Abstract
Programmed necrosis induced by DNA alkylating agents, such as MNNG, is a caspase-independent mode of cell death mediated by apoptosis-inducing factor (AIF). After poly(ADP-ribose) polymerase 1, calpain, and Bax activation, AIF moves from the mitochondria to the nucleus where it induces chromatinolysis and cell death. The mechanisms underlying the nuclear action of AIF are, however, largely unknown. We show here that, through its C-terminal proline-rich binding domain (PBD, residues 543–559), AIF associates in the nucleus with histone H2AX. This interaction regulates chromatinolysis and programmed necrosis by generating an active DNA-degrading complex with cyclophilin A (CypA). Deletion or directed mutagenesis in the AIF C-terminal PBD abolishes AIF/H2AX interaction and AIF-mediated chromatinolysis. H2AX genetic ablation or CypA downregulation confers resistance to programmed necrosis. AIF fails to induce chromatinolysis in H2AX or CypA-deficient nuclei. We also establish that H2AX is phosphorylated at Ser139 after MNNG treatment and that this phosphorylation is critical for caspase-independent programmed necrosis. Overall, our data shed new light in the mechanisms regulating programmed necrosis, elucidate a key nuclear partner of AIF, and uncover an AIF apoptogenic motif.
Keywords: AIF, DNA damage, H2AX, PARP-1, programmed necrosis
Introduction
Cell death was initially divided into two generic categories: apoptosis, which involves the activation of caspases and necrosis, which is considered as a fortuitous and uncontrolled event. Nevertheless, recent pharmacological and genetic evidences show that, similar to apoptosis, necrosis could be a tightly regulated form of caspase-independent cell death (Golstein et al, 2003; Festjens et al, 2006). RIP1, RIP3, poly(ADP-ribose) polymerase 1 (PARP-1), and apoptosis-inducing factor (AIF) are involved in the execution of the most studied necrotic pathways. RIP1 and RIP3 kinase activities are required in necroptosis (Hitomi et al, 2008) and TNFα-mediated necrosis (Cho et al, 2009; He et al, 2009). PARP-1 and AIF regulate parthanatos and programmed necrosis (Moubarak et al, 2007; Wang et al, 2009a).
AIF is a FAD-dependent oxidoreductase that has a vital role in oxidative phosphorylation (Vahsen et al, 2004). After a caspase-independent cell death insult, AIF is cleaved by calpains and/or cathepsins to yield truncated AIF (tAIF), the pro-apoptotic AIF form (∼57 kDa) (Polster et al, 2005; Yuste et al, 2005). tAIF relocates from the mitochondria to the cytosol and nucleus, where it provokes chromatinolysis and programmed cell death (PCD) (Susin et al, 1999). Thorough studies on AIF have increased our knowledge of its structure/function relationship (Mate et al, 2002; Ye et al, 2002; Sevrioukova, 2009), the molecular mechanisms regulating its mitochondrial release (Otera et al, 2005; Polster et al, 2005; Yuste et al, 2005), and the genetic characteristics of five isoforms: AIF, AIF2 (AIFexB), AIFsh, AIFsh2, and AIFsh3 (Susin et al, 1999; Loeffler et al, 2001; Delettre et al, 2006a, 2006b; Hangen et al, 2010). It is also known that AIF cytosolic/nuclear transit is regulated by Heat Shock Protein-70 and cyclophilin A (CypA) (Gurbuxani et al, 2003; Zhu et al, 2007). A complementary study indicates that, through a yet undefined mechanism, AIF associates in the nucleus with CypA to induce chromatinolysis (Cande et al, 2004).
The understanding of the dual physiological role of AIF, which deals with life in the mitochondria and death in the nucleus, has been largely illustrated (Lorenzo and Susin, 2007; Joza et al, 2009). In this sense, the Harlequin mouse strain, which exhibits an 80% reduction in AIF expression, has been particularly useful (Klein et al, 2002). Concerning the downstream effectors of AIF, the data obtained in Caenorhabditis elegans, Saccharomyces cereviseae, and Drosophila melanogaster are of particular interest. The C. elegans AIF ortholog WAH-1 interacts with CSP-6 (the mammalian endonuclease G ortholog) to mediate nuclear DNA degradation (Wang et al, 2002). AIF also associates with SCRM-1 to regulate externalization of phosphatidylserine (PS) on the surface of apoptotic cells (Wang et al, 2007). In S. cereviseae, CypA is also essential to AIF-proapoptotic activity (Wissing et al, 2004). Finally, in D. melanogaster, downregulation of DmTrx-2 suppresses AIF-mediated PCD (Joza et al, 2008). In mammals, given that the translocation of AIF from the mitochondria to the nucleus occurs in nearly all caspase-independent PCD paradigms, AIF is considered as a main effector of this mode of PCD (Dawson and Dawson, 2004; Lorenzo and Susin, 2007; Joza et al, 2009). Strong evidence for a role of AIF in caspase-independent PCD has come from studies performed with exocitotoxins or alkylating DNA damage agents, such as the nitrosourea MNNG (Yu et al, 2002; Wang et al, 2004, 2009a, 2009b; Zong et al, 2004; Cheung et al, 2005; Ishihara et al, 2005; Moubarak et al, 2007; Hegedus et al, 2008).
In spite of the considerable amount of work performed on AIF, key questions remain to be addressed. What happens when AIF is in the nucleus? How does this protein induce chromatinolysis? Are there unknown nuclear partners associated with AIF chromatinolytic activity? By resorting to a combination of biophysical, cellular, genetic, and molecular biology methodologies, we identify here histone H2AX as a key nuclear partner of AIF in MNNG-mediated programmed necrosis.
H2AX is a member of the histone H2A family, which is part of the histone nucleosome core. H2AX, which represents about 2–25% of total H2A, is a sensitive marker for DNA double-strand breaks (DSB) (Fernandez-Capetillo et al, 2004). On exposure of cells to a DSB DNA damage inducer, H2AX is phosphorylated at Ser139 in the nucleosomes surrounding the break point (Thiriet and Hayes, 2005). Phosphorylated H2AX (γH2AX) modifies here the structure of chromatin to render damaged DNA sites accessible to repair factors (Paull et al, 2000; Redon et al, 2002; Celeste et al, 2003; Pilch et al, 2003). The function of H2AX is therefore primarily associated with DNA damage repair. γH2AX is also implicated in the regulation of apoptosis occurring through the caspase-3/caspase-activated DNase (CAD) pathway (Rogakou et al, 2000; Lu et al, 2006). However, little is known about the implication of H2AX in caspase-independent PCD. We show here that (i) H2AX phosphorylates on Ser139 after MNNG treatment, (ii) γH2AX is essential for necrotic PCD, and (iii) the genetic ablation of H2AX prevents the effect of AIF in the nucleus, the AIF/CypA association, and caspase-independent programmed necrosis.
Results
AIF interacts with histone H2AX in caspase-independent programmed necrosis
Treatment with DNA alkylating agents, such as MNNG, provokes breaks in the DNA. When the DNA damage is extensive, the cell undertakes a caspase-independent type of PCD called programmed necrosis. This type of PCD is regulated by the sequential activation of PARP-1, calpains, Bax, and AIF. The apoptogenic form of AIF redistributes from the mitochondria to the nucleus where it provokes, by an unknown mechanism, chromatinolysis and PCD (Figure 1A; Supplementary Figure 1) (Moubarak et al, 2007).
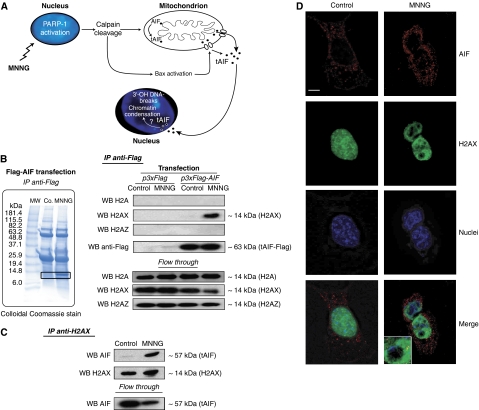
Figure 1.
AIF interacts with histone H2AX in MNNG-induced programmed necrosis. (A) Model for alkylating DNA damage-mediated death. MNNG induces death through PARP-1, calpains, and Bax activation. These proteins facilitate the cleavage and release of tAIF (the pro-apoptotic AIF form) from the mitochondrion to the cytosol and nucleus where tAIF generates 3′-OH DNA breaks and chromatin condensation through a yet unknown mechanism. (B) Representative SDS–PAGE and Coomassie blue staining of Flag-immunoprecipitates (IP anti-Flag) obtained from MEFs transfected with Flag-AIF and either untreated (Co.) or treated with MNNG (9 h). MW, molecular weight marker. After MNNG treatment, a 14 kDa protein (in the square) was co-purified with AIF. Flag-immunoprecipitates (IP anti-Flag) obtained from MEFs transfected with Flag-AIF and either untreated (Co.) or treated with MNNG (9 h) were analysed by western blotting (WB) using anti-H2A, anti-H2AX, anti-H2AZ, and anti-Flag antibodies. WB on lysates after IP is also shown (flow through). (C) H2AX immunoprecipitation on MEFs incubated or not with MNNG (9 h). AIF and H2AX were detected by immunoblot. WB on lysates after IP is also shown (flow through). (D) MEFs left untreated (control) or treated with MNNG as above were subjected to confocal immunofluorescent detection of AIF (red) and H2AX (green). Nuclei were stained with Hoechst 33342 (blue). This experiment was repeated five times, yielding similar results. Bar: 10 μm.
