Abstract
DNA-dependent protein kinase (DNA-PK) is a double-strand breaks repair complex, the subunits of which (KU and DNA-PKcs) are paradoxically present at mammalian telomeres. Telomere fusion has been reported in cells lacking these proteins, raising two questions: how is DNA–PK prevented from initiating classical ligase IV (LIG4)-dependent non-homologous end-joining (C-NHEJ) at telomeres and how is the backup end-joining (EJ) activity (B-NHEJ) that operates at telomeres under conditions of C-NHEJ deficiency controlled? To address these questions, we have investigated EJ using plasmid substrates bearing double-stranded telomeric tracks and human cell extracts with variable C-NHEJ or B-NHEJ activity. We found that (1) TRF2/RAP1 prevents C-NHEJ-mediated end fusion at the initial DNA–PK end binding and activation step and (2) DNA–PK counteracts a potent LIG4-independent EJ mechanism. Thus, telomeres are protected against EJ by a lock with two bolts. These results account for observations with mammalian models and underline the importance of alternative non-classical EJ pathways for telomere fusions in cells.
Keywords: DNA repair, genome instability, non-homologous end-joining, telomeres
Introduction
Loss of chromosome fragments can lead to apoptosis or initiate carcinogenesis by means of improper expression or silencing of key genes controlling cell proliferation. Thus, signalling and repair of DNA double-strand breaks (DSB) are essential for genome stability (O'Driscoll and Jeggo, 2006). Mammals have evolved two main DSB repair mechanisms, homologous recombination (HR) and non-homologous end-joining (NHEJ) (Pardo et al, 2009). HR uses strand exchange at the break site mainly with the sister chromatid during S and G2 phases of the cell cycle. NHEJ, on the other hand, rejoins the two ends of the break throughout the cell cycle and is the predominant mechanism in G1. The main NHEJ reaction, hereafter named classical NHEJ (C-NHEJ), relies on recognition, protection and bridging of the DNA ends by the DNA-dependent protein kinase complex (DNA–PK). This complex is composed of the DNA binding KU70/KU80 heterodimer, which recruits the serine-threonine kinase catalytic subunit (DNA–PKcs) (for a review, see Mahaney et al, 2009). DNA–PK also activates end-processing enzymes such as the Artemis nuclease (Goodarzi et al, 2006) and is required for the stable recruitment of the XRCC4/DNA ligase IV (LIG4)/Cernunnos–XLF complex that catalyses the final ligation step (Drouet et al, 2005; Wu et al, 2007). Recently, evidence has accumulated for an alternative or backup NHEJ pathway (hereafter named B-NHEJ), which accounts for residual end-joining (EJ) of DSB in cells that are deficient in components of C-NHEJ (for reviews, see Nussenzweig and Nussenzweig, 2007; Haber, 2008; McVey and Lee, 2008). B-NHEJ is repressed by C-NHEJ and preferentially uses DNA microhomology for EJ (Guirouilh-Barbat et al, 2007; Schulte-Uentrop et al, 2008). Data from our and other laboratories have implicated XRCC1/DNA ligase III and PARP-1 in B-NHEJ (Audebert et al, 2004; Wang et al, 2005; Robert et al, 2009); in addition these proteins are involved in the repair of base damage and single-strand breaks.
Efficient cellular mechanisms for coping with genomic DSB present a challenge to linear chromosomes that must prevent unwanted signalling, joining or recombination at their ends. In most eukaryotes, chomosome ends are composed of special nucleoprotein structures called telomeres. In humans, the telomeric DNA comprises typically 10–15 kb of T2AG3 duplex repeats oriented 5′ to 3′ towards the chromosome end, followed by a 3′ single-stranded extension composed of these same repeats over 50–500 nucleotides. The T2AG3 repeats associate with shelterin, a complex of proteins: Tin2 bridges the TRF1 and TRF2/RAP1 complex that is bound to the double-stranded (ds) telomeric repeats whereas the POT1/TPP1 complex is attached to the G-rich tail (reviewed in Palm and de Lange, 2008). The 3′ overhang folds back and invades the duplex telomeric repeat to form the so-called T loop (Griffith et al, 1999) most likely because of the DNA unwinding properties of TRF2 (Amiard et al, 2007). A minimal telomere length is needed, probably required for shelterin assembly. This is maintained either by the telomerase activity or alternative recombination-based mechanisms (reviewed in Verdun and Karlseder, 2007; Cesare and Reddel, 2008).
Some of these proteins participate in DSB signalling avoidance at telomeres; it has been shown that TRF2 is necessary to prevent ATM activation whereas POT1 in association with TPP1 is responsible for ATR inhibition (Denchi and de Lange, 2007). They also have a role in preventing end fusion; TRF2 is essential for inhibition of telomere fusion in cells (van Steensel et al, 1998; Celli and de Lange, 2005; Celli et al, 2006), most likely by anchoring RAP1 on telomeres (Sarthy et al, 2009)). The extent of end fusion on loss of TRF2 is far more pronounced than for POT1 deficiency (Hockemeyer et al, 2006). In addition, the G-rich tail is not required for TRF2/RAP1-mediated inhibition of EJ on telomeric DNA in vitro (Bae and Baumann, 2007). Some human telomeres whose length is incompatible with T-loop formation also appear to escape fusion in cells (Baird et al, 2003; Xu and Blackburn, 2007). These results suggest that duplex telomeric repeats and the associated proteins are a major contribution to EJ avoidance at telomeres.
Surprisingly, both KU and DNA–PKcs components of the C-NHEJ machinery are present on telomeres (Hsu et al, 1999; d'Adda di Fagagna et al, 2001). This observation underlines the paradoxical protection and fusion functions of these proteins at telomeres (reviewed in Fisher and Zakian, 2005; Riha et al, 2006). The near complete absence of chromosome fusions on inhibition or loss of TRF2 in an LIG4-deficient background clearly argues for a predominant role of the C-NHEJ mechanism for telomeric fusions under these shelterin destabilization conditions (Smogorzewska et al, 2002; Celli and de Lange, 2005). Under these conditions, telomeric fusion has been shown to predominate in G1 phase of the cell cycle (Konishi and de Lange, 2008). Similarly, LIG4 is responsible for the fusion of sister telomeres in cells deficient in the telomeric poly(adenosine-diphosphate ribose) polymerase tankyrase-1 (Hsiao and Smith, 2009). However, significant chromosome end fusions occur on TRF2 loss in the absence KU, although 10-fold less frequently than in wild-type cells (Celli et al, 2006). In addition, spontaneous chromosomal end fusions are also promoted by a deficiency in either KU (Bailey et al, 1999; Hsu et al, 1999, 2000; Samper et al, 2000; d'Adda di Fagagna et al, 2001; Espejel et al, 2004; Li et al, 2007) or DNA–PKcs (Gilley et al, 2001; Goytisolo et al, 2001), where, at least in the absence of KU, the core shelterin complex appears broadly normal (Celli et al, 2006). Moreover, both DNA–PKcs and LIG4 have been shown to be dispensable for chromosomal fusions arising in telomerase-deficient mouse cells (Maser et al, 2007), in line with the report of KU- and LIG4-independent telomere fusions on attrition in fission yeast (Wang and Baumann, 2008). However, another group has reported that KU and DNA–PKcs are necessary for fusions in telomerase-deficient mouse cells (Espejel et al, 2002a, 2002b).
These observations raise several questions: (1) which of the end-recognitions or the DNA ligation steps in the C-NHEJ mechanism is blocked at telomeres? (2) Does a DNA–PK-independent and LIG4-mediated or a B-NHEJ alternative EJ mechanism operate at chromosome ends under special conditions? (3) What is the connection between both the shelterin and the NHEJ proteins at telomeres and this DNA–PK-independent EJ mechanism?
To address these questions, we have used an EJ assay with plasmid substrate bearing at one end ds telomeric tracks, defined as the minimal structure protecting from EJ (Bae and Baumann, 2007). The joining reaction was catalysed by human cell extracts under controlled conditions enabling either C-NHEJ or B-NHEJ. We show that TRF2/RAP1 and DNA–PK complexes protect telomeres by two complementary mechanisms. The former prevents C-NHEJ-mediated end fusion at the initial DNA–PK end-binding step whereas the latter counteracts a potent LIG4-independent EJ mechanism that promotes ligation in the absence of DNA–PK. This double-lock protection accounts for observations with mammalian models and underlines the importance of alternative non-classical EJ pathways for telomeres fusion in cells.
Results
Inhibition of C-NHEJ-mediated joining of telomeric ds DNA ends relies on an impaired DNA–PKcs activation
An in vitro assay with cell extracts and DNA substrates was used to mimick the effect of telomeric DNA on EJ mediated by the NHEJ apparatus. As ligation substrate, the pUCtelo2 plasmid containing 648 bp of 5′-T2AG3 repeats was used (Amiard et al, 2007), which closely matches the natural mean telomere length (Figure 1A). Appropriate restrictions of pUCtelo2 produced linear plasmids with a ds telomeric tract plus 1 bp at one end, at the 5′ end (pT5′), at the 3′end (pT3′) or ending with a 31 bp non-telomeric sequence (pT3′H). The EJ reaction obtained with the HeLa extracts was sensitive to the addition of a DNA–PKcs-specific inhibitor or antibodies directed against XRCC4, showing that it was true C-NHEJ (Supplementary Figure S1A, lanes 1 and 3, respectively). When introduced into the EJ assay, 30–31% of a control pBS substrate digested with the same restriction enzyme as pUCtelo2 was converted into dimers and multimers, whatever the DNA end (Figure 1B, lanes 1 and 5; Supplementary Figure S1B, lanes 1–3). In contrast, only 15–18% of the pT3′ was rejoined and only dimers were observed (Figure 1B, lanes 2 and 6; Supplementary Figure S1B, lane 5). However, the pT5′ and the pT3′H substrates were as efficiently rejoined as the control pBS (Supplementary Figure S1B, lanes 4 and 6, respectively). All the ligation events were sensitive to NU7026 (Figure 1B, lane 9; Supplementary Figure S1B, lanes 7–9). These results show that the C-NHEJ inhibition mediated by the telomeric tract was restricted to the T2AG3-3′ orientation (pT3′) and was released by the addition of non-telomeric 31 bp at this end.
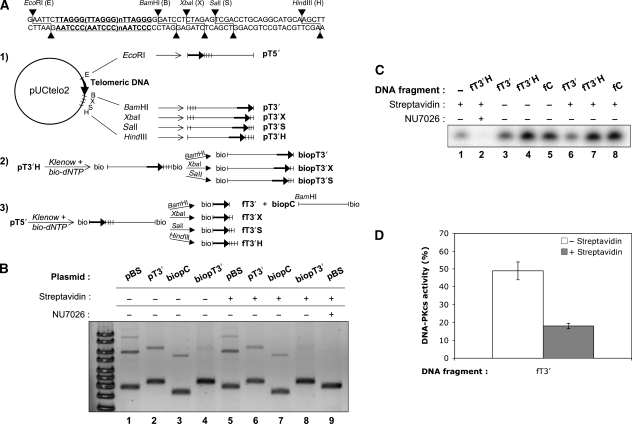
Figure 1.
DNA substrates construction and EJ and kinase assays with telomeric DNA. (A) Construction scheme of the plasmids and DNA fragments used. pUCtelo2 plasmid contains a 648 bp telomeric sequence inserted between EcoRI and BamHI sites as detailed in the upper part of the figure. Track 1: non-biotinylated plasmids. pUtelo2 was digested with the indicated enzymes to produce linearized plasmid bearing a telomeric sequence at the 5′ end (pT5′), the 3′ end (pT3′) or moved at various distance inward from the 3′ end (pT3′X, pT3′S, pT3′H). Track 2: biotinylated plasmids. Biotinylation followed by appropriate restriction produced the same 3′ended-telomeric plasmids but containing biotin at the opposite end (biopT3′, biopT3′X, biopT3′S). Track 3: biotinylated fragments. Biotinylation followed by appropriate restriction produced fragments with the telomeric sequence at various distance inward from the 3′ end (fT3′, fT3′X, fT3′S, fT3′H) and containing biotin at the opposite end. The control plasmid (biopC) corresponds to the pUCtelo2 plasmid without telomeric sequence and biotinylated at the 5′ end. A control non-telomeric 502 bp fragment biotinylated or not at the 5′-end was amplified by PCR from pBluescript-KS-II(−) (see Materials and methods). (B) EJ assay catalysed under standard reaction conditions with the indicated plasmids and HeLa extracts, in the presence or not of streptavidin or DNA–PK-specific inhibitor NU7026. DNA ligation products were separated by agarose gel electrophoresis followed by SYBR-Green staining. pBS stands for pBluescript-KS-II(−). Ligation efficiency (% of multimers versus monomer) was 30.2, 17.9, 13.7, 2.7 for lanes 1–4 (without streptavidin) and 31, 15.2, 12.7, 1.6 for lanes 5–8 (with streptavidin), respectively (C) DNA–PK assay catalysed under standard conditions with the indicated DNA fragments and HeLa extracts, in the presence or not of streptavidin or DNA–PK-specific inhibitor NU7026. DNA–PK peptide substrate was isolated by polyacrylamide denaturing gel electrophoresis followed by auto-radiography of the gel. (D) Quantification of independent experiments as shown in (C) (n=3). Relative DNA–PKcs activity was calcultated as the % of radiolabel incorporation in the peptide substrate obtained with fT3′ fragment as activating DNA compared with the incorporation obtained with fT3′H fragment, after subtraction in each case of the background incorporation obtained without DNA. Error bars correspond to s.e.m.
Having validated our in vitro assay for EJ inhibition by telomeric DNA as reported earlier (Bae and Baumann, 2007), we restricted the accessibility of ligation only to one end by constructing another series of substrates, bearing a biotin residue at the end opposite to the telomeric end (Figure 1A, biopT plasmids). A biotinylated non-telomeric plasmid with a smilar length and free end was used as control (Figure 1A, biopC plasmid, track 3). As shown in Figure 1B, blocking the non-telomeric end by a biotin residue was sufficient to prevent the residual dimer formation on the pT3′ substrate and led to an extensive EJ inhibition, which was very similar with or without streptavidin in the reaction (87 and 80%, respectively, as quantified from Figure 1B). This shows that a long T2AG3-3′ tract mimicking a natural ds chromosome end is refractory to NHEJ-mediated ligation in vitro.
DNA–PKcs activity is necessary for efficient EJ of DSB in vitro and in cells (Meek et al, 2008). Therefore, we checked the effect of telomeric tracts at a DNA end on the activation of DNA–PKcs in vitro. The telomeric fragment was isolated from pUCTelo2 and biotin was incorporated at one end to restrain the possibility of DNA–PKcs activation either at the 3′ telomeric end (fT3′) or the 3′ telomeric end extended with a 31 bp non-telomeric sequence (fT3′H) (Figure 1A). DNA–PKcs activity on a standard peptide substrate was assessed in the NHEJ competent HeLa extracts under the same buffer conditions as the EJ reaction and with the various activating DNA fragments biotinylated at one end, with or without streptavidin (Figure 1C). Peptide phosphorylation was completely abolished with NU7026 showing that this reaction requires DNA–PKcs activity. In the absence of streptavidin, all added fragments promoted an activity above the background obtained with the extracts alone, indicating that DNA–PKcs activation was dependent on the exogenous DNA ends provided in the assay. On streptavidin addition, the DNA–PKcs activity promoted by the fT3′ fragment dropped markedly whereas it remained unchanged with the other DNA fragments. As quantified in Figure 1D, DNA–PKcs activation targeted on the T2AG3-3′ extremity (fT3′+streptavidin) was reduced by more than 80% when compared with the activation obtained with a 31 bp non-telomeric DNA extension added to the telomeric tract (fT3′H). The latter fragment yielded the same DNA–PKcs activation level as a control DNA fragment (Figure 1C), indicating that 31 non-telomeric bps were sufficient to completely overcome the inhibitory effect of telomeric repeats at the 3′ end on DNA–PKcs.
