Abstract
Amassments of heterochromatin in somatic cells occur in close contact with the nuclear envelope (NE) but are gapped by channel- and cone-like zones that appear largely free of heterochromatin and associated with the nuclear pore complexes (NPCs). To identify proteins involved in forming such heterochromatin exclusion zones (HEZs), we used a cell culture model in which chromatin condensation induced by poliovirus (PV) infection revealed HEZs resembling those in normal tissue cells. HEZ occurrence depended on the NPC-associated protein Tpr and its large coiled coil-forming domain. RNAi-mediated loss of Tpr allowed condensing chromatin to occur all along the NE's nuclear surface, resulting in HEZs no longer being established and NPCs covered by heterochromatin. These results assign a central function to Tpr as a determinant of perinuclear organization, with a direct role in forming a morphologically distinct nuclear sub-compartment and delimiting heterochromatin distribution.
Keywords: heterochromatin, nuclear compartmentalization, nuclear pore complex, Tpr
Introduction
The interphase nucleus of eukaryotic cells is compartmentalized into distinct territories, including areas occupied by transcriptionally active euchromatin and those with highly condensed, transcriptionally more inert heterochromatin. Whereas only small amounts of heterochromatin occur in proliferating cultures of permanent cell lines, it can occupy much of the nucleus in terminally differentiated cells (e.g., Francastel et al, 2000). In transmission electron microscopy (TEM) such heterochromatin appears as dense patches deep within the nucleus and aligned along the inner surface of the nuclear envelope (NE).
However, even conspicuous amassments of condensed nuclear-peripheral chromatin are known since long to be gapped by electron-lucent zones free of heterochromatin (e.g., Swift, 1959; Watson, 1959). Such heterochromatin exclusion zones (HEZs) can differ in length and expansion, sometimes appear to segment large areas of the nuclear interior (e.g., Davies et al, 1974), and are often found associated with the nuclear pore complexes (NPCs) that serve as gateways of nucleo-cytoplasmic transport. However, even though different models exist of how nuclear compartmentalization might be accomplished (e.g., Jackson, 2003; Misteli, 2005, 2007; Cremer et al, 2006; Branco and Pombo, 2007; Hancock, 2007; Lanctôt et al, 2007; Rippe, 2007; Schneider and Grosschedl, 2007; Richter et al, 2008), the cellular factors or mechanisms that establish the perinuclear HEZs have remained unknown. While these zones are sometimes also referred to as euchromatic, chromatin-free, or interchromatin channels, or as part of a reticular inter-chromosomal compartment traversing the nucleus, the designation HEZ used in this study refers to the NPC-proximal parts of these zones.
The NPC is a macromolecular structure of eightfold rotational symmetry that perforates the NE. Fibrillar appendices have been found attached to the NPC's nuclear side that vary significantly in length in different cell types, sometimes projecting deep into the nuclear interior as in vertebrate oocytes (e.g., Franke and Scheer, 1974; Scheer et al, 1988; Ris and Malecki, 1993; Goldberg et al, 1997). There, as well as in insect salivary gland cells and in the protozoan Dictyostelium, eight NPC-attached fibrils interdigitate with each other laterally, forming a structure called the nuclear basket (NB) (Ris, 1989; Jarnik and Aebi, 1991; Kiseleva et al, 1996; Beck et al, 2004). NB-reminiscent structures have also been described in yeast (Kiseleva et al, 2004) and proposed to exist in mammalian cells (e.g., Iborra et al, 2000; Frosst et al, 2002; Krull et al, 2004). In fact, an NPC-attached ‘fish trap'-like assembly of nuclear fibrils in monkey cells was reported as early as 1976 (Maul, 1976). However, due to the delicate nature of these fibrils and difficulties in visualizing them by conventional TEM, it has remained uncertain whether these findings reflect the occurrence of an NB common to all NPCs. Moreover, the protein composition of fibrils and NBs has not yet been unambiguously determined, and different proteins are considered as their potential components (e.g., Krull et al, 2004; Prunuske et al, 2006; Brown and Silver, 2007; Köhler and Hurt, 2007).
Two of the candidate proteins formerly proposed as fibrillar appendices of the NPC are called Nup153 and Nup98 (e.g., Sukegawa and Blobel, 1993; Powers et al, 1995; Radu et al, 1995; Fahrenkrog and Aebi, 2002; Frosst et al, 2002). Both belong to a subfamily of NPC proteins (nucleoporins) that contain large, natively unfolded domains with FG-tandem repeats (Denning et al, 2003). In human HeLa cells though, both the FG-repeat and the NPC-anchor domains of Nup98 were found at the NPC proper (Griffis et al, 2003; Krull et al, 2004), indicating that they are not very likely components of fibrillar material that may be peripherally attached to the NPCs in this particular cell type. The NPC-targeting domain of Nup153 too was found located at the NPC in HeLa and in other cell types (e.g., Walther et al, 2001; Fahrenkrog et al, 2002; Krull et al, 2004), but the location of its large FG-repeat domain remains enigmatic. Found within the NPC proper in fission yeast (Balasundaram et al, 1999), the FG-repeat domain of the vertebrate orthologue might form extended and highly flexible fibrils capable of projecting into the nuclear interior (e.g., Fahrenkrog et al, 2002, 2004).
Another constituent of fibrillar NPC appendices is a 267-kDa protein called Tpr (e.g., Mitchell and Cooper, 1992; Cordes et al, 1997; Frosst et al, 2002). It forms long coiled-coil homodimers of rod-like shape via its N-terminal domain of 190 kDa, which also harbours a short segment required for anchorage to the NPC (Bangs et al, 1998; Cordes et al, 1998; Hase et al, 2001). By contrast, its C-terminal ‘tail' domain does not homodimerize but appears largely unfolded and flexible (Hase et al, 2001). In different mammalian cell types Tpr's NPC-binding segment was indeed found positioned at the NPC, whereas other parts of the rod domain and the C-terminal tail were located deeper within the nucleus (Cordes et al, 1997; Frosst et al, 2002; Krull et al, 2004; Hüve et al, 2008).
To investigate whether such a protein plays a role in HEZ establishment, one could attempt eliminating it in a cellular background in which HEZs are prevalent. However, in tissue cells with clearly contoured HEZs, target protein elimination is generally difficult to achieve. On the other hand, target protein levels can be easily knocked down by RNA interference (RNAi) in permanent cell lines (Elbashir et al, 2001). In such cells, however, condensed chromatin is usually scarce, and HEZs are imperceptible at most NPCs. Therefore, a suitable model system would need to concurrently allow rapid chromatin condensation and RNAi-mediated protein knockdown to possibly visualise HEZs and then study their fate after RNAi.
Hyper-condensation of chromatin can be induced by infecting cultured cells with picornaviruses (Bienz et al, 1973). Within a well-defined timeframe, such condensation rapidly spreads throughout the nucleus of the infected cell, whereas HEZs are not trespassed even late after infection (Belov et al, 2004; Lidsky et al, 2006). This results in the contouring of HEZs at essentially all NPCs, thereby revealing shapes similar to those in terminally differentiated somatic cells. Here, we have exploited this possibility of visualizing HEZs by infecting HeLa cells with poliovirus (PV). Using this in combination with RNAi, we found Tpr to be an element essential for HEZ establishment and for delimiting perinuclear heterochromatin distribution.
Results
NPC-associated HEZs in PV-infected cells
In cell lines such as HeLa, only small amounts of heterochromatin are aligned along the NE between neighbouring NPCs (Figure 1A). Local amassments of such heterochromatin occur only sporadically (Supplementary Figure S1A). Furthermore, the frequency of NPCs in these contact regions between heterochromatin clusters and the NE can be notably lower than that in the neighbouring areas (see also Maul, 1977; Garcia-Segura et al, 1989). Therefore, ultrathin sections of such cells only infrequently include perpendicular sections through NPCs juxtaposed to heterochromatin amassments. The few present, however, generally reveal an NPC-associated heterochromatin-free zone, often with seemingly hyperbolic contours or shaped like an isosceles triangle or trapezium with its basis next to the NPC. Similar clearance zones are also seen when the granular, pre-ribosomal material of the nucleolus is positioned directly in front of an NPC (Supplementary Figure S1A). Again though, NPCs are comparatively rare in such contact regions (Maul, 1977).
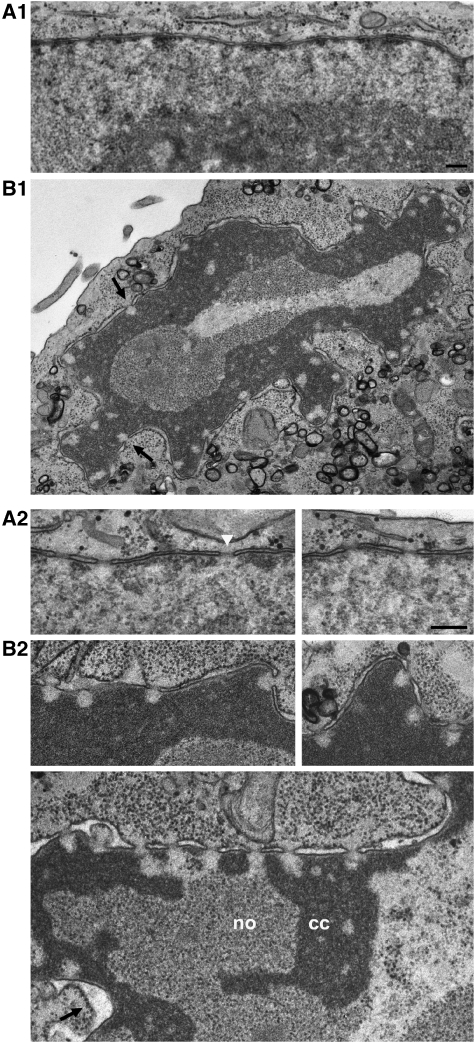
Figure 1.