AIF redistributes from the mitochondria to the nucleus 6/9 h post-MNNG treatment (Moubarak et al, 2007). Thus, we used this timeframe to identify nuclear components downstream of AIF in programmed necrosis. Lysates obtained from mouse embryo fibroblasts (MEFs) that were transfected with AIF tagged with a Flag epitope, and treated or not with MNNG, were immunoprecipitated with an anti-Flag antibody. As shown in Figure 1B, SDS–PAGE and colloidal Coomassie blue stain of the immunoprecipitates showed a protein of ∼14 kDa, which was only detected in samples obtained from MNNG-treated cells. To identify this protein, the band was in-gel digested and analysed by MALDI-TOF (Supplementary Figure 2). We visualized a single 944.5 m/z ion, in which we performed a complementary post-source decay analysis. This approach yielded an amino acidic sequence matching with histones H2A, H2AX, or H2AZ (AGLQFPVGR). We clarified the identity of this histone through a complementary immunoblot approach. Indeed, though H2A or H2AZ-specific antibodies failed to recognize the ∼14 kDa band immunoprecipitated from MNNG-treated MEFs, an H2AX-specific antibody reacted against this band (Figure 1B). It therefore appears that H2AX associates with AIF in programmed necrosis.
To confirm this assumption and to assess whether this interaction could be observed with endogenous proteins, we performed a reverse immunoprecipitation. As depicted in Figure 1C, AIF and H2AX did not co-immunoprecipitate in control MEFs. However, treatment with MNNG led to an AIF/H2AX association. Similar co-precipitations performed with H2A and H2AZ antibodies yielded negative results (not shown). The AIF/H2AX interaction was corroborated by confocal microscopy. In control cells, AIF was confined to the mitochondria, whereas H2AX was localized in the nucleus. On induction of programmed necrosis, H2AX and AIF co-localized in defined nuclear areas (Figure 1D). Altogether, these data show that, after MNNG treatment, AIF interacts with H2AX in the nucleus.
Characterization of the physical interaction between AIF and H2AX by surface plasmon resonance and molecular modelling
We resorted to surface plasmon resonance (SPR) assays to obtain further confirmation of the direct interaction between AIF and H2AX, and to determine its characteristics. His-tagged recombinant tAIF, the nuclear apoptogenic form of AIF, was generated and captured on an NTA sensor chip. Recombinant H2AX was brought into contact with tAIF by a continuous flow. A specific SPR resonance signal was detected in the flowcell, indicating that H2AX was bound to tAIF. This signal was reproducibly proportional to the concentration of H2AX that was injected (Figure 2A). The dissociation of the tAIF/H2AX complex showed two phases, one fast (koff(1)=0.065 s−1) and the other slow (koff(2)=1.29 × 10−3 s−1). The concentration-dependence of the steady-state SPR signals indicated a tAIF/H2AX dissociation constant (KD) of 8.7±2.2 × 10−7 M (Figure 2A). These results show that the tAIF/H2AX interaction is direct and does not require additional proteins.
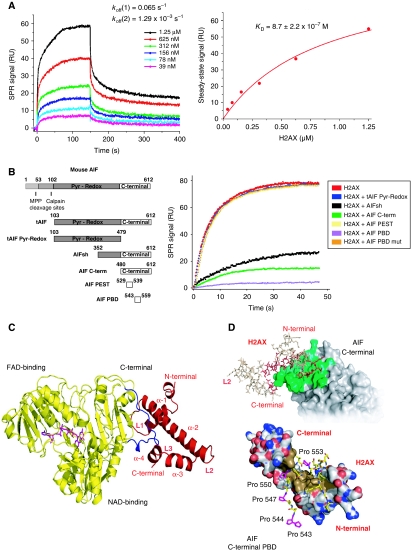
Figure 2.
AIF interacts with H2AX through its C-terminal PBD. (A) SPR sensorgrams resulting from the injection of the indicated concentrations of soluble H2AX onto immobilized tAIF. koff, dissociation rate constants. Right: H2AX concentration dependence of the steady-state response. KD, equilibrium dissociation constant. (B) Mouse AIF, tAIF, tAIF Pyr-Redox, AIFsh, AIF C-term, AIF PEST, and AIF PBD protein organization (Left). Numbers designate aminoacids. Pyr-Redox and C-terminal AIF domains are indicated. Right: SPR sensorgrams showing the association of H2AX with immobilized tAIF, in the absence or presence of tAIF Pyr-Redox, AIFsh, AIF C-term, AIF PEST, AIF PBD, or AIF PBD mut. (C) Model of the tAIF/H2AX complex obtained by homology with the known crystal structures of the individual partners. Left: ribbon structure of tAIF (yellow) and H2AX (red). AIF interacts with H2AX through its C-terminal residues 543–559 (in blue). FAD (the AIF cofactor) is drawn in magenta. H2AX folding consists of four α helices (α-1 to α-4) linked by three short loops (L1–L3). (D) tAIF/H2AX molecular docking. Upper: surface of the tAIF binding interface (green). Residues of H2AX interacting with tAIF surface are drawn in red. Lower: tAIF C-terminal proline-rich residues interact with H2AX through a hydrophobic cluster (brown). Acidic and basic residues are, respectively, depicted in red and blue. AIF C-terminal Pro543, 544, 547, 550, and 553 are drawn in magenta.
As the C-terminal domain of AIF is the pro-apoptotic region of the molecule (Cheung et al, 2006; Delettre et al, 2006a; Zhang et al, 2009), we tested whether this domain was implicated in the association with H2AX. We set up an SPR-based competition assay in which, before being injected into the tAIF-captured chip, H2AX was mixed with different AIF forms (Figure 2B), including two motifs enclosed in this C-terminal domain: a predicted PEST sequence (amino acids 529–539) and a proline-rich binding domain (PBD; amino acids 543–559) (Mate et al, 2002). The PEST and PBD sequences are usually involved in protein–protein interactions. As shown in Figure 2B, the amount of H2AX available for binding tAIF was significantly lower when H2AX was pre-incubated with AIF forms containing the C-terminal domain of the protein (AIFsh and the entire C-terminus). This indicated that, unlike the tAIF Pyr-Redox N-terminal region, the C-terminal domain of AIF interacts with H2AX. Remarkably, in contrast to what was observed in the PEST sequence, the tAIF/H2AX binding signal was reduced when pre-incubating H2AX with the PBD sequence (AIF PBD) (Figure 2B). This indicated that H2AX specifically associates with this PBD. We confirmed this finding by directed mutagenesis: substitution of the five prolines of the PBD motif by alanines (AIF PBD mut) eliminated the interaction of this domain with H2AX.
Molecular modelling, based in our above-described SPR-based competition assay (e.g. relevance of AIF's C-terminal domain), illustrated the AIF/H2AX interaction (Figure 2C and D). As reported earlier, AIF comprises three structural domains: FAD binding, NADH binding, and C-terminal domain (Mate et al, 2002). Regarding H2AX, our model includes a short N-terminal pre-helix (4 residues), an 11-residue helix (α-1), followed by a short loop (L1), a long 27-residue helix (α-2), a second loop (L2), an 8-residue helix (α-3), a third loop (L3), and a final C-terminal α-4 helix (6 residues). This core is followed by a C-terminal tail, which is not involved in the interaction with AIF (residues 85–142). The AIF/H2AX complex is formed by an H2AX hydrophobic region (residues 51–64), an additional leucine cluster located in α-4 helix (Leu93, Leu96, and Leu97), and the PBD enclosed in the C-terminal domain of AIF, which includes the critical binding prolines 543, 544, 547, 550, and 553. According to this model, the interface surface area is 871.18 Å2 (Figure 2D).
H2AX is phosphorylated on Ser139 during programmed necrosis
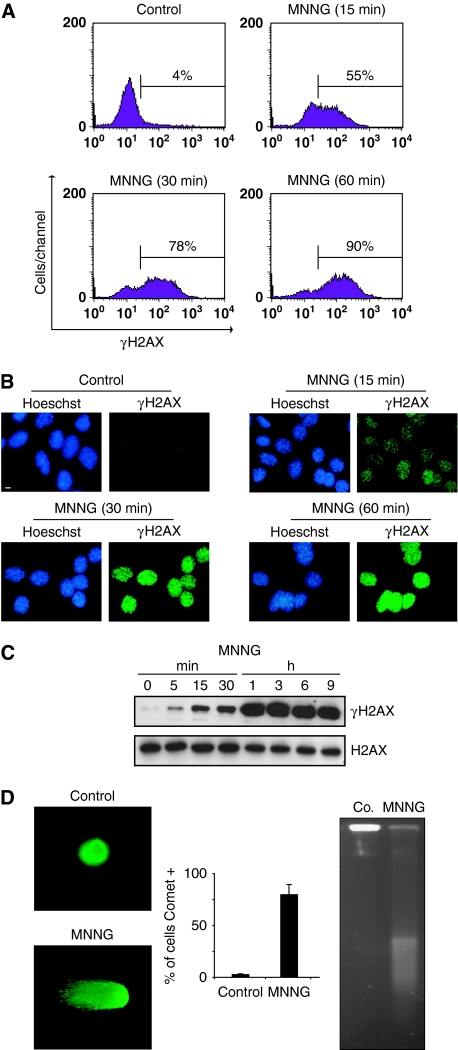
Phosphorylation of H2AX on Ser139 is a key event in caspase-dependent PCD (Rogakou et al, 2000; Lu et al, 2006; Solier et al, 2009). Thus, to investigate the biological role of H2AX (and the above-described AIF/H2AX association) in programmed necrosis, we first assessed whether H2AX was here phosphorylated. Flow cytometry detection successfully revealed a rapid and time-dependent H2AX phosphorylation on Ser139 in MNNG-treated cells (Figure 3A). This result was confirmed by immunofluorescence, which detected formation of γH2AX nuclear foci and immunoblotting (Figure 3B and C). Detection of γH2AX nuclear foci, which usually characterizes DSB generation (Banath and Olive, 2003), led us to assess the DNA damage provoked by MNNG. Two independent tests were applied: neutral comet assay and field inversed gel electrophoresis (FIGE). Both methods corroborated that high doses of MNNG generate DSB in DNA (Figure 3D).
Figure 3.
MNNG induces H2AX phosphorylation and DNA double-strand breaks. (A) Kinetics of the H2AX phosphorylation induced by MNNG in MEFs. Cells were left untreated (control) or treated with MNNG, and H2AX Ser139 phosphorylation was assessed by flow cytometry. Representative cytofluorometric plots are shown. Percentages refer to γH2AX-positive staining. This experiment was repeated five times with a lower experimental variability (⩽5%). (B) Immunofluorescent staining of γH2AX detected in MEFs left untreated (control) or treated with MNNG and stained with Hoechst 33342 (to visualize nuclei) or a specific γH2AX antibody. Representative cells are shown. This experiment was repeated eight times with similar results. Bar: 10 μm. (C) γH2AX immunoblotting detection in total extracts obtained at different times from MEFs untreated or treated with MNNG. The membrane was reblotted for total H2AX to control protein loading. (D) Analysis of DNA DSB by a neutral comet assay performed in MEFs untreated or treated with MNNG (1 h). Representative comet images and percentage of cells with a tail (comet +) are shown. Data are means±s.d. (n=6). Alternatively, MEFs were analysed by field inversion gel electrophoresis (FIGE). The smear observed in MNNG-treated cells indicated accumulation of DSB (Saintigny et al, 2001). A full-colour version of this figure is available at The EMBO Journal Online.
γH2AX regulates MNNG-mediated PCD
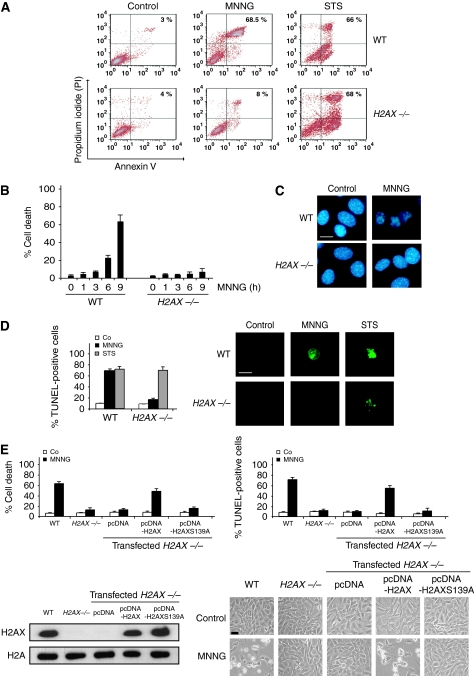
Through the analysis of γH2AX and DSB caused by UVA exposure or etoposide treatment, a recent work has reported that H2AX−/− MEFs were less sensitive to these two PCD inducers than control cells (Lu et al, 2006). This shows that H2AX controls UVA and etoposide-mediated death. γH2AX and DSB are also hallmarks of programmed necrosis. Thus, to assess the relevance of H2AX in this type of PCD, we tested the responsiveness of H2AX−/− cells to MNNG treatment. As depicted in Figure 4A and B, MNNG-mediated PCD was delayed in H2AX−/− MEFs. In these cells, the percentage of PS exposure and loss of viability reached ∼8% 9 h after MNNG treatment. At the same time, MNNG induced death in about 70% of H2AX-wt (WT) cells. As an internal control, we verified that WT and H2AX−/− MEFs treated with the apoptosis inducer staurosporine (STS) showed similar PCD levels (Figure 4A), confirming that H2AX−/− MEFs remained sensitive to STS-mediated death (Mukherjee et al, 2006). Strikingly, MNNG-treated H2AX−/− cells did not present TUNEL positivity or chromatin condensation: the hallmarks characterizing AIF-mediated chromatinolysis (Figure 4C and D). Thus, it seems that the presence of H2AX in the nucleus is required for both AIF-mediated chromatinolysis and programmed necrosis. To confirm that the blockade of these PCD hallmarks was due to the specific lack of H2AX, we reintroduced the H2AX gene into the H2AX−/− cells. As was to be expected, H2AX transfection resensitized H2AX−/− cells to MNNG-induced death (Figure 4E). Finally, to investigate the importance of γH2AX in programmed necrosis, we mutated Ser139 to Ala (S139A). Then, we transfected H2AXS139A into H2AX−/− cells. Strikingly, contrary to H2AX, H2AXS139A did not restore the MNNG ability to induce programmed necrosis in H2AX−/− cells (Figure 4E). The generation of γH2AX is thus critical in this type of PCD.
Figure 4.
γH2AX is essential for programmed necrosis. (A) WT and H2AX−/− cells were untreated (control) or treated with MNNG (9 h) or staurosporine (STS), labelled with AnnexinV-FITC and PI, and analysed by flow cytometry. Representative cytofluorometric plots are shown. Percentages refer to double-positive staining. (B) Kinetic analysis of PS exposure and cell viability loss induced by MNNG in WT and H2AX−/− MEFs. After the indicated times, cells were stained as in (A) and the frequency of double-positive labelling was recorded and expressed as a percentage. Data are the means of 10 independent experiments±s.d. (C) WT and H2AX−/− MEFs were treated with MNNG (9 h) and stained with Hoechst 33342 to visualize nuclei. Representative nuclei from untreated (control) or MNNG-treated cells are shown. Bar: 10 μm. (D) WT and H2AX−/− MEFs were untreated (Co) or treated with MNNG (9 h) or STS as in (A), stained for the detection of 3′-OH DNA breaks, and analysed by flow cytometry. Data are the means of six independent experiments±s.d. Right: representative microphotographs from each treatment are shown. Bar: 10 μm. (E) H2AX−/− MEFs were transfected with the indicated expression plasmids and selected as described in ‘Materials and methods' section. Then, cells were untreated (Co) or treated with MNNG (9 h). Cell death was analysed by AnnexinV-FITC/PI and TUNEL staining. Data are means±s.d. (n=8). The expression level of H2AX was assessed by immunoblotting. Equal loading was confirmed by histone H2A assessment. Representative microphotographs of each treatment are shown. Bar: 40 μm. A full-colour version of this figure is available at The EMBO Journal Online.
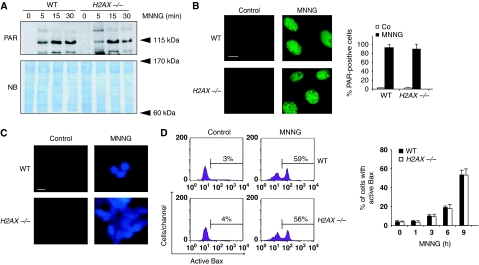
PARP-1, calpains, Bax, and AIF are activated in MNNG-treated H2AX−/− MEFs
Genetic deletion of H2AX prevents programmed necrosis. Therefore, it could be anticipated that the absence of H2AX in the nucleus would inactivate a key programmed necrotic effector (e.g. PARP-1, calpains, Bax, or AIF). We thus verified whether these proteins were activated or not in H2AX−/− MEFs after MNNG treatment. PARP-1 activation is assessed by the inverse relationship existing between poly(ADP-ribose) (PAR) and NAD+ and ATP cellular levels. As shown by the formation of PAR polymers, PARP-1 was equally activated in WT and H2AX−/− MEFs (Figure 5A and B). This showed that, even in the absence of H2AX, PARP-1 is an active enzyme. In line with that, MNNG caused a rapid and similar decrease in NAD+ and ATP cellular levels in both WT and H2AX−/− MEFs (Supplementary Figure 3). The specificity of the PARP-dependent NAD+ loss associated with MNNG treatment was confirmed using a pharmacological PARP inhibitor, PJ34. We next focused on calpains and Bax. Calpain activity was first measured with the help of a cell-permeable calpain substrate, which reports calpain activity in live cells. This approach showed that MNNG induced rapid calpain activation in WT and H2AX−/− MEFs (Figure 5C). These results were corroborated by a fluorogenic substrate-based assay (Supplementary Figure 4A). Finally, we showed a time-dependent increase in activated Bax in WT and H2AX−/− MNNG-treated cells (Figure 5D). This was confirmed using an immunofluorescence approach (Supplementary Figure 4B).
Figure 5.