To check more precisely the distance from the end that is required for the inhibitory effect on NHEJ of telomeric repeats at the 3′ end, the 5′ biotinylated substrates biopT3′X and biopT3′S were constructed, bearing 7 or 13 non-telomeric bps added 3′ to the telomeric sequence, respectively (Figure 1A). As shown in Supplementary Figure S2A, NHEJ was repressed on 3′X ends as strongly as on 3′ telomeric plasmids (lanes 1, 2 and 4, 5) but was efficient on 3′S substrates, leading to multimers without biotin (pT3′S plasmid, lane 3) and to dimers with biotin (biopT3′S plasmid, lane 6). Thus, under these conditions, the 3′ telomeric tract did not inhibit NHEJ-mediated EJ at this end beyond a 13 bp distance. We then assessed the effect of the telomeric tract distance from the 3′ end on DNA–PKcs activity. Supplementary Figure S2B shows that the strong kinase inhibition observed at the fT3′ fragment (lane 2) was already absent when the telomeric tract was moved 7 bp inward from the 3′ end (lane 3, fT3′X substrate).
TRF2/RAP1 complex mediates C-NHEJ inhibition at telomeric ends through hindrance at the KU loading and DNA–PK activation steps
Binding of TRF2/RAP1 complex to ds telomeric DNA has a major role in EJ inhibition (Bae and Baumann, 2007). To investigate the molecular basis of NHEJ inhibition at ds telomeric DNA ends under our in vitro conditions, we immuno-depleted HeLa extracts for TRF2 and RAP1 proteins (Figure 2A). After immuno-depletion, RAP1 was undetectable whereas a faint amount of TRF2 was still present and TRF1 concentration remained unchanged. We failed to selectively immuno-deplete TRF1 from the extracts because of a cross-reactivity with TRF2 of immuno-precipitant anti-TRF1 antibodies (Supplementary Figure S3), as reported (Bae and Baumann, 2007). When immuno-depleted extracts were assayed with the telomeric ends and control substrates, EJ was largely re-established at the telomeric DNA end of the biopT3′ plasmid (Figure 2B and C). A decrease in the overall EJ activity on the control substrate was observed, probably because of extract manipulations during immuno-depletion. EJ remained sensitive to NU7026 (Figure 2B, lane 5) and was undetectable in the absence of LIG4 (Supplementary Figure S4B, lane 8), indicating that telomeric ends ligation still relied on C-NHEJ in the absence of TRF2/RAP1 complex.
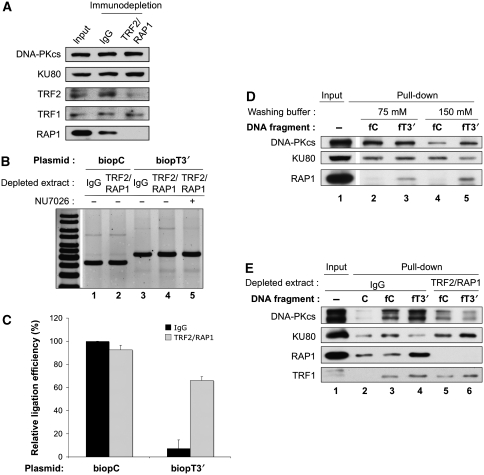
Figure 2.
TRF2/RAP1complex mediates C-NHEJ inhibition at telomeric ends through hindrance at the KU loading and DNA–PK activation steps. (A) Western blotting analysis of HeLa protein extracts after immuno-depletion as indicated. Protein samples were denatured and separated on 8% SDS–PAGE gel followed by electrotransfer on membrane and blotting with the antibodies as indicated. (B) EJ assay catalysed under standard reaction conditions with the indicated plasmids and HeLa extracts depleted as specified, in the presence or not of NU7026. DNA ligation products were separated by agarose gel electrophoresis followed by SYBR-Green staining. (C) Quantification of independent experiments as shown in (B) (n=3). Relative ligation efficiency was calcultated as the % of ligation obtained in each case related to the ligation obtained on the control biopC plasmid with the control IgG-depleted extracts. Error bars correspond to s.e.m. Black bars correspond to control IgG-depleted extracts and grey bars to TRF2/RAP1-depleted extracts. Absolute ligation efficiency was 4.6±0.8% s.e.m. for IgG-depleted extracts on bioPC plasmid. (D) Pull-down experiment under standard conditions with HeLa extracts mixed with the indicated biotinylated DNA fragments. Salt molarity during washing of the beads is specified. The initial amount of protein used during the pull-down experiment was loaded as input. Protein samples were denatured and separated on 8% SDS–PAGE gel followed by electrotransfer on membrane and blotting with the antibodies as indicated. (E) Pull-down experiment as in (D) but with immuno-depleted HeLa extracts as indicated and washing under high salt conditions. C stands for the 502 bp non-biotinylated control DNA fragment.
Under the salt conditions used here, KU binding to DNA ends is necessary for DNA–PK activation (Hammarsten and Chu, 1998). As DNA–PK activity was impaired at telomeric ends, we have perfomed pull-down experiments with streptavidin beads and biotinylated DNA fragments to assess KU loading at these ends with HeLa extracts proficient or deficient in TRF2/RAP1. The reaction contained ATP to allow DNA–PKcs activation, auto-phosphorylation (Chan and Lees-Miller, 1996) and detachment from the probe in case of proper activation, as shown earlier (Calsou et al, 1999). As binding of KU to the telomeric probe might occur indirectly through interaction with TRF2 or TRF1 proteins (Hsu et al, 1999; Song et al, 2000), we first tested the effect of the salt molarity during the washing step (Figure 2D). As expected, RAP1 was specifically pulled down with the telomeric probe and resisted high salt washing (lanes 3 and 5). At the lower salt concentration, no difference was observed for KU and DNA–PK pull-down between non-telomeric and telomeric probes (lanes 2 and 3). In contrast, a marked difference between the two probes was observed for the two proteins under a higher salt molarity: KU failed to accumulate at the telomeric probe whereas DNA–PKcs was mainly pulled down on this probe (lanes 4 and 5). This pattern is reminiscent to what we have observed under conditions of kinase inhibition that led to a blocked DNA–PK complex at the DNA end (Calsou et al, 1999). The higher salt conditions were therefore chosen to perform a pull-down with both probes and control and TRF2/RAP1-depleted extracts in parallel (Figure 2E). Again, RAP1 was specifically pulled down with the telomeric probe but, as expected, was absent in pull-down from the depleted extracts. In contrast, TRF1 was equally pulled down on the telomeric probes, whether TRF2/RAP1 was present or not (lanes 4 and 6). Regarding KU and DNA–PKcs, the same pattern as in Figure 2D was found with the control extract, consistent with the presence of a blocked DNA–PK complex at the telomeric-ended probe (lanes 3 and 4). In contrast, KU was fully pulled down on the telomeric probe from TRF2/RAP1-depleted extracts (lane 6) and DNA–PKcs was equally pulled down with both the probes in the absence of TRF2/RAP1, suggesting equal kinase activity on both probes. At ds telomeric ends, TRF2/RAP1 complex, but not TRF1 alone, is likely responsible for an hindrance of the DNA–PK loading on DNA necessary for proper DNA–PKcs activation that in turn impairs LIG4-dependent EJ at these ends.
KU and DNA–PKcs are the main factors that prevent B-NHEJ from operating on telomeric ends
Various instances of telomere fusions independent of C-NHEJ have been described in cells (Bailey et al, 1999; Hsu et al, 1999, 2000; Samper et al, 2000; Gilley et al, 2001; Goytisolo et al, 2001; d'Adda di Fagagna et al, 2001; Espejel et al, 2004; Li et al, 2007; Maser et al, 2007). We, therefore, decided to characterize this EJ activity in vitro. To prevent C-NHEJ, we used extracts from a human pre-B-cell line, N114P2, with targeted disruption in both LIG4 alleles (Grawunder et al, 1998), and from its parental line, Nalm-6 as control. As expected, no LIG4 was detected in N114P2 extracts when compared with extracts from the parental Nalm-6 cell (Figure 3A; Supplementary Figure S4A). Likewise as shown in Supplementary Figure S4B, no ligation activity was observed with LIG4− extracts on control plasmid (lanes 2 and 4), whereas LIG4+ extracts ligated the control plasmid but not the telomeric substrate (lanes 1 and 5). As already observed with HeLa extracts (Figure 2B), immuno-depletion of TRF2/RAP1 proteins from LIG4+ extracts overcame the ligation inhibition on telomeric plasmid (Supplementary Figure S4B, compare lanes 5 and 7), whereas TRF2/RAP1-depleted LIG4− extracts still did not ligate this substrate (compare lanes 6 and 8), indicating that C-NHEJ is involved.
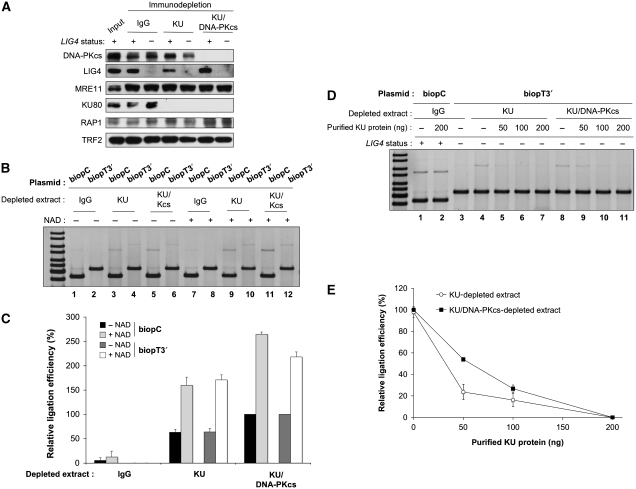
Figure 3.
KU and DNA–PKcs prevent B-NHEJ at telomeric ends. (A) Western blotting analysis of Nalm-6 and N114P2 protein extracts after immuno-depletion as indicated. Protein samples were denatured and separated on 8% SDS–PAGE gel followed by electrotransfer on membrane and blotting with the antibodies as indicated. (B) EJ assay catalysed under standard reaction conditions with the indicated plasmids and N114P2 LIG4- extracts depleted as specified, in the presence or not of β-NAD. DNA ligation products were separated by agarose gel electrophoresis followed by SYBR-Green staining. (C) Quantification of independent experiments as shown in (B) (n=3). For each plasmid series (biopC control or biopT3′ telomeric plasmid), relative ligation efficiency was calcultated as the % of ligation obtained in each case related to the ligation obtained with the KU/DNA–PKcs-depleted extracts without NAD. Error bars correspond to s.e.m. (D) EJ assay catalysed under standard reaction conditions with the indicated plasmids and Nalm-6 LIG4+ or N114P2 LIG4− extracts depleted as specified, in the presence or not of the indicated amount of purified KU protein. DNA ligation products were separated by agarose gel electrophoresis followed by SYBR-Green staining. (E) Quantification of independent experiments as shown in (D) (n=3). Relative ligation efficiency was calcultated as the % of ligation obtained in each case related to the ligation obtained without KU added to the extracts. Error bars correspond to s.e.m.
KU has been described as a major inhibitor of alternative routes for EJ in vitro (Wang et al, 2006) and in vivo (Guirouilh-Barbat et al, 2007; Schulte-Uentrop et al, 2008). Therefore, extracts were immuno-depleted for KU or both KU and DNA–PKcs proteins and EJ capacity was assessed with LIG4-deficient extracts to prevent C-NHEJ activity (Figure 3A). As shown in Figure 3B, KU depletion showed an EJ activity in the absence of LIG4, operating on non-telomeric and telomeric-ended plasmid (lanes 3 and 4). Strikingly, the addition of 2 mM β-NAD stimulated more than two-fold the ligation of both substrates (Figure 3C), indicating that this LIG4-independent ligation relied in part on PARP activation (Audebert et al, 2004; Wang et al, 2006). A two-fold stimulation of ligation in KU-depleted LIG4− extracts was also observed by increasing magnesium concentration up to 3 mM (data not shown) in agreement with published results (Wang et al, 2001). The ligation observed in the absence of KU was bona fide B-NHEJ because it was efficient in the absence of LIG4 (Figure 3B) and it was resistant to both the DNA–PKcs inhibitor, NU7026, and the anti-serum against XRCC4, which otherwise strongly inhibited C-NHEJ (Supplementary Figure S4C and S4D). In addition, we have observed a preferential ligation of compatible plasmid DNA ends versus incompatible ends under our B-NHEJ conditions (Supplementary Figure S5), in agreement with the microhomology preference of LIG4-independent ligation (McVey and Lee, 2008). Compared with the effect of only KU depletion, co-depletion of both components of DNA–PK significantly increased the ligation yield (Figure 3C; Supplementary Figure S6), suggesting a facilitating effect of DNA–PKcs in addition to KU for inhibition of B-NHEJ at ds telomeric ends.
As co-depletion of some unknown component in addition to KU could have occurred, we added back purified human KU dimer to the depleted extracts. A dose-dependent inhibition of ligation was obtained using the telomeric-ended substrate over the range of KU concentrations used, and complete inhibition was observed for the maximal amount (200 ng; Figure 3D), although it remained far below the amount present in the original extracts (Supplementary Figure S7). Interestingly, the inhibition obtained was greater for extracts still containing DNA–PKcs than for extracts lacking both DNA–PK components (Figure 3E). This suggests again a facilitating effect of DNA–PKcs in addition to KU for B-NHEJ repression at telomeric ds ends.
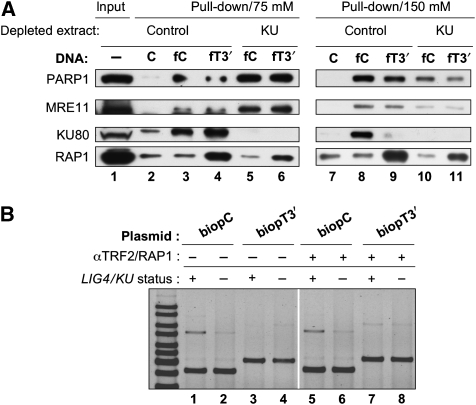
Several other proteins have been proposed to participate in alternative EJ pathways (McVey and Lee, 2008). Our and other groups have shown that in biochemical assays with cell extracts KU competes with PARP-1 for DNA end binding, that PARP-1 can perform a synapsis activity and that it is required for a subsequent LIG3-dependent joining step (Audebert et al, 2004; Wang et al, 2006; Liang et al, 2008). In addition, recent results argue for a role of the MRN complex in this alternative repair route (Dinkelmann et al, 2009; Rass et al, 2009; Xie et al, 2009; Zhuang et al, 2009). We, therefore, investigated the recovery of PARP-1 and MRE11 as representative candidate proteins in pull-down experiments from LIG4-deficient extracts proficient or deficient in KU and with either telomeric or non-telomeric probes. As salt sensitivity of B-NHEJ protein/DNA complexes could not be anticipated, we washed beads under two salt concentration conditions. As shown in Figure 4A, the same pattern was observed for KU as in HeLa extracts in Figure 2D and E; at the lower salt concentration, there was no difference for KU pull-down for non-telomeric or telomeric probes (lanes 3 and 4) whereas at a higher concentration, KU failed to accumulate on the telomeric probe (lanes 8 and 9). RAP1 was specifically pulled down with the telomeric probe and resisted high salt washing (lanes 4, 6, 9 and 11). Strikingly, PARP-1 and MRE11 exhibited the same pattern, whether the DNA end was telomeric or not; under low salt conditions, there was a marked increase in both proteins pulled down from KU-depleted extracts (lanes 5, 6) when compared with control extracts (lanes 3, 4), whereas this difference was lost under higher salt molarity. This result indicates that KU competes with PARP-1 and MRE11 for loading at DNA ends; in addition, PARP-1 and MRE11 binding at telomeric ds seems to be insensitive to the shelterin complex.
Figure 4.
TRF2/RAP1complex does not impair loading of candidate B-NHEJ proteins or B-NHEJ catalysed EJ at telomeric ends. (A) Pull-down experiment under standard conditions with control or Ku-depleted N114P2 extracts mixed with the indicated DNA fragments. Salt molarity during washing of the beads is specified. The initial amount of protein used during the pull-down experiment was loaded as input. Protein samples were denatured and separated on 8% SDS–PAGE gel followed by electrotransfer on membrane and blotting with the antibodies as indicated. C stands for the 502 bp non-biotinylated control DNA fragment. (B) EJ assay catalysed under standard reaction conditions with the indicated plasmids and Nalm-6 LIG4+ or N114P2 LIG4− extracts depleted as specified, in the presence or not of a mixture of antibodies against TRF2 and RAP1. DNA ligation products were separated by agarose gel electrophoresis followed by SYBR-Green staining. The ratio of ligation efficiency with TRF2/RAP1 antibodies versus without antibodies was 0.9, 1, 8.4 and 1.2 for pair of lanes 5/1, 6/2, 7/3 and 9/4, respectively.
TRF2/RAP1 complex does not impair B-NHEJ at telomeric ends
As TRF2 and RAP1 proteins were still present in KU-depleted extracts (Figure 3A), we addressed their effect on B-NHEJ. A combination of RAP1 and TRF2 antibodies was able to overcome the C-NHEJ inhibition observed using the telomeric substrate, mimicking immuno-depletion of both proteins (Supplementary Figure S8). As shown in Figure 4B, this mixture of antibodies clearly stimulated C-NHEJ on telomeric plasmid with the control LIG4+/KU+ extracts (eight-fold stimulation of ligation efficiency between lanes 3 and 7); strikingly, it had no significant effect on the B-NHEJ activity operating on telomeric plasmid with KU-depleted LIG4− extracts (ratio 1.2 of ligation efficiency between lanes 8 and 4). Together, these results indicate that KU/DNA–PKcs rather than TRF2/RAP1 complex has a major role for B-NHEJ inhibition at ds telomeric ends.
Discussion
Interplay between shelterin and C-NHEJ proteins at ds telomeric end
Bae and Baumann (2007) have observed an inhibition of LIG4-dependent EJ at short ds telomere tracks with cell extracts. Here, we have analysed the first steps of this reaction by using a similar EJ reaction assay with human cell extracts, longer telomeric tracks and restricting the accessibility of ligation to only one end of the plasmid substrate. Under these conditions, we measured 80–90% EJ inhibition at ds 3′-telomeric ends. We show for the first time that TRF2/RAP1 impairs the proper DNA–PK binding at ds telomeric ends that are necessary for DNA–PKcs activation, which is in turn indispensable for LIG4-dependent EJ. When binding of TRF2/RAP1 complex to telomeric ends was impeded by immuno-depletion, DNA–PK binding and activation were restored and allowed subsequent LIG4-dependent ligation.
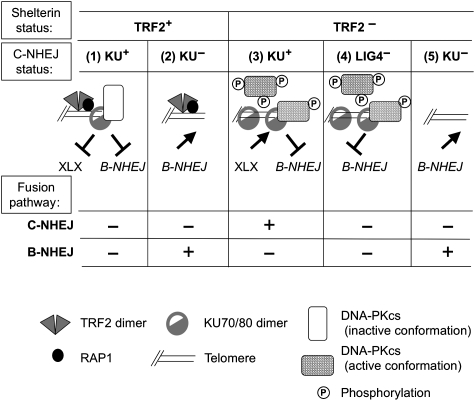
In Figure 5, we propose a model integrating our pull-down results from control and TRF2/RAP1-depleted extracts with the telomeric probe (Figure 2D and E). It is known that DNA–PKcs loading on KU, which is bound to a DNA end, induces an inward translocation of KU (Yoo and Dynan, 1999). This translocation is probably required for the melting of DNA termini needed for DNA–PKcs activation (Hammarsten et al, 2000; Jovanovic and Dynan, 2006). The presence of the shelterin DNA complex at the telomeric terminus likely causes steric hindrance (dependent on TRF2/RAP1) that impairs a proper inward sliding of KU on the DNA helix (Figure 5, column 1), as shown with extracts from two different cell lines (Figures 2E and 4A). Under such conditions, DNA–PK remains at the DNA end in an inactive form, with a protein/DNA end molar ratio likely close to one, as shown elsewhere (Calsou et al, 1999). This explains the DNA–PKcs accumulation at telomeric ends compared with freely activating DNA ends, despite a low amount of pull-down KU (Figure 2D and E). In contrast, TRF2/RAP1 depletion allows KU inward translocation, DNA–PKcs activation and detachment by auto-phosphorylation (Chan and Lees-Miller, 1996) (Figure 5, column 3). As demonstrated earlier under similar pull-down conditions (Calsou et al, 1999), iterative binding and activation events then occur and promote KU accumulation along the DNA telomeric probe (Figure 5, column 3), thus increasing KU/DNA-end molar ratio (Figure 2D and E). The polarity of the EJ inhibition suggests that the conformation of the shelterin complex at the telomeric end facing KU may be different in the 3′ and 5′ orientation.
Figure 5.
Model for the interplay between shelterin and EJ proteins at telomeres. See ‘Discussion' section for details. XLX refers to the ligation complex of the C-NHEJ pathway (XRCC4/LIG4/Cernunnos–XLF).
Interestingly, when the telomeric tract was moved inward from the 3′ end, DNA–PK activation was completely restored when the telomeric sequence was 7 bp from the 3′ end while ligation activity required 13 bp. KU inward sliding from the DNA end likely covers a distance shorter than that necessary for proper positioning of the ligation complex (Modesti et al, 1999; Lu et al, 2007).
Negative control of B-NHEJ at DNA ends by DNA–PK
Biochemical data with purified components have indicated that KU stimulates other mammalian ligases than LIG4 (Ramsden and Gellert, 1998). However, the rescue of the embryonic lethality of LigIV−/− mice (Karanjawala et al, 2002) or radiosensitivity of LIG4-deficient cells by a simultaneous abrogation of KU (Adachi et al, 2001) rather supports a dominant negative inhibition exerted by KU on backup EJ in the absence of LIG4. In agreement, we observe here that KU immuno-depletion from LIG4− extracts mediates EJ and back complementation with purified protein abolishes it. This clearly shows that KU is the main contributor to B-NHEJ inhibition at ds telomeric ends. In addition, the DNA–PKcs subunit exerts a facilitating effect on this B-NHEJ repression.
What is the basis of KU-dependent inhibition of B-NHEJ at telomeres? It has been shown that a DNA end bearing a blocked DNA–PK complex is protected from further processing by either DNA polymerization, degradation or ligation (Calsou et al, 1999). As DNA–PK complex is likely blocked at ds telomeric ends because of TRF2/RAP1-dependent hindrance, the simplest model is that this DNA–PK prevents, in turn, loading of key B-NHEJ proteins (Figure 5, column 1), in agreement with the observation that DNA–PK suppresses B-NHEJ in vitro (Perrault et al, 2004). Furthermore, we and others have reported evidence for the involvement of PARP-1 in alternative EJ (this work and Audebert et al, 2004; Wang et al, 2006; Robert et al, 2009). Although under normal conditions, DNA–PK competes with PARP-1 for binding to DSB, PARP-1 becomes more efficient than DNA–PK in the absence of KU (Wang et al, 2006), in agreement with our results. Additionally, the shelterin-driven KU localization at telomeres may have a role. Indeed, KU localizes at telomeres most likely through interactions with the TRF2 and TRF1 components of the shelterin apparatus (Hsu et al, 2000; Song et al, 2000). As TRF2/RAP1 depletion in LIG4− extracts did not promote ligation of the telomeric-ended substrate (Supplementary Figure S3), it is unlikely that TRF2–KU interaction affords protection against B-NHEJ. However, a TRF1–KU interaction may persist under these conditions, and cannot be excluded as a participant in protection from EJ, as proposed (Hsu et al, 2000).
