Abstract
Nonsense-mediated mRNA decay (NMD) is a translation-linked process that destroys mRNAs with premature translation termination codons (PTCs). In mammalian cells, NMD is also linked to pre-mRNA splicing, usually PTCs trigger strong NMD only when positioned upstream of at least one intron. The exon junction complex (EJC) is believed to mediate the link between splicing and NMD in these systems. Here, we report that in Schizosaccharomyces pombe splicing also enhances NMD, but against the EJC model prediction, an intron stimulated NMD regardless of whether it is positioned upstream or downstream of the PTC and EJC components are not required. Still the effect of splicing seems to be direct—we have found that the important NMD determinant is the proximity of an intron to the PTC, not just the occurrence of splicing. On the basis of these results, we propose a new model to explain how splicing could affect NMD.
Keywords: NMD, S. pombe, splicing, translation, yeast
Introduction
Alleles carrying nonsense mutations often yield very low mRNA levels. This phenomenon has been observed in all investigated organisms, from bacteria to mammalian cells. In eukaryotic cells, the phenomenon is called nonsense-mediated mRNA decay (NMD) and it seems to be an active mechanism that selectively destroys mRNAs carrying premature translation termination codons (PTCs) (reviewed in Maquat, 2004; Amrani et al, 2006; Muhlemann et al, 2008; Brogna and Wen, 2009). NMD requires active translation and specific trans-acting factors. Three trans-acting factors are the evolutionary conserved proteins UPF1, UPF2 and UPF3; typically deletion or silencing of the corresponding genes prevents NMD in all tested organisms (Maquat, 2004; Amrani et al, 2006; Muhlemann et al, 2008). Additional factors have also been implicated in NMD in multicellular organisms (Maquat, 2004; Muhlemann et al, 2008). Perhaps NMD serves as an mRNA surveillance mechanism that prevents the synthesis of truncated proteins that potentially could have toxic effects, but its full physiological importance is not yet clear. UPF1, UPF2 and UPF3 are not essential for viability in Saccharomyces cerevisiae or Caenorhabditis elegans (Culbertson et al, 1980; Hodgkin et al, 1989). Some NMD factors are essential for viability in other organisms (Medghalchi et al, 2001; Metzstein and Krasnow, 2006; Yoine et al, 2006; Wittkopp et al, 2009); but it is not yet known whether NMD or some other important function of these proteins is required for survival (Brumbaugh et al, 2004; Azzalin and Lingner, 2006; Azzalin et al, 2007; Luke et al, 2007; Ajamian et al, 2008). A definite class of NMD substrates are immunoglobulin and T-cell receptor mRNAs that acquired PTCs as a consequence of V(D)J rearrangements during lymphocyte maturation (Li and Wilkinson, 1998; Muhlemann et al, 2008). However, in the typical cell, the main NMD substrates are probably abnormal transcripts that acquired PTCs as a consequence of errors during transcription or pre-mRNA processing (He et al, 1993; Jaillon et al, 2008; Sayani et al, 2008). The discovery that unspliced pre-mRNAs accumulate in NMD mutants in S. cerevisiae and other organisms also suggests that NMD may have a proof-reading function in gene expression, eliminating transcripts that have not been spliced due to suboptimal splice signals (Jaillon et al, 2008; Sayani et al, 2008).
NMD is clearly linked to translation termination; the important question is how PTCs are distinguished from normal stop codons. Surprisingly, in mammalian cells, the distinction seems to be linked to splicing: strong mRNA reduction typically occurs when the nonsense mutation is located upstream of an intron (Carter et al, 1996; Thermann et al, 1998; Zhang et al, 1998). Moreover, in some instances, the artificial insertion of an intron downstream of a normal stop codon can make it behave like a PTC (Carter et al, 1996; Thermann et al, 1998); similar observations were also reported in plants (Kerenyi et al, 2008). It is proposed that in these systems, the exon junction complex (EJC)—a multiprotein complex that is deposited on the mRNA during splicing in the nucleus and remains associated with the mRNA during export to the cytoplasm—mediates the link between splicing and NMD (Le Hir et al, 2001; Le Hir and Seraphin, 2008). The current model is that upon translation termination, a downstream EJC promotes recruitment and activation of UPF1 on the mRNA, initiating NMD (Kashima et al, 2006; Chamieh et al, 2008). Stop codons located in the last exon do not induce NMD, probably because the translating ribosome displaces the EJC before reaching the stop codon. In addition to its function in NMD, the EJC is believed to be the important explanation for why pre-mRNA processing impinges on later events such as subcellular mRNA localization and translation (Tange et al, 2004). However, the mechanism of mammalian NMD remains poorly understood; in fact, recent studies have reported examples of EJC-independent NMD and have questioned that splicing and the EJC have an important function in NMD (Buhler et al, 2006; Eberle et al, 2008; Singh et al, 2008).
In S. cerevisiae, ∼4% of the genes have introns and there has been no evidence of a link between PTC recognition and splicing (Goffeau et al, 1996; Amrani et al, 2006). Instead, the current view is that in this organism, the distance between PTC and the 3′ end is the important determinant (Amrani et al, 2006). This mechanism is conceptualized by the so-called faux (false) 3′-UTR model, which says that premature termination is intrinsically abnormal because it takes place faraway from the normal 3′ UTR, and this prevents the normal interaction between cytoplasmic poly(A)-binding protein (PABPC) and the terminating ribosome. Instead, it allows the interaction with NMD factors that shunt the mRNA for destruction (Amrani et al, 2004, 2006). The faux 3′-UTR model might explain the observation that often PTCs trigger strong NMD only when located early in the coding region—this polarity effect of NMD has been previously observed in many organisms, including S. cerevisiae, Schizosaccharomyces pombe, C. elegans and Drosophila melanogaster and Drosophila S2 cells (Losson and Lacroute, 1979; Peltz et al, 1993; Brogna, 1999; Mendell et al, 2000; Behm-Ansmant et al, 2007; Longman et al, 2007). The faux 3′-UTR model might explain a number of NMD features in mammalian and Drosophila cells as well (Behm-Ansmant et al, 2007; Eberle et al, 2008; Ivanov et al, 2008; Muhlemann et al, 2008; Silva et al, 2008; Singh et al, 2008). However, contrary to the prediction of the faux 3′-UTR model, recent studies have reported that neither the poly(A) tail nor PABPC are required for NMD in S. cerevisiae (Meaux et al, 2008). We have argued that neither the faux 3′-UTR nor the EJC model explains all the available data satisfactorily in any organism (Brogna and Wen, 2009).
Here, we have investigated the connection between NMD and splicing in S. pombe, in which some 43% of genes have introns (Wood et al, 2002). We found that introns strongly enhance NMD, but contrary to the prediction of the EJC model, an intron enhanced NMD that was positioned either before or after the PTC and deletion of orthologs of EJC components did not affect NMD. We found that it is the proximity of an intron to the PTC that triggers the mRNA reduction, indicating that the effect of pre-mRNA splicing on NMD is direct, not the secondary consequence of splicing enhancing mRNA translation. In the absence of introns, only PTCs located at the beginning of the gene caused NMD; however, against the faux 3′-UTR model prediction, we found only a modest correlation with the distance of the PTC from the 3′ end.
