Abstract
Recent studies have shown a critical function for the ubiquitin-proteasome system (UPS) in regulating the signalling network for DNA damage responses and DNA repair. To search for new UPS targets in the DNA damage signalling pathway, we have carried out a non-biased assay to identify fast-turnover proteins induced by various types of genotoxic stress. This endeavour led to the identification of Rad17 as a protein exhibiting a distinctive pattern of upregulation followed by subsequent degradation after exposure to UV radiation in human primary cells. Our characterization showed that UV-induced Rad17 oscillation is mediated by Cdh1/APC, a ubiquitin-protein ligase. Studies using a degradation-resistant Rad17 mutant demonstrated that Rad17 stabilization prevents the termination of checkpoint signalling, which in turn attenuates the cellular re-entry into cell-cycle progression. The findings provide an insight into how the proteolysis of Rad17 by Cdh1/APC regulates the termination of checkpoint signalling and the recovery from genotoxic stress.
Keywords: Cdh1/APC, checkpoint regulation and carcinogenesis, proteolysis, Rad17
Introduction
Cellular responses to DNA damage are tightly controlled by checkpoints. After DNA damage or replication disruption, checkpoint signalling would arrest the progression of the cell cycle at G1 or G2/M, thus enabling DNA repair (Kastan and Bartek, 2004; Harper and Elledge, 2007). Failure at the checkpoint would result in genomic instability and carcinogenesis (Huang and D'Andrea, 2006; Halazonetis et al, 2008; Helleday et al, 2008; Nouspikel, 2008; Hannah and Zhou, 2009). Targeting the circuitry of DNA damage responses and DNA repair has been an important focus in the development of anti-cancer drug as well as novel cancer therapeutic strategies (Helleday et al, 2008). On exposure of cells to genotoxic stress, ATR and ATM protein kinases of the PI-3 protein-like kinase family would activate the DNA damage signalling circuitry. ATR is central to cellular responses to UV and other agents, whereas ATM principally responds to double-strand breaks in DNA (Zhou and Elledge, 2000; Abraham, 2001; Kastan and Bartek, 2004). Studies in yeast and mammal have revealed that several Rad proteins, including Rad17, Rad1, Rad9 and Hus1, function as important components during the activation of both the DNA damage and replication checkpoints (Abraham, 2001; Harper and Elledge, 2007). Rad17 is structurally similar to subunits of the replication factor C (RFC) bearing Walker A-type and B-type nucleotide-binding motifs and potentially acts as an important component of the PCNA-like checkpoint-loading complex, which loads the 9-1-1 checkpoint-sliding clamp (Siede et al, 1996; Paulovich et al, 1997; Dean et al, 1998; Venclovas and Thelen, 2000). On the activation of DNA damage response machinery, binding of replication protein A (RPA) to the damaged single-strand DNA recruits ATR through its regulatory subunit ATRIP (Zou and Elledge, 2003; Cimprich and Cortez, 2008), which leads to phosphorylation of Rad17 (Bao et al, 2001; Wang et al, 2006). Recent report implied a function for phosphorylated Rad17 in uploading Claspin, a recruiter of Chk1 to the chromatin, whereas earlier studies also suggested a possible function for phosphorylated Rad17 in loading 9-1-1 (Bao et al, 2001; Wang et al, 2006). Rad17-mediated activation of Chk1 in turn is thought to initiate downstream withdraw of cell-cycle progression (Abraham, 2001).
Rad17 was initially identified in fission yeast as a DNA damage checkpoint protein (Siede et al, 1996; Paulovich et al, 1997). Yeast genetic analyses implicated the importance of Rad17 in cell-cycle arrest and cellular survival in response to DNA damage and DNA replication blockage (Paulovich et al, 1997). Rad17-knockout mouse displayed embryonic lethality with numerous developmental defects (Budzowska et al, 2004). Targeted deletion of Rad17 in HCT116 and DT40 cells inhibits mitotic as well as S-phase damage checkpoint functions and further results in chromosomal aberration and endoreduplication (Wang et al, 2003; Kobayashi et al, 2004). Biochemical study of the mechanism by which Rad17 is involved in the activation of DNA damage checkpoints in human cell has demonstrated a functional relationships between Rad17 and ATR, in which ATR, in response to DNA damage would phosphorylate Rad17 at two SQ motifs (Ser 635 and Ser 645) at the C-terminus (Dean et al, 1998; Li et al, 1999; Bao et al, 2001). Analyses of Rad17 gene structure has demonstrated an essential functional domain termed P-loop, a chaperone-like ATPase domain, which facilitates Rad17-mediated checkpoint activation by connecting Rad17 to DNA (Dean et al, 1998; Venclovas and Thelen, 2000). In addition, an oscillating pattern of phosphorylation of Rad17 is observed at different stages of the cell cycle, where periodic interaction between phosphorylated Rad17 and DNA polymerase ɛ is detected (Post et al, 2001). Although a function for Rad17 phosphorylation has been delineated by the above studies, the exact mechanism by which phosphorylated Rad17 associates with ATR remains unclear and controversial (Bao et al, 2001; Zou et al, 2002). The study of Rad17 in DNA damage responses has focused extensively on the initial 2 h after cellular exposure to genotoxic stress (Bao et al, 2001; Zou et al, 2002), but this focus has limited our understanding of the kinetics of Rad17 regulation during the entire process of DNA damage responses.
We have carried out a non-biased assay to identify fast-turnover proteins induced by various types of genotoxic stress. This effort led to the identification of Rad17 as a highly fluctuating protein after exposure to UV radiation in human primary cells. We have demonstrated that the UV-induced Rad17 oscillation is mediated by a ubiuqitin-protein ligase, Cdh1/APC. We have further identified a degron region that mediates Rad17 proteolysis and have engineered a degradation-resistant Rad17 mutant. Studies using the Rad17 stable mutant have unveiled that a failure in Rad17 degradation disrupts the termination of checkpoint signalling, which in turn attenuates the cellular re-entry into cell-cycle progression. The results suggest that proteolytic regulation of Rad17 is a mechanism for turning off checkpoint signalling after the completion of checkