal PCM1 immunoreactivity was not observed. (G) Undifferentiated SH-SY5Y cell, showing centrosomal localization (arrow). The counter stain used in (A), (B), (D) and (E) is methyl green (a nuclear stain), with cresyl violet used in (C). Scale bar = 10 µm. Scale bar in (A) also applies to (B), (D) and (E); scale bar in (F) also applies to (G).
Effects of DISC1 genotype and schizophrenia on centrosomal PCM1 immunoreactivity
As neuronal centrosomal PCM1 immunoreactivity could not be identified, the measurements of centrosomal PCM1 immunoreactive area were carried out over glia. The resulting data were normally distributed within each diagnostic and genotypic group, and there were no correlations of PCM1 immunoreactivity with potential confounding variables (post mortem interval, age, brain pH; all P > 0.210). In subjects with schizophrenia, PCM1 immunoreactive area did not correlate with any measure of antipsychotic exposure nor was it influenced by smoking or substance abuse histories (data not shown).
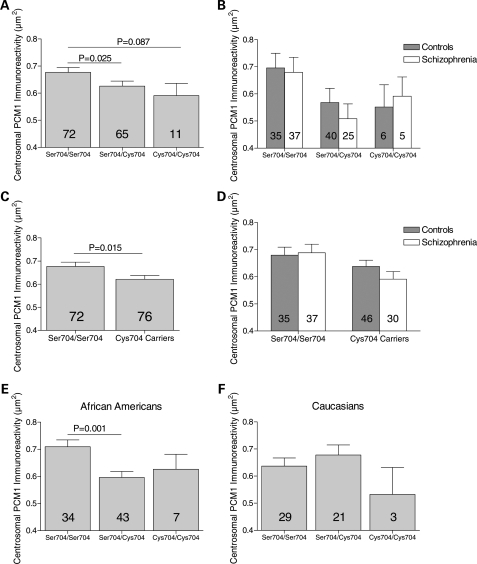
A significant effect of Ser704Cys genotype upon centrosomal PCM1 immunoreactive area was detected (F2, 142 = 3.303, P = 0.040; Fig. 3A and B), with planned comparisons revealing decreased immunoreactive area in heterozygotes compared with Ser704 homozygotes (P = 0.025), and a trend difference between Cys704 homozygotes and Ser704 homozygotes (P = 0.087; Fig. 3A). Owing to the low frequency of the minor allele, Cys704 carriers were also grouped together and compared with Ser704 homozygotes; centrosomal PCM1 immunoreactive area was smaller in subjects carrying a Cys704 allele (F1, 144 = 6.122, P = 0.015); Fig. 3C and D). In both ANOVAs described above, no interactions between genotype and diagnosis were detected (P > 0.268). Given the heterogeneous ethnicity of the sample, we also explored whether race impacted on the genetic association. This analysis was restricted to African Americans and Caucasians since these comprised by far the largest groups. We found a significant interaction between race and Ser704Cys genotype (F3, 131 = 2.737, P = 0.046), with a significant effect of Ser704Cys genotype on PCM1 immunoreactivity in African Americans (F2, 78 = 6.013, P = 0.004; Fig. 3E), but not in Caucasians (F2, 47 = 1.047, P = 0.359; Fig. 3F). A similar result was seen when subjects carrying a Cys704 allele were compared with Ser704 homozygotes (African Americans: F1, 80 = 11.398, P = 0.001; Caucasians: F1, 49 = 0.235, P = 0.630; data not shown).
Figure 3.
Association between Ser704Cys substitution and centrosomal PCM1 immunoreactive area in white matter glia of human superior temporal gyrus. (A) There was an overall effect of genotype (P = 0.04), with PCM1 centrosomal immunoreactive area being decreased in heterozygotes when compared with Ser704 homozygotes, with a similar trend between Ser704 homozygotes and Cys704 homozygotes. (B) Data in (A) parsed by diagnosis. No effect of diagnosis nor genotype by diagnosis interaction was detected. (C) Centrosomal PCM1 immunoreactive area was significantly lower in Cys704 carriers compared with Ser704 homozygotes. (D) Data in (C) parsed by diagnosis. No effect of diagnosis nor genotype by diagnosis interaction was detected. (E) Ser704Cys affected centrosomal PCM1 immunoreactive area in African Americans (P = 0.004), being decreased in heterozygotes compared with Ser704 homozygotes. (F) No influence of Ser704Cys upon PCM1 centrosomal immunoreactive area was detected in Caucasians.
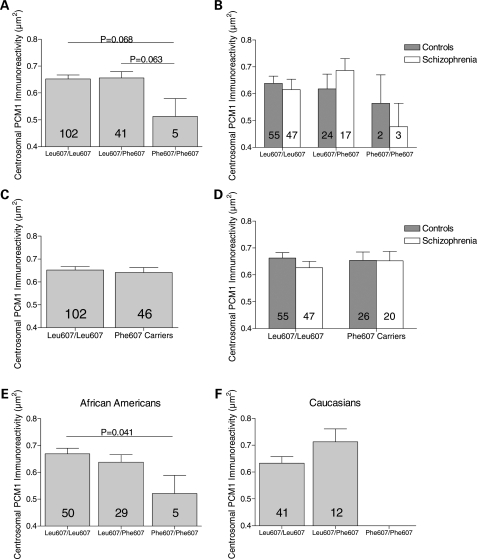
For the Leu607Phe substitution, there was no overall effect of genotype (F2, 142 = 1.794, P = 0.170). However, planned comparisons revealed a trend for smaller PCM1 centrosomal immunoreactive area in Phe607 homozygotes compared with heterozygotes (P = 0.063), and compared with Leu607 homozygotes (P = 0.068; Fig. 4A and B). No differences were seen when Phe607 carriers were compared with Leu607 homozygotes (F2, 144 = 0.154, P = 0.696; Fig. 4C and D), and no interaction between genotype and diagnosis was detected (P ≥ 0.71). A post hoc analysis revealed a trend interaction between genotype and race (F3, 131 = 3.164, P = 0.078), and although there was not an overall effect of genotype for any racial group (data not shown), in African Americans, a significant decrease in centrosomal PCM1 immunoreactive area was detected in Phe607 homozygotes compared with Leu607 homozygotes (P = 0.041; Fig. 4E).
Figure 4.
Association between Leu607Phe substitution and centrosomal PCM1 immunoreactive area in white matter glia of human superior temporal gyrus. (A) No overall effect of genotype was observed for the Leu607Phe substitution. However, there was a trend towards smaller centrosomal immunoreactive area in Phe607 homozygotes when compared with heterozygotes and Leu607 homozygotes. (B) Data in (A) parsed by diagnosis. No effect of diagnosis, nor genotype by diagnosis interaction was detected. (C) Phe607 carriers did not differ from Leu607 homozygotes. (D) Data in (C) parsed by diagnosis. No effect of diagnosis nor genotype by diagnosis interaction was detected. (E) Although no overall effect of genotype was found in African Americans, centrosomal PCM1 area was lower in Phe607 homozygotes than in Leu607 homozygotes. (F) No effect of Leu607Phe genotype in Caucasians, although note the absence of Phe607 homozygotes.
In an exploratory analysis we examined whether the two polymorphisms might have a cumulative effect on centrosomal PCM1 immunoreactivity. Individuals who were Phe607 homozygotes and Cys704 carriers (i.e. the two genotypes associated separately with lower centrosomal PCM1 immunoreactivity; Fig. 3C and 4A) were compared with those who had only one of the two ‘risk genotypes’. Centrosomal PCM1 immunoreactivity in the Phe607Phe/Cys704 carriers (n = 3; 0.390 ± 0.06 µm2, mean ± SD) was less than in subjects who were either Phe607Phe/Ser704Ser or Leu607 carriers/Cys704 carriers (combined n = 75; 0.632 ± 0.145 µm2; Mann–Whitney, P = 0.006).
No difference in centrosomal PCM1 immunoreactive area was found between subjects with schizophrenia (0.638 ± 0.155 µm2, mean ± SD) and controls (0.657 ± 0.150 µm2; F1, 142 < 0.50, P > 0.50). The schizophrenia and control groups were not well matched for age, post mortem delay or brain pH (see Table 1). However, as none of these variables correlated with centrosomal PCM1 immunoreactive area, differences between the two diagnostic groups for these potential confounding variables is unlikely to account for our negative finding. Furthermore, when subjects were excluded to form a subset matched for these variables (66 controls, 48 with schizophrenia), centrosomal PCM1 immunoreactive area remained unaltered in schizophrenia (data not shown).
Table 1.
Demographic details of subjects included in the study, stratified by diagnosis and by genotype
| Ser704Cys (rs826161) | |||
| Genotype | Ser704/Ser704 (n = 72) | Ser704/Cys704 (n = 65) | Cys704/Cys704 (n = 11) |
| Age (years) | 48 ± 15 | 44 ± 16 | 46 ± 15 |
| Post mortem delay (h) | 37 ± 16 | 35 ± 19 | 29 ± 17 |
| Brain pH | 6.5 ± 0.3 | 6.5 ± 0.3 | 6.4 ± 0.2 |
| Sex (male:female) | 49:23 | 39:26 | 7:4 |
| Racea | 34AA, 29C, 6H, 3A | 43AA, 21C | 7AA, 3C, 1H |
| Leu607Phe (rs6675281) | |||
| Genotype | Leu607/Leu607 (n = 102) | Leu607/Phe607 (n = 41) | Phe607/Phe607 (n = 5) |
| Age (years) | 47 ± 16 | 46 ± 15 | 45 ± 20 |
| Post mortem delay (h) | 34 ± 17 | 41 ± 17 | 27 ± 18 |
| Brain pH | 6.5 ± 0.3 | 6.4 ± 0.3 | 6.4 ± 0.4 |
| Sex (male:female) | 68:34 | 22:19 | 5:0 |
| Race | 50AA, 41C, 7H, 3A | 29AA, 12C | 5AA |
| Diagnosis | Control (n = 81) | Schizophrenia (n = 67) | |
| Age (years) | 42 ± 15 | 52 ± 16 | |
| Post mortem delay (h) | 32 ± 15 | 40 ± 19 | |
| Brain pH | 6.6 ± 0.3 | 6.4 ± 0.3 | |
| Sex (male:female) | 54:27 | 41:26 | |
| Race | 46AA, 28C, 4H, 3A | 38AA, 25C, 3H | |
| Age at onset (years) | — | 23 ± 8 | |
Values are mean ± standard deviation.
A, Asian Pacific; AA, African American; C, Caucasian; H, Hispanic.
aRace was unknown for one subject.
Effects of differentiation on centrosomal localization of PCM1 in SH-SY5Y cells
The finding that PCM1 immunoreactivity was not centrosomal in neurons (Fig. 2A and D) was unexpected, given the earlier observation in the SH-SY5Y neuronal cell line (22). We wondered whether this might relate to the state of neuronal differentiation, since the transfected SH-SY5Y cells we used were undifferentiated (22). Here, we confirmed the centrosomal localization of PCM1 in undifferentiated SH-SY5Y cells (Fig. 2G). In contrast, in differentiated SH-SY5Y cells, PCM1 was observed throughout the cell body and extended into neuritic processes, and centrosomal staining was not readily detected (Fig. 2F), thus resembling the neurons seen in human and rat brain sections.
DISCUSSION
Previously we reported that transfection of undifferentiated SH-SY5Y cells with a DISC1 risk variant, Phe607, resulted in decreased centrosomal localization of its binding partner, PCM1 (22), compared with untransfected cells and those transfected with Leu607 DISC1. Here we extended our study of DISC1 genetic variants to adult human brain. We investigated PCM1 centrosomal localization in cells of the STG, explored its association with two DISC1 coding variants within the PCM1 binding region, and whether this is altered in schizophrenia. The main findings are the following: (i) PCM1 immunoreactivity was concentrated at the centrosome in glia, but was widespread in neurons, such that neuronal centrosomal PCM1 immunostaining could not be readily detected. For this reason, measurements of centrosomal PCM1 were carried out over glia. (ii) DISC1 Ser704Cys (rs821616) genotype was associated with centrosomal PCM1 immunoreactive area, the latter being decreased in subjects carrying one or two Cys704 alleles. For the Leu607Phe (rs6675281) substitution, a similar trend was seen in Phe607 homozygotes. (iii) Post hoc analyses demonstrated that significant influences of these SNPs were restricted to African Americans, and also suggested a possible cumulative effect of the two variants. (iv) Centrosomal PCM1 immunoreactive area did not differ between controls and subjects with schizophrenia. (v) Differentiated SH-SY5Y cells lost the centrosomal localization of PCM1 present in undifferentiated SH-SY5Y cells. Our data provide additional evidence that Ser704Cys and Leu607Phe are functional DISC1 variants, and suggest that the altered centrosomal localization of the DISC1 binding partner, PCM1, may be part of the underlying cellular mechanism. The findings also highlight the neglected role of glia in the biology of DISC1.
Expression and subcellular distribution of PCM1
To date, the expression of PCM1 has not been characterized in the human brain, and western blotting for PCM1 using human protein has not been performed. In our hands, although the antibody utilized targets a region common to full-length (228 kDa) PCM1 (23) and two other isoforms identified to date (UniProt), the main band detected in human STG protein was 60–65 kDa (corresponding to PCM1 isoform 3; Q15154–3, UniProt). The fact that the antibody was able to detect GST-tagged PCM1 fusion protein, and full-length PCM1 in HEK293 cells, confirmed the specificity of the antibody. Our inability to detect the full-length PCM1 in human STG protein is in keeping with an observation that this isoform is not detected in differentiated tissue (Andreas Merdes, Centre National de la Recherche Scientifique-Pierre Fabre, France, personal communication), and our data suggest that isoform 3 may be the main PCM1 protein in human STG.
PCM1 immunoreactivity was observed in both neurons and glia of human and rat brain (Fig 2A–E). It was differentially distributed in these two cells types, and as reported previously for mouse (24), PCM1 was not readily observed at the centrosome in neurons. Of note, in the adult mouse brain the distribution of ninein, a protein regulated by PCM1 (16,19) is similar to that reported here for PCM1, being found in the soma and dendrites in neurons, while being restricted to the centrosome in glia (25). These findings suggest that centrosomes differ significantly between neurons and glia in their protein composition. Given the importance of PCM1 at the centrosome in the regulation of cell cycle progression (23,26,27), it is tempting to speculate that this dissimilarity might reflect, to some extent, differences between neurons and glia in the mature brain in terms of their ability to further differentiate or divide. Our findings in SH-SY5Y cells are consistent with this possibility, in that the undifferentiated cells have centrosomal PCM1 immunoreactivity, whereas the differentiated cells show a more diffuse, ‘neuron-like’ immunoreactive profile. DISC1 itself undergoes a similar redistribution in SH-SY5Y cells (28), as does ninein during neuronal differentiation in vivo (25) and PCM1 during muscle cell differentiation (29). It would be of interest to examine fetal brain to see whether the subcellular distribution of PCM1 in neurons is different from that in the adult, utilizing markers of cell type and maturation to clarify the characteristics of cells which have centrosomal versus non-centrosomal PCM1 immunoreactivity. In this context, it is notable that developmental perturbation of PCM1 or DISC1 function results in altered neurogenesis and cell migration (8,30).
As most glia within the white matter are oligodendrocytes, the majority of our quantitative data probably come from this glial cell population and speak to a role for DISC1, as well as PCM1, in oligodendrocyte function. Notably, Seshadri et al. (31) recently reported the first clear evidence that DISC1 is expressed in glia, including oligodendrocytes, in human brain; previously, although DISC1 had been detected in glia (32), interest in the role of DISC1 had generally been limited to neurons. Together, these results indicate that further studies of DISC1 biology in glia are warranted. Equally, as PCM1 is found in neurons in the mature brain (albeit in different subcellular locations than the centrosome; Fig. 2A), the study of the relationship between Leu607Phe and Ser704Cys substitutions and PCM1 in neurons is also warranted.
Centrosomal PCM1 immunoreactivity is influenced by genetic variation in DISC1
Evidence that the DISC1 Ser704Cys substitution is functional has been based primarily on its association with disease risk and with brain structure and function. The SNP has been associated with schizophrenia (33–35) and major depression (36), and in addition, has been reported to influence hippocampal grey matter volume and function (33,37), cingulate grey matter volume (36), pre-frontal cortical function (38) and cognitive decline during ageing (39). Although less widely studied, the Leu607Phe substitution has been associated with schizoaffective disorder (21), decreased cortical grey matter (40) and a greater severity of positive symptoms in schizophrenia (40); in each case, Phe607 is the risk variant. Separate evidence for functionality of Ser704Cys and Leu607Phe is provided from recent data—in many of the same brains as studied here—showing that they impact on DISC1 gene expression (41). Alternative splicing produces more than 50 DISC1 transcripts in human brain, and the expression of one truncated variant is influenced by the Leu607Phe and Ser704Cys substitutions, with Phe607 carriers and Cys704 homozygotes expressing a greater proportion of this truncated transcript in the hippocampus (41). Whether our findings are related to the impact of the two SNPs on differential expression of DISC1 isoforms, or to the other possible mechanisms discussed below, is unknown.
The studies of Ser704Cys vary as to which is the associated risk allele [for reviews see (2,3,42)]. For example, in a North American Caucasian population, Ser704 was over-transmitted in schizophrenia and associated with reduced hippocampal grey matter volume (33), whereas Cys704 was associated with decreased hippocampal grey matter volume and poorer hippocampal function in an Italian Caucasian population (37). Such discrepancies might have several explanations, other than chance, including the ‘flip–flop’ phenomenon and haplotype background (see 37). In this respect, it is interesting that a significant interaction was detected here between race and genotype, with the influence of genotype upon centrosomal PCM1 localization, reaching significance only in African Americans (Fig. 3E); the effect of Ser704Cys on DISC1 transcript expression was also limited to African Americans (41). Our finding of an additive effect of the two DISC1 SNPs on centrosomal PCM1, though very preliminary, provides another indication that individual SNPs should not be considered in isolation, with increasing evidence that intragenic (and intergenic) interactions are relevant in the association of DISC1 and other genes in the genetic architecture of schizophrenia (3,43). Equally, future studies will benefit from inclusion of other DISC1 SNPs, both coding and non-coding, that have been associated with brain phenotypes and disorders.
We found no differences in centrosomal PCM1 immunoreactivity between cases and controls, nor a diagnosis-by-genotype interaction. Our data therefore do not extend the evidence for involvement of oligodendrocytes in schizophrenia (44,45).
How may DISC1 genetic variants influence centrosomal PCM1 localization?
Our results do not speak to the mechanism by which the two coding variants in DISC1 affect localization of PCM1 to the centrosome in glia. One possibility is that it results from their influence on DISC1 transcript expression mentioned earlier, but it may also reflect an alteration in the direct or indirect interaction between DISC1 and PCM1 proteins. Since isoform 3 does not contain the middle portion of full-length PCM1 (amino acids 741–1420) demonstrated to be important for DISC1–PCM1 interaction (16), our data suggesting that this is the major PCM1 isoform in human STG suggests that an ‘indirect’ scenario may be more likely. For example, Ser704Cys has been associated with extracellular signal related kinase (ERK) activity (36), and it may be that the influence of this kinase on cytoskeletal remodelling (36) could indirectly contribute to the influence of Ser704Cys upon PCM1 localization. Ser704Cys has also been demonstrated to influence the binding of nuclear distribution element-like (NDEL1) and its homologue NDE1 to DISC1, with increased NDEL1 binding being associated with Cys704 DISC1 (42,46). As NDEL1 plays a role in the transport of PCM1 to the centrosome (47), it may be that changes in centrosomal PCM1 immunoreactivity associated with Ser704Cys may occur indirectly via this SNP's influence on the binding of NDEL1 to DISC1.
Mechanisms involving a direct DISC1–PCM1 interaction are also possible, even for PCM1 isoform 3, since the N-terminal region of PCM1 does interact with DISC1, albeit weakly (16). As Leu607Phe and Ser704Cys occur within the region of DISC1 to which PCM1 binds, the SNPs may affect the extent of centrosomal PCM1 localization by impacting on the affinity of DISC1 for its binding partners; a recent in vitro study supports this possibility, at least for Ser704Cys (48). Alternatively, it may be that DISC1–PCM1 binding is unchanged, but the SNPs influence the recruitment and localization of DISC1 to the centrosome, resulting in concomitantly decreased PCM1 localization. For the Leu607Phe substitution, we previously proposed (22) that the latter situation may occur, via the disruption of a putative leucine zipper and coiled-coil domain, protein motifs important in regulating the localization of proteins to the centrosome (49). Finally, the C-terminal region of DISC1 (including the PCM1 binding region) contains four putative coiled-coil domains, which contribute to self-association, and indeed, a section of DISC1 (amino acids 403–504) has been demonstrated to be a functional self-association domain (50). Based on these structural predictions and an earlier study in which subjects with chronic psychiatric illness were found to exhibit an increase in insoluble DISC1 aggregates (51), Leliveld et al. (48) investigated the role of Ser704Cys in DISC1 dimerization and oligomerization, and found that Cys704 DISC1 displayed a higher oligomerization propensity, which could culminate in an alteration in binding to its protein partners. Whether an increase in DISC1 oligomerization changes its affinity for PCM1, which in turn alters the centrosomal localization of the latter, remains to be determined.
Distinguishing between these various potential explanations for the current data will require considerable further study. A critical issue is whether the effects on PCM1 are specific, or whether the DISC1 SNPs also influence the localization of other centrosomal DISC1 binding partners, such as NDEL1, LIS1, PDE4B or pericentrin (14,15), or affect centrosomal proteins that are not direct DISC1 binding partners. Similarly, the effects and interactions of other DISC1 SNPs, and SNPs in PCM1 and other centrosomal protein genes, require evaluation. Only when these issues are clarified will it be possible to interpret the significance of the current findings in terms of their implications for centrosomal function, and how this in turn contributes to the roles which genetic variation in DISC1 and PCM1 play in brain function and psychiatric disorders.
MATERIALS AND METHODS
Study subjects
Post mortem tissue samples from the STG were collected at the Clinical Brain Disorders Branch, NIMH from 81 normal control subjects and 67 subjects with schizophrenia (Table 1). Determination of diagnoses and details of neuroleptic medication exposure, smoking and substance abuse histories were as described previously (20). Frozen sections (14 µm) were collected onto Superfrost Plus slides (VWR, Lutterworth, UK) and stored at −80°C until use.
PCM1 immunohistochemistry
The antibody utilized in this study (sc-67204, Insight Biotechnology, Wembley, UK) was designed against amino acids 1–262 of human PCM1 (and common to all three PCM1 isoforms listed in UniProt). Sections were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) for 5 min, dehydrated in increasing concentrations of industrial methylated spirits (IMS) and then placed in methanol containing 3% hydrogen peroxide for 30 min. After rehydration through decreasing concentrations of IMS and two 10 min washes in PBS, non-specific binding sites were blocked by incubating the sections for 30 min in PBS containing 0.3% Triton-X100 (PBS-T) and 10% normal goat serum. Sections were incubated overnight at 4°C with the anti-PCM1 antibody diluted 1:200 in PBS-T and 1% normal goat serum. After washes in PBS, bound antibody was visualized using a Vectastain ABC Elite kit (Vector Laboratories, Peterborough, UK) and diaminobenzidine. Sections were counterstained with either methyl green (for our initial studies of normative PCM1 cellular distribution) or cresyl violet (for quantitative centrosomal PCM1 immunoreactivity analyses, see below) and cover slipped using DPX mountant and ‘0’ thickness coverslips (VWR, Lutterworth, UK).
Experimental controls
To confirm the specificity of the antibody utilized above, several experimental controls were performed. Protein from human STG, rat frontal cortex and HEK293 cells (gift from Philip Burnet) was extracted using standard methods, and 30 µg of the protein was loaded and fractionated by sodium dodecyl sulphate (SDS)–polyacrylamide gel electrophoresis (PAGE) before being transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% milk and probed with the anti-PCM1 antibody diluted to 1:5000, followed by goat anti-rabbit IgG horse radish peroxidase conjugate (Bio-Rad, Hemel Hempstead, UK) at 1:5000. Antibody binding was visualized using ECL Plus Western Blotting Detection System (GE Healthcare, Chalfont, UK).
Additional western blots were performed using PCM1 fusion protein. Briefly, bacterial cells (KRX competent cells, Promega, Southampton, UK) were transfected with pGEX4T-1 vector, containing amino acids 1–620 of full-length human PCM1 and an amino terminal GST tag (gift from Andreas Merdes) and fusion protein expression induced with 0.5 mm IPTG. After 3 h, cells were lysed in RIPA buffer (Sigma, Poole, Dorset) and PCM1 western blotting performed as described above. Westerns were also performed in the absence of primary antibody. Finally, immunohistochemistry utilizing a second commercially available antibody (diluted 1:200; H00005108-B01, Abnova, Taipei, Taiwan) was performed as described above.
Measurement of human PCM1 centrosomal immunoreactivity
Cell staining was examined using a Nikon Eclipse 3600 microscope and a ×60 oil objective, and images captured using an MCID Elite version 7.0 image analysis system (Interfocus, Haverhill, UK). Pilot studies established that counterstaining with cresyl violet produced a higher contrast image than that obtained using methyl green, enabling better distinction and measurement of centrosomal PCM1 immunoreactivity under the black and white camera conditions utilized for image analysis (chosen in preference to a colour camera due to the quicker image capture times and ease of image capture). As PCM1 at the centrosome was only readily identifiable in glia but not neurons (see Results), measurements of centrosomal PCM1 immunoreactive area were made over glia in the white matter. A total of 50 of these PCM1 immunoreactive cells were selected randomly, and the centrosomal PCM1 immunoreactive area outlined by hand using the cursor, and calibrated to square micrometres using a microscale and the MCID Elite system. The test–retest variability in mean PCM1 immunoreactive area per subject was <2%. All assessments were carried out blinded to diagnosis and genotype.
DISC1 genotype determination
Genomic DNA was extracted from each subject using a Nucleon Genomic DNA extraction kit (Tepnel Life Sciences, Manchester, UK). Genotype was determined using Applied Biosystems (Warrington, UK), pre-designed Taqman SNP genotyping assays (Leu607Phe: C_1650667_10; Ser704Cys: C_1433135_20) in accordance with manufacturer's instructions using a 7900HT real-time PCR system and SDS 2.2.2 software. Genotype reproducibility was 100%. The SH-SY5Y cells were also genotyped, and were Leu607/Leu607 and Ser704/Ser704.
Differentiation and staining of SH-SY5Y cells
The differentiation protocol utilized was based on that established by Encinas et al. (52) which results in fully differentiated neuron-like cells. Initially, SH-SY5Y cells (European Collection of Cell Cultures, Porton Down, UK) were cultured in Dulbecco's Modified Eagle Medium (DMEM; Sigma, Poole, UK) supplemented with 10% foetal calf serum (Sigma), 2 mm l-glutamine (Sigma) and 1% non-essential amino acids (Sigma). Cells were maintained in a humidified incubator at 37°C and 5% CO2. To initiate the differentiation process, cells were seeded at a density of 10 000 cells per well into 4 well chamber slides (Lab-Tek II chamber slides, CC2 treated, VWR). After settling overnight, the media was changed to DMEM supplemented with 5% foetal calf serum and 10 µm all-trans retinoic acid (Sigma). After culturing for 10 days in the presence of retinoic acid, the medium was substituted with DMEM (not supplemented with foetal calf serum) containing 50 ng/ml of recombinant human BDNF (Universal Biologicals, Cambridge, UK) for 7 days.
Differentiated and undifferentiated SH-SY5Y cells were fixed with methanol at −20°C for 6 min, given two 10 min washes with PBS and blocked for 1 h at room temperature (RT) with 10% normal goat serum (Sigma) diluted in PBS-T and 2% bovine serum albumin (BSA; Sigma). Cells were then incubated for 1 h at RT with rabbit anti-PCM1 diluted at 1:200 in PBS-T, containing 1% normal goat serum and 2% BSA. After three 10 min washes in PBS, cells were incubated for 1 h at RT with goat anti-rabbit Alexa Fluor 568 (at 1:1000; Invitrogen, Paisley, UK) in PBS-T containing 1% normal goat serum and 2% BSA. Cells were given three 10 min washes in PBS, dipped into distilled water and cover slipped using Vectashield (Vector Laboratories).
Statistical analyses
All statistical analyses were performed using SPSS version 16. One sample Kolmogorov–Smirnov tests were used to verify that the data were normally distributed. Potential influences of continuous variables (e.g. brain pH, post mortem interval, age) upon centrosomal PCM1 immunoreactive area were examined using Spearman correlations. Effects of diagnosis, genotype and race (and their interaction) were analysed using ANOVA, with planned comparisons between diagnostic groups and genotypes explored using LSD tests. Owing to the low frequency of the minor alleles for each SNP, additional ANOVAs were performed in which subjects carrying the minor allele were grouped together and compared with subjects homozygous for the higher frequency allele.
FUNDING
This work was funded by the UK Medical Research Council. The brain specimens utilized in this study were acquired with funding provided by the Intramural Research Program of the National Institute of Mental Health, National Institutes of Health, Department of Health and Human Services, United States Government.
ACKNOWLEDGEMENTS
We thank Katherine Mountain for her contributions, Li Chen for technical help, Phil Burnet for HEK293 protein extracts, Andreas Merdes for the PCM1 construct and Atsushi Kamiya for advice.
Conflict of Interest statement. P.J.H. reports receiving in the past 3 years honoraria for educational lectures, chairing scientific meetings or advisory boards, from Bristol Myers Squibb, Janssen, Merck, Sanofi, and Wyeth, and an unrestricted educational grant from GlaxoSmithKline.
REFERENCES
- 1.Millar J.K., Wilson-Annan J.C., Anderson S., Christie S., Taylor M.S., Semple C.A., Devon R.S., Clair D.M., Muir W.J., Blackwood D.H., et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. doi:10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 2.Chubb J.E., Bradshaw N.J., Soares D.C., Porteous D.J., Millar J.K. The DISC locus in psychiatric illness. Mol. Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. doi:10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 3.Hennah W., Thomson P., McQuillin A., Bass N., Loukola A., Anjorin A., Blackwood D., Curtis D., Deary I.J., Harris S.E., et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol. Psychiatry. 2009;14:865–873. doi: 10.1038/mp.2008.22. doi:10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- 4.Ishizuka K., Paek M., Kamiya A., Sawa A. A review of disrupted-in-schizophrenia-1 (DISC1): neurodevelopment, cognition and mental conditions. Biol. Psychiatry. 2006;59:1189–1197. doi: 10.1016/j.biopsych.2006.03.065. doi:10.1016/j.biopsych.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 5.Porteous D.J., Thomson P., Brandon N.J., Millar J.K. The genetics and biology of DISC1—an emerging role in psychosis and cognition. Biol. Psychiatry. 2006;60:123–131. doi: 10.1016/j.biopsych.2006.04.008. doi:10.1016/j.biopsych.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Camargo L.M., Collura V., Rain J.C., Mizuguchi K., Hermjakob H., Kerrien S., Bonnert T.P., Whiting P.J., Brandon N.J. Disrupted in schizophrenia 1 interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. doi:10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 7.Jaaro-Peled H., Hayashi-Takagi A., Seshadri S., Kamiya A., Brandon N.J., Sawa A. Neurodevelopmental mechanisms of schizophrenia: understanding disturbed postnatal brain maturation through neuregulin-1-ErbB4 and DISC1. Trends Neurosci. 2009;32:485–495. doi: 10.1016/j.tins.2009.05.007. doi:10.1016/j.tins.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi-Takagi A., Takaki M., Graziane N., Seshadri S., Murdoch H., Dunlop A.J., Makino Y., Seshadri A.J., Ishizuka K., Srivastava D.P., et al. Disrupted-in-schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via RAC1. Nat. Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. doi:10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enomoto A., Asai N., Namba T., Wang Y., Kato T., Tanaka M., Tatsumi H., Taya S., Tsuboi D., Kuroda K., et al. Roles of disrupted-in-schizophrenia-1-interacting protein Girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. doi:10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Kim J.Y., Duan X., Liu C.Y., Jang M.H., Guo J.J.U., Pow-Anpongkul N., Kang E.C., Song H.J., Ming G.L. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. doi:10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer K.D., Morris J.A. Disc1 regulates granule cell migration in the developing hippocampus. Hum. Mol. Genet. 2009;18:3286–3297. doi: 10.1093/hmg/ddp266. doi:10.1093/hmg/ddp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Y.W., Ge X.C., Frank C.L., Madison J.M., Koehler A.N., Doud M.K., Tassa C., Berry E.M., Soda T., Singh K.K., et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3 beta-Catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. doi:10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris J.A., Kandpal G., Ma L., Austin C.P. DISC1 (disrupted-in-schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003;13:1591–1608. doi: 10.1093/hmg/ddg162. doi:10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi K., Asanuma M., Miyazaki I., Diaz-Corrales F.J., Katayama T., Tohyama M., Ogawa N. DISC1 localizes to the centrosome by binding to kendrin. Biochem. Biophys. Res. Commun. 2004;317:1195–1199. doi: 10.1016/j.bbrc.2004.03.163. doi:10.1016/j.bbrc.2004.03.163. [DOI] [PubMed] [Google Scholar]
- 15.Bradshaw N.J., Ogawa F., Antolin-Fontes B., Chubb J.E., Carlyle B.C., Christie S., Claessens A., Porteous D.J., Millar J.K. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem. Biophys. Res. Commun. 2008;377:1091–1096. doi: 10.1016/j.bbrc.2008.10.120. doi:10.1016/j.bbrc.2008.10.120. [DOI] [PubMed] [Google Scholar]
- 16.Kamiya A., Tan P.L., Kubo K., Engelhard C., Ishizuka K., Kubo A., Tsukita S., Pulver A.E., Nakajima K., Cascella N.G., et al. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch. Gen. Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. doi:10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datta S.R., McQuillin A., Rizig M., Blaveri E., Thirumalai S., Kalsi G., Lawrence J., Bass N.J., Puri V., Choudhury K., et al. A threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia. Mol. Psychiatry. 2008 doi: 10.1038/mp.2008.128. DOI 10.1038/mp.2008.128. 10.1038/mp.2008.128. [DOI] [PubMed] [Google Scholar]
- 18.Gurling H.M., Critchley H., Datta S.R., McQuillin A., Blaveri E., Thirumalai S., Pimm J., Krasucki R., Kalsi G., Quested D., et al. Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch. Gen. Psychiatry. 2006;63:844–854. doi: 10.1001/archpsyc.63.8.844. doi:10.1001/archpsyc.63.8.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dammermann A., Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. doi:10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipska B.K., Peters T., Hyde T.M., Halim N., Horowitz C., Mitkus S., Weickert C.S., Matsumoto M., Sawa A., Straub R.E., et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum. Mol. Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. doi:10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 21.Hodgkinson C.A., Goldman D., Jaeger J., Persaud S., Kane J.M., Lipsky R.H., Malhotra A.K. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet. 2004;75:862–872. doi: 10.1086/425586. doi:10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eastwood S.L., Hodgkinson C.A., Harrison P.J. DISC-1 Leu607Phe alleles differentially affect centrosomal PCM1 localization and neurotransmitter release. Mol. Psychiatry. 2009;14:556–557. doi: 10.1038/mp.2009.13. doi:10.1038/mp.2009.13. [DOI] [PubMed] [Google Scholar]
- 23.Balczon R., Bao L., Zimmer W.E. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J. Cell Biol. 1994;124:783–793. doi: 10.1083/jcb.124.5.783. doi:10.1083/jcb.124.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubo A., Tsukita S. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 2003;116:919–928. doi: 10.1242/jcs.00282. doi:10.1242/jcs.00282. [DOI] [PubMed] [Google Scholar]
- 25.Ohama Y., Hayashi K. Relocalization of a microtubule-anchoring protein, ninein, from the centrosome to dendrites during differentiation of mouse neurons. Histochem. Cell Biol. 2009;132:515–524. doi: 10.1007/s00418-009-0631-z. doi:10.1007/s00418-009-0631-z. [DOI] [PubMed] [Google Scholar]
- 26.Balczon R., Simerly C., Takahashi D., Schatten G. Arrest of cell cycle progression during first interphase in murine zygotes microinjected with anti-PCM-1 antibodies. Cell Motil. Cytoskeleton. 2002;52:183–192. doi: 10.1002/cm.10043. doi:10.1002/cm.10043. [DOI] [PubMed] [Google Scholar]
- 27.Srsen V., Gnadt N., Dammermann A., Merdes A. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J. Cell Biol. 2006;174:625–630. doi: 10.1083/jcb.200606051. doi:10.1083/jcb.200606051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James R., Adams R.R., Christie S., Buchanan D.J., Porteous D.J., Millar J.K. Disrupted in schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol. Cell. Neurosci. 2004;26:112–122. doi: 10.1016/j.mcn.2004.01.013. doi:10.1016/j.mcn.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Srsen V., Fant X., Heald R., Rabouille C., Merdes A. Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 2009;10:28. doi: 10.1186/1471-2121-10-28. doi:10.1186/1471-2121-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge X., Frank C.L., de Anda F.C., Tsai H. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. doi:10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seshadri S., Kamiya A., Yokota Y., Prikulis I., Kano S., Hayashi-Takagi A., Stanco A., Eom T.Y., Rao S., Ishizuka K., et al. Disrupted-in-schizophrenia-1 expression is regulated by {beta}-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc. Natl Acad. Sci. USA. 2010;107:5622–5627. doi: 10.1073/pnas.0909284107. doi:10.1073/pnas.0909284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkpatrick B., Xu L., Cascella N., Ozeki Y., Sawa A., Roberts R.C. DISC1 immunoreactivity at the light and ultrastructural level in the human neocortex. J. Comp. Neurol. 2006;497:436–450. doi: 10.1002/cne.21007. doi:10.1002/cne.21007. [DOI] [PubMed] [Google Scholar]
- 33.Callicott J.H., Straub R.E., Pezawas L., Egan M.F., Mattay V.S., Hariri A.R., Verchinski B.A., Meyer-Lindenberg A., Balkissoon R., Kolachana B., et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc. Natl Acad. Sci. USA. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. doi:10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu M., Tang F., Yue W., Ruan Y., Lu T., Liu Z., Zhang H., Han Y., Zhang D., Wang F., Zhang D. Positive association of the disrupted-in-schizophrenia-1 gene (DISC1) with schizophrenia in the Chinese Han population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:266–270. doi: 10.1002/ajmg.b.30322. [DOI] [PubMed] [Google Scholar]
- 35.Song W., Li W., Feng J., Heston L.L., Scaringe W.A., Sommer S.S. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem. Biophys. Res. Commun. 2008;367:700–706. doi: 10.1016/j.bbrc.2007.12.117. doi:10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto R., Numakawa T., Ohnishi T., Kumamaru E., Yagasaki Y., Ishimoto T., Mori T., Nemoto K., Adachi N., Izumi A., et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum. Mol. Genet. 2006;15:3024–3033. doi: 10.1093/hmg/ddl244. doi:10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- 37.Di Giorgio A., Blasi G., Sambataro F., Rampino A., Papazacharias A., Gambi F., Romano R., Caforio G., Rizzo M., Latorre V., et al. Association of the SerCys DISC1 polymorphism with human hippocampal formation gray matter and function during memory encoding. Eur. J. Neurosci. 2008;28:2129–2136. doi: 10.1111/j.1460-9568.2008.06482.x. doi:10.1111/j.1460-9568.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prata D.P., Mechelli A., Fu C.H.Y., Picchioni M., Kane F., Kalidini S., McDonald C., Kravariti E., Toulopoulou T., Miorelli A., et al. Effect of disrupted-in-schizophrenia-1 on pre-frontal cortical function. Mol. Psychiatry. 2008;13:915–917. doi: 10.1038/mp.2008.76. doi:10.1038/mp.2008.76. [DOI] [PubMed] [Google Scholar]
- 39.Thomson P.A., Harris S.E., Starr J.M., Whalley L.J., Porteous D.J., Deary I.J. Association between genotype at an exonic SNP in DISC1 and normal cognitive aging. Neurosci. Lett. 2005;389:41–45. doi: 10.1016/j.neulet.2005.07.004. doi:10.1016/j.neulet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Szeszko P.R., Hodgkinson C.A., Robinson D.G., Derosse P., Bilder R.M., Lencz T., Burdick K.E., Napolitano B., Betensky J.D., Kane J.M., et al. DISC1 is associated with prefrontal cortical gray matter and positive symptoms in schizophrenia. Biol. Psychol. 2008;79:103–110. doi: 10.1016/j.biopsycho.2007.10.011. doi:10.1016/j.biopsycho.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakata K., Lipska B.K., Hyde T.M., Ye T., Newburn E.N., Morita Y., Vakkalanka R., Barenboim M., Sei Y., Weinberger D.R., Kleinman J.E. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc. Natl Acad. Sci. USA. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. doi:10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burdick K.E., Kamiya A., Hodgkinson C.A., Lencz T., Derosse P., Ishizuka K., Elashvili S., Arai H., Goldman D., Sawa A., Malhotra A.K. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum. Mol. Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. doi:10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicodemus K.K., Law A.J., Radulescu E., Luna A., Kolachana B., Vakkalanka R., Rujescu D., Giegling I., Straub R.E., McGee K., et al. NRG1, ERBB4 and AKT1 epistasis increases schizophrenia risk and is biologically validated via functional neuroimaging in healthy controls. Arch. Gen. Psychiatry. 2010 doi: 10.1001/archgenpsychiatry.2010.117. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal D., Koschnick J.R., Siegers L.H., Hof P.R. Oligodendrocyte pathophysiology: a new view of schizophrenia. Int. J. Neuropsychopharmacol. 2007;10:503–511. doi: 10.1017/S146114570600722X. doi:10.1017/S146114570600722X. [DOI] [PubMed] [Google Scholar]
- 45.Martins-de-Souza D. Proteome and transcriptome analysis suggests oligodendrocyte dysfunction in schizophrenia. J. Psychiatr. Res. 2010;44:149–156. doi: 10.1016/j.jpsychires.2009.07.007. doi:10.1016/j.jpsychires.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Kamiya A., Tomoda T., Chang J., Takaki M., Zhan C., Morita M., Cascio M.B., Elashvili S., Koizumi H., Takanezawa Y., et al. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outqrowth, is modulated by genetic variations of DISC1. Hum. Mol. Genet. 2006;15:3313–3323. doi: 10.1093/hmg/ddl407. doi:10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- 47.Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell. 2006;17:680–689. doi: 10.1091/mbc.E05-04-0360. doi:10.1091/mbc.E05-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leliveld S.R., Hendriks P., Michel M., Sajnani G., Bader V., Trossbach S., Prikulis I., Hartman R., Jonas E., Willbold D., Requena J.R., Korth C. Oligomer assembly of the C-terminal DISC1 domain (640–685) is controlled by self-association motifs and disease-associated polymorphism S704C. Biochemistry. 2009;48:7746–7755. doi: 10.1021/bi900901e. doi:10.1021/bi900901e. [DOI] [PubMed] [Google Scholar]
- 49.Park H.J., Seo H.J., Kim H.W., Kim J.S., Hwang S.Y., Seong Y.S. The centrosomal localization of KM-HN-1 (MGC33607) depends on that leucine zipper motif and the C-terminal coiled-coil domain. Exp. Mol. Med. 2009;39:828–838. doi: 10.1038/emm.2007.90. [DOI] [PubMed] [Google Scholar]
- 50.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 51.Leliveld S.R., Bader V., Hendriks P., Prikulis I., Sajnani G., Requena J.R., Korth C. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J. Neurosci. 2008;28:3839–3845. doi: 10.1523/JNEUROSCI.5389-07.2008. doi:10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Encinas M., Iglesias M., Liu Y., Wang H., Muhaisen A., Cena V., Gallego C., Comella J.X. Sequential treatment of SH-SY5Y cells with retoinic acid and brain derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. doi:10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]