Results
NMD depends on the position of the PTC along the coding region
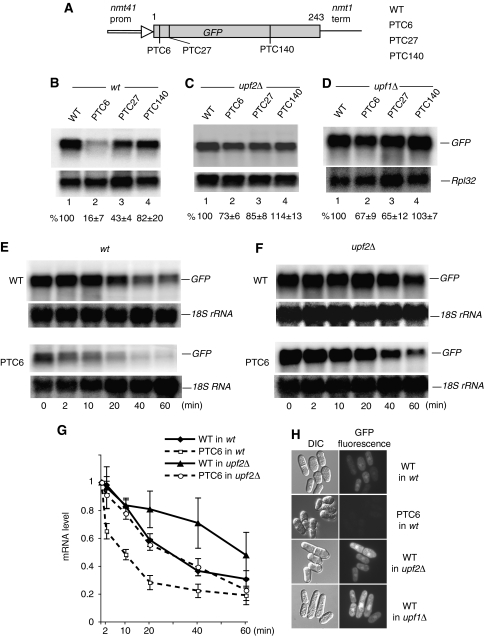
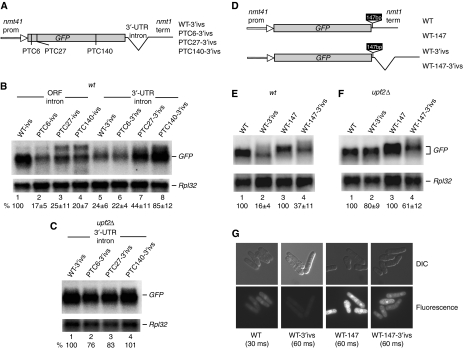
To investigate NMD, we generated GFP reporters with or without a PTC, under the control of the promoter (nmt41) and terminator of the nmt1 gene of S. pombe (Maundrell, 1993). Three nonsense mutations were introduced in the open reading frame (ORF) at codon positions 6, 27 and 140 (Figure 1A). The plasmid constructs were transformed in a wild-type strain of S. pombe (Supplementary Table 1) and RNA levels were analysed by Northern blotting. A reduced amount of mRNA was apparent with the PTC6 and PTC27 reporters; however, the reporter with PTC140 showed minimal reduction (Figure 1B). The mRNA levels were restored in upf2Δ and upf1Δ strains, suggesting that the reduction is caused by canonical NMD (Figure 1C and D).
Figure 1.
NMD depends on PTC position. (A) Diagram of the GFP reporters. Nonsense mutations (TAA) were introduced at codon positions 6, 27 and 140. (B–D) Northern blotting analysis of total RNA from wild-type (B), upf2Δ (C) and upf1Δ (D) cells, transformed with the plasmids indicated. Top panels show hybridization with a GFP-specific probe; the bottom panel shows hybridization with a probe specific for the large ribosomal protein 32 (Rpl32) mRNA, as a loading control. The values below each lane are percentages with s.d., of the GFP mRNA present in the PTC-less control, lane 1. Bands intensities were quantified with a phophorimager and standardized to the RpL32 signal (see Materials and methods). Calculations are based on three independent experiments. (E–G) Stability of the GFP mRNA in wild-type (E) and upf2Δ (F) cells following transcription inhibition with 1,10-phenanthroline (see Materials and methods). The PTC-less control (top panel) and PTC6 mRNA (bottom panels) were independently monitored and (G) quantified by Northern blotting as above, except that values were standardized to the 18S rRNA signal (this RNA does not show visible decay over the time period of the experiment). Total RNA was extracted at six different time points after adding the drug: 0, 2, 10, 20, 40 and 60 min. Error bars are s.d. from three different experiments. (H) Micrographs of cells transformed with the indicated reporters, showing GFP fluorescence. The fluorescent images were acquired with the same microscope and camera settings.
The presence of the mutations probably lowered mRNA stability. We assessed this for the PTC6 mRNA. Transcript levels were assayed at regular time intervals after adding 1,10-phenanthroline to inhibit transcription. This drug was used previously to inhibit transcription and to measure genome-wide mRNA stability in S. cerevisiae and S. pombe—the effects of 1,10-phenanthroline were similar to using rpb1-1, a temperature-sensitive allele of RNA polymerase II (Rodriguez-Gabriel et al, 2003; Grigull et al, 2004; Lackner et al, 2007). We found that the PTC6 mRNA decays faster; however, the decay rate is not gradual like that of the PTC-less mRNA: it is initially faster but then becomes slower, comparable to that of the control (Figure 1E and G). In upf2Δ cells, the PTC6 mRNA was stabilized, the decay kinetic was similar to that of the PTC-less mRNA in the wild-type strain (Figure 1G). Surprisingly, deletion of upf2 seems to stabilize the PTC-less mRNA as well (Figure 1F and G). Therefore, deletion of upf2 might also have a slight non-NMD-specific mRNA stabilization phenotype; this is not a feature peculiar to this mRNA reporter or organism: examples of PTC-less mRNAs that increase in level in NMD mutants have previously been reported in other well-characterized NMD systems (see Discussion). Moreover, the PTC-less reporter produced more GFP in upf1Δ and upf2Δ cells than in wild type (Figure 1H); the increase is reminiscent of the phenotype of loss-of-function mutations (photoshop mutations) in NMD factors of D. melanogaster, which increase expression of GFP and other transgenes with no apparent NMD features (Metzstein and Krasnow, 2006).
In summary, in this system, NMD depends on the position of the PTC along the coding region and requires the core NMD components UPF1 and UPF2—the system displays NMD features seen in other organisms.
Splicing enhances NMD by an EJC-independent mechanism
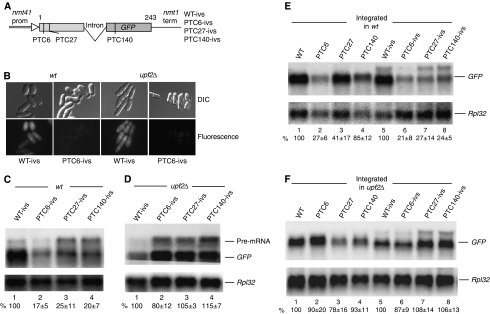
To examine whether splicing affects NMD, we generated additional reporters carrying an intron derived from the endogenous gene ubc4; the intron was inserted within codon 110, approximately in the middle of the GFP-coding region (Figure 2A). The intron is correctly spliced: an mRNA of the correct size is produced and GFP fluorescence is readily detected in the PTC-less control (Figure 2B); splicing fidelity was also verified by sequencing RT–PCR-amplified cDNA clones of the coding region (data not shown). We found that the mRNA level was drastically reduced in all PTC-containing reporters (Figure 2C). In particular, PTC140 caused a strong mRNA reduction, whereas the same mutation in the intron-less reporter caused only a minor decrease (Figure 2C versus 1B). The level of the PTC27 mRNA was also lower than that seen with the intron-less reporters (Figure 2C versus 1B). In summary, all PTC mutations resulted in a strong mRNA reduction in the intron-containing reporters, regardless of the position along the coding region; the presence of the intron abolished the ‘polar' NMD phenotype seen with the intron-less constructs. Similarly to the intron-less reporters, mRNA levels were restored in the upf2Δ strain (Figure 2D). Notably, the level of unspliced pre-RNA was also increased in the upf2Δ strain, consistent with the view that NMD normally degrades transcripts that fail to be spliced (Figure 2D). Some pre-mRNA accumulation is also apparent in wild-type cells; probably because the intron is not very efficiently spliced. Notably, in wild-type cells, the level PTC6 pre-mRNA (Figure 2C, lane 2) is much lower than that with PTC27 or PTC140 (lanes 3–4) or that of the control that carries only intron-encoded PTCs (lane 1); this behavior suggests that pre-mRNAs that fail to be spliced are rapidly degraded if they carry an early PTC, because like PTC6 they are also subjected to splicing-independent NMD, similar to mRNAs that have not undergone splicing. In several Northern blots, we found more pre-mRNA accumulation with the PTC27 and PTC140 reporters than with the PTC-less control (for example in Figure 2C and D), its significance remains to be investigated.
Figure 2.
An intron can enhance NMD. (A) Diagram of the intron-containing GFP reporters. The intron is placed in the middle of codon 110. (B) GFP fluorescence of cells transformed with the indicated intron-containing reporters. No GFP fluorescence was observed in cells transformed with PTC6, PTC27 and PTC140 reporters (data not shown for PTC27 and PTC140). (C, D) Northern blotting analysis of total RNA from wild-type (C) and upf2Δ (D) cells transformed with the indicated reporters. The top panel shows hybridization with the GFP probe (the slower migrating band corresponds to unspliced pre-mRNA); the bottom panel shows hybridization with the Rpl32 probe. Values are standardized percentages of the GFP signal in the PTC-less control, lane 1. Quantification as in Figure 1; based on three independent experiments. (E, F) Northern blot analysis of total RNA of wild-type (E) and upf2Δ (F) cells carrying an integrated copy of the indicated reporters.