NPC-associated HEZs of distinct size and shape withstand the expansion of condensed chromatin after PV infection. (A1) TEM of a perpendicular NE cross-section of a non-infected HeLa cell in mid-interphase, with a thin layer of NE-associated heterochromatin between the NPCs. (B1) A PV-infected HeLa cell (assembled from two micrographs) at 12 h post-infection, illustrating the characteristic nuclear distortion and emergence of NPC-associated HEZs (arrows). (A2, B2) Higher magnification views of NE cross-sections of non-infected HeLa cells (A2) and of cells 12 h after PV infection (B2), illustrating the contouring of NPC-associated HEZs by condensed chromatin (cc). Nucleolar materials (no) were excluded from these zones too. Chromatin hyper-condensation was also accompanied by gradual loss of the electron-dense NPC midplanes seen in non-infected cells (white arrowhead), and often by dilation of the NE lumen (arrow) late in the infection process. Bars: 200 nm; same magnification for panels A1 and B1, and panels A2 and B2.
However, PV infection of HeLa causes amassments of condensed chromatin, first at the NE and later also deep in the nucleus, without trespassing ostensive boundaries in front of most, if not all, NPCs. This happens within a few hours post-infection and results in the contouring of HEZs of different lengths and shapes, with a seemingly temporal order of predominant appearance.
Longer HEZ section areas of rectangular shape or with contours reminiscent of parabolic or hyperbolic curves as from sections of elongate cones, were more often observed in the earlier stages of chromatin condensation. Although notably varying in length, the majority of measured section heights (hy) were between 100 and 150 nm, some also reaching 150–250 nm (Supplementary Figure S1B). Very rarely, exceptionally long rectangular HEZ sections up to 460 nm in length were observed (Supplementary Figure S1B3), reminiscent of channel-like HEZs seen in other cell types (e.g., Visser et al, 2000; Rego et al, 2008). Analysis of consecutive cell sections showed that some of the long HEZs were longitudinally surrounded by condensed chromatin, indicative that they can be of cylindrical shape.
Later in the infection process, the elongate types of HEZ sections were less frequently observed and shorter HEZ sections of triangular and trapezoid-like appearance or with hyperbola-like contours were prevalent. Although also observed in the earlier stages of chromatin condensation, these shorter HEZ section shapes were strikingly predominant in cells late in PV infection, with highly distorted nucleus and amassments of condensed chromatin. Their mean size did not appear to diminish further, suggestive of a rather steady residual HEZ ‘core' region (Figure 1B and Supplementary Figure S1B).
To estimate the size of such minimal HEZs, we chose a stereometric approach, on the basis of the simplifying assumption that the underlying HEZ shape can be approximatively described as a cone-like space. To this end, we measured the 2D-shape parameters of longitudinally sectioned HEZs next to perpendicularly sectioned NPCs and normalized for non-diametric section planes. These values were used for trigonometric calculations and approximation of HEZ dimensions and shape (Figure 2 and Supplementary Figure S2).
Figure 2.
Stereometric size approximation of the NPC-associated minimal HEZ. (A) A perpendicularly but non-diametrically sectioned NPC-associated HEZ enclosed by condensed chromatin, from the late stage of PV infection. Bar: 50 nm. (B1) Schematic depiction of a non-diametrically sectioned HEZ and measuring tracks (double-headed arrows) of intra-membrane NPC channel diameter (2rn), of HEZ-section diameter at the base (2rb) and parallel to the base at 40 nm distance (2r40), and of HEZ-section height (hy). A total of 121 perpendicularly cross-sectioned HEZs from the late stages of PV infection were measured and normalized for non-diametric section planes as described in Supplementary Figure S2. (B2) Schematic depiction of the mean normalized diametric HEZ section, with a basis diameter 2Rb of 119 nm and heights hyfr and hapex of 74 and 96 nm, respectively. (C) The 3D shape of the corresponding mean HEZ, showing a cone with flattened top. Schemes are drawn in scale to an NPC with an intra-membrane channel diameter set to 84 nm; for possibly smaller channel diameters in PV-infected cells and the effect on calculated HEZ dimensions, see Supplementary Figure S2.
Even though the resulting model was likely to be slightly smaller than the authentic size (Supplementary Figure S2), it provided an estimate of the minimal dimensions of the HEZ ‘core' region, showing a cone with flattened top, with a diameter (2Rb) of 119 nm at its basis and an approximated height (hapex) of 96 nm. This was slightly smaller than the calculated means of the rare HEZs found in non-infected HeLa cells, yielding a truncated cone with a 2Rb value of 129 nm and a hapex of 107 nm (Supplementary Figure S2D). Noteworthy, the shapes and dimensions of the trapezoid-like HEZ sections from PV- and non-infected HeLa cells closely resembled those from terminally differentiated cells (e.g., Tokuyasu et al, 1968; Maul, 1977).
Maintenance of NPC-associated HEZs after PV infection correlates with the proteolytic insensitivity of Tpr's coiled-coil domain
PV infection causes silencing of host cell transcription and specific degradation of a small number of nuclear proteins (e.g., Crawford et al, 1981; Davies et al, 1991; Rubinstein et al, 1992; Shen et al, 1996; Yalamanchili et al, 1997). Two nucleoporins, Nup153 and Nup62, were found degraded in PV-infected HeLa cells too (Gustin and Sarnow, 2001). Whereas Nup62 is an FG-repeat protein of the NPC core, the structurally unfolded C-terminal half of Nup153 possibly projects into the nucleus. Its degradation, though, would indicate that it is dispensable for the HEZs seen in such cells. Moreover, NPC anchorage of the coiled-coil protein Tpr is mediated by a short segment of Nup153 located within its N-terminal half (Hase and Cordes, 2003). Complete Nup153 degradation might therefore prevent or destabilize NPC binding of Tpr and thus also exclude a role for Tpr in HEZ establishment. However, Tpr's actual fate during PV infection, and that of other FG-repeat nucleoporins, such as Nup98, was unknown at the beginning of this work.
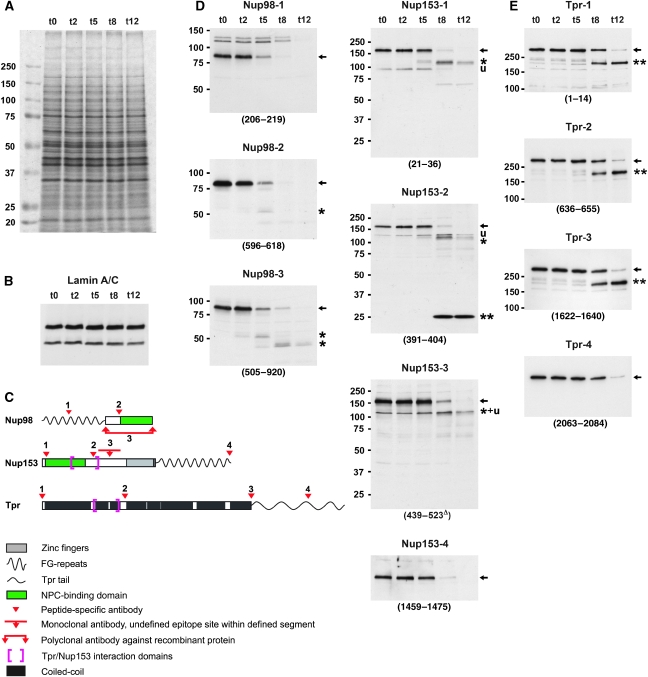
We hence studied these and other proteins by immunoblotting of total HeLa proteins, collected at different time points after PV infection. SDS–PAGE showed that the bulk of cellular proteins remained unaffected by PV-induced proteolysis (Figure 3A). Furthermore, immunoblotting showed that lamins (Figure 3B) and most NPC-core proteins were left unharmed (Supplementary Figure S3). This included those of the Nup160 subcomplex, which represent the fundament for direct or indirect NPC anchorage of other NPC-associated proteins such as Nup98, Nup153, and Tpr (e.g., Vasu et al, 2001). By contrast, most FG-repeat nucleoporins (Figure 3C and D, and Supplementary Figure S3) had their FG-repeat domains removed. Tpr was also partially degraded (Figure 3C and E).
Figure 3.
Nup153 and Nup98 appear largely degraded upon PV infection whereas the coiled-coil rod domain of protein Tpr remains intact. (A) SDS–PAGE and Coomassie staining of whole-protein extracts from HeLa cells before and at 2–12 h post-infection, showing the bulk of cellular proteins unaffected by PV-induced proteolysis. The extracts were from the same HeLa cell cultures analysed by TEM in Figure 1. (B) Immunoblotting of extracts used in panel A, showing that lamin A and lamin C, like lamin B (not shown), remain unaffected. This indicates that rearrangements within the nuclear lamina or its NE attachments, but not lamin proteolysis, are required for the NE upfolding observed. (C) Epitope sites of Nup153, Nup98, and Tpr antibodies indicated by arrowheads. Different antibodies targeting the same protein are numbered as in panels D and E. (D, E) Immunoblotting of Nup153, Nup98, and Tpr, using cell extracts shown in panel A; (see also Supplementary Figure S3). Target regions are given in parentheses; Δ indicates mAbs for which actual epitopes within defined protein segments are unknown. (D) At 8–12 h post-infection, when HEZs are visible at almost all NPCs, Nup153 (full-length proteins marked by arrows) appears largely degraded, except for a small segment (double-asterisk) comprising at least part of the Tpr-binding region. Additionally, only minor amounts of a 120-kDa degradation product (asterisk) and some unspecific cross-reactions (u) are seen with some Nup153 antibodies. Nup98 is degraded more rapidly, with only its C-terminal domain (asterisk) resisting proteolysis slightly longer. (E) Whereas Tpr's C-terminal domain is being degraded around 8 h post-infection, its entire rod domain (double-asterisk) withstands proteolysis. The membrane marked 1622–1640 was first incubated with rb-anti-Tpr-4 (2063–2084), then stripped and re-incubated with gp-anti-Tpr-3 (1622–1640).