We observed that B-NHEJ inhibition at ds telomeric ends was released by KU depletion even in TRF2/RAP1-proficient extracts. Additionally, B-NHEJ proteins were pulled down despite the presence of TRF2/RAP1 on the DNA probe and antibody-mediated inactivation of this complex did not impact on the B-NHEJ efficiency. Togther, these results indicate that, contrary to C-NHEJ, loading and activation of B-NHEJ proteins are not significantly impaired by the shelterin at the ds telomeric ends (Figure 5, column 2). This may be a good reason for the involvement of a second KU-dependent mechanism against improper joining at these ends. Further experiments are now needed in cellulo to decipher the interference at telomeric ends between shelterin and B-NHEJ candidate proteins reported here.
Relevance to in vivo situations
In humans, the telomeric DNA comprises T2AG3 duplex repeats oriented 5′ to 3′ toward the chromosome end, followed by a 3′ T2AG3 single-stranded extension. Although the G-tail may participate in EJ inhibition at telomeres, for example through impairment of DNA–PK activation (Tsai et al, 2007), our results together with a published report (Bae and Baumann, 2007) clearly emphasize a specific role of the duplex repeats for this inhibition. Telomeric ends devoid of the 3′G-tail may be relevant to certain situations in cells such as blunt telomeres generated by leading strand synthesis (Gilson and Geli, 2007) or processing by the nuclease activity of ERCC1/XPF (Zhu et al, 2003) or MRE11 (Deng et al, 2009). Indeed, our results account for various results obtained in cellulo. We found that on TRF2/RAP1 depletion, EJ was mainly LIG4 dependent, which agrees with the LIG4 dependency of telomere fusion consecutive to either TRF2 inhibition by expression of a dominant negative variant (Smogorzewska et al, 2002), expression of a thermosensitive mutant of TRF2 (Konishi and de Lange, 2008) or deletion by Cre-dependent recombination in mouse cells (Celli and de Lange, 2005; Celli et al, 2006) (Figure 5, column 3). On TRF2 inactivation, it was observed that although LIG4 deficiency lowered 100-fold the telomere fusion frequency, this frequencey was only lowered 10-fold by KU deficiency, leading the authors to suggest that ‘a subset of telomere fusions require DNA LIG4 but not KU' (Celli et al, 2006). According to our results, we rather believe that KU deficiency released the inhibition of an alternative LIG4-independent EJ route, which is otherwise blocked by KU in LIG4− cells (Figure 5, columns 4 and 5), although it might be partially restrained in the former case by persistent 3′ overhangs (Celli et al, 2006). It would be interesting to perform TRF2 deletion with Cre in LIG4−/− versus LIG4+/+ MEFs together with KU inactivation that may promote alternative EJ to some extent. The LIG4-independent alternative EJ mechanism that we report here may also account for spontaneous chromosomal end fusions in the absence of KU or DNA–PKcs (Bailey et al, 1999; Hsu et al, 1999, 2000; Samper et al, 2000; Gilley et al, 2001; Goytisolo et al, 2001; d'Adda di Fagagna et al, 2001; Espejel et al, 2004; Li et al, 2007) (Figure 5, column 2).
From our data, we speculate that chromosome stability benefits from a local high density of DNA–PK at telomeres. In case of fortuitous exposure of a blunt telomere end like at post-replicative leading strand, DNA–PK could instantly recognize and bind to this end; maintained in a kinase inactive configuration by the shelterin components, it may not initiate C-NHEJ while preventing aberrant EJ by alternative shelterin-insensitive mechanisms. This post-replicative blockade of DNA–PK at blunt leading strand may only be transient and reversed by some unknown mechanism leading to DNA–PKcs release by phosphorylation and G-tail generation by end processing. Indeed, DNA–PKcs activity is required to maintain telomere stability (Bailey et al, 2004; Williams et al, 2009).
Chromosome ends may be particularly at risk from ligation by B-NHEJ as candidate components such as PARP-1 and MRE11 also interact with TRF2 (Zhu et al, 2000; Gomez et al, 2006). This competition model could explain the DNA–PKcs- and LIG4-independent fusions arising after telomere attrition in telomerase-deficient mouse cells (Maser et al, 2007). In this case, a lower local density of shelterin-associated DNA–PK at the shortened telomeres may compete less efficiently with B-NHEJ proteins, which additionally may be insensitive to shelterin hindrance at the telomere end, as shown here. Accordingly, PARP-1 has been shown to accumulate at eroded telomeres caused by telomerase deficiency (Gomez et al, 2006).
In summary, we have used an EJ assay with plasmid DNA and cell extracts under controlled conditions enabling either C-NHEJ or B-NHEJ. We show that TRF2/RAP1 and DNA–PK complexes sustain a dual protection at telomeres: TRF2/RAP1 complex prevents C-NHEJ-mediated end fusion at the initial DNA–PK end-binding step, whereas the DNA–PK complex counteracts a potent B-NHEJ mechanism.
Materials and methods
Cell culture
All culture media were from Gibco-Invitrogen and were supplemented with 10% foetal calf serum, 2 mM glutamine, 125 U/ml penicillin and 125 μg/ml streptomycin. All cells were grown at 37°C in a humidified atmosphere with 5% CO2. HeLa cells, from the American Type Culture Collection, were grown in Dulbecco's modified Eagle's medium (Invitrogen). The LIG4-defective cell line, N114P2 (Grawunder et al, 1998), and its parental cell line, Nalm-6 (gifts from MR Lieber, University of Southern California, Los Angeles, CA, USA) were maintained in RPMI 1640 medium.
Antibodies
Mouse monoclonal antibodies anti-TRF2 (4A794), anti-KU80 (S10B1) and anti-DNA–PKcs (clone 18.2) were from Abcam, anti-RAP1 (4C8/1) was from Sigma, anti-PARP (4C10-5) was from BD Pharmingen and anti-KU (clone Ab3-162) and anti-DNA–PKcs (clone 25.4) used for immuno-depletion were from Thermo Scientific. DNA LIG4 (AHP554) polyclonal antibody was obtained from Serotec. Rabbit serum anti-TRF2 used for immuno-depletion was raised against full-length protein and was purified with HiTrap Protein A HP (GE Healthcare). Rabbit serum anti-XRCC4 was raised against full-length recombinant protein, produced in baculovirus and was affinity purified.
Plasmid substrates and DNA probes
The scheme for the production of the plasmids and DNA fragments used in this study is shown in Figure 1A. All the enzymes used for pUCtelo2 (Amiard et al, 2007) or pBluescript-KS-II(−) restriction were from Biolabs and used according to the manufacturer's instructions. Linearized plasmids (Figure 1A, tracks 1 and 2) were purified by PCR and Gel Clean-Up System (Promega). Biotinylated substrates (Figure 1A, tracks 2 and 3) were obtained by filling linearized pUCtelo2 with Klenow exo-enzyme (Stratagene) and biotinylated analogs biotin-21-dUTP and biotin-14-dATP (Clontech). For generation of telomeric fragments (Figure 1A, track 3) plasmids were further restricted, gel separated and fragments of interest were purified from the gel slices with the Gel Clean-Up System (Promega) followed by phenol/chloroform extraction and precipitation. Control non-biotinylated C and biotinylated fC 502 bp fragments were amplified by PCR in a total volume of 20 μl of HF Buffer (Finnzymes), from 20 ng of pBluescript-KS-II(−), with 0.5 μM non-biotinylated (fragment C) or biotinylated (fragment fC) reverse primer (GCGTTATCCCCTGATTCTGTGG), respectively, and 0.5 μM forward primer (GGGCGAATTGGAGCTCCACC), 400 μM dNTP (Fermentas), 0.4 U Phusion Hot Start (Finnzymes) with annealing temperature at 69°C and 35 cycles of amplification, and denaturation and polymerization conditions as recommended by the manufacturer.