To assess whether the effect of splicing on NMD depends on whether the reporters are expressed from plasmids or from single copy chromosomal genes, we integrated the constructs at the leu1 locus in both wild-type and upf2Δ strains. As for plasmid expression, we found that NMD was strongly enhanced in the intron-containing reporters (Figure 2E); and mRNA levels were restored in the upf2Δ strain (Figure 2F). The integrated reporters produce about 10-fold less mRNA than plasmid expression; thus NMD efficiency does not seem to depend on substrate level—the NMD capacity of the cell probably is not affected by high substrate levels; in agreement with this view, we found that NMD is efficient also when a reporter is expressed with a promoter that yields seven to eight times more mRNA than the plasmid reporters we have used throughout this study (see Supplementary Figure 2).
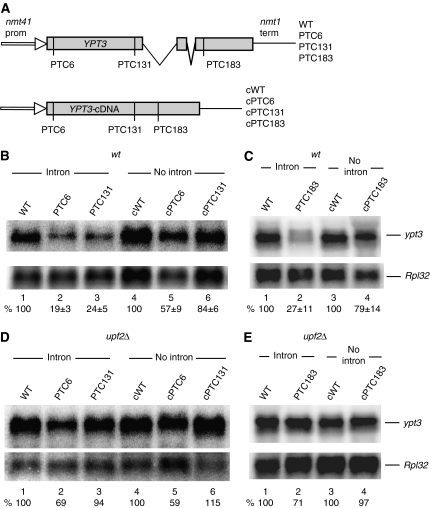
Next, we tested whether splicing affects NMD in the endogenous gene ypt3 (SPAC18G6.03). This evolutionary conserved gene contains two introns in the middle of the coding region. Three nonsense mutations were introduced at either codon 6, 131 or 183 (Figure 3A). We found that all mutations reduced the mRNA level (Figure 3B and C) and the reduction was clearly suppressed in the upf2Δ strain (Figure 3D and E). To test whether the introns are required for the mRNA reduction, we generated similar constructs with a cDNA version of the gene (Figure 3A, bottom panel). We found that such PTC-containing mRNAs were only modestly reduced; the PTC6 mRNA was reduced to about 50% of the PTC-less control, but the PTC131 and PTC183 mRNAs were reduced by only 16 and 21%, respectively (Figure 3B, lanes 1–3 versus 4–6; and Figure 3C, lanes 1–2 versus 3–4). In the upf2Δ strain, the levels of PTC131 and PTC183 mRNAs were increased, but that of PTC6 was not affected. The data further indicate that mRNAs derived from intron-less reporters are not subjected to NMD as much as spliced mRNAs.
Figure 3.
Introns are necessary for strong NMD in the ypt3 gene. (A) Diagram of the reporters expressing either the wild-type endogenous ypt3 gene or cDNA derivatives lacking both introns. PTCs are at codon positions 6, 131 and 183. (B–E) Northern blotting analysis of total RNA from wild-type (B, C) and upf2Δ (D, E) cells transformed with the indicated reporters. Top panels show hybridization with an ypt3-specific probe; bottom panels with the RpL32 probe. Values are standardized percentages of the ypt3 mRNA signal in the PTC-less control, lane 1; s.d. are from three independent experiments.
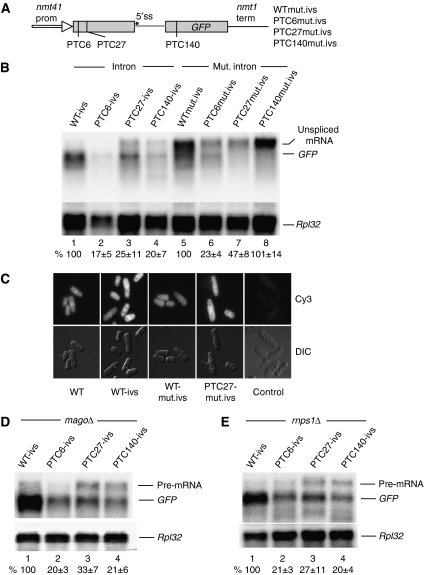
To further examine the function of splicing in NMD, we mutated the 5′ splicing site (ss) of the intron from GUAG to AUAA in our GFP reporters (Figure 4A). The mutation abolished splicing almost completely, resulting in the accumulation of a larger transcript of the expected size (Figure 4B, lanes 5–8)—four putative cryptic 5′ ss located very close to the 5′ end probably account for the weak band migrating as the spliced GFP mRNA. The relative levels of the unspliced transcripts were similar to those of the intron-less reporters: the PTC6 and PTC27 mutations caused modest NMD, whereas PTC140 did not seem to affect the transcript level (Figure 4B). The retention of the intron gives rise to additional intron-encoded PTCs (the first of which is embedded in the mutated 5′ ss); therefore, in the PTC140 reporter, the earliest PTC is actually at position 111. It is feasible that the unspliced transcripts escape NMD because they are retained in the nucleus (Legrain and Rosbash, 1989). To investigate the issue further, we assayed the subcellular distribution of the RNAs by fluorescent in situ hybridization (FISH). We found that the unspliced RNA with the mutated 5′ ss accumulates throughout the cell, similar to the fully spliced mRNA (Figure 4C).
Figure 4.
Intron-dependent NMD requires splicing but not the EJC. (A) Diagram of GFP reporters carrying mutation in the intron 5′ splice site (5′ ss). (B) Northern blotting analysis of total RNA from wild-type cells transformed with the indicated reporters. Top panel shows hybridization with the GFP probe; bottom panel shows hybridization with the Rpl32 probe. Values are percentages of the standardized intensity of the GFP mRNA in lane 1 (lanes 1–4) or of the unspliced transcript in lane 5 (lanes 5–8). Calculations are based on three independent experiments. (C) FISH analysis showing the subcellular distribution of the GFP transcript in cells expressing different NMD reporters. Cells were transformed with either the original intron-less reporter (WT), or with one carrying the wild-type intron (WT-ivs), or the mutated intron (WTmut.ivs), or with the mutated intron and a PTC at position 27 (PTC27mut.ivs). The panels show micrographs of cells viewed with an epifluorescence microscope; top panels show Cy3 fluorescence, bottom panels DIC images of the same field of view. The transcripts seem to be distributed throughout the cell in all strains. The right column shows cells not expressing GFP, which show no FISH signal. (D, E) Northern blotting analysis of total RNA extracted from Mago or RNPS1 deletion strains transformed with the indicated reporters. The top panel shows hybridizations with the GFP probe, the bottom panels with the Rpl32 probe. Values are percentages of the intensity of the GFP mRNA in the PTC-less control in lane 1; s.d. are from three independent experiments.
Given that the PTC140 mutation is located downstream of the intron, it is probably not the EJC that enhances NMD. We are not aware of any experimental evidence that suggests that splicing deposits an EJC in S. pombe. However, the genome of this organism contains orthologs of some EJC components: we identified the core proteins MAGO, eIF4AIII and RNPS1, whereas the putative Y14 homolog lacks the well-conserved N-terminal domain found in other organisms and MLN51 is missing altogether (Supplementary Figure 1; Fribourg et al, 2003). RNAi depletion or mutations of EJC components stabilize PTC-containing mRNAs in mammalian cells (Lykke-Andersen et al, 2001; Gehring et al, 2003, 2005; Shibuya et al, 2004). However, we analysed MAGO and RNPS1 deletion mutants and found that neither affected NMD: the levels of the PTC-containing mRNAs were reduced as in the wild-type strain (Figure 4D and E). We fail to knockout eIF4AIII; the gene is probably essential like in other organisms (Palacios et al, 2004).