PARP-1, calpains, and Bax are activated in the absence of H2AX. (A) Poly(ADP-ribose) (PAR) immunoblotting detection in total extracts from WT and H2AX−/− MEFs untreated or treated with MNNG at different times. The membrane was stained with naphtol blue (NB) to assess protein loading. (B) WT and H2AX−/− MEFs were untreated (control) or treated with MNNG (15 min), immunostained for PAR detection, and visualized by fluorescent microscopy. Representative micrographs of each cell type are shown. Data are means±s.d. (n=5). Bar: 10 μm. (C) Fluorescent assessment of calpain activity measured in WT and H2AX−/− MEFs untreated (control) or treated with MNNG (1 h). Representative micrographs of each treatment are shown. This experiment was repeated four times, yielding similar results. Bar: 10 μm. (D) WT and H2AX−/− MEFs were treated with MNNG and Bax activation was measured by flow cytometry. Representative cytofluorometric plots of untreated (control) and MNNG-treated (9 h) cells are shown. Data are the means±s.d. (n=6). A full-colour version of this figure is available at The EMBO Journal Online.
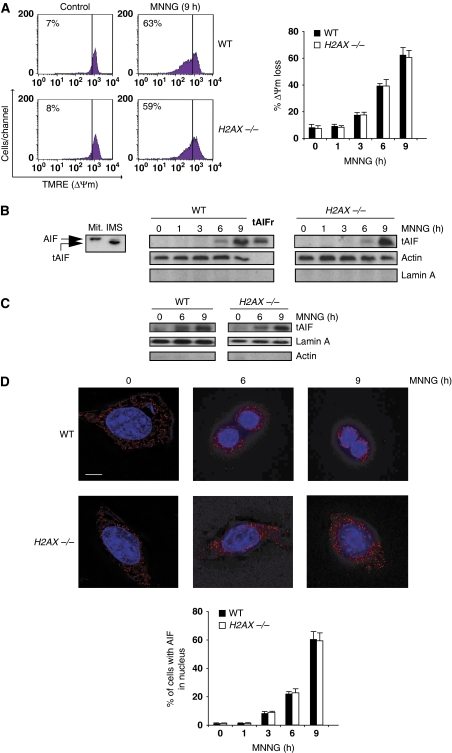
The activation of PARP-1, calpains, and Bax measured in WT and H2AX−/− MEFs would be expected to provoke AIF processing, mitochondrial damage, and tAIF translocation from the mitochondria to the nucleus. This is indeed the case. MNNG-treated WT and H2AX−/− MEFs underwent mitochondrial transmembrane potential (ΔΨm) dissipation (Figure 6A). This alteration is associated with the mitochondrial release of tAIF and its translocation to the cytosol and nucleus (Figure 6B and C). As supported by confocal analysis (Figure 6D), in untreated cells AIF was excluded from the nucleus and showed the typical mitochondrial pattern. However, after MNNG treatment, a portion of AIF was found within the nucleus. Overall, these results show that H2AX−/− MNNG-treated MEFs activate PARP-1, calpains, Bax, and AIF in exactly the same way as WT cells.
Figure 6.
MNNG-induced ΔΨm loss and tAIF relocalization from the mitochondria to the cytosol and nucleus in H2AX−/− MEFs. (A) After the indicated time post-MNNG treatment, WT and H2AX−/− MEFs were labelled with TMRE, and the frequency of cells with ΔΨm loss was recorded and illustrated as a plot. Data presented in the bar chart are from the means of five independent experiments±s.d. In cytometry panels, percentages refer to cells with ΔΨm loss. (B) Mitochondrial AIF (Mit.; 62 kDa) is cleaved into a lower molecular weight tAIF (57 kDa) in the mitochondrial intermembrane space (IMS) upon atractyloside treatment (Yuste et al, 2005) (left panel). Cytosolic fractions recovered from WT and H2AX−/− MEFs after MNNG treatment at different times were blotted for tAIF detection. Compared to the mitochondria isolated from WT MEFs and tAIF recombinant protein (tAIFr), MNNG treatment induces both AIF cleavage into tAIF and time-dependent tAIF release to cytosol. Lamin A and actin were used to control fractionation quality and protein loading. (C) tAIF was also detected in nuclear extracts obtained from WT and H2AX−/− MEFs treated as in (B). Actin and lamin A were used to control fractionation quality and protein loading. (D) WT and H2AX−/− MEFs were untreated or treated with MNNG (6 and 9 h), immunostained for AIF detection (red), and examined by confocal microscopy. Hoechst 33342 (blue) was used to visualize nuclei. Representative microphotographs are shown. Bar: 10 μm. Cells presenting AIF in the nucleus are quantified and plotted as a percentage of total cells. This experiment was repeated four times, yielding similar results.
These data raised a complementary question: how did MNNG-treated H2AX−/− MEFs survive with activated PARP-1, calpains, Bax, and AIF, consumed NAD+ and ATP pools, and dissipated ΔΨm? To answer this question, we performed a long MNNG treatment in H2AX−/− MEFs. As depicted in Supplementary Figure 5A, in H2AX−/− MEFs we only detected a significant positive PS exposure/loss of viability labelling 24 h post-MNNG incubation. Note that WT MEFs reached a similar per cent of PCD 9 h after MNNG treatment (Figure 4B). Interestingly, MNNG-treated H2AX−/− cells never presented the hallmarks characterizing AIF-mediated chromatinolysis: TUNEL positivity or peripheral chromatin condensation. Indeed, after 24 h of MNNG treatment, H2AX−/− MEFs exploded, as in uncontrolled or secondary necrosis (Supplementary Figure 5B and C). These results strongly suggest a key role for the above-described AIF/H2AX link in the management of the MNNG necrotic programme. Indeed, without this link, it seems that the cell is unable to entirely accomplish a ‘PCD' event.
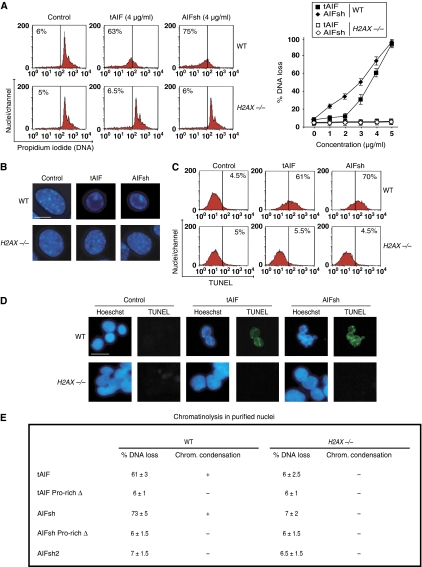
Cooperation between AIF and H2AX is required to induce chromatinolysis
After MNNG treatment, H2AX knockout cells did not present the hallmarks characteristic of AIF-mediated chromatinolysis, even if the active form of AIF was in the nucleus. Thus, it seems that AIF/H2AX interaction is required to provoke chromatinolysis and PCD. To verify this possibility, we used mouse tAIF and AIFsh recombinant proteins (the two natural AIF pro-apoptotic forms) in a cell-free in vitro system in which they were added to highly purified WT and H2AX−/− activated nuclei. After 90 min, nuclear chromatinolysis was quantified by (i) cytofluorometry, to assess nuclei presenting DNA breaks; (ii) Hoechst immunofluorescence, to analyse nuclear morphology; (iii) cytofluorometry and immunofluorescence, to visualize TUNEL detectable DNA breaks. Using these methodologies, we observed that recombinants tAIF and AIFsh induced DNA loss (Figure 7A), peripheral chromatin condensation (Figure 7B), and TUNEL positivity (Figure 7C and D) in WT nuclei. These nuclear modifications were not evident after addition of tAIF or AIFsh to nuclei purified from H2AX−/− MEFs (Figure 7A–D). The presence of H2AX in the nucleus is thus essential to AIF-mediated chromatin condensation and DNA degradation. Besides, we found that AIFsh2 (a natural AIF isoform that lacks the C-terminal AIF domain) (Delettre et al, 2006b) did not induce chromatinolysis in WT nuclei (Figure 7E), confirming the pro-apoptotic functional hierarchy of the C-terminal domain of AIF. This result prompted us to further investigate the relevance of the AIF/H2AX link in AIF-mediated chromatinolysis. As indicated in Figure 2, the region of AIF interacting with H2AX is the C-terminal PBD motif. Thus, to verify the significance of the AIF/H2AX link in AIF-mediated chromatin condensation and DNA degradation, we deleted this PBD in tAIF and AIFsh, and added these mutated proteins to WT purified nuclei. As indicated in Figure 7E, tAIF Pro-rich Δ and AIFsh Pro-rich Δ lost their chromatinolytic properties. Therefore, in the absence of the C-terminal PBD (the AIF/H2AX interacting region), the pro-apoptotic AIF forms are inefficient in triggering chromatinolysis. Overall, these data show that the presence of H2AX in the nucleus is critical for AIF-mediated chromatin condensation and TUNEL positivity. The interaction between the C-terminal PBD of AIF and H2AX regulates this chromatinolytic action.
Figure 7.