EJ extracts
Cell extract preparation was carried out as described earlier (Buck et al, 2006). Briefly, exponentially growing cells were lysed through three freeze/thaw cycles in lysis buffer (25 mM Tris–HCl pH 7.5, 333 mM KCl, 1.5 mM EDTA pH 8.0, 4 mM DTT) containing protease inhibitor cocktail (Roche Diagnostics), and phosphatase inhibitor cocktails I and II (Sigma-Aldrich). Lysates were incubated at 4°C for 20 min, cleared by centrifugation, and dialysed against dialysis buffer (20 mM Tris–HCl pH 8.0, 100 mM potassium acetate, 20% glycerol, 0.5 mM EDTA pH 8.0, 1 mM DTT). Protein concentration was determined using the Bradford assay (Bio-Rad) and EJ extracts were stored at −80°C.
EJ assay
When necessary, extracts were pre-incubated with Nu7026 (Sigma-Aldrich), blocking antibodies or purified KU protein as indicated for 10 min at 4°C. Pretreated or mock-treated extracts (40 μg) were incubated for 2 h at 25°C in 10 μl reaction mixture containing 5 ng linearized plasmid in EJ buffer (50 mM triethanolamine pH 8.0, 0.5 mM magnesium acetate, 1 mM dithiothreitol, 0.1 mg/ml BSA, 60 mM potassium acetate) with 1 mM ATP added at last to initiate the reaction in the presence or not of 2 mM β-NAD (Sigma-Aldrich) when necessary. Standard conditions for B-NHEJ refer to conditions as above except the presence of 3 mM magnesium acetate and 2 mM β-NAD. Samples were then treated with 100 μg/ml RNAse A for 10 min at 37°C and deproteinized. DNA ligation products were separated in 0.7% agarose gels together with GeneRuler DNA ladder mix (0.5–10 kb, Fermentas) in one lane and stained with SYBR-Green (Invitrogen). Fluorescence was detected and analysed on a Typhoon fluorimager (Molecular Dynamics). Quantitative analysis of the gel was performed with the ImageJ software (version 1.4).
Kinase assay
When necessary, extracts were pre-incubated with Nu7026 (Sigma-Aldrich) for 10 min at 4°C. DNA fragment as indicated (0.1 pmol) was mixed with 2 pmol streptavidin (Sigma) in EJ buffer, incubated for 10 min at RT and then mixed in 10 μl reaction volume with 10 μg pretreated or mocked-treated EJ extract, 2.5 μg DNA–PK peptide substrate (Promega) and then 50 μM ATP and 1 μCi γ32P-ATP (Perkin-Elmer) added at last to initiate the reaction. Incubation was for 20 min at 25°C. Samples were boiled in loading sample buffer for 10 min and run on a 22% polyacrylamide denaturing gel. Auto-radiography of the gel was processed with a PhosphorImager (Molecular Dynamics, Storm System TM). Quantitative analysis of the gel was performed with the ImageQuant software (version 5.2).
Immuno-depletion
Immuno-depletion was carried out in a 65 μl reaction volume containing 20 μl of protein A/G-PLUS agarose wet beads (Santa Cruz Biotechnology), 1 mg of EJ extract in EJ buffer, 10 μg of antibody or IgG control or 10 μl of serum. After an overnight incubation at 4°C under constant rotation, supernatant was recovered by spinning, mixed with 20 μl of beads as above, incubated for 1 h at 4°C and spun down again to remove the beads. Sample aliquots were processed for western blotting and the remaining extracts were frozen at −80°C until use.
Pull-down experiment
DNA fragment as indicated was incubated with high capacity streptavidin agarose beads (Thermo-scientific) at the ratio 1 pmol/10 μl wet beads in two-fold diluted phosphate-buffered saline (PBS) buffer under agitation for 25 min at RT before washing in the same buffer. Then, 40 μl wet beads were mixed with 80 μg EJ extract immuno-depleted with the indicated antibody in 80 μl reaction volume under kinase buffer conditions as above (except that the peptide substrate and γ32P-ATP were omitted) and incubated for 25 min at 25°C under agitation. The beads were washed rapidly twice with either two-fold diluted or undiluted PBS buffer and then processed for western blotting.
Western blotting
Protein samples were mixed with concentrated loading sample buffer to 1X final concentration (50 mM Tris–HCl pH 6.8, 10% glycerol, 1% SDS, 300 mM 2-mercaptoethanol, 0.01% bromophenol blue), boiled, separated by SDS–PAGE (8% polyacrylamide) and blotted onto Immobilon-P polyvinylidene difluoride membranes (Millipore). Membranes were blocked for 1 h with 5% dry milk in PBS-T (PBS, 0.1% Tween-20 (Sigma-Aldrich)) and incubated for 1 h with primary antibody diluted in PBS containing 0.02% Tween-20 and 1% bovine serum albumin (fraction V, Sigma-Aldrich). After three washes with PBS-T, membranes were incubated for 1 h with secondary antibodies in PBS containing 0.02% Tween-20 and 5% dry milk. Membranes were washed five times with PBS-T and immuno-blots were visualized by enhanced chemiluminescence (ImmunofaxA, Yelen). When indicated, successive rounds of immuno-blotting were performed on the same membranes after stripping (Restore Western Blot Stripping Buffer, Pierce).
Purification of KU protein complex
The cDNAs of KU80 and the fusion construct His-KU70 were cloned into pFastBac1 vector (Invitrogen). Baculoviruses were produced and amplified in Sf9 cells according to manufacturer's procedures (Invitrogen). For purification of KU70/KU80 complex, 1 l of Sf9 cells were grown in suspension in Insect-Xpress medium (BioWhittaker) and inoculated with both His-KU70 and KU80 viruses at MOI of 10 and 15, respectively. After 72 h incubation at 27°C, cells were collected by centrifugation at 1500 g for 5 min at 4°C. The pellet was washed twice with ice cold 1X PBS. Cells were lysed at 4°C for 30 min in Buffer A (50 mM Hepes-KOH pH 7.5, 10% glycerol, 2 mM EDTA, 0.01% NP-40) containing 1 M NaCl. The lysate was clarified by centrifugation at 175 000 g for 1 h at 4°C and adjusted to 100 mM NaCl before loading onto a HiTrap-Q HP column (GE Healthcare). Proteins were eluted with buffer A containing increasing concentration of KCl. The presence of KU proteins was assessed by western blot. Positive fractions were pooled, diluted in buffer A without EDTA and adjusted to 500 mM NaCl before loading onto a Ni−HiTrap Chelating HP column (GE Healthcare). After washing, proteins were eluted with buffer A without EDTA and supplemented with 100 mM NaCl and 150 mM imidazole. The presence and purity of KU proteins was monitored by Coomassie blue staining. Positive fractions were pooled, and proteins were concentrated and equilibrated in storing buffer (50 mM Tris–HCl pH 7.8, 100 mM NaCl 10% glycerol) using Centricon-100 microconcentrator column (Millipore). Protein concentration was estimated with Bradford protein assay (Bio-Rad) and samples were aliquoted and stored at −80°C.