The distance between PTC and intron determines NMD efficiency
As mentioned in Introduction, it has been reported that the insertion of an intron in the 3′ UTR can make the normal stop codon behave like a PTC, causing a strong mRNA reduction, which requires NMD factors. To assess whether this applies to S. pombe, we inserted an intron just after the normal GFP stop codon (Figure 5A). This caused strong mRNA reduction in the PTC-less reporter with just the GFP stop codon; but surprisingly, no further reduction in the reporters with PTC27 and PTC140, compared with the corresponding intro-less reporters (Figure 5B versus Figure 1B). The mRNA levels were restored in the upf2Δ strain (Figure 5C). We reasoned that the PTC27 and PTC140 transcripts were not affected probably because the mutations are too far from the intron—the PTC27 transcript is more affected than PTC140 most likely because it is more sensitive to splicing-independent NMD, like we have seen for mRNAs that have not undergone splicing (Figure 1B). On the basis of these results, we reasoned that splicing could perhaps affect NMD only when termination occurs close to the splice site. To test this possibility further, we inserted a spacer between stop codon and intron in a reporter with a 3′-UTR intron (Figure 5D, this reporter is slightly different from that shown in 5A, see figure legend). We found that the 3′-UTR intron causes strong NMD when close to the stop codon, as before (Figure 5E and F); instead, in the reporter with the insert we found that the mRNA level is partially restored (5E, lane 2 versus 4)—in this experiment, the mRNA levels were also estimated by real-time RT–PCR, which confirmed that the mRNA with the spacer is about 2.5-fold more abundant (see Supplementary Figure 3). In agreement with the mRNA quantifications, the reporter with the spacer yields more GFP (Figure 5G, compare WT-3′ivs and WT-147-3ivs), suggesting that lengthening the distance between stop codon and intron not only suppresses NMD, but it also does so without impairing steady-state translation efficiency.
Figure 5.
A 3′-UTR intron induces stronger NMD when it is closer to the stop codon. (A) Diagram of the reporters with a 3′-UTR intron. The intron is located 9 nt after the GFP ORF (see Materials and methods). Three of the constructs (PTC6-3′ivs, PTC27-3′ivs and PTC140-3′ivs) carry a PTC mutation at codon positions 6, 27 and 140, respectively; WT-3′ivs carries no PTC mutations. (B, C) Northern blot analysis of total RNA extracted from wild-type (B) and upf2Δ (C) strains transformed with the reporters with a 3′-UTR intron. The top panel shows hybridization with the GFP probe, the bottom panel with the Rpl32 probe. The values in (B) are percentages of the GFP mRNA signal of the PTC-less control; s.d. are from three independent experiments. (D) Diagram of the reporters with a 147 nt insert in the 3′ UTR; the bottom panel shows a reporter with the intron cloned immediately after the insert (see Materials and methods). WT and WT-3′ivs are similar to the reporters described in Figures 1A and 5A, but contain a 6 nt insertion after the stop codon (an AvrII site used for cloning). (E, F) Northern blotting analysis of total RNA extracted from wild-type (E) and upf2Δ cells (F) transformed with the reporters indicated. The top panel shows hybridization with the GFP probe, bottom panel with the Rpl32 probe. Values are percentages with s.d. (from three experiments) of the intensity of the corresponding PTC-less GFP mRNA control, lanes 1 and 3. (G) Visualization of GFP fluorescence in cells transformed with the indicated reporters. Cells transformed with the WT-147-3′ivs reporter show more GFP fluorescence than those transformed with WT-3′ivs. Fluorescence images were taken with the same camera setting and exposure time (60 ms), except for the GFP control, which was taken with a 30 ms exposure.
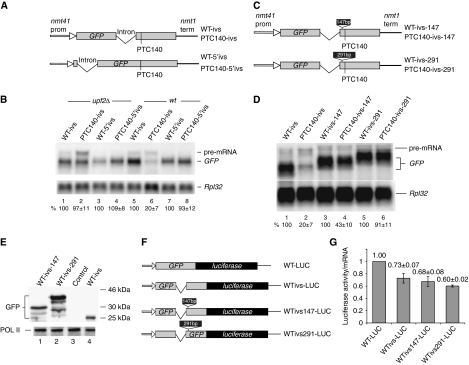
To further assess the importance of the distance between intron and PTC, in another construct, we have moved the intron to the very beginning of the GFP-coding region (Figure 6A, bottom diagram). We found that at this position (417 nt from the PTC) the intron does not enhance NMD of the PTC140 mRNA—yet the intron is spliced very efficiently, we see no pre-mRNA accumulation (Figure 6B, lane 6 versus 8). In addition, we tested whether artificial lengthening of the distance between the intron and the PTC prevents NMD in the initial reporter with the intron before the PTC (PTC140-ivs). We inserted either a 147-bp or a 291-bp fragment between the intron and PTC140 (Figure 6C). We expected the 147-bp insertion to partially prevent NMD and the 291-bp to restore the mRNA level to that of the PTC-less control—the 291-bp insertion places PTC140 at a similar distance from the intron as in the reporter with a 3′-UTR intron (PTC140-3′ivs), which was not subjected to NMD. We found that both insertions suppress NMD; in particular, the 147-bp insertion restored the mRNA level about two-fold, whereas the 291-bp insertion recovered the mRNA level almost to that of the PTC-less control (Figure 6D). The insertion of these spacers in the PTC-less reporter did not noticeably change the mRNA level, arguing against the inserts per se having major effects on transcription or processing. Moreover, the mRNAs protein yields were either comparable or increased, suggesting that the insertions also do not inhibit translation (Figure 6E). Thus, NMD reduction is probably not due to less efficient steady-state translation of the mRNAs with the insertions. To further test this issue, we have made some more reporters in which a GFP construct is joined at the 3′ end with the coding region of firefly luciferase (Figure 6F); these reporters allowed us to compare translation yields more accurately. In agreement with the western blotting result, we found that the reporters with the inserts (WTivs147-LUC and WTivs291-LUC) are not translated less efficiently than the reporters without (Figure 6G). Thus, the insertions do not suppress NMD indirectly, by inhibiting steady-state translation for example. Surprisingly, we also found that the presence of an intron in the middle of GFP appreciably reduced translation efficiency rather than enhancing it (Figure 6G). Therefore, we conclude that the effects of splicing on NMD are probably direct despite being EJC independent and that splicing enhances NMD more when the intron is close to the PTC.
Figure 6.
NMD depends on the distance between PTC and intron. (A) Diagram of two reporters (with or without PTC140) carrying an intron immediately after start codon of GFP. The top panel shows the reporter with the intron in the middle of codon 110 (similar to that in Figure 1A, but for the presence of a 6 nt insertion encoding an AvrII site after the start codon, which was used for cloning). The intron is the same in the two constructs (see Materials and methods). (B) Northern blot analysis of total RNA extracted from wild-type and upf2Δ strains transformed with the reporters indicated in (A). The top panel shows hybridization with the GFP probe, the bottom panel with the Rpl32 probe. The values are percentages of the GFP mRNA signal of the PTC-less control (lanes 1, 3, 5 and 7); s.d. are from three independent experiments. (C) Diagram of the reporters in which the distance between the intron and the PTC was expanded by placing a 147- or 291-bp insert between PTC140 and the intron in the WT-ivs construct (see panel A); both inserts maintain the GFP ORF intact. The 147 or 291 bp inserts are after codon 120 (see Materials and methods). (D) Northern blotting analysis of total RNA extracted from wild-type cells transformed with the reporters with indicated reporters. The top panel shows hybridization with the GFP probe, bottom panel with the Rpl32 probe. Values are percentages with s.d. (from three experiments) of the intensity of the corresponding PTC-less control (lanes 1, 3 and 5). (E) Western blotting analysis of whole-cell protein extracts of cells transformed with the indicated reporters. The top panel shows a blot probed with anti-GFP, bottom panel shows the blot re-probed with anti-Pol II, as loading control. (F) Diagram of four GFP reporters fused with firefly luciferase at the C terminal. WT-LUC contains the wild-type GFP-coding region; WTivs-LUC carry GFP with the same intron as WT-ivs (see Figure 2A). WTivs147-LUC and WTivs291-LUC are derivatives of the constructs shown in (C). (G) Luciferase activities measured in cells expressing the reporters described in (F). Light units were normalized by mRNA levels, which were quantified with a phosphorimager following Northern blotting (data not shown). Values are averages of three measurements.
The distance of the PTC from the 3′ end and the nature of the 3′ UTR do not greatly affect NMD
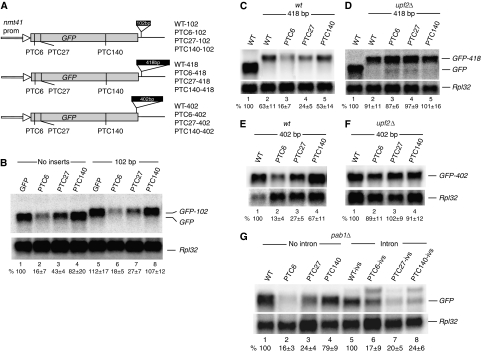
The faux 3′-UTR model predicts that NMD should increase with the distance of the PTC from the 3′ end. Our observation that NMD is less apparent when the PTC is located in the second half of the gene seems to be, at least in intron-less reporters, consistent with the model. To test it directly, first we inserted either a 102 or 418 bp fragment just downstream of the stop codon (Figure 7A). The expectation was that both insertions would enhance NMD because termination occurs further away from the 3′ end. Lengthening the 3′ UTR by 102 bases should make PTC27 behave like PTC6, enhancing NMD, whereas lengthening the 3′ UTR by 418 bases would trigger NMD of the PTC140 transcript; the insertion places the mutation at the same distance from the 3′ end as PTC6 in the original reporter. We found that the 102-base insertion reproducibly enhanced NMD of the mRNA with PTC27 (Figure 7B, lane 3 versus 7), but did not affect that with PTC6 (lane 2 versus 6). Surprisingly, the level of the PTC140 mRNA was constantly higher than that with the original 3′ UTR rather than being reduced (Figure 7B, lane 4 versus 8). In the reporters with the 418-base insertion, we found that the mRNA without PTCs was reduced to about 63% of that with the normal 3′ UTR (Figure 7C); the mRNA level was restored in the upf2Δ strain (Figure 7D), consistent with the view that mRNAs with artificially long 3′ UTR become NMD substrates. However, we found no linear relationship between mRNA levels and distance of the PTC from the 3′ end. For example, it was expected that the 418-bp insertion would make PTC140 behave like PTC6 in the initial reporter, triggering strong NMD. Instead, we found that the PTC140 mRNA level was only half of that of the reporter with the original 3′ UTR (Figure 7C, lane 1 versus 5), and it was barely reduced relative to the PTC-less control (Figure 7C, lane 2 versus 5). To further assess to what extent the distance from the 3′ end determines NMD, we inserted an in-frame 402 bp fragment just before the normal stop codon (Figure 7A)—we reasoned that an insertion in the coding region is less likely to influence mRNA stability features that might be present in the original 3′ UTR. We found that PTCs at positions 6 and 27 caused as strong NMD as in the earlier reporters. However, the PTC140 mRNA was only modestly reduced, even though the PTC is at the same distance from the 3′ end as the NMD inducing PTC6 was in the constructs with the original 3′ UTR (Figure 7E and F).
Figure 7.
Lengthening the 3′ UTR and lack of PABPC only modestly affect NMD. (A) Diagram of reporters with different length 3′ UTRs. The WT-102 reporter carries a 102 bp insert 9 bp after the normal stop codon, WT-418, a 418 bp at the same position. WT-402 contains a 402 bp insert 6 bp upstream of the normal stop codon, in frame with the GFP ORF. (B–F) Northern blot analysis of total RNA from wild-type (B, C, E) and upf2Δ (D, F) cells transformed with the indicated reporters. The top panels show hybridizations with the GFP probe, bottom panels with the Rpl32 probe. Values are percentages with s.d. based on three independent experiments. (G) Northern blot analysis of total RNA from pab1Δ cells transformed with the indicated reporters. The top panel shows hybridization with the GFP probe, bottom panel with the Rpl32 probe. Quantifications of the GFP mRNA are as above, based on two independent experiments.
We also investigated whether the sequence of the 3′ UTR affects NMD. We generated reporter constructs in which the nmt41 terminator sequencer was replaced by that of either the ade6 or adh gene. The ade6 3′ UTR was selected because it has been previously reported that nonsense mutations in the ade6 gene cause strong NMD, indicating that the 3′ UTR of this gene is NMD permissive (Mendell et al, 2000). The other 3′ UTR was selected because the adh gene is highly expressed. We found the levels of the PTC6 and PTC27 mRNA were reduced, but the PTC140 mRNA was not: the same pattern seen with the reporters carrying the nmt1 3′ UTR (Figure 2E and F, and data not shown).
We also assessed whether NMD is affected by PABPC, as predicted by the original faux 3′-UTR model (Amrani et al, 2004). We found that PTCs induced mRNA reduction in the pab1Δ strain as they did in the wild-type strain (Figure 7G). pab1 is the only PABC homolog in the genome of S. pombe but is not essential.
We conclude that there is not a clear correlation between NMD and distance of the PTC from the mRNA 3′ end, which could be predictive of how much premature translation termination will affect the mRNA level. We also conclude that PABPC is not necessary for NMD in fission yeast.
Discussion
It has been reported that nonsense mutations located at the beginning of the S. pombe ADE6 gene induce NMD even though the gene does not have introns (Mendell et al, 2000). Here, we have also found that PTCs can induce strong NMD in intron-less reporter genes, but only when located at the beginning of the coding region, whereas PTCs in the second half of the coding region do not induce strong NMD. We discovered that in the presence of a nearby intron, PTCs also cause strong NMD when located in the second half of the gene. In addition, inserting an intron in the 3′ UTR made the normal stop codon behave like a PTC and caused a strong mRNA reduction, as previously reported in mammalian systems (Carter et al, 1996; Thermann et al, 1998). Similarly, we found that removal of the introns from an endogenous gene drastically reduced NMD, arguing that introns are likely to have a general function in S. pombe NMD.
These results initially suggested the involvement of the EJC, as in mammalian cells. However, contrary to the prediction of the EJC model, the intron enhanced NMD when positioned either before or after the PTC. Moreover, NMD was not affected by deletion of the orthologs of the EJC components MAGO and RNPS1—of which MAGO seems to be a core component (Chamieh et al, 2008). Although we cannot exclude that eIF4AIII alone can assemble an EJC-like complex in S. pombe, this is unlikely: in S. cerevisiae there is a protein very similar to eIF4AIII yet no evidence of an EJC (Supplementary Figure 1). The absence of an MLN51 homolog also argues against the formation of a stable EJCs in S. pombe—this protein is probably necessary to stabilize the EJC on the RNA in mammalian cells (Le Hir and Andersen, 2008). In addition, as the intron also stimulated NMD when positioned before the PTC, we conclude that the EJC model cannot explain why splicing enhances NMD in fission yeast. It is difficult to explain how the ribosome could recognize the PTC without scanning through preceding exon junctions, the ribosome tunnel through which the mRNA moves is just large enough to accommodate naked single-stranded RNA, so that the mRNA is stripped off before scanning (Yusupova et al, 2001). Similar results have been reported for the β-globin gene in mammalian cells: it was observed that an upstream intron is required for NMD when downstream introns are absent (Matsuda et al, 2007). However, this mode of NMD seems to require some of the EJC proteins, RNPS1 was also dispensable (Matsuda et al, 2007).