Proteolysis of the FG-repeat proteins, including Nup153 and Nup98, first affected their unfolded FG-repeat domains. The one from Nup98 was eliminated earliest, in line with another recent study of PV-mediated Nup98 degradation (Park et al, 2008). The NPC-binding domains of these nucleoporins withstood degradation longer but when the HEZs became visible, the anchor domains appeared largely degraded too, except for those of Nup358, an FG-repeat nucleoporin located at the NPC's cytoplasmic side (Supplementary Figure S3), and Nup153 (Figure 3D). Whereas most of Nup153 appeared degraded at 12 h post-infection, a small segment of ∼26 kDa withstood degradation. This segment likely harbours the sequences essential for NPC anchorage and Tpr binding, both residing within a region comprising aa 228–439 (Enarson et al, 1998; Vasu et al, 2001; Hase and Cordes, 2003; Griffis et al, 2004).
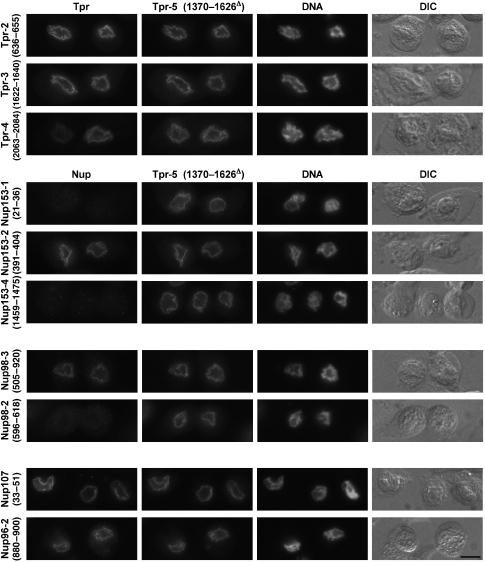
Tpr degradation, in contrast, was restricted to its unfolded C-terminal tail domain whereas its large coiled-coil domain (aa 1–1630), including Tpr's NPC- and Nup153-binding region, remained unharmed. Moreover, immunofluorescence microscopy (IFM) showed that these Tpr rods remained bound to the NE even late after PV infection. Similarly, the NPC-anchor segment of Nup153 remained attached to the NE, as did residual copies of Nup98's NPC-anchor domain. By contrast, already earlier after infection, neither other parts of Nup153, nor the FG-repeat domains of Nup98 and other nucleoporins, were detectable at the NE any longer (Figure 4 and Supplementary Figure S4).
Figure 4.
The coiled-coil rod domain of Tpr remains anchored to the NPC even late after PV infection. IFM of HeLa cells at 10 h post-infection, showing that antibodies against Tpr's coiled-coil domain (anti-Tpr-2 (636–655), -3 (1622–1640), -5 (1370–1626Δ)) and the Tpr-binding domain of Nup153 (anti-Nup153-2 (391–404)) still label the NE. At this time point, and earlier ones (not shown), the other parts of Nup153, and the FG-repeat domain of Nup98, are no longer detectable whereas the NPC anchor of Nup98 and Tpr's C-terminal tail are still present at some NEs. Nucleoporins of the Nup107 subcomplex (also Supplementary Figure S3), representing direct and indirect anchor sites for Tpr, Nup153, and Nup98, remain bound to the NPC. DNA staining and differential interference contrast (DIC) micrographs show nuclear-peripheral chromatin accumulation and cell rounding, characteristic for later stages of PV infection. Bar: 10 μm; same magnification for all the micrographs.
NPC-associated HEZs can no longer be established after in vivo depletion of Tpr
If fibrillar NPC appendices mark the periphery of the NPC-associated HEZ, a largely degraded Nup153, and Tpr's tail domain could not be regarded as prime candidates for maintaining the HEZs still visible late in PV infection. By contrast, Tpr's rod domain, distinguished by its integrity and persisting attachment to the NPC, remained a potential candidate.
To test this further, we attempted to PV-infect cells after having depleted them of Tpr. In principle, elimination of Tpr was known to be achievable in HeLa cells by RNAi. Such Tpr-depleted cells are still capable of cell-cycle progression and nucleo-cytoplasmic transport of proteins and mRNAs (e.g., Hase, 2003; Hase and Cordes, 2003; Lee et al, 2008; and data not shown; see also Shibata et al, 2002). Furthermore, chromosome positioning as monitored by telomere and centromere detection, as well as the patterns of several epigenetic histone modifications, appeared unaffected in most Tpr-deficient cells (Supplementary Figure S5).
Furthermore, it had been shown that activation of the RNAi machinery does not necessarily trigger antiviral responses, and siRNA-transfected cells can remain susceptible for subsequent PV infection (Gitlin et al, 2002). Nonetheless, to avoid innate cellular immune responses that could impair PV infection (e.g., Ida-Hosonuma et al, 2005), we first screened different Tpr siRNAs for high knockdown efficiencies without off-target effects unrelated to Tpr deficiency (data not shown). Several pre-selected siRNAs, complementary to non-overlapping ORF segments of the Tpr mRNA, caused a clear knockdown of Tpr protein levels at day 4 after transfection (Figure 5A). The mean intensities of residual Tpr staining in the transfected cells could be as low as 4–6% as determined by IFM, whereas residual Tpr levels in immunoblots of total cell extracts commonly ranged between 10% and less than 20% (Figure 5B). TEM of such cell populations allowed sporadic detection of perpendicularly sectioned NPCs that were juxtaposed to heterochromatic or nucleolar material but now mostly lacked distinct exclusion zones. This indicated that Tpr might at least play a role in the establishment of the few HEZs seen in non-infected HeLa cells. By contrast, even though correspondingly positioned NPCs were similarly sparse in simultaneous controls, they were still characterized by distinct HEZs (Figure 5C; see also Supplementary Text to Figure 5, and Supplementary Figure S6).
Figure 5.
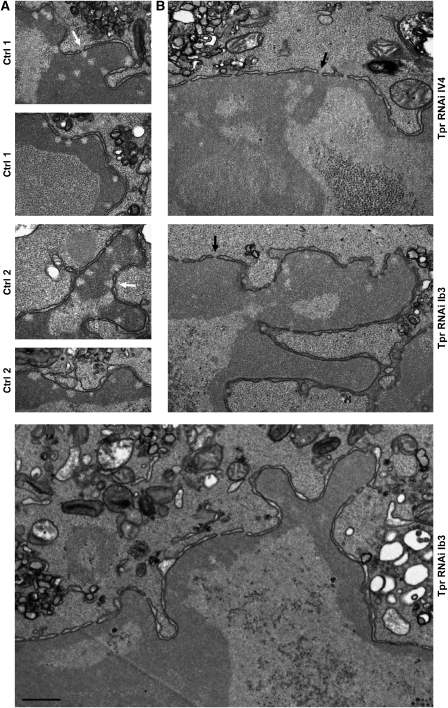
RNAi-mediated Tpr knockdown in HeLa cells. (A) Confocal IFM of Tpr at day 4 after transfection with different Tpr siRNAs (Ib3, IV2, IV4) or target-less control siRNAs. Only traces of Tpr staining are seen in most cells after Tpr RNAi; bright nuclear rim staining shown as reference is visible only in cells that remained untransfected. For occasionally observed dots of residual Tpr staining at otherwise largely Tpr-deficient NEs, see Supplementary Figure S6. Bar: 20 μm. (B1) SDS–PAGE and Coomassie staining of serial dilutions of whole-protein extracts from non-transfected cells (Ctrl 2), and cells treated with Tpr siRNAs (Ib3, III4, III5, IV2, IV4) or transfection reagent alone (Ctrl 1), showing that the bulk of cellular proteins remains unaffected by siRNA treatment. (B2) Immunoblotting of identical loadings as in panel B1. Incubations with anti-Tpr and anti-Nup98 (asterisks: unrelated cross-reactions) were on different halves of the same membrane. Efficient Tpr knockdown was achieved with all Tpr siRNAs without eliciting distinct effects on other NPC proteins, including Nup93, Nup107, Nup133, and gp210 (not shown). Cells transfected with III4 and III5, however, were later found to not allow for a normal PV-infection process, so that these siRNAs were not used further. Of the infection-compatible siRNAs, Ib3 and IV4 were used for all subsequent PV-infection experiments in parallel. (C) TEM of non-transfected cells, and of cell populations at day 4 after treatment with transfection reagent alone, and transfection with Tpr siRNAs Ib3, or non-target control siRNAs (Ctrl 3). NPCs juxtaposed to heterochromatic or nucleolar material (arrows) in control cells remained characterized by HEZs but mostly lacked such exclusion zones after Tpr RNAi. Bars: 200 nm; same magnification for all the images.
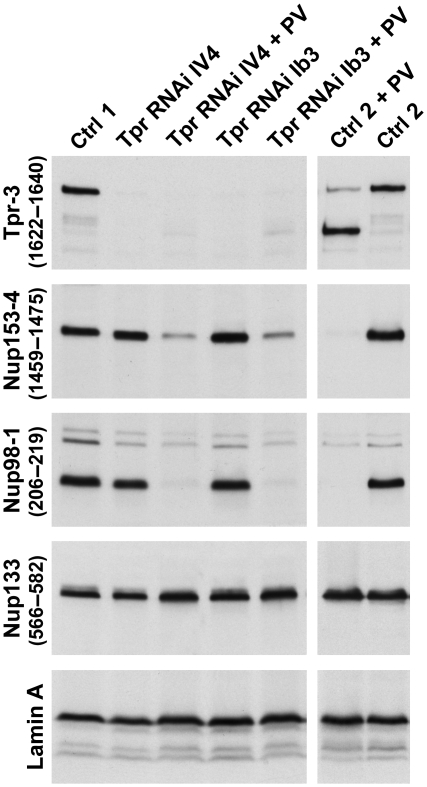
Tpr-deficient cell populations were then tested for susceptibility to PV infection, paralleled by infecting control cells that had been transfected with a target-less siRNA, treated with transfection reagent only, or had been left untreated. For several Tpr siRNAs, PV infection of transfected cells was found to appear normal. Characteristic changes in morphology, such as cell rounding, and their time points of occurrence were similar to that in the controls. At the molecular level, the characteristic NPC-protein degradation patterns were basically the same, except that the time points of complete degradation were slightly delayed in the siRNA-transfected cells (Figure 6).
Figure 6.
Post-transcriptional tpr gene silencing by RNAi does not impair subsequent PV infection and degradation of nucleoporins. Four days after transfection with Tpr siRNAs or mock transfection with non-target control siRNAs (Ctrl 1), or after incubation with transfection reagent alone (Ctrl 2), cells were either infected with PV or not, and harvested 10 h later. Total cell proteins were analysed by immunoblotting. Regardless of whether Tpr had already been eliminated by RNAi before PV infection or not, Nup153 and Nup98 were again degraded in the infected cells whereas the NPC core protein Nup133 and lamin A remained unaffected. PV-induced degradation of nucleoporins such as Nup35, and stability of others such as Nup107 and gp210, was also similar in control and Tpr-RNAi cells (not shown).
At the cytological level, progression of chromatin condensation and its final expansion throughout the nucleus appeared similar too. However, whereas NPC-associated HEZs were omnipresent in the infected control cells, the majority of cells in the Tpr siRNA-treated populations lacked HEZs (Figure 7 and Supplementary Figure S7). In fact, in nuclei in which nuclear-peripheral chromatin had started to condense, such material was already found distributed laterally along the NE's inner face, concealing the NPCs' nuclear entrances. Also at time points when the condensed chromatin had filled larger areas of the nucleus, the NPCs of Tpr-deficient cells remained devoid of HEZs.
Figure 7.
NPC-associated HEZs are no longer maintained after in vivo depletion of Tpr, resulting in NPC coating by heterochromatin. (A) After mock transfection (Ctrl 1) or treatment with transfection reagent alone (Ctrl 2), HeLa controls were PV-infected, harvested 9.5–10 h post-infection, and then analysed by TEM, in parallel to the specimens shown in panel B. The hyper-condensed chromatin masses contoured but did not trespass the borderlines of the NPC-associated HEZs (white arrows) that persisted in these cells. (B) Four days after transfection with Tpr siRNAs, HeLa cells were infected with PV and harvested 9.5–10 h later. Amassments of condensed chromatin were found across the entrance of most NPCs (black arrows). Beside HEZ loss in the Tpr-knockdown cells, the staining of the hyper-condensed chromatin often appeared slightly lighter than in parallel controls. Whether such decreased affinity for heavy metal stains correlates with altered chromatin condensation, and how this might be caused by Tpr deficiency, remains unknown. Bar: 500 nm; same magnification for all the images.
Discussion
NPC-associated HEZs occur as the distal parts of an interconnected system of a largely chromatin-free space traversing the nucleus of many somatic cell types. Observed since five decades as perinuclear sub-compartments of either channel-like appearance (e.g., Swift, 1959; Watson, 1959; Davies, 1961; Visser et al, 2000; Rego et al, 2008) or possibly cone-like shape (e.g., Tokuyasu et al, 1968; Maul, 1977), the factors that establish these HEZs have remained largely unknown. Here we show that their formation and maintenance requires the large coiled-coil protein Tpr.
Current model concepts of nuclear compartmentalization
Current propositions about how nuclear compartmentalization is accomplished in most somatic cells include different but not necessarily mutually exclusive notions. A common starting point is that chromosomes are in fact non-randomly arranged within the interphase nucleus, with each chromosome residing within its own spatially restricted chromosome territory (CT; e.g., Cremer et al, 1993). However, different views exist of how such a spatial organization is achieved and how the borderlines between these CTs are established.
In one model, intrinsic self-organizing properties and the sum of cohesive interactions within the chromosomes themselves define each CT. The borderlines between the CTs and the resulting inter-chromosomal space (ICS) on their outside would thus primarily stem from intra-chromosomal self-stabilization, with no need for static boundaries formed by structural elements (e.g., Cremer et al, 2006; Misteli, 2007). Transiently bound extra-chromosomal factors, such as components of the transcription- and pre-mRNA processing machineries, could help reinforce these borderlines in a dynamic manner (e.g., Zirbel et al, 1993). Cargo flux density and macromolecular crowding, both within the territories and the ICS, could represent additional factors maintaining such spatial arrangements (e.g., Hancock, 2004, 2007; Rippe, 2007; Richter et al, 2008; Bancaud et al, 2009).
In this scenario, the only major boundary forced upon the chromosomes by an extra-chromosomal structural framework would be the one formed by the NE and its nuclear lamina, to which the nuclear-peripheral chromosomes are tethered. The NPCs, often non-randomly distributed and sometimes arranged in clusters or lines within the NE of differentiated cells (e.g., Maul, 1977), would thereby be mostly segregated from the NE-attached chromatin areas and sometimes even contour the underlying interface between neighbouring CTs (Murray and Davies, 1979; Lopez-Velazquez et al, 1996). In such cases, most of the long, NPC-associated inter-chromosomal zones seen in ultrathin sections would need to be interpreted as orthogonally sectioned ditches rather than tubular channels.
In a conceptually different scenario, not only the nuclear lamina but also dedicated architectural elements deep within the nucleus would play a role in chromatin organization. These would partition nuclear space by forming scaffolds, thereby determining the spatial arrangement of chromosomes and delimiting their distribution in a static manner. However, plausible arguments have been adduced against such a concept and an extensive intra-nuclear scaffold network. Moreover, no structural elements that could account for such large-scale nuclear partitioning have been identified so far (e.g., Misteli, 2007).
Early evidence for scaffold-surrounded zones spatially confined to the nuclear periphery
Even though a formerly proposed extensive nucleoskeleton (e.g., Nickerson, 2001; Jackson, 2003) is not supported by recent data, it remains possible that structural elements contribute to locally restricted nuclear sub-compartments. Indications that the NPC-associated HEZs might represent one type of scaffold-defined zone came from different studies. For example, TEM of serial cell sections (Visser et al, 2000; also Ghadially et al, 1985) showed that NPC-associated HEZs do not only locate between neighbouring CTs but also occur as tube-like and essentially chromatin-free channels that project deep into and even pass through individual CTs. A recent TEM study of the inactivated X chromosome (Xi) in female fibroblasts (Rego et al, 2008) further showed that such HEZ channels could penetrate up to a depth of 400 nm into the core of the Barr body that contains the silenced and non-genic sequences of the Xi heterochromatin (Clemson et al, 2006), indicating that these channels do not need to be maintained by dynamic interactions with transcription complexes. Furthermore, some HEZs in differentiated cells were found to occur as isolated zones completely encapsulated by heterochromatin (Maul, 1977), indicative that they need not be in continuum with the ICS and maintained by high cargo flux density. Such findings prompted the question as to how intra-chromosomal interactions alone could create cylindrical channels and which intrinsic chromatin properties could determine such distinct boundaries.
As an alternative explanation, the NPC-proximal parts of these channels reflect the existence of a distinct scaffold. Supporting the notion of a scaffold whose maintenance does not necessarily depend on surrounding chromatin, a TEM study of monkey kidney cells infected with simian virus 40 (Maul, 1976) found SV40 particles of 45 nm in diameter to accumulate within the nucleus and displace both euchromatin and heterochromatin from the nuclear periphery. Yet even in the seeming absence of neighbouring chromatin in the late stages of infection, the particles remained partially excluded from an NPC-associated zone extending into the nucleus for ∼200 nm, and completely restrained from an ∼80nmlong zone directly in front of the NPC proper. Sporadically observed fibrous material of fish trap-like appearance was proposed to have a role in excluding the virus particles from these zones.
The relationship between HEZ and an NB-like scaffold, and the contribution of Tpr
To investigate which proteins might have a role in also excluding heterochromatin from such NPC-associated zones, we visualized HEZs in PV-infected HeLa cells. There, most if not all NPCs were associated with HEZs, including cone-like HEZs omnipresent in the late stages of PV infection, as well as longer ones more frequently seen at earlier time points. This approach of contouring HEZs, in combination with RNAi, resulted in identifying the coiled-coil protein Tpr as an element required for defining the borderlines between HEZs and the surrounding heterochromatin. Accordingly, loss of both cone-shaped and tube-like HEZs was found to go along with Tpr deficiency.
Stereometric size estimation of the cone-like HEZ suggested that its borderlines could be equivalent to those formed by the fish trap in monkey cells or the NBs in other organisms. Even though fibrils could not be depicted, the minimal HEZ dimensions were reminiscent of the NB-enclosed space in amphibian oocytes. In such cells, in which the inner NE side can be studied by three-dimensional EM imaging, the eight NB fibrils enclose a space reminiscent of a truncated pyramid with an octangular base. The model of the residual HEZ from the late stages of PV infection would allow fitting such an average-sized oocyte NB into it. The latter has a mean diameter of 110–120 nm at its basis and reported heights of 50–100 nm (e.g., Ris, 1989, 1997; Jarnik and Aebi, 1991).
Furthermore, in immunogold-TEM studies of cells with little or no perinuclear chromatin, or when chromatin had been detached from the NE during sample preparation, Tpr was located at NPC-attached fibrillar material (Cordes et al, 1997; Zimowska et al, 1997; Frosst et al, 2002; Soop et al, 2005) possibly comprising the fibrils building an NB or fish trap. In cultured cells with preserved perinuclear chromatin, such fibrils were usually not discernible, but there too Tpr was located in proximity to NPCs (Krull et al, 2004).
If the cone-shaped HEZ would reflect the existence of an NB-like structure in HeLa cells, the presented TEM and RNAi results would hence point to Tpr as one of its essential components. Consequently, one could assign a central function in perinuclear organization not only to Tpr but also to the macromolecular NB as a whole. Visualization of cone-shaped HEZs at essentially all NPCs further indicates that such NBs are common in HeLa cells. Moreover, as Tpr is attached to the NPCs of many vertebrate cell types of different germ-layer origin (Cordes et al, 1997; Kuznetsov et al, 2002), these cells may likewise possess NB-like structures, thus representing a near-ubiquitous tool of nuclear-peripheral compartmentalization.
In such NBs, Tpr's large coiled coil-dominated N-terminal ‘rod' domain (Hase et al, 2001) would be an essential architectural element (Figure 8). By contrast, the structural contribution to an NB-like scaffold by Nup153 or even Nup98, largely degraded in the PV-infected cells, could be regarded as supportive at most. In this case, small proteolytic fragments that might have escaped detection could still have a scaffold-stabilizing role in the PV-infected cell. It also cannot be ruled out that those parts of Nup153 degraded during PV infection normally play some role in the initial steps of HEZ formation, or contribute to the slightly larger HEZs seen in non-infected cells. Furthermore, our data do not exclude the possibility that additional, yet unidentified proteins contribute to the formation of NBs and HEZs.
Figure 8.
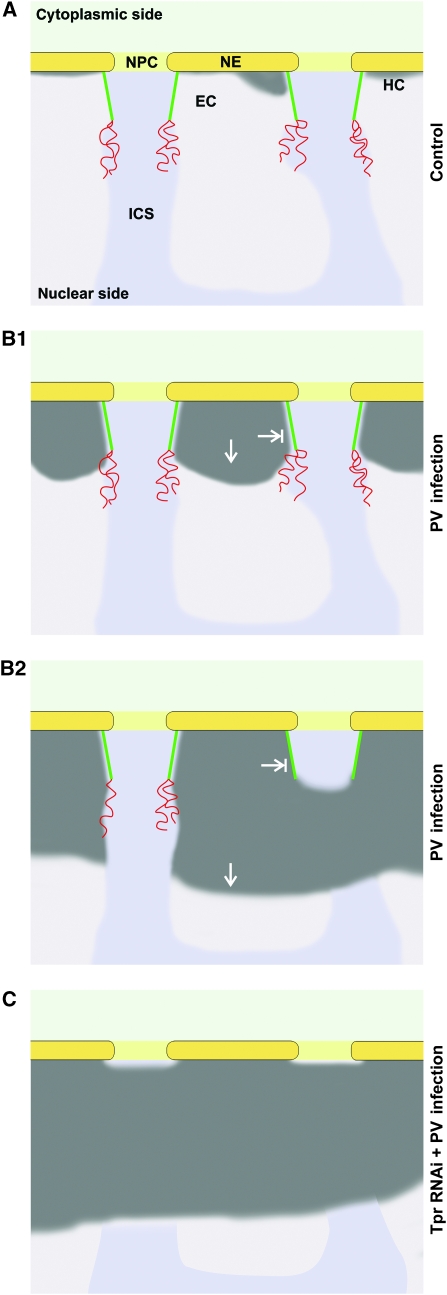
A model for Tpr as an essential structural element of perinuclear compartmentalization. Schemes summarizing the spatial relationships between NPCs and nuclear-peripheral heterochromatin (HC) in HeLa control cells (A), and between NPCs and hyper-condensed chromatin in PV-infected cells with (B) and without Tpr (C). NEs, NPCs, and HEZs are shown in diametric cross-section and approximately to scale. The light and medium grey areas represent the euchromatic regions (EC) and the inter-chromosomal space (ICS). The coiled-coil rod domains of Tpr contribute to forming a fibrillar HEZ scaffold (green) that corresponds to the NB seen in other cell types. The unfolded Tpr tail domains (red), or other yet unidentified appendices, may project deeper into the nucleus. (A) In HeLa cells, nuclear-peripheral heterochromatin is limited to a thin layer only a few nanometres thick. Whether such heterochromatin is actually excluded from the NPC entrance zones is unclear at this point. (B1) Early upon PV infection, a process of chromatin compaction appears to spread towards the nuclear interior, thereby outlining the NPC-proximal parts of HEZs reminiscent of those observed in different types of tissue cells. (B2) Later after infection, the HEZs can be completely engirded by condensed chromatin excluded from this region. By then, the Tpr tails or any other appendices are either largely degraded or have collapsed onto the residual HEZ scaffold comprising the intact Tpr rods. Similarly shaped HEZs are observed in terminally differentiated cells with high heterochromatin content. (C) In Tpr-deficient cells, the nuclear entrances of the NPCs are covered by condensed chromatin after PV infection. It remains unknown though whether the HEZs are in fact trespassed by expanding masses of heterochromatin, or whether euchromatin possibly present in these regions after loss of Tpr is condensed upon PV infection.
Tpr's largely unfolded C-terminal ‘tail' domain of ∼730 aa would in this model not be needed for maintaining the NB-like scaffold. Instead, it could project into the NB-engirded HEZ or towards the nuclear interior as a flexible appendix. However, located rather close to the conjectured distal end of an NB (Krull et al, 2004), the tail alone is unlikely to form the very long HEZs that markedly exceed the dimensions of an NB scaffold. However, as Tpr deficiency also precludes their occurrence upon PV infection, their formation or maintenance too is either directly or indirectly dependent on Tpr. Whether other components though need to be tethered to an NB-like scaffold to form longer HEZs remains to be investigated. The potential conversion of such long HEZs into smaller, cone-shaped ones during progression of PV infection might correlate with the detachment of additional components from the NB. The latter might be degraded themselves or released by proteolysis of an NB component such as Tpr's tail domain.
A possible role for an NPC-associated NB-like scaffold in heterochromatin exclusion
The mechanism by which compounds as different as condensed chromatin, the granular material of the nucleolus, and SV40 particles are all segregated from the cone-shaped HEZ remains unknown. If the HEZ scaffold in somatic cells were indeed an NB-like structure of octagonal symmetry, the maximal width between neighbouring fibrils would be ∼40 nm. However, if these fibrils would act as a physical barrier, the nucleolar granules of ∼15–20 nm diameter could pass through them, if not retained by the nucleolus itself or prevented by an NB-mediated sorting process. By contrast, the large SV40 particles could indeed be excluded due to their size.
Regarding chromatin, simple size exclusion could be imagined for a folded euchromatic 30nm fibre and even more so for the condensed heterochromatic material. However, as fibrillar connections observed between the NB-like fish trap and the surrounding chromatin (Maul, 1971, 1976) could suggest, chromatin might not merely be excluded by size but kept in place by being tethered to the HEZ scaffold.
The reason why an HEZ-forming scaffold should exclude chromatin from an NPC-proximal zone could simply be that access of large cargo complexes to the NPC translocation channel would be impeded by a meshwork of dense chromatin fibres, and facilitated by HEZs. Especially in differentiated cells with a high content of very dense heterochromatin but still active gene expression, clogging the paths to the NPCs should variably affect the export of different cargo molecules. However, without experimental evidence from such cells, the proposition that heterochromatin could impede the accessibility of their NPCs needs to be treated with caution.
In cultured cells, heterochromatic regions are in fact accessible for tracer molecules of certain size, as shown for fluorescently labelled dextrans injected into nuclei. Whereas regions of perinuclear heterochromatin were found inaccessible for 282-kDa dextrans, dextrans of 42 kDa were reported to have unlimited access. Dextrans of 77 kDa were excluded from the densest heterochromatin in such cells, but less dense regions appeared partially accessible for dextrans of 70–148 kDa (Verschure et al, 2003; Görisch et al, 2005a, 2005b).
The sizes of dextrans in solution can be described by their radii of gyration RG (e.g., Seksek et al, 1997; Lénárt and Ellenberg, 2006). These can be used to calculate the approximate sizes of differently shaped particles that would have similar hydrodynamic properties (e.g., van Holde, 1971; Verschure et al, 2003). For macromolecular complexes of spherical shape, approximate RG values of 7.5 nm (42-kDa dextrans), 9.5 nm (77 kDa), and 18 nm (282 kDa) would correspond to particle diameters of ∼19, 25, and 46 nm, respectively. For a prolate rotational ellipsoid with an axial ratio of 4, representing the approximate shape and axial proportions characteristic for the mRNPs that comprise the bulk of moderately sized mRNAs of only a few kilobases (kb) (Batisse et al, 2009), the above-mentioned RG values would correspond to particle lengths of 32, 40, and 76 nm.
Therefore, if size were the only criterion for exclusion, export cargos such as the ribosomal 40S and 60S subunits of 23–25 by 13–23 nm in size (e.g., Haga et al, 1970; Nonomura et al, 1971; Frank et al, 1981), with corresponding RG values of 7–9 nm, might be too small to be prevented in a cultured cell from reaching an NPC coated by a layer of moderately dense heterochromatin. The latter should then also not prevent NPC access of the bulk of mRNPs, with particle lengths of 15–30 nm (Batisse et al, 2009), and a calculated RG of 7.1 nm for the 30-nm particle.