Cooperative effect of AIF and H2AX on chromatinolysis. (A) Purified WT and H2AX−/− MEFs nuclei were incubated in the absence (control) or presence of mouse recombinant proteins tAIF and AIFsh and stained with PI. The percentage of DNA loss was measured by flow cytometry and illustrated as a plot. Data are shown as mean values±s.d. (n=4). In cytometry panels, numbers indicate the % of hypoploid nuclei. (B) WT and H2AX−/− MEFs nuclei were treated with tAIF and AIFsh (4 μg/ml) and stained with Hoescht 33342 to visualize chromatin condensation by fluorescent microscopy. Representative micrographs of each treatment are shown. This experiment was done three times, yielding similar results. (C) Nuclei were treated with tAIF and AIFsh as in (B), stained for the detection of 3′-OH DNA breaks, and analysed by flow cytometry. Representative cytofluorometric plots are shown. (D) In a similar experiment to (C), nuclei were stained with Hoechst 33342, and blue and green (TUNEL positive) fluorescence was visualized. Representative microphotographs from untreated (control), tAIF, or AIFsh-treated nuclei are shown. Bar: 20 μm. (E) Table compiling the results obtained in WT and H2AX−/− MEFs nuclei incubated in the presence of tAIF, tAIF Pro-rich Δ, AIFsh, AIFsh Pro-rich Δ, and AIFsh2 (4 μg/ml). Data are represented as % of DNA loss measured as in (A) (X±s.d.; n=5), +: induction of chromatin condensation, or −: absence of chromatin condensation. A full-colour version of this figure is available at The EMBO Journal Online.
AIF/H2AX association promotes DNA degradation through CypA
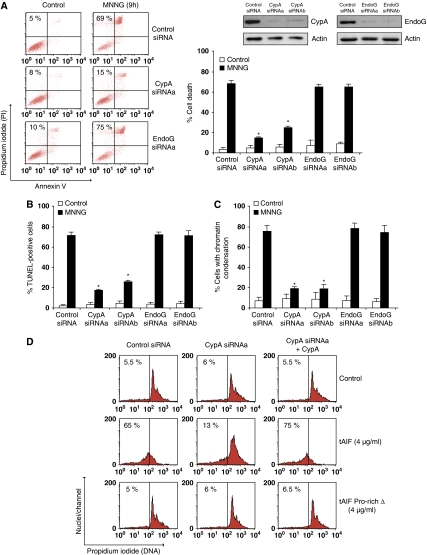
The interaction between AIF and H2AX is critical for the chromatinolytic action that controls caspase-independent programmed necrosis. However, AIF and H2AX have no DNAase activity. Therefore, how does the association between AIF and H2AX promote DNA degradation? In apoptotic caspase-dependent PCD, H2AX improves the DNA-degrading ability of the CAD endonuclease (Lu et al, 2006). Hence, it seems plausible to consider that, in caspase-independent PCD, the AIF/H2AX link could improve DNA accessibility to CypA or EndoG, two endonucleases that have been previously related to the AIF apoptogenic action (Wang et al, 2002; Cande et al, 2004; Wissing et al, 2004; Zhu et al, 2007). To verify this possibility, we first tested whether these endonucleases are implicated in programmed necrosis. As depicted in Figure 8A, silencing of CypA abrogated MNNG toxicity. Moreover, CypA downregulation significantly eliminated the hallmarks of AIF-mediated chromatinolysis (Figure 8B and C). No effect was detected in similar experiments performed with two different siRNAs designed against EndoG. This indicates that CypA, but not EndoG, participates in MNNG-mediated PCD.
Figure 8.
The AIF/H2AX link promotes DNA degradation by activating CypA. (A) WT MEFs were transfected with a scramble siRNA (control siRNA) or two different siRNAs against mouse CypA (CypAsiRNAa and CypAsiRNAb) or mouse EndoG (EndoGsiRNAa and EndoGsiRNAb). Total cell lysates from siRNA cells were prepared and the expression levels of CypA or EndoG were assessed by immunoblotting. Actin was used as a loading control; 48 h after the indicated transfection, cells were untreated (control) or treated with MNNG (9 h), labelled with AnnexinV-FITC and PI, and analysed by flow cytometry. Representative cytofluorometric plots are shown. Percentages refer to double-positive staining. Kinetic analysis of PS exposure and cell viability loss induced by MNNG. After the indicated times, cells were stained as above and the frequency of double-positive labelling was recorded and expressed as a percentage. Data are the means of eight independent experiments±s.d. (B) MEFs were untreated or treated with MNNG as in (A), stained for the detection of 3′-OH DNA breaks, and analysed by flow cytometry. Data are the means of six independent experiments±s.d. (C) MEFs were untreated or treated with MNNG (9 h) and stained with Hoechst 33342 to visualize nuclei. The number of cells presenting chromatin condensation were quantified and plotted as a percentage of total cells. Data are the means±s.d. (n=6). The asterisks in (A–C) indicate a significant effect (P<0.01). (D) Nuclei purified from the MEFs described in (A) were incubated in the absence (control) or presence of mouse recombinant proteins tAIF or tAIF Pro-rich Δ and stained with PI. The percentage of DNA loss was measured by flow cytometry, numbers indicate the % of hypoploid nuclei. In selected experiments, nuclei purified from WT CypAsiRNAa MEFs were reconstituted with recombinant CypA (10 nM). A full-colour version of this figure is available at The EMBO Journal Online.
CypA partially locates in the nucleus, has a latent nuclease activity, and generates the DNA loss characterizing the AIF nuclear action (e.g. TUNEL positivity) (Montague et al, 1997). To investigate whether the association between AIF and H2AX induces DNA degradation by promoting the activation of the latent nuclear CypA, we used a cell-free system assay in which tAIF, AIFsh, tAIF Pro-rich Δ, and AIFsh Pro-rich Δ recombinant proteins were added to nuclei purified from WT MEFs transfected with a control siRNA or an siRNA against CypA. Using this assay, we observed that recombinants tAIF and AIFsh induced DNA loss in WT control siRNA nuclei (Figure 8D; Supplementary Figure 6). These nuclear modifications were absent in nuclei purified from WT siRNA CypA. As in CypA downregulated cells, these results prove that the presence of CypA in the nucleus is crucial to AIF-mediated chromatinolysis. In this way, reconstitution of WT siRNA CypA nuclei with recombinant CypA resensitized the tAIF/AIFsh apoptogenic action (Figure 8D; Supplementary Figure 6). Note that CypA alone (without tAIF or AIFsh) does not sensitize WT siRNA CypA nuclei. Importantly, tAIF Pro-rich Δ and AIFsh Pro-rich Δ, the two AIF forms deleted in the AIF/H2AX interacting region are inefficient in triggering chromatinolysis. And that is the case even in WT siRNA CypA nuclei reconstituted with recombinant CypA (Figure 8D; Supplementary Figure 6). Thus, the AIF/H2AX link promotes the CypA chromatinolytic action that is associated to AIF and caspase-independent programmed necrosis. As a control experiment, we corroborated that the addition of the active recombinant proteins tAIF and CypA to nuclei purified from H2AX−/− MEFs did not provoke DNA degradation (Supplementary Figure 6). Together with the results included in Figure 7, these data indicate that the synchronized presence of AIF, H2AX, and CypA in the nucleus is required to provoke DNA degradation.
Discussion
Despite the substantial amount of work performed on AIF, 10 years after its discovery little is known about the means used by this protein to promote chromatinolysis and caspase-independent PCD. In this study, we provide a key insight into the mechanisms of AIF's pro-apoptotic action. We show that alkylating DNA damage-mediated PCD requires the nuclear interaction between AIF and H2AX. This association is critical in the generation of a DNA-degrading complex that also implicates CypA. Together with our previous results (Moubarak et al, 2007), our new data establish a novel sequence of molecular events for caspase-independent programmed necrosis: MNNG induces in turn the activation of PARP-1, calpain, Bax, and AIF. Upon transfer from the mitochondria to the nucleus in its pro-apoptotic form, AIF interacts with H2AX to promote chromatinolysis in cooperation with CypA.
Given its similar features (e.g. γH2AX, PARP-1, or Bax activation), our new results indicate that the apoptotic and the programmed necrotic pathways could represent alternate outcomes of a similar PCD programme (see also Moubarak et al, 2007). Indeed, the main differences between these two PCD modalities seem to be the different mode of activation of PARP-1 and the implication of either caspases or AIF. Interestingly, the discovery of the AIF/H2AX link reveals that DNA chromatinolysis is a major step in programmed necrosis. Without the DNA degradation promoted by this link, the MNNG necrotic program remains uncomplete and the cells burst.
One of the first responses to the production of DSB in DNA is the rapid generation of phosphorylated H2AX near the DNA break point. This feature distinguishes H2AX from other members of the H2A family (Redon et al, 2002; Thiriet and Hayes, 2005). Traditionally, phosphorylated H2AX (γH2AX) has been associated to chromatin restructuration and recruitment of DNA repair factors (Celeste et al, 2003; Fernandez-Capetillo et al, 2004). However, the introduction of DSB into DNA triggers a large panel of cellular responses, which include DNA repair, but also PCD (Bonner et al, 2008). Thus, theoretically, γH2AX might serve as a signalling platform to control the nuclear PCD features. This has been demonstrated for caspase-dependent PCD (Rogakou et al, 2000; Lu et al, 2006). γH2AX is here critical for DNA fragmentation triggered by the caspase-3 downstream target CAD (Lu et al, 2006), by altering the chromatin conformation to improve DNA accessibility to the CAD endonuclease (Sluss and Davis, 2006). Our data indicate that H2AX is also phosphorylated at Ser139 after MNNG treatment and that this phosphorylation is crucial for caspase-independent programmed necrosis. Indeed, the particular redistribution of γH2AX within the nuclear volume, which helps in organizing specialized protein complexes (Bewersdorf et al, 2006), and the DNA restructuration induced by γH2AX (Fernandez-Capetillo et al, 2004; Sluss and Davis, 2006) appear essential to the AIF/CypA action that promotes caspase-independent DNA degradation. Thus, chromatinolysis mediated by γH2AX is a common feature in caspase-dependent and caspase-independent PCD. The difference in the nature of the DNA degradation representing these two types of death, oligonucleosomal for caspase-dependent PCD and large scale for caspase-independent PCD, seems related to the actors implicated: CAD or AIF/CypA.
Our results strongly favour the hypothesis that AIF/H2AX interaction promotes chromatinolysis and PCD by generating an active DNA-degrading complex involving CypA: (i) a specific deletion in the AIF/H2AX interaction site (the C-terminal PBD of AIF) abolishes both AIF and CypA activity; (ii) H2AX and CypA-deficient cells exhibit a major defect in AIF-mediated DNA fragmentation and programmed necrosis. Indeed, our study on H2AX and CypA-deficient cells shows for the first time that the presence of the apoptogenic form of AIF in the nucleus is required but not sufficient for caspase-independent chromatinolysis. Together with our previous work (Moubarak et al, 2007), we show that AIF, H2AX, and CypA-deficient cells present lower responsiveness to MNNG-mediated programmed necrosis than WT cells. This indicates that H2AX, AIF, and CypA are involved in a single cell death pathway; (iii) our cell-free in vitro system corroborates the cellular data: the individual lack of H2AX, AIF, or CypA abolishes the nuclear DNA-degrading activity characterizing caspase-independent PCD; and (iv) the H2AX interaction site is located in residues 543–559 of AIF, and is different from the AIF/CypA binding domain (amino acids 366–398) (Cande et al, 2004) (Supplementary Figure 7). Thus, theoretically, AIF could simultaneously associate with H2AX and CypA. This is the case in MNNG-treated WT MEFs (Supplementary Figure 8). Remarkably, AIF interacts with H2AX even in the absence of CypA. In contrast, AIF needs H2AX in the nucleus to associate with CypA (Supplementary Figure 8). These results seem to indicate that the nuclear AIF/H2AX association favours the interaction of AIF with CypA. Then, providing a strategic bridge between H2AX and CypA (illustrated in Supplementary Figure 9), AIF generates the lethal DNA-degrading activity characterizing programmed necrosis. In this sense, an AIF-derived peptide containing both the H2AX and CypA binding residues provokes DNA degradation. In contrast, AIF-derived peptides containing either the PBD or the CypA binding sites are unable to induce chromatinolysis (Supplementary Figure 10). AIF needs therefore its H2AX and CypA binding sites to promote chromatinolysis. This explains the absence of chromatinolytic activity in some of the previously described AIF forms: AIFsh2 (Delettre et al, 2006b), which lacks the H2AX and CypA binding domains; AIF Δ 263–399 (Cande et al, 2004), which lacks the CypA binding domain; or tAIF Pro-rich Δ and AIFsh Pro-rich Δ (this study), which lack the H2AX binding domain. On the contrary, tAIF and AIFsh, which possess both the H2AX and CypA binding sites, are active apoptogenic proteins. Overall, our data led us to conclude that the specific characteristics of H2AX (phosphorylation, nuclear redistribution, and generation of DNA accessibility), CypA (DNase), and AIF (specialized binding domains) generate a nuclear chromatinolytic complex that controls programmed necrosis.
Two signal transducers of DNA damage regulate programmed necrosis: PARP-1 and H2AX. Indeed, PARP-1 and H2AX share common physiological features. They are activated in response to DNA damage generated by exogenous treatments, participate in DNA repair, and are implicated in PCD (Haince et al, 2005; Bonner et al, 2008). However, in apoptotic and necrotic PCD, these two proteins could share different properties. In caspase-dependent apoptosis, PARP-1 and H2AX seem to regulate each other. Moreover, H2AX-deficient cells are more sensitive to apoptotic PCD than WT cells (Meador et al, 2008). In caspase-independent PCD, both PARP-1 and H2AX-deficient MEFs are less sensitive than WT cells (Moubarak et al, 2007). Additionally, in contrast to apoptotic PCD, PARP-1 activity is independent from H2AX (Figure 5A and B; Supplementary Figure 3).
We show here that nitrosoureas engage a highly regulated necrotic pathway that can be modulated independently of classical apoptotic pathways. This offers new possibilities for the activation of PCD in tumour resistant cells (e.g. targeting caspase-independent effectors). AIF is therefore a promising drug target. First, AIF is implicated in most of the caspase-independent PCD paradigms tested in tumour cells (Galluzzi et al, 2006; Lorenzo and Susin, 2007; Constantinou et al, 2009). Second, based on our current results, it seems conceivable to activate the caspase-independent PCD pathway by using AIF-derived peptides, such as the PBD/CypA peptide described here. In this sense, our new results might pave the way for the development of novel pharmacological tools in cancer research.
Materials and methods
Cell culture and cell death induction
MEFs were cultured in DMEM supplemented with 10% FCS, 2 mM L-glutamine, and 100 U/ml penicillin/streptomycin (Invitrogen), and maintained at 37°C in a 5% CO2 atmosphere. To induce death, cells were treated either with MNNG (500 μM) or with STS (1 μM; 6 h). In MNNG experiments, the treating medium was replaced after 20 min of incubation by fresh medium devoid of MNNG, and cells (80% confluence) were cultured for the indicated times.
Immunoprecipitation and cell transfection
Cells were transfected in a Nucleofector® system (Amaxa) with p3xFlag-AIF or p3xFlag empty vectors; 24 h after transfection, cells were untreated or treated with MNNG (9 h). Cells were then collected in IP buffer (20 mM Tris/HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 10% Glycerol, 1% Triton X-100) supplemented with protease inhibitors (Roche). One milligram of protein was taken and adjusted with the IP buffer to achieve a final concentration of 1 μg/μl. In all, 40 μl of anti-Flag M2-agarose-coupled antibody were added and incubated overnight at 4°C. Beads were washed with IP buffer and eluted for 30 min at 4°C with 50 μl of TBS containing 150 ng/ml of 3xFlag competitor peptide. Alternatively, 1 mg of supernatant from control or MNNG-treated MEFs was subjected to immunoprecipitation with 2 μg of anti-H2AX antibody (Novus, NB100-383). H2AX−/− MEFs were transfected as above with pcDNA3.1-H2AX, pcDNA3.1-H2AXS139A, or pcDNA3.1 empty vector. Then, they were treated with Zeocin and cultured for 4 weeks before analysis of H2AX expression by western blot and limited dilution selection of individual clones. Clones with similar H2AX levels to H2AX-wt were expanded and analysed.
SPR measurements
A BIAcore 2000 SPR biosensor was used to assay the interaction of soluble H2AX (Abnova) with His-tagged recombinant tAIF. 400–500 RU (1 RU=1 pg/mm2) of tAIF were immobilized on an NTA sensor chip as described (Jomain et al, 2007). The interaction between H2AX (39 nM–1.25 μM) and immobilized tAIF was assayed at 25°C at 100 μl/min using the following buffer: 50 mM Tris pH 7.4, 1 mM EDTA, 450 mM NaCl, 0.01% Tween 20, and 0.1 mg/ml of BSA. For competition experiments, H2AX (1.25 μM) was mixed with tAIF Pyr-Redox, AIFsh, AIF C-term, AIF 543–559, and AIF 543–559 mut (1.25 μM) and equilibrated for 30 min at RT before injecting on the tAIF surface.
Protein modelling
A model of the AIF/H2AX complex was obtained with GRAMM-X (Tovchigrechko and Vakser, 2005) using pdb 1GV4 and the histone H2AX from nucleosome core particles (NCP, pdb 1P3A chain C) as templates for AIF and H2AX, respectively. H2AX structure was modelled from NCP with DALI (Holm and Sander, 1993). Refinement was performed in SwissPDBViewer and the resulting model was edited with Pymol (Guex et al, 1999).
Flow cytometry
We used Annexin V (0.1 μg/ml) for the assessment of PS exposure, propidium iodide (PI, 0.5 μg/ml) for cell viability, and TMRE (20 nM) for ΔΨm quantification. Cell death was recorded in an FACSCanto II (BD Biosciences) in total population (10 000 cells/nuclei).
Immunofluorescence
For AIF redistribution and AIF/H2AX colocalization analysis, MEFs seeded on coverslips were washed in PBS and fixed in PFA (4%). Cells were quenched 10 min in 50 mM glycin, saturated in 1 mg/ml BSA, and permeabilized in 0.05% saponin, 1 mg/ml BSA (incubation buffer, IB). Cells were incubated with anti-AIF and anti-H2AX (M-20; Sta Cruz) diluted in IB, washed three times, and incubated with anti-rabbit and anti-goat IgG conjugated with Alexa® Fluor 594 and 488 (Invitrogen). Cells were mounted in DABCO medium, and fluorescence observed in a Leica DM-RXA23D microscope, equipped with a piezo z-drive (Physik Instrument). We used a × 100 1.4NA-PL-APO objective. Multicolour image stacks (0.2 μm) were acquired using Metamorph software (MDS) through a cooled CDD-camera (Photometrics Coolsnap HQ). Alternatively, cells were stained with Hoechst 33342 before assessment of AIF localization. The quantification of AIF nuclear redistribution by fluorescence microscopy was performed on 100 cells for each data point. For PAR immunofluorescence, cells were fixed in methanol, incubated with an anti-PAR antibody (clone 10H, Alexis), and detected by anti-rabbit IgG conjugated with Alexa® Fluor 488. Fluorescence was observed in a Nikon Eclipse TE2000-U microscope, and analysed using Nikon ACT-1 software. To visualize chromatin condensation, cells were stained with Hoechst 33342. Quantification in Figures 3D, 5B, 6D, 7E, and 8C was performed on 150 cells/nuclei for each data point. For γH2AX and Bax detection, MEFs were fixed in 1% PFA, permeabilized with 0.1% saponin, incubated with an anti-γH2AX (Cell Signaling) or an anti-Bax (clone 6A7, BD Biosciences), and detected by an anti-mouse IgG conjugated with Alexa® Fluor 488. Cells were analysed by immunofluorescence or cytofluorometry as indicated above. In selected assays, cells were co-stained with Hoechst 33342.
Protein extraction, cell fractionation, and western blotting
For whole protein extracts, 1 × 106 cells were lysed in a buffer containing 20 mM Tris–HCl, pH 7.6, 150 mM NaCl, and 1% Triton X-100. For cytosolic purification, cells were resuspended in 220 mM mannitol, 70 mM sucrose, 50 mM Hepes-KOH (pH 7.2), 10 mM KCl, 5 mM EGTA, 2 mM MgCl2, and 0.025% digitonine, and kept on ice for 5 min. Lysed cells were centrifuged and the supernatant was retained as a cytosolic fraction. Nuclear fractions were obtained as reported (Artus et al, 2006). Mitochondrial fractions were obtained using a kit from Pierce. In vitro assays of AIF mitochondrial release on atractyloside treatment were performed as described (Yuste et al, 2005). Histones were extracted with 0.1 M HCl (Lu et al, 2006). For poly(ADP ribose) detection, 1 × 106 cells were lysed as described (Xu et al, 2006); 20 μg of protein was loaded on a SDS–PAGE gel and transferred into a nitrocellulose membrane (Millipore). Membranes were probed with antibodies against PAR (clone LP98-10, Alexis), Lamin A (Cell Signaling), H2AX (Novus), H2A, H2AZ, γH2AX, CypA (Upstate, Millipore), AIF, EndoG, FlagM2, actin.
siRNA
MEFs were transfected as above with siRNA double-stranded oligonucleotides designed against mouse CypA (CypA siRNAa, 5′-CUGAAUGGCUGGAUGGCAA-3′ and CypA siRNAb, 5′-UCUGAGCACUGGAGAGAAA-3′) or mouse EndoG (EndoG siRNAa, 5′-GGACCGAGGCUGAUGGGAA-3′ and EndoG siRNAb, 5′-CUGGAAACCUCAAGGCUAU-3′). As a control, we used an irrelevant siRNA (Control siRNA, 5′-GCGAUAAGUCGUGUCUUAC-3′).
COMET assay
Neutral comet assay was performed as described (Olive and Banath, 2006). Single cell images were captured and analysed using a Nikon Eclipse TE2000-U microscope as indicated above.
Field inversed gel electrophoresis
DNA was prepared from agarose plugs (2 × 106 cells), followed by electrophoresis in a Bio-Rad Laboratories Fige Mapper cell. Running conditions were 180 V (forward pulse), 120 V (reverse pulse), and 1.5 and 3.5 s for the initial and final switch times (forward and reverse pulses, linear ramp) for 20 h.
Recombinant proteins and peptides
Mouse tAIF (AIFΔ1–102), AIFsh (AIFΔ1–351), tAIF Pyr-Redox (AIFΔ1–102; Δ480–612), AIF C-term (AIFΔ1–479), AIFsh2 (AIFΔ352–612), tAIF Pro-rich Δ (AIFΔ1–102; Δ543–559), and AIFsh Pro-rich Δ (AIFΔ1–351; Δ543–559) recombinant proteins were produced as described (Yuste et al, 2005). CypA was from Abnova. NeoMPS synthesized AIF PEST (AIF529–539), AIF PBD (AIF543–559), AIF PBD mut (AASAAAVAQVAVEGEDY), AIF CypA (AIF366–398), and AIF PBD/CypA (AIF366–559).
TUNEL assay and cell-free system
Detection of 3′-OH DNA breaks was performed as described earlier (Moubarak et al, 2007). Alternatively, cells were examined in a Nikon Eclipse TE2000-U microscope.
Cell-free system was performed as reported (Susin et al, 1999). Briefly, nuclei purified from different types of MEFs treated 20 min with MNNG were incubated in the presence of the indicated recombinant protein or peptide for 90 min at 37°C. Nuclei were used for TUNEL and Hoechst 33342 labelling, and examined by fluorescence microscopy and flow cytometry as above. Alternatively, nuclei were stained with PI (0.5 μg/ml) and analysed by flow cytometry.
Calpain activity
Calpain activity in live cells was detected using the cell-permeable calpain substrate Boc-Leu-Met-CMAC (Invitrogen). Cells spread on coverslides were incubated for 30 min with 50 μM of calpain substrate in DMEM culture medium. Cells were then treated or not with MNNG and observed in a Nikon Eclipse TE2000-U microscope.
Statistical analysis
The significance of data included in Figure 8 was determined using the Student's t-test for unpaired observations.
Unless specified, all reagents used were from Sigma-Aldrich.
Supplementary Material
Acknowledgments
We thank A Nussenzweig (NIH, NCI, Bethesda) for immortalized H2AX-wt and H2AX−/− MEFs, NV Tomilin (Institute of Cytology, St Petersburg) for the H2AX and H2AXS139A cDNA, and M Segade and S Krantic for critical comments. SAS is supported by INSERM, UPMC-Paris 6, CNRS, ANR (contract ANR-09-BLAN-0247-01), ARC (contracts 4043, 5104, and 7987), and Fondation de France.
Footnotes
The authors declare that they have no conflict of interest.
References
- Artus C, Maquarre E, Moubarak RS, Delettre C, Jasmin C, Susin SA, Robert-Lezenes J (2006) CD44 ligation induces caspase-independent cell death via a novel calpain/AIF pathway in human erythroleukemia cells. Oncogene 25: 5741–5751 [DOI] [PubMed] [Google Scholar]
- Banath JP, Olive PL (2003) Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks. Cancer Res 63: 4347–4350 [PubMed] [Google Scholar]
- Bewersdorf J, Bennett BT, Knight KL (2006) H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy. Proc Natl Acad Sci USA 103: 18137–18142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y (2008) GammaH2AX and cancer. Nat Rev Cancer 8: 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande C, Vahsen N, Kouranti I, Schmitt E, Daugas E, Spahr C, Luban J, Kroemer RT, Giordanetto F, Garrido C, Penninger JM, Kroemer G (2004) AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene 23: 1514–1521 [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Cheung EC, Joza N, Steenaart NA, McClellan KA, Neuspiel M, McNamara S, Maclaurin JG, Rippstein P, Park DS, Shore GC, McBride HM, Penninger JM, Slack RS (2006) Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J 25: 4061–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung EC, Melanson-Drapeau L, Cregan SP, Vanderluit JL, Ferguson KL, McIntosh WC, Park DS, Bennett SA, Slack RS (2005) Apoptosis-inducing factor is a key factor in neuronal cell death propagated by BAX-dependent and BAX-independent mechanisms. J Neurosci 25: 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK (2009) Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137: 1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou C, Papas KA, Constantinou AI (2009) Caspase-independent pathways of programmed cell death: the unraveling of new targets of cancer therapy? Curr Cancer Drug Targets 9: 717–728 [DOI] [PubMed] [Google Scholar]
- Dawson VL, Dawson TM (2004) Deadly conversations: nuclear-mitochondrial cross-talk. J Bioenerg Biomembr 36: 287–294 [DOI] [PubMed] [Google Scholar]
- Delettre C, Yuste VJ, Moubarak RS, Bras M, Lesbordes-Brion JC, Petres S, Bellalou J, Susin SA (2006a) AIFsh, a novel Apoptosis-inducing Factor (AIF) pro-apoptotic isoform with potential pathological relevance in human cancer. J Biol Chem 281: 6413–6427 [DOI] [PubMed] [Google Scholar]
- Delettre C, Yuste VJ, Moubarak RS, Bras M, Robert N, Susin SA (2006b) Identification and characterization of AIFsh2, a mitochondrial Apoptosis-inducing Factor (AIF) isoform with NADH oxidase activity. J Biol Chem 281: 18507–18518 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of the genome. DNA Repair (Amst) 3: 959–967 [DOI] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Vandenabeele P (2006) Necrosis, a well-orchestrated form of cell demise: signalling cascades, important mediators and concomitant immune response. Biochim Biophys Acta 1757: 1371–1387 [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Larochette N, Zamzami N, Kroemer G (2006) Mitochondria as therapeutic targets for cancer chemotherapy. Oncogene 25: 4812–4830 [DOI] [PubMed] [Google Scholar]
- Golstein P, Aubry L, Levraud JP (2003) Cell-death alternative model organisms: why and which? Nat Rev Mol Cell Biol 4: 798–807 [DOI] [PubMed] [Google Scholar]
- Guex N, Diemand A, Peitsch MC (1999) Protein modelling for all. Trends Biochem Sci 24: 364–367 [DOI] [PubMed] [Google Scholar]
- Gurbuxani S, Schmitt E, Cande C, Parcellier A, Hammann A, Daugas E, Kouranti I, Spahr C, Pance A, Kroemer G, Garrido C (2003) Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene 22: 6669–6678 [DOI] [PubMed] [Google Scholar]
- Haince JF, Rouleau M, Hendzel MJ, Masson JY, Poirier GG (2005) Targeting poly(ADP-ribosyl)ation: a promising approach in cancer therapy. Trends Mol Med 11: 456–463 [DOI] [PubMed] [Google Scholar]
- Hangen E, De Zio D, Bordi M, Zhu C, Dessen P, Caffin F, Lachkar S, Perfettini JL, Lazar V, Benard J, Fimia GM, Piacentini M, Harper F, Pierron G, Vicencio JM, Benit P, de Andrade A, Hoglinger G, Culmsee C, Rustin P et al. (2010) A brain-specific isoform of mitochondrial apoptosis-inducing factor: AIF2. Cell Death Differ (advance online publication 29 January 2010; doi:10.1038/cdd.2009.211) [DOI] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X (2009) Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 137: 1100–1111 [DOI] [PubMed] [Google Scholar]