An alternative explanation for why introns enhance NMD might be that spliced mRNAs are translated more efficiently and as a result PTCs are recognized more frequently, enhancing NMD indirectly. This view is based on several studies that have reported that splicing can enhance translation (Nott et al, 2003, 2004; Wiegand et al, 2003; Sanford et al, 2004; Ma et al, 2008; Michlewski et al, 2008). In particular, it has been reported that strengthening of splice signals enhances both translation and NMD, whereas mutations that weakened splicing have the opposite effect (Gudikote et al, 2005). It has been suggested that the EJC enhances initiation of the first pioneer round of translation directly, but the efficiency of the first round might positively affect subsequent rounds and steady-state translation (Ishigaki et al, 2001; Ma et al, 2008). Splicing might also enhance translation in S. cerevisiae and in other yeast species including S. pombe (Juneau et al, 2006; Man and Pilpel, 2007); however, we have found that splicing slightly decreases translation yield of our reporters rather than enhancing it.
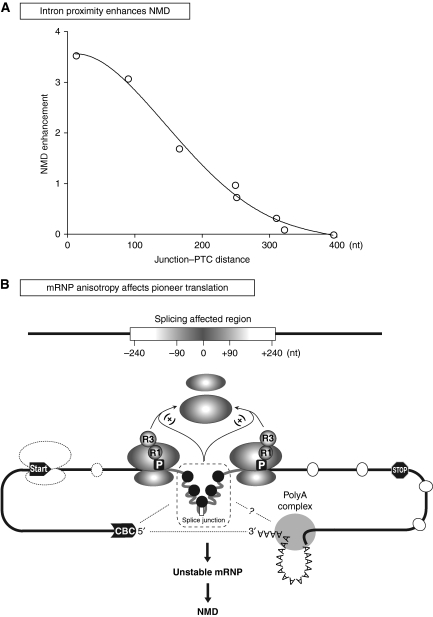
Notably, we found that splicing enhances NMD only when the PTC is not more than 240 bases away from the intron (Figure 8A). Reporters in which we have artificially increased the distance between intron and PTC are no longer subjected to NMD, yet our control experiments indicate that steady-state translation efficiency is not affected. Similarly, lengthening the distance between stop codon and a 3′-UTR intron suppresses NMD but clearly not translation yield. These data indicate that pre-mRNA splicing impinges on NMD directly, yet they exclude that an EJC-like protein complex mediates the effect. Although we still postulate that proteins that remain associated with the mRNA after splicing influence NMD, we propose that the effect is not due to a specific factor deposited at a particular distance before the splice junction (Le Hir et al, 2000, 2001). Instead, we postulate that splicing simply changes the composition or conformation of the mRNP at either side of the junction, and that the local mRNP structure influences translation termination or the release of the post-termination ribosome when the PTC is near but not when it is at a distance from the splice region (Figure 8B). The model predicts that NMD occurs because such perturbations on the ribosome destabilize the mRNP, possibly by preventing establishment of a stable translation circuit as suggested earlier (Brogna and Wen, 2009). It is also possible that there are connections between the spliced region and mRNP components outside the coding region, which could affect the local mRNP; some of the physical interactions of splicing with capping and 3′-end formation, which occur during pre-mRNA processing, may be maintained in the mature mRNP (Proudfoot et al, 2002). In particular, interactions between cap-binding complex (CBC) and splicing have been reported in S. cerevisiae (Colot et al, 1996; Fortes et al, 1999). Notably the yeast CBC can interact with the translation initiation factor eIF4G, and the CBP80 subunit of CBC has structural similarity with eIF4G (Fortes et al, 2000; Marintchev and Wagner, 2005); furthermore, in mammalian cells, NMD substrates are preferentially associated with CBC rather than the translation initiation factor eIF4E (Ishigaki et al, 2001). In summary, we propose that splicing enhances NMD because it influences the structure of the mRNP around the spliced region (the mRNP inisotropy model, Figure 8B). An important assumption of the model is that such anisotropy in mRNP composition and structure is only transient; the prediction is that splicing might only enhance NMD of newly synthesized transcripts before disassociation of the ‘processing' factors. Instead—provided that the PTC is in a region not subjected to splicing-independent NMD—the transcripts are stable afterward. The observation that steady-state mRNA is immune to splicing-dependent NMD in mammalian cells is in agreement with this model (reviewed in Maquat, 2004).
Figure 8.
The mRNP anisotropy model. (A) An intron enhances NMD only when it is close to the PTC. Plot showing the relationship between the distance of the intron from the PTC and NMD enhancement. The numbers are based on the data reported in this paper; from left to right (close to distant): WT3′ivs (Figure 5B, lane 5), PTC140ivs (Figure 2C, lane 4), PTC140ivs-147 (Figure 6D, lane 4), WT-147-3′ivs (Figure 5E, lane 4), PTC27ivs (Figure 2C, lane 3), PTC140-3′ivs (Figure 5B, lane 8), PTC140ivs-291 (Figure 6D, lane 6) and PTC140-5′ivs (Figure 6B, lane 8). The NMD level r corresponds to the fold mRNA reduction relative to the intron-less reporter. (B) Local mRNP anisotropy triggers splice-dependent NMD. Pre-mRNA splicing deposits proteins around either side of the splice junction. These proteins remain associated with the mRNA and change the mRNP local configuration, which in turn affect translation termination or the fate of the post-termination ribosome (e.g. ribosome release). The proteins remain associated with the mRNA only transiently, possibly until the first round of translation. The local mRNP configuration involves interactions of the splice region with the cap-binding complex (CBC) and other proteins associated with the 3′ end. White ovals are nonspecific mRNA-binding proteins; black circles are proteins deposited by splicing. R1, translation release factor 1; R3, translation release factor 3, P, PTC.
Our data clearly indicate that splicing enhances NMD in S. pombe. However, PTCs at the beginning of the coding region can trigger strong NMD also in genes without introns; similarly to what has been reported in other systems. Do these observations indicate that there are two separate NMD mechanisms? Given that deletion of UPF1 and UPF2 suppresses NMD in both cases, the process is probably not entirely distinct. However, the genetic analysis alone may be misleading because NMD mutants might have a nonspecific mRNA stabilization phenotype: we found that in upf1Δ and upf2Δ strains PTC-less mRNAs are also stabilized. This is not an idiosyncrasy of our experimental system, similar observations have previously been reported in Drosophila, both in cell culture and in flies (Gatfield et al, 2003; Metzstein and Krasnow, 2006); examples of over-stabilization of control PTC-less mRNAs upon knockdown of NMD factors were also reported in mammalian cells yet not explained (Figures 1 and 4; Eberle et al, 2008). Furthermore, early studies in budding yeast also reported cases of over-stabilization of wild-type mRNAs in UPF1 mutants (Table 1 in Peltz et al, 1994). In addition, microarray expression studies have clearly indicated that mRNAs with no obvious NMD features are also stabilized in NMD mutants (Rehwinkel et al, 2005). Therefore, we cannot conclude that the two mRNA reduction mechanisms are similar just because lack of UPF1 or UPF2 suppresses them both. Generally, NMD is viewed as a single well-defined biochemical process, which similar to other cellular mechanisms, should be conserved across organisms. Against this view, there are cases in which UPF2 and UPF3 are not required for NMD (Gehring et al, 2005; Chan et al, 2007). On the basis of these conflicting observations, it is plausible that the nonsense-mediated mRNA reduction, typically attributed to NMD, is in fact the compound effect of diverse mechanisms that destroy mRNPs that are not efficiently translated (Brogna and Wen, 2009).