In principle though, size-dependent exclusion by heterochromatin would be well conceivable for very large mRNPs. Such exist, for example, in the polytene salivary gland cells of some insects, where transcripts of 35–40 kb are assembled into spherical mRNP particles ∼50 nm in diameter (e.g., Stevens and Swift, 1966; Mehlin et al, 1991). With an according RG value of >19 nm, these mRNPs would outreach the proposed size-exclusion limit for heterochromatin (Görisch et al, 2005b). Yet in this cell type, devoid of perinuclear heterochromatin, HEZs would not be needed.
However, vertebrate mRNPs containing the 20- to 100-kb mRNAs for giant myofibrillar proteins (e.g., Sanger and Sanger, 2001) may be of similar size or even larger, and in this case noticeable amounts of heterochromatin with HEZs exist in different muscle cell types (e.g., Rhodin, 1968; Bloom, 1970; Smetana et al, 1970; Sabatelli et al, 2001). It appears to be obvious that for such cargos the heterochromatin would need to be excluded from their paths to the NPC.
Other functions
In addition to preventing heterochromatic material from entering the HEZ, a scaffold might also confine the diffusive movement of export cargos. In fact, it has been found that large RNP complexes can virtually move freely within the ICS but appear to be excluded from the chromatin-rich regions (e.g., Politz et al, 1999; Bridger et al, 2005). However, if the NB-like scaffold, and any fibrillar extensions, would primarily consist of thin and rather widely spaced fibrils, it appears unlikely that they could corral the bulk of the cargo complexes within the HEZ merely by size exclusion. The fact that mRNP particles are nonetheless largely confined to the ICS indicates that this involves other mechanisms. These might include transient interactions between cargos and components stably associated to the HEZ scaffold, and even play a role in the NPC directedness of cargo movement near the nuclear periphery. The C-terminal domain of Tpr, perhaps dispensable for HEZ formation and apparently not required for its maintenance, could be one component that directs export cargos to the NPC. Accordingly, various RNP-associated components were found to bind to Tpr's tail domain (e.g., Hase, 2003; our unpublished data). Interestingly, some nuclear import receptors also bind to this domain (Shah et al, 1998; Ben-Efraim et al, 2009) but relevance of these interactions in the context of Tpr's NPC attachment still remains unclear. Furthermore, the ERK2 kinase binds stably to Tpr's tail and was proposed to have a role in cargo modification at this location (Vomastek et al, 2008).
Facilitating particle access to the NPC by chromatin exclusion may therefore not have been the primary purpose of NPC-attached appendices. The heterochromatin could simply have been expelled from this region as a consequence imposed by the NB's design and dimensions, if prerequisite for proper nuclear-peripheral positioning of Tpr's C-terminal domain for other functions. Indeed, NBs in insect salivary gland cells act as intermediate docking and rearrangement sites for large RNPs during nuclear export (Kiseleva et al, 1996). Likewise, the yeast Tpr homologue Mlp1 has been found to act as a docking site for hnRNPs (Green et al, 2003; Vinciguerra et al, 2005) and a retention site for unspliced mRNAs (e.g., Galy et al, 2004). Moreover, presence of NPC-attached Tpr in proliferating cells with sparse heterochromatin, and in cell types with no or little perinuclear chromatin at all, as in insect salivary gland cells and in fully grown amphibian oocytes, further illustrates that heterochromatin exclusion is not Tpr's only function.
Whether Tpr has a role in establishing or maintaining a certain condensation status of the surrounding chromatin, as might be deduced from subtle differences in perinuclear chromatin appearance noted between Tpr-deficient and control cells (not shown), remains to be investigated. If so, this could attribute a role in perinuclear transcription regulation to mammalian Tpr, as has been found for its orthologue in insects (Mendjan et al, 2006) and yeast (e.g., Casolari et al, 2005; Vinciguerra et al, 2005; Dieppois et al, 2006; Luthra et al, 2007). In such a scenario, Tpr could tether chromatin to the vicinity of the NPC in order to keep it in a distinct state. One could also envision Tpr as a recruitment and operational platform for factors that modulate chromatin properties and thereby contribute to a distinct NB-to-chromatin interface. This could even act as a transcriptionally permissive microenvironment (e.g., Sexton et al, 2007; Skaggs et al, 2007; Zhao et al, 2009) and thereby contribute to HEZ formation and maintenance. The factors involved, as well as transiently bound RNPs, might be lost during the PV-infection process, along with degradation of peripheral NPC proteins like Nup153, and Tpr's tail domain. This might explain why the dimensions of the cone-shaped HEZs late in PV infection appear slightly smaller than those found in non-infected cells. Identification of proteins that specifically bind to either Tpr's rod or tail domain should not only provide further insight into Tpr's functional repertoire but also into that of the NB-like scaffold as a whole. Moreover, whereas Tpr deficiency and loss of its probably diverse functions at the NE, as well as those in mitosis after NPC disassembly (Lee et al, 2008; De Souza et al, 2009; Lince-Faria et al, 2009), appear tolerable in highly aneuploid tumour cells like HeLa, Tpr loss in diploid tissue cells might cause more pronounced phenotypes and perhaps also uncover cell type-specific functions of this protein.
Materials and methods
Antibodies, viruses, and cells
Mouse mAbs against Tpr and Nup153 were described earlier (Cordes et al, 1997; Hase and Cordes, 2003) and are here referred to as Tpr-5 (1370–1626Δ) and Nup153-3 (439–523Δ). Most guinea pig (gp) and rabbit (rb) antibodies against Tpr and NPC proteins were raised against synthetic peptides (Hase and Cordes, 2003; Krull et al, 2004; Patre et al, 2006). Rb antibodies against hNup98 aa 505–920 (Griffis et al, 2002) and mAb X167 against lamin A/C (Höger et al, 1991) were provided by Maureen Powers and Georg Krohne, respectively. Rabbit antibodies against lamin A were from Covance (Emeryville, CA, USA). Secondary antibodies were from Jackson ImmunoResearch (West Grove, PA, USA) and Dako (Hamburg, Germany). For PV infections (Belov et al, 2004), near-confluent HeLa cultures were, if not stated otherwise, inoculated with 50 PFUs (plaque forming units) per cell of PV type 1, strain Mahoney, propagated on Green Monkey kidney cells before use on HeLa cells. The culture conditions for HeLa cells of high passage numbers were as described (Kuznetsov et al, 2002).
SDS–PAGE and immunoblotting
Preparation of total protein samples from HeLa cells and immunoblotting was performed as described (Hase et al, 2001; Kuznetsov et al, 2002). SDS–PAGE was according to Thomas and Kornberg (1975). Some filters were reused after stripping bound antibodies by incubation in 100 mM glycine (pH 2.4) at RT for 15 min, subsequent washes in H2O, incubation in 62.5 mM Tris–HCl (pH 6.7) with 2% SDS and 50 mM DTT at 60°C for 30 min, further washes in H2O, and re-equilibration in TBST at 4°C o/n.
Post-transcriptional gene silencing
Transfection of HeLa cells with siRNAs and Oligofectamine (Invitrogen, Karlsruhe, Germany) was as described by Elbashir et al (2001) and Hase and Cordes (2003). Unmodified 19-mer RNA oligonucleotides finally selected in the course of this study were from Dharmacon Research (Lafayette, CO, USA) and Qiagen (Hilden, Germany). Antisense strands were complementary to nt 3176–3194 (here termed pair Ib3), 6040–6058 (IV2), and 6241–6259 (IV4) of the hTpr cDNA sequence (U69668). siRNAs III4 and III5 (Figure 5B) were 25-mer Stealth siRNAs (Invitrogen). For control transfections, Dharmacon's RISC-Free siRNA#1 and non-targeting siRNA#2 were used, as well as other 19-mer siRNAs with at least three mismatches to human mRNAs and <50% GC content (Dharmacon).
Immunofluorescence microscopy
IFM of formaldehyde (FA)-fixed HeLa cells was performed as described (Hase and Cordes, 2003). Conventional IFM was performed with a Leica DM RXA, equipped with the Openlab Software. For confocal laser-scanning microscopy, a Zeiss LSM 510 and a Leica SP5 were used. For quantitative analysis of Tpr signal intensities in siRNA-transfected versus non-transfected cells on the same coverslip, images were acquired with the Leica SP5 with offset levels of the corresponding PMT channel set to zero, as in Figure 5A. Tpr-signal intensities of non-transfected (n⩾20 per coverslip) and neighbouring transfected cells (n⩾40) of similar nuclear diameter and DNA-staining intensity were then determined by ImageJ (http://rsb.info.nih.gov/ij/).
Electron microscopy
HeLa cells adherent to culture dishes were washed with PBS and fixed in 3% glutaraldehyde in 0.1 M sodium cacodylate–HCl buffer (pH 7.3) with 0.05 M sucrose o/n. They were then scraped off and sedimented at 200 g for 10 min. After rinsing in cacodylate buffer, the cells were post-fixed in cacodylate buffer with 1.5% OsO4 and 0.7% potassium ferrocyanate at 4°C for 90 min, again rinsed in cacodylate buffer, dehydrated in ethanol (70, 95, and 100%; each for 1 h), and then stained with 2% uranyl acetate in ethanol for 30 min. The cells were consecutively incubated in a 2:1 and then 1:2 (v/v) mixture of ethanol and Spurr resin (Electron Microscopy Sciences, Hatfield, PA, USA) at 20°C for 1 h each, followed by infiltration with pure resin at 20°C for 3 h, and polymerization at 70°C for 15 h. Sections of approximately 40–60 nm thickness were transferred onto 200-mesh copper grids without supporting film and stained with lead citrate for 2 min. Specimens were examined using a Philips CM120 EM or a Hitachi H-7600 at 80 kV, both equipped with a MegaView 3 CCD camera (SIS, Soft Imaging Solutions, Münster, Germany). The iTEM software from SIS was used for distance measurements.