Supplementary Material
Acknowledgments
We thank MR Lieber (USC, Los Angeles, CA, USA) for the gift of cell lines. We thank Neil Johnson for the reading of the manuscript. This work was partly supported by grants from the Ligue Nationale Contre le Cancer (‘équipe labellisée') and a grant from the Institut National Contre le Cancer (TELINCA program). O Bombarde was supported by a PhD fellowship from La Ligue Nationale Contre le Cancer. P Calsou is a scientist from INSERM, France. OB, CB, DG and PF performed the experiments; M-JGP and EG provided materials; PC designed the experiments; M-JGP, EG, BS and PC analysed the data; PC wrote the paper; OB, DG, PF, M-JGP, EG and BS corrected the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adachi N, Ishino T, Ishii Y, Takeda S, Koyama H (2001) DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: implications for DNA double-strand break repair. Proc Natl Acad Sci USA 98: 12109–12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, Paoletti J, Gilson E, Giraud-Panis MJ (2007) A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol 14: 147–154 [DOI] [PubMed] [Google Scholar]
- Audebert M, Salles B, Calsou P (2004) Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 279: 55117–55126 [DOI] [PubMed] [Google Scholar]
- Bae NS, Baumann P (2007) A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell 26: 323–334 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Brenneman MA, Halbrook J, Nickoloff JA, Ullrich RL, Goodwin EH (2004) The kinase activity of DNA-PK is required to protect mammalian telomeres. DNA Repair (Amst) 3: 225–233 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci USA 96: 14899–14904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D (2003) Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet 33: 203–207 [DOI] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P (2006) Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299 [DOI] [PubMed] [Google Scholar]
- Calsou P, Frit P, Humbert O, Muller C, Chen DJ, Salles B (1999) The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J Biol Chem 274: 7848–7856 [DOI] [PubMed] [Google Scholar]
- Celli GB, de Lange T (2005) DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 7: 712–718 [DOI] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T (2006) Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 8: 885–890 [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR (2008) Telomere uncapping and alternative lengthening of telomeres. Mech Ageing Dev 129: 99–108 [DOI] [PubMed] [Google Scholar]
- Chan DW, Lees-Miller SP (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem 271: 8936–8941 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP (2001) Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol 11: 1192–1196 [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Deng Y, Guo X, Ferguson DO, Chang S (2009) Multiple roles for MRE11 at uncapped telomeres. Nature 460: 914–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, Ferguson DO (2009) Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol 16: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet J, Delteil C, Lefrancois J, Concannon P, Salles B, Calsou P (2005) DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J Biol Chem 280: 7060–7069 [DOI] [PubMed] [Google Scholar]
- Espejel S, Franco S, Rodriguez-Perales S, Bouffler SD, Cigudosa JC, Blasco MA (2002a) Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J 21: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel S, Franco S, Sgura A, Gae D, Bailey SM, Taccioli GE, Blasco MA (2002b) Functional interaction between DNA-PKcs and telomerase in telomere length maintenance. EMBO J 21: 6275–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel S, Klatt P, Menissier-de Murcia J, Martin-Caballero J, Flores JM, Taccioli G, de Murcia G, Blasco MA (2004) Impact of telomerase ablation on organismal viability, aging, and tumorigenesis in mice lacking the DNA repair proteins PARP-1, Ku86, or DNA-PKcs. J Cell Biol 167: 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Zakian VA (2005) Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 4: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Gilley D, Tanaka H, Hande MP, Kurimasa A, Li GC, Oshimura M, Chen DJ (2001) DNA-PKcs is critical for telomere capping. Proc Natl Acad Sci USA 98: 15084–15088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Gomez M, Wu J, Schreiber V, Dunlap J, Dantzer F, Wang Y, Liu Y (2006) PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol Biol Cell 17: 1686–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP (2006) DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J 25: 3880–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytisolo FA, Samper E, Edmonson S, Taccioli GE, Blasco MA (2001) The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol Cell Biol 21: 3642–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR (1998) DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell 2: 477–484 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS (2007) Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA 104: 20902–20907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE (2008) Alternative endings. Proc Natl Acad Sci USA 105: 405–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarsten O, Chu G (1998) DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA 95: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarsten O, DeFazio LG, Chu G (2000) Activation of DNA-dependent protein kinase by single-stranded DNA ends. J Biol Chem 275: 1541–1550 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T (2006) Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S (2009) Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J Cell Biol 184: 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Blackburn EH, Chen DJ (1999) Ku is associated with the telomere in mammals. Proc Natl Acad Sci USA 96: 12454–12458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev 14: 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Dynan WS (2006) Terminal DNA structure and ATP influence binding parameters of the DNA-dependent protein kinase at an early step prior to DNA synapsis. Nucleic Acids Res 34: 1112–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanjawala ZE, Adachi N, Irvine RA, Oh EK, Shibata D, Schwarz K, Hsieh CL, Lieber MR (2002) The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair (Amst) 1: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Konishi A, de Lange T (2008) Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev 22: 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Vogel H, Holcomb VB, Gu Y, Hasty P (2007) Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol Cell Biol 27: 8205–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Deng L, Nguyen SC, Zhao X, Maulion CD, Shao C, Tischfield JA (2008) Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res 36: 3297–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Pannicke U, Schwarz K, Lieber MR (2007) Length-dependent binding of human XLF to DNA and stimulation of XRCC4: DNA ligase IV activity. J Biol Chem 282: 11155–11162 [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Meek K, Lees-Miller SP (2009) Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 417: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Wong KK, Sahin E, Xia H, Naylor M, Hedberg HM, Artandi SE, DePinho RA (2007) DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol Cell Biol 27: 2253–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Lee SE (2008) MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet 24: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Dang V, Lees-Miller SP (2008) DNA-PK the means to justify the ends? Adv Immunol 99: 33–58 [DOI] [PubMed] [Google Scholar]
- Modesti M, Hesse JE, Gellert M (1999) DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J 18: 2008–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC (2007) A backup DNA repair pathway moves to the forefront. Cell 131: 223–225 [DOI] [PubMed] [Google Scholar]
- O'Driscoll M, Jeggo PA (2006) The role of double-strand break repair—insights from human genetics. Nat Rev Genet 7: 45–54 [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Pardo B, Gomez-Gonzalez B, Aguilera A (2009) DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci 66: 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault R, Wang H, Wang M, Rosidi B, Iliakis G (2004) Backup pathways of NHEJ are suppressed by DNA-PK. J Cell Biochem 92: 781–794 [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M (1998) Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J 17: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS (2009) Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 16: 819–824 [DOI] [PubMed] [Google Scholar]
- Riha K, Heacock ML, Shippen DE (2006) The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet 40: 237–277 [DOI] [PubMed] [Google Scholar]
- Robert I, Dantzer F, Reina-San-Martin B (2009) Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med 206: 1047–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA (2000) Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep 1: 244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy J, Bae NS, Scrafford J, Baumann P (2009) Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J 28: 3390–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J (2008) Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res 36: 2561–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T (2002) DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12: 1635–1644 [DOI] [PubMed] [Google Scholar]
- Song K, Jung D, Jung Y, Lee SG, Lee I (2000) Interaction of human Ku70 with TRF2. FEBS Lett 481: 81–85 [DOI] [PubMed] [Google Scholar]
- Tsai YC, Qi H, Liu LF (2007) Protection of DNA ends by telomeric 3′ G-Tail sequences. J Biol Chem 282: 18786–18792 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J (2007) Replication and protection of telomeres. Nature 447: 924–931 [DOI] [PubMed] [Google Scholar]
- Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G (2005) DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 65: 4020–4030 [DOI] [PubMed] [Google Scholar]
- Wang H, Zeng ZC, Perrault AR, Cheng X, Qin W, Iliakis G (2001) Genetic evidence for the involvement of DNA ligase IV in the DNA-PK-dependent pathway of non-homologous end joining in mammalian cells. Nucleic Acids Res 29: 1653–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G (2006) PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34: 6170–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Baumann P (2008) Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol Cell 31: 463–473 [DOI] [PubMed] [Google Scholar]
- Williams ES, Klingler R, Ponnaiya B, Hardt T, Schrock E, Lees-Miller SP, Meek K, Ullrich RL, Bailey SM (2009) Telomere dysfunction and DNA-PKcs deficiency: characterization and consequence. Cancer Res 69: 2100–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Frit P, Malivert L, Revy P, Biard D, Salles B, Calsou P (2007) Interplay between cernunnos-XLF and NHEJ proteins at DNA ends in the cell. J Biol Chem 282: 31937–31943 [DOI] [PubMed] [Google Scholar]
- Xie A, Kwok A, Scully R (2009) Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 16: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Blackburn EH (2007) Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol Cell 28: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Dynan WS (1999) Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res 27: 4679–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet 25: 347–352 [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell 12: 1489–1498 [DOI] [PubMed] [Google Scholar]
- Zhuang J, Jiang G, Willers H, Xia F (2009) The exonuclease function of human Mre11 promotes deletional non-homologous end-joining. J Biol Chem 284: 30565–30573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.