Finally, in agreement with earlier studies, we also found cases in which lengthening of the 3′ UTR induced an UPF1/UPF2-dependent reduction of the mRNA. However, against the faux 3′-UTR model prediction, we found no clear correlation with the distance of the PTC from the 3′ end (we also found no correlation with the distance from the 5′ end, data not shown). Furthermore, we found that PABPC is not required for NMD of either spliced or unspliced mRNAs, in agreement with the report that neither the poly(A) tail nor PABPC are required for NMD in S. cerevisiae (Meaux et al, 2008). Our data argue against the notion that it is a long distance from either the 3′ or 5′ end, which primarily triggers splicing-independent NMD; instead the data suggest that some sequence elements flanking the PTC might affect NMD; this view is in agreement with earlier studies in S. cerevisiae, which concluded that downstream sequence elements located after the PTC modulate NMD (Peltz et al, 1993; Zhang et al, 1995). In conclusion, the studies presented here clearly establish that current models cannot explain NMD in S. pombe, future work shall investigate the molecular mechanisms and establish if NMD is different in this organism or whether existing models are generally not accurate.
Materials and methods
Plasmid constructs
The GFP reporters were generated by cloning the GFP-coding region into the BamH1 sites of the pREP41 plasmid vector. The GFP fragment was amplified from pRS316-GFP, which contains the original S65T GFP, accession number: BD062761 (Heim et al, 1995; Kuperwasser et al, 2004). Mutations were introduced by overlapping PCR. The intron in the WT-ivs constructs derives from the ubc4 gene, which was amplified from S. pombe genomic DNA and inserted into the PmlI site located at position 328 in the GFP-coding region, by blunt-end ligation. The WT-3′ivs constructs carry the same intron as above, cloned in the XmaI site, which is located 9-bases after the GFP stop codon. The WT-102 and WT-418 constructs were generated by inserting 102 and 418 bp DNA fragment in the Xma I site, as above. The fragments correspond to overlapping regions of the firefly luciferase gene; they were amplified using primers:
5′-AACGACCCGGGCTGAATACAAATCACA-3′ (F-luc.sense)
5′-ATATTCCCGGGCGCAACTGCAACTCC-3′ (102 bp.reverse)
5′-ACTATCCCGGGCCACACCCTTAGGTAACC-3′ (418 bp.reverse)
The WT-402 constructs were generated by inserting 402 bp DNA fragment into an AvrII site (introduced by PCR) located 6 bp before the native stop codon. The fragment was PCR amplified from a firefly luciferase clone using primers:
5′-ACTGCCCTAGGAATCACAGAATCGTCGTATG-3′ (402 bp.sense) and
5′-CCACTCCTAGGCACACCCTTAGGTAACCCA-3′ (402 bp.reverse)
The WT-ivs-147 and WT-ivs-291 constructs (and PTC-containing derivatives) were generated by inserting either a 147 or 291 bp DNA fragment in an AvrII site, which was introduced by overlapping PCR between codons 120 and 121. The fragments were PCR amplified from EGFP (pEGFP-C3, CLONTECH) using primers:
5′-CTTGAGCCTAGGTACCCATACGATGTTCCTGA-3′ (EGFP.sense),
5′-TTGGTTCCTAGGGGTCAGGGTGGTCACGAG-3′ (EGFP291 bp.reverse),
5′-TTTGTACCTAGGCTCGACCAGGATGGGCAC-3′ (EGFP147 bp.reverse).
The WT-147-3′ivs construct was generated by inserting a 147 bp fragment in an AvrII site located immediately downstream of the normal stop codon in a derivative of WT-3′ivs; the AvrII site was introduced by PCR. The GFP-luciferase fusion constructs were generated by joining the GFP and firefly luciferase sequences by overlapping PCR, followed by cloning of the fusion PCR fragments into the Bam HI site in pREP41, as above. The WT-5′ivs was generated by inserting the intron sequence into an AvrII site (introduced by PCR) located immediately downstream of ATG. All plasmids were verified by sequencing and restriction digestion.
Transformation, gene disruption and plasmid integration
Fission yeast transformations were as described earlier with minor modifications (Okazaki et al, 1990). Yeast cultures were grown to 1 OD600, washed with water and resuspended in lithium acetate buffer; 1 μg plasmids or 10 μg PCR products were mixed with 20 μg ssDNA and added to 100 μl of cells and incubated at room temperature for 30 min. Later, 260 μl of 50% PEG4000 were added and incubated for a further 60 min at 30°C, followed by heat shock at 42°C for 30 min. The RNPS1 homolog was deleted by homologous recombination using a PCR-amplified fragment containing the kanMX6 cassette, flanked targeting sequences (Bahler et al, 1998). The following primers were used:
5′-TCCTCTCAAAAGTTTAGTAATAAAATATAAAACATTCTCAATTATGCTTTTTGTAATACTcggatccccgggttaattaa-3′, and
5′-TTCAAACTGGAAATTTGTTTTCACTTTTCTTTTTTATTCAGTAGGCCACAATTTGCGGGAgaattcgagctcgtttaaac-3′—uppercase are
the RNPS1 homologous sequences, lowercase are the sequences complementary to the KanMX6 cassette. PCR products were gel purified and 5–10 μg of DNA was transformed into SPJK039. Transformants were selected by 100 μg/ml G418 and screened by yeast colony PCR and confirmed by PCR of purified genomic DNA. Plasmid integration was as described earlier using the pDUAL multicopy vector (Matsuyama et al, 2004). Briefly, the GFP reporters were cloned into the BamH1 site of pDUAL1. Plasmids constructs were linearized by SacI digestion and transformed into either the wild-type or upf2Δ strain. Recombinants were selected EMM plates lacking leucine.
RNA extraction and RNA analysis
Total RNA was extracted with hot acid phenol as described (Ausubel et al, 1996); typically from 10 ml cell cultures grown to 1 OD600. The RNA was separated on 1.2% agarose gels in the presence of formaldehyde. The RNA was transferred by overnight capillary transfer onto a nylon membrane (Hybond-N, GE Healthcare) and hybridized with random-primer 32P-labelled probes as described (Yang et al, 1993). Probes were PCR amplified from plasmid clones (GFP) or from genomic DNA (RpL32-2 and 18S rRNA). The intensity of the radioactive bands was calculated with a phosphorimager (Molecular Image-FX, Bio-Rad). For RNA half-life analysis, a 50 ml cell culture was grown until 1 OD600, treated with 150 μg/ml 1,10-phenanthroline (t0), and then 7 ml aliquots were transferred into a tube with about 1 volume of crushed ice after 0, 2, 10, 20, 40 and 60 min. RNA was extracted with acid phenol as described above.
FISH in fission yeast
FISH was performed as described earlier for S. cerevisae with minor modifications (Kuperwasser et al, 2004). Briefly, cells were harvested from 5 ml cultures, OD600∼0.8, fixed in 4% formaldehyde and spread on polylysine treated slides. Novazyme (50 μg/ml, Interspex Products Inc) and zymolyase 20T (100 μg/ml, Europa Bioproducts Ltd) were used to make spheroplasts by incubating the cells for 30 min at 37°C in 0.1 M potassium phosphate, pH 6.5). The GFP probes consisted of four amino-modified oligonucleotides, as described earlier (Dower and Rosbash, 2002). The oligos were a gift from Torben Heick Jensen (University of Aarhus) and were labelled with Cy3 using the Cy 3-mono-Reactive Dye Pack (GE Healthcare). Slides were viewed with a Nikon ECLIPSE E600 epifluorescence microscope. Pictures were taken with a CCD camera (C4742-95, Hamamatsu).
Protein extraction, western blotting and luciferase assay
Proteins were typically extracted from 10 ml cultures, 1–2 OD600; cell number was equalized following counting with a hemocytometer. Cells were spanned down and pellets resuspended gently in 450 μl of sterile ddH2O, 50 μl of 3.5 N NaOH and incubated for 5 min at room temperature. Cells were centrifuged at 3500 r.p.m. for 2 min and resuspended in 100 μl and loaded into 10% SDS–PAGE. Following SDS–PAGE, proteins were transferred to a nitrocellulose membrane, blocked and incubated with a goat polyclone anti-GFP (AbD Serotec). The secondary antibody was a rabbit HRP-conjugated anti-goat IgG, which was detected with chemiluminescent HRP substrate (Immobilon Western, Millipore). The chemiluminescent signal was analysed with Quantity One (Bio-rad). The monoclonal 8WG16 (Covance) was used to detect the RNA polymerase II. Luciferase expression was measured with the Steady-Glo Luciferase Assay System (Promega) directly from an aliquot of cell culture: 10 μl of culture (in EMM) were mixed with 15 μl of Passive Lysis Buffer (Promega) and 25 μl of Steady-Glo substrate solution (Promega) in a 96-well microtiter plate; luminescence was measured with a plate reader (Tecan).