Supplementary Material
Acknowledgments
We thank Birgitta Björkroth and Marie-Louise Spångberg for EM sectioning, Anne Cordes for help with stereometric analyses, and Heinz-Jürgen Dehne for expert technical support. We greatly appreciate Dirk Görlich for financial support of SR and thank Valerie Doye, Wolfgang Fischle, Christer Höög, Ralph Kehlenbach, Georg Krohne, and Maureen Powers for kindly providing research reagents. Part of this work was supported by the Swedish Research Council.
Footnotes
The authors declare that they have no conflict of interest.
References
- Balasundaram D, Benedik MJ, Morphew M, Dang VD, Levin HL (1999) Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol Cell Biol 19: 5768–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J (2009) Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J 28: 3785–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs P, Burke B, Powers C, Craig R, Purohit A, Doxsey S (1998) Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J Cell Biol 143: 1801–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batisse J, Batisse C, Budd A, Bottcher B, Hurt E (2009) Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem 284: 34911–34917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O (2004) Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306: 1387–1390 [DOI] [PubMed] [Google Scholar]
- Belov GA, Lidsky PV, Mikitas OV, Egger D, Lukyanov KA, Bienz K, Agol VI (2004) Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J Virol 78: 10166–10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I, Frosst PD, Gerace L (2009) Karyopherin binding interactions and nuclear import mechanism of nuclear pore complex protein Tpr. BMC Cell Biol 10: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz K, Egger D, Wolff DA (1973) Virus replication, cytopathology, and lysosomal enzyme response of mitotic and interphase Hep-2 cells infected with poliovirus. J Virol 11: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom S (1970) Structural changes in nuclear envelopes during elongation of heart muscle cells. J Cell Biol 44: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A (2007) Chromosome organization: new facts, new models. Trends Cell Biol 17: 127–134 [DOI] [PubMed] [Google Scholar]
- Bridger JM, Kalla C, Wodrich H, Weitz S, King JA, Khazaie K, Kräusslich H-G, Lichter P (2005) Nuclear RNAs confined to a reticular compartment between chromosome territories. Exp Cell Res 302: 180–193 [DOI] [PubMed] [Google Scholar]
- Brown CR, Silver PA (2007) Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev 17: 100–106 [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA (2005) Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev 19: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB (2006) The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci USA 103: 7688–7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Hase ME, Müller L (1998) Molecular segments of protein Tpr that confer nuclear targeting and association with the nuclear pore complex. Exp Cell Res 245: 43–56 [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW (1997) Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol 136: 515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N, Fire A, Samuels M, Sharp PA, Baltimore D (1981) Inhibition of transcription factor activity by poliovirus. Cell 27: 555–561 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S (2006) Chromosome territories—a functional nuclear landscape. Curr Opin Cell Biol 18: 307–316 [DOI] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schröck E, Speicher MR, Mathieu U, Jauch A, Emmerich P, Scherthan H, Ried T, Cremer C, Lichter P (1993) Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb Symp Quant Biol 58: 777–792 [DOI] [PubMed] [Google Scholar]
- Davies HG (1961) Structure in nucleated erythrocytes. J Biophys Biochem Cytol 9: 671–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies HG, Murray AB, Walmsley ME (1974) Electron-microscope observations on the organization of the nucleus in chicken erythrocytes and a superunit thread hypothesis for chromosome structure. J Cell Sci 16: 261–299 [DOI] [PubMed] [Google Scholar]
- Davies MV, Pelletier J, Meerovitch K, Sonenberg N, Kaufman RJ (1991) The effect of poliovirus proteinase 2Apro expression on cellular metabolism. Inhibition of DNA replication, RNA polymerase II transcription, and translation. J Biol Chem 266: 14714–14720 [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA (2009) Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell 20: 2146–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DP, Patel SS, Uversky V, Fink AL, Rexach M (2003) Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci USA 100: 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F (2006) Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 26: 7858–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Enarson P, Enarson M, Bastos R, Burke B (1998) Amino-terminal sequences that direct nucleoporin Nup153 to the inner surface of the nuclear envelope. Chromosoma 107: 228–236 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Aebi U (2002) The vertebrate nuclear pore complex: from structure to function. Results Probl Cell Differ 35: 25–48 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Koser J, Aebi U (2004) The nuclear pore complex: a jack of all trades? Trends Biochem Sci 29: 175–182 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Maco B, Fager AM, Köser J, Sauder U, Ullman KS, Aebi U (2002) Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol 140: 254–267 [DOI] [PubMed] [Google Scholar]
- Francastel C, Schubeler D, Martin DI, Groudine M (2000) Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol 1: 137–143 [DOI] [PubMed] [Google Scholar]
- Frank J, Verschoor A, Boublik M (1981) Computer averaging of electron micrographs of 40S ribosomal subunits. Science 214: 1353–1355 [DOI] [PubMed] [Google Scholar]
- Franke WW, Scheer U (1974) Structures and functions of the nuclear envelope. In The Cell Nucleus, Busch H (ed), pp 219–347. New York: Academic Press Inc [Google Scholar]
- Frosst P, Guan T, Subauste C, Hahn K, Gerace L (2002) Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol 156: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Garcia-Segura LM, Lafarga M, Berciano MT, Hernandez P, Andres MA (1989) Distribution of nuclear pores and chromatin organization in neurons and glial cells of the rat cerebellar cortex. J Comp Neurol 290: 440–450 [DOI] [PubMed] [Google Scholar]
- Ghadially FN, Senoo A, Fuse Y (1985) A serial section study of nuclear pockets containing nuclear material. J Submicrosc Cytol 17: 687–694 [PubMed] [Google Scholar]
- Gitlin L, Karelsky S, Andino R (2002) Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418: 430–434 [DOI] [PubMed] [Google Scholar]
- Goldberg MW, Solovei I, Allen TD (1997) Nuclear pore complex structure in birds. J Struct Biol 119: 284–294 [PubMed] [Google Scholar]
- Görisch SM, Lichter P, Rippe K (2005a) Mobility of multi-subunit complexes in the nucleus: accessibility and dynamics of chromatin subcompartments. Histochem Cell Biol 123: 217–228 [DOI] [PubMed] [Google Scholar]
- Görisch SM, Wachsmuth M, Toth KF, Lichter P, Rippe K (2005b) Histone acetylation increases chromatin accessibility. J Cell Sci 118: 5825–5834 [DOI] [PubMed] [Google Scholar]
- Green DM, Johnson CP, Hagan H, Corbett AH (2003) The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci USA 100: 1010–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA (2002) Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell 13: 1282–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA (2004) Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell 15: 1991–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Xu S, Powers MA (2003) Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol Biol Cell 14: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin KE, Sarnow P (2001) Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J 20: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga JY, Hamilton MG, Petermann ML (1970) Electron microscopic observations on the large subunit of the rat liver ribosome. J Cell Biol 47: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R (2004) Internal organisation of the nucleus: assembly of compartments by macromolecular crowding and the nuclear matrix model. Biol Cell 96: 595–601 [DOI] [PubMed] [Google Scholar]
- Hancock R (2007) Packing of the polynucleosome chain in interphase chromosomes: evidence for a contribution of crowding and entropic forces. Semin Cell Dev Biol 18: 668–675 [DOI] [PubMed] [Google Scholar]
- Hase ME (2003) Molecular and Ultrastructural Analysis of Tpr, a Nuclear Pore Complex-Attached Coiled-Coil Protein. Stockholm: Karolinska University Press. ISBN 91-7349-525-5 [Google Scholar]
- Hase ME, Cordes VC (2003) Direct interaction with Nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14: 1923–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Kuznetsov NV, Cordes VC (2001) Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol Biol Cell 12: 2433–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höger TH, Grund C, Franke WW, Krohne G (1991) Immunolocalization of lamins in the thick nuclear lamina of human synovial cells. Eur J Cell Biol 54: 150–156 [PubMed] [Google Scholar]
- Hüve J, Wesselmann R, Kahms M, Peters R (2008) 4Pi microscopy of the nuclear pore complex. Biophys J 95: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Jackson DA, Cook PR (2000) The path of RNA through nuclear pores: apparent entry from the sides into specialized pores. J Cell Sci 113: 291–302 [DOI] [PubMed] [Google Scholar]
- Ida-Hosonuma M, Iwasaki T, Yoshikawa T, Nagata N, Sato Y, Sata T, Yoneyama M, Fujita T, Taya C, Yonekawa H, Koike S (2005) The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J Virol 79: 4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA (2003) The principles of nuclear structure. Chromosome Res 11: 387–401 [DOI] [PubMed] [Google Scholar]
- Jarnik M, Aebi U (1991) Toward a more complete 3D structure of the nuclear pore complex. J Struct Biol 107: 291–308 [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Allen TD, Rutherford S, Bucci M, Wente SR, Goldberg MW (2004) Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J Struct Biol 145: 272–288 [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Daneholt B, Allen TD (1996) RNP export is mediated by structural reorganization of the nuclear pore basket. J Mol Biol 260: 304–311 [DOI] [PubMed] [Google Scholar]