- Hegedus C, Lakatos P, Olah G, Toth BI, Gergely S, Szabo E, Biro T, Szabo C, Virag L (2008) Protein kinase C protects from DNA damage-induced necrotic cell death by inhibiting poly(ADP-ribose) polymerase-1. FEBS Lett 582: 1672–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J (2008) Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Ishihara N, Takagi N, Niimura M, Takagi K, Nakano M, Tanonaka K, Funakoshi H, Matsumoto K, Nakamura T, Takeo S (2005) Inhibition of apoptosis-inducing factor translocation is involved in protective effects of hepatocyte growth factor against excitotoxic cell death in cultured hippocampal neurons. J Neurochem 95: 1277–1286 [DOI] [PubMed] [Google Scholar]
- Jomain JB, Tallet E, Broutin I, Hoos S, van Agthoven J, Ducruix A, Kelly PA, Kragelund BB, England P, Goffin V (2007) Structural and thermodynamic bases for the design of pure prolactin receptor antagonists: X-ray structure of Del1-9-G129R-hPRL. J Biol Chem 282: 33118–33131 [DOI] [PubMed] [Google Scholar]
- Joza N, Galindo K, Pospisilik JA, Benit P, Rangachari M, Kanitz EE, Nakashima Y, Neely GG, Rustin P, Abrams JM, Kroemer G, Penninger JM (2008) The molecular archaeology of a mitochondrial death effector: AIF in Drosophila. Cell Death Differ 15: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joza N, Pospisilik JA, Hangen E, Hanada T, Modjtahedi N, Penninger JM, Kroemer G (2009) AIF: not just an apoptosis-inducing factor. Ann NY Acad Sci 1171: 2–11 [DOI] [PubMed] [Google Scholar]
- Klein JA, Longo-Guess CM, Rossmann MP, Seburn KL, Hurd RE, Frankel WN, Bronson RT, Ackerman SL (2002) The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature 419: 367–374 [DOI] [PubMed] [Google Scholar]
- Loeffler M, Daugas E, Susin SA, Zamzami N, Metivier D, Nieminen AL, Brothers G, Penninger JM, Kroemer G (2001) Dominant cell death induction by extramitochondrially targeted apoptosis-inducing factor. FASEB J 15: 758–767 [DOI] [PubMed] [Google Scholar]
- Lorenzo HK, Susin SA (2007) Therapeutic potential of AIF-mediated caspase-independent programmed cell death. Drug Resist Updat 10: 235–255 [DOI] [PubMed] [Google Scholar]
- Lu C, Zhu F, Cho YY, Tang F, Zykova T, Ma WY, Bode AM, Dong Z (2006) Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell 23: 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mate MJ, Ortiz-Lombardia M, Boitel B, Haouz A, Tello D, Susin SA, Penninger J, Kroemer G, Alzari PM (2002) The crystal structure of the mouse apoptosis-inducing factor AIF. Nat Struct Biol 9: 442–446 [DOI] [PubMed] [Google Scholar]
- Meador JA, Zhao M, Su Y, Narayan G, Geard CR, Balajee AS (2008) Histone H2AX is a critical factor for cellular protection against DNA alkylating agents. Oncogene 27: 5662–5671 [DOI] [PubMed] [Google Scholar]
- Montague JW, Hughes FM Jr, Cidlowski JA (1997) Native recombinant cyclophilins A, B, and C degrade DNA independently of peptidylprolyl cis-trans-isomerase activity. Potential roles of cyclophilins in apoptosis. J Biol Chem 272: 6677–6684 [DOI] [PubMed] [Google Scholar]
- Moubarak RS, Yuste VJ, Artus C, Bouharrour A, Greer PA, Menissier-de Murcia J, Susin SA (2007) Sequential activation of Poly(ADP-Ribose) polymerase 1, calpains, and Bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Mol Cell Biol 27: 4844–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B, Kessinger C, Kobayashi J, Chen BP, Chen DJ, Chatterjee A, Burma S (2006) DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair (Amst) 5: 575–590 [DOI] [PubMed] [Google Scholar]
- Olive PL, Banath JP (2006) The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 1: 23–29 [DOI] [PubMed] [Google Scholar]
- Otera H, Ohsakaya S, Nagaura Z, Ishihara N, Mihara K (2005) Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J 24: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10: 886–895 [DOI] [PubMed] [Google Scholar]
- Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM (2003) Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol 81: 123–129 [DOI] [PubMed] [Google Scholar]
- Polster BM, Basanez G, Etxebarria A, Hardwick JM, Nicholls DG (2005) Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J Biol Chem 280: 6447–6454 [DOI] [PubMed] [Google Scholar]
- Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W (2002) Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev 12: 162–169 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM (2000) Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem 275: 9390–9395 [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS (2001) Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J 20: 3861–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF (2009) Redox-linked conformational dynamics in apoptosis-inducing factor. J Mol Biol 390: 924–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluss HK, Davis RJ (2006) H2AX is a target of the JNK signaling pathway that is required for apoptotic DNA fragmentation. Mol Cell 23: 152–153 [DOI] [PubMed] [Google Scholar]
- Solier S, Sordet O, Kohn KW, Pommier Y (2009) Death receptor-induced activation of the Chk2- and histone H2AX-associated DNA damage response pathways. Mol Cell Biol 29: 68–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G (1999) Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397: 441–446 [DOI] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ (2005) Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol Cell 18: 617–622 [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA (2005) Development and testing of an automated approach to protein docking. Proteins 60: 296–301 [DOI] [PubMed] [Google Scholar]
- Vahsen N, Cande C, Briere JJ, Benit P, Joza N, Larochette N, Mastroberardino PG, Pequignot MO, Casares N, Lazar V, Feraud O, Debili N, Wissing S, Engelhardt S, Madeo F, Piacentini M, Penninger JM, Schagger H, Rustin P, Kroemer G (2004) AIF deficiency compromises oxidative phosphorylation. EMBO J 23: 4679–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yu SW, Koh DW, Lew J, Coombs C, Bowers W, Federoff HJ, Poirier GG, Dawson TM, Dawson VL (2004) Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci 24: 10963–10973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang J, Gengyo-Ando K, Gu L, Sun CL, Yang C, Shi Y, Kobayashi T, Shi Y, Mitani S, Xie XS, Xue D (2007) C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat Cell Biol 9: 541–549 [DOI] [PubMed] [Google Scholar]
- Wang X, Yang C, Chai J, Shi Y, Xue D (2002) Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science 298: 1587–1592 [DOI] [PubMed] [Google Scholar]
- Wang Y, Dawson VL, Dawson TM (2009a) Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol 218: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kim NS, Li X, Greer PA, Koehler RC, Dawson VL, Dawson TM (2009b) Calpain activation is not required for AIF translocation in PARP-1-dependent cell death (parthanatos). J Neurochem 110: 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Ludovico P, Herker E, Buttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, Corte-Real M, Frohlich KU, Manns J, Cande C, Sigrist SJ, Kroemer G, Madeo F (2004) An AIF orthologue regulates apoptosis in yeast. J Cell Biol 166: 969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Huang S, Liu ZG, Han J (2006) Poly(ADP-ribose) polymerase-1 signaling to mitochondria in necrotic cell death requires RIP1/TRAF2-mediated JNK1 activation. J Biol Chem 281: 8788–8795 [DOI] [PubMed] [Google Scholar]
- Ye H, Cande C, Stephanou NC, Jiang S, Gurbuxani S, Larochette N, Daugas E, Garrido C, Kroemer G, Wu H (2002) DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nat Struct Biol 9: 680–684 [DOI] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL (2002) Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297: 259–263 [DOI] [PubMed] [Google Scholar]
- Yuste VJ, Moubarak RS, Delettre C, Bras M, Sancho P, Robert N, d′Alayer J, Susin SA (2005) Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. Cell Death Differ 12: 1445–1448 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Han T, Zhu Q, Zhang W, Bao W, Fu HJ, Yang J, Huang XJ, Wei JX, Meng YL, Zhao J, Cao YX, Jia LT, Yangi AG (2009) The proapoptotic activity of C-terminal domain of apoptosis-inducing factor (AIF) is separated from its N-terminal. Biol Res 42: 249–260 [PubMed] [Google Scholar]
- Zhu C, Wang X, Deinum J, Huang Z, Gao J, Modjtahedi N, Neagu MR, Nilsson M, Eriksson PS, Hagberg H, Luban J, Kroemer G, Blomgren K (2007) Cyclophilin A participates in the nuclear translocation of apoptosis-inducing factor in neurons after cerebral hypoxia-ischemia. J Exp Med 204: 1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB (2004) Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev 18: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.