Supplementary Material
Acknowledgments
We thank Stephen Dove for help and support. We also thank Jiannan Guo for technical help with real-time PCR. In addition, we thank Dr Jurg Bahler, Dr John Davey, Dr Anabelle Decottignies, Dr Ravi Dhar, Dr Harry Dietz, Dr Iain Hagan, Dr Torben Heick Jensen, Dr Akihisa Matsuyama, Dr Michael Rosbash, Dr Paul Russell and Dr Gerald Smith for providing reagents. This work was supported by a Royal Society URF fellowship and a Wellcome Trust project grant to SB and a Darwin Trust of Edinburgh scholarship to JW. We make the following declaration: SB and JW have conceived and designed the experiments, and wrote the paper. JW performed all the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ (2008) Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14: 914–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, Jacobson A (2006) Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7: 415–425 [DOI] [PubMed] [Google Scholar]
- Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, K S (1996) Current Protocols in Molecular Biology. New York: John Wiley & Sons [Google Scholar]
- Azzalin CM, Lingner J (2006) The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16: 433–439 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast (Chichester, England) 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E (2007) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S (1999) Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA 5: 562–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S, Wen J (2009) Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol 16: 107–113 [DOI] [PubMed] [Google Scholar]
- Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT (2004) The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell 14: 585–598 [DOI] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O (2006) EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat Struct Mol Biol 13: 462–464 [DOI] [PubMed] [Google Scholar]
- Carter MS, Li SL, Wilkinson MF (1996) A splicing dependent regulatory mechanism that detects translation signals. EMBO J 15: 5965–5975 [PMC free article] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H (2008) NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93 [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA II, Wilkinson MF (2007) An alternative branch of the nonsense-mediated decay pathway. EMBO J 26: 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M (1996) The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20 the small subunit of the nuclear cap-binding complex. Genes Dev 10: 1699–1708 [DOI] [PubMed] [Google Scholar]
- Culbertson MR, Underbrink KM, Fink GR (1980) Frameshift suppression in Saccharomyces cerevisiae II. Genetic properties of group II suppressors. Genetics 95: 833–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower K, Rosbash M (2002) T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O (2008) Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Bilbao-Cortes D, Fornerod M, Rigaut G, Raymond W, Seraphin B, Mattaj IW (1999) Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev 13: 2425–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Inada T, Preiss T, Hentze MW, Mattaj IW, Sachs AB (2000) The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol Cell 6: 191–196 [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E (2003) A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol 10: 433–439 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E (2003) Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 22: 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE (2005) Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE (2003) Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949 [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 genes. Science 274: 546, 563–547 [DOI] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR (2004) Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudikote JP, Imam JS, Garcia RF, Wilkinson MF (2005) RNA splicing promotes translation and RNA surveillance. Nat Struct Mol Biol 12: 801–809 [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A (1993) Stabilization and ribosome association of unspliced pre-mRNAs in a yeast Upf1- mutant. Proc Natl Acad Sci USA 90: 7034–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY (1995) Improved green fluorescence. Nature 373: 663–664 [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P (1989) A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Bouhouche K, Gout JF, Aury JM, Noel B, Saudemont B, Nowacki M, Serrano V, Porcel BM, Segurens B, Le Mouel A, Lepere G, Schachter V, Betermier M, Cohen J, Wincker P, Sperling L, Duret L, Meyer E (2008) Translational control of intron splicing in eukaryotes. Nature 451: 359–362 [DOI] [PubMed] [Google Scholar]
- Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW (2006) Introns regulate RNA and protein abundance in yeast. Genetics 174: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D (2008) Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 27: 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser N, Brogna S, Dower K, Rosbash M (2004) Nonsense-mediated decay does not occur within the yeast nucleus. RNA 10: 1907–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Andersen GR (2008) Structural insights into the exon junction complex. Curr Opin Struct Biol 18: 112–119 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ (2001) The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev 14: 1098–1108 [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Seraphin B (2008) EJCs at the heart of translational control. Cell 133: 213–216 [DOI] [PubMed] [Google Scholar]
- Legrain P, Rosbash M (1989) Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583 [DOI] [PubMed] [Google Scholar]
- Li SL, Wilkinson MF (1998) Nonsense surveillance in lymphocytes? Immunity 8: 135–141 [DOI] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF (2007) Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 21: 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R, Lacroute F (1979) Interference of nonsense mutations with eukaryotic messanger RNA stability. Proc Natl Acad Sci USA 76: 5134–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Azzalin CM, Hug N, Deplazes A, Peter M, Lingner J (2007) Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res 35: 7688–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA (2001) Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839 [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J (2008) SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133: 303–313 [DOI] [PubMed] [Google Scholar]
- Man O, Pilpel Y (2007) Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet 39: 415–421 [DOI] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Marintchev A, Wagner G (2005) eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry 44: 12265–12272 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Hosoda N, Kim YK, Maquat LE (2007) Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol 14: 974–979 [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Shirai A, Yashiroda Y, Kamata A, Horinouchi S, Yoshida M (2004) pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast (Chichester, England) 21: 1289–1305 [DOI] [PubMed] [Google Scholar]
- Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130 [DOI] [PubMed] [Google Scholar]
- Meaux S, van Hoof A, Baker KE (2008) Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell 29: 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 10: 99–105 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol 20: 8944–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzstein MM, Krasnow MA (2006) Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet 2: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Caceres JF (2008) The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell 30: 179–189 [DOI] [PubMed] [Google Scholar]
- Muhlemann O, Eberle AB, Stalder L, Zamudio Orozco R (2008) Recognition and elimination of nonsense mRNA. Biochim Biophys Acta 1779: 538–549 [DOI] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ (2004) Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev 18: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Meislin SH, Moore MJ (2003) A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H (1990) High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res 18: 6485–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E (2004) An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757 [DOI] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A (1993) Messenger RNA destabilization triggered by premature translational termination depends on at least 3 cis-acting sequence elements and one trans-acting factor. Genes Dev 7: 1737–1754 [DOI] [PubMed] [Google Scholar]
- Peltz SW, He F, Welch E, Jacobson A (1994) Nonsense-mediated mRNA decay in yeast. Prog Nucleic Acid Res Mol Biol 47: 271–298 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E (2005) Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11: 1530–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gabriel MA, Burns G, McDonald WH, Martin V, Yates JR III, Bahler J, Russell P (2003) RNA-binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J 22: 6256–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Caceres JF (2004) A novel role for shuttling SR proteins in mRNA translation. Genes Dev 18: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF (2008) Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell 31: 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ (2004) eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol 11: 346–351 [DOI] [PubMed] [Google Scholar]
- Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L (2008) Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J (2008) A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ (2004) The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol 16: 279–284 [DOI] [PubMed] [Google Scholar]
- Thermann R, NeuYilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR (2003) Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA 100: 11327–11332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E (2009) Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol 29: 3517–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
- Yang H, McLeese J, Weisbart M, Dionne JL, Lemaire I, Aubin RA (1993) Simplified high throughput protocol for northern hybridization. Nucleic Acids Res 21: 3337–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Nishii T, Nakamura K (2006) Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol 47: 572–580 [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF (2001) The path of messenger RNA through the ribosome. Cell 106: 233–241 [DOI] [PubMed] [Google Scholar]
- Zhang J, Sun XL, Qian YM, LaDuca JP, Maquat LE (1998) At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol 18: 5272–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SA, Ruizechevarria MJ, Quan Y, Peltz SW (1995) Identification and characterization of a sequence motif involved in nonsense-mediated messenger RNA decay. Mol Cell Biol 15: 2231–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.