- Köhler A, Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Krull S, Thyberg J, Björkroth B, Rackwitz HR, Cordes VC (2004) Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell 15: 4261–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov NV, Sandblad L, Hase ME, Hunziker A, Hergt M, Cordes VC (2002) The evolutionarily conserved single-copy gene for murine Tpr encodes one prevalent isoform in somatic cells and lacks paralogs in higher eukaryotes. Chromosoma 111: 236–255 [DOI] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8: 104–115 [DOI] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F (2008) Tpr directly binds to Mad1 and Mad2 and is important for the Mad1–Mad2-mediated mitotic spindle checkpoint. Genes Dev 22: 2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P, Ellenberg J (2006) Monitoring the permeability of the nuclear envelope during the cell cycle. Methods 38: 17–24 [DOI] [PubMed] [Google Scholar]
- Lidsky PV, Hato S, Bardina MV, Aminev AG, Palmenberg AC, Sheval EV, Polyakov VY, van Kuppeveld FJ, Agol VI (2006) Nucleocytoplasmic traffic disorder induced by cardioviruses. J Virol 80: 2705–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M, Maffini S, Orr B, Ding Y, Florindo C, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H (2009) Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol 184: 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Velazquez G, Marquez J, Ubaldo E, Corkidi G, Echeverria O, Vazquez Nin GH (1996) Three-dimensional analysis of the arrangement of compact chromatin in the nucleus of G0 rat lymphocytes. Histochem Cell Biol 105: 153–161 [DOI] [PubMed] [Google Scholar]
- Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH (2007) Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem 282: 3042–3049 [DOI] [PubMed] [Google Scholar]
- Maul GG (1971) On the octagonality of the nuclear pore complex. J Cell Biol 51: 558–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG (1976) Fibrils attached to the nuclear pore prevent egress of SV40 particles from the infected nucleus. J Cell Biol 70: 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG (1977) The nuclear and the cytoplasmic pore complex: structure, dynamics, distribution, and evolution. Int Rev Cytol Suppl 6: 75–186348630 [Google Scholar]
- Mehlin H, Skoglund U, Daneholt B (1991) Transport of Balbiani ring granules through nuclear pores in Chironomus tentans. Exp Cell Res 193: 72–77 [DOI] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A (2006) Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21: 811–823 [DOI] [PubMed] [Google Scholar]
- Misteli T (2005) Concepts in nuclear architecture. BioEssays 27: 477–487 [DOI] [PubMed] [Google Scholar]
- Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Cooper CS (1992) The human tpr gene encodes a protein of 2094 amino acids that has extensive coiled-coil regions and an acidic C-terminal domain. Oncogene 7: 2329–2333 [PubMed] [Google Scholar]
- Murray AB, Davies HG (1979) Three-dimensional reconstruction of the chromatin bodies in the nuclei of mature erythrocytes from the newt Triturus cristatus: the number of nuclear envelope-attachment sites. J Cell Sci 35: 59–66 [DOI] [PubMed] [Google Scholar]
- Nickerson J (2001) Experimental observations of a nuclear matrix. J Cell Sci 114: 463–474 [DOI] [PubMed] [Google Scholar]
- Nonomura Y, Blobel G, Sabatini D (1971) Structure of liver ribosomes studied by negative staining. J Mol Biol 60: 303–323 [DOI] [PubMed] [Google Scholar]
- Park N, Katikaneni P, Skern T, Gustin KE (2008) Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J Virol 82: 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patre M, Tabbert A, Hermann D, Walczak H, Rackwitz HR, Cordes VC, Ferrando-May E (2006) Caspases target only two architectural components within the core structure of the nuclear pore complex. J Biol Chem 281: 1296–1304 [DOI] [PubMed] [Google Scholar]
- Politz JC, Tuft RA, Pederson T, Singer RH (1999) Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr Biol 9: 285–291 [DOI] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz FR, Forbes DJ (1995) Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol 128: 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunuske AJ, Liu J, Elgort S, Joseph J, Dasso M, Ullman KS (2006) Nuclear envelope breakdown is coordinated by both Nup358/RanBP2 and Nup153, two nucleoporins with zinc finger modules. Mol Biol Cell 17: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G (1995) The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81: 215–222 [DOI] [PubMed] [Google Scholar]
- Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS (2008) The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci 121: 1119–1127 [DOI] [PubMed] [Google Scholar]
- Rhodin JA (1968) Ultrastructure of mammalian venous capillaries, venules, and small collecting veins. J Ultrastruct Res 25: 452–500 [DOI] [PubMed] [Google Scholar]
- Richter K, Nessling M, Lichter P (2008) Macromolecular crowding and its potential impact on nuclear function. Biochim Biophys Acta 1783: 2100–2107 [DOI] [PubMed] [Google Scholar]
- Rippe K (2007) Dynamic organization of the cell nucleus. Curr Opin Genet Dev 17: 373–380 [DOI] [PubMed] [Google Scholar]
- Ris H (1989) Three-dimensional imaging of cell ultrastructure with high resolution low voltage SEM. Inst Phys Conf Ser 98: 657–662 [Google Scholar]
- Ris H (1997) High-resolution field-emission scanning electron microscopy of nuclear pore complex. Scanning 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Ris H, Malecki M (1993) High-resolution field emission scanning electron microscope imaging of internal cell structures after Epon extraction from sections: a new approach to correlative ultrastructural and immunocytochemical studies. J Struct Biol 111: 148–157 [DOI] [PubMed] [Google Scholar]
- Rubinstein SJ, Hammerle T, Wimmer E, Dasgupta A (1992) Infection of HeLa cells with poliovirus results in modification of a complex that binds to the rRNA promoter. J Virol 66: 3062–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli P, Lattanzi G, Ognibene A, Columbaro M, Capanni C, Merlini L, Maraldi NM, Squarzoni S (2001) Nuclear alterations in autosomal-dominant Emery–Dreifuss muscular dystrophy. Muscle Nerve 24: 826–829 [DOI] [PubMed] [Google Scholar]
- Sanger JW, Sanger JM (2001) Fishing out proteins that bind to titin. J Cell Biol 154: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Dabauvalle MC, Merkert H, Benevente R (1988) The nuclear envelope and the organization of the pore complexes. Cell Biol Int Rep 12: 669–689 [DOI] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R (2007) Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21: 3027–3043 [DOI] [PubMed] [Google Scholar]
- Seksek O, Biwersi J, Verkman AS (1997) Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J Cell Biol 138: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Schober H, Fraser P, Gasser SM (2007) Gene regulation through nuclear organization. Nat Struct Mol Biol 14: 1049–1055 [DOI] [PubMed] [Google Scholar]
- Shah S, Tugendreich S, Forbes D (1998) Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol 141: 31–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Igo M, Yalamanchili P, Berk AJ, Dasgupta A (1996) DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol Cell Biol 16: 4163–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Matsuoka Y, Yoneda Y (2002) Nucleocytoplasmic transport of proteins and poly(A)+ RNA in reconstituted Tpr-less nuclei in living mammalian cells. Genes Cells 7: 421–434 [DOI] [PubMed] [Google Scholar]
- Skaggs HS, Xing H, Wilkerson DC, Murphy LA, Hong Y, Mayhew CN, Sarge KD (2007) HSF1-TPR interaction facilitates export of stress-induced HSP70 mRNA. J Biol Chem 282: 33902–33907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana K, Gyorkey F, Gyorkey P, Busch H (1970) Studies on the ultrastructure of nucleoli in human smooth muscle cells. Exp Cell Res 60: 175–184 [DOI] [PubMed] [Google Scholar]
- Soop T, Ivarsson B, Björkroth B, Fomproix N, Masich S, Cordes VC, Daneholt B (2005) Nup153 affects entry of messenger and ribosomal ribonucleoproteins into the nuclear basket during export. Mol Biol Cell 16: 5610–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens BJ, Swift H (1966) RNA transport from nucleus to cytoplasm in Chironomus salivary glands. J Cell Biol 31: 55–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G (1993) A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 72: 29–38 [DOI] [PubMed] [Google Scholar]
- Swift H (1959) Studies on nuclear fine structure. Brookhaven Symp Biol 12: 134–152 [PubMed] [Google Scholar]
- Thomas JO, Kornberg RD (1975) An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci USA 72: 2626–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K, Madden SC, Zeldis LJ (1968) Fine structural alterations of interphase nuclei of lymphocytes stimulated to growth activity in vitro. J Cell Biol 39: 630–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde KE (1971) Scattering. In Physical Biochemistry, Hager L, Wold F (eds), pp 180–202. Englewood Cliffs, New Jersey, USA: Prentice-Hall Inc. [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ (2001) Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol 155: 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschure PJ, van der Kraan I, Manders EMM, Hoogstraten D, Houtsmuller AB, van Driel R (2003) Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep 4: 861–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F (2005) Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J 24: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser AE, Jaunin F, Fakan S, Aten JA (2000) High resolution analysis of interphase chromosome domains. J Cell Sci 113: 2585–2593 [DOI] [PubMed] [Google Scholar]
- Vomastek T, Iwanicki MP, Burack WR, Tiwari D, Kumar D, Parsons JT, Weber MJ, Nandicoori VK (2008) Extracellular signal-regulated kinase 2 (ERK2) phosphorylation sites and docking domain on the nuclear pore complex protein Tpr cooperatively regulate ERK2–Tpr interaction. Mol Cell Biol 28: 6954–6966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW (2001) The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J 20: 5703–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML (1959) Further observations on the nuclear envelope of the animal cell. J Biophys Biochem Cytol 6: 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalamanchili P, Weidman K, Dasgupta A (1997) Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology 239: 176–185 [DOI] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL (2009) Nuclear neighborhoods and gene expression. Curr Opin Genet Dev 19: 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR (1997) A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci 110: 927–944 [DOI] [PubMed] [Google Scholar]
- Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P (1993) Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res 1: 93–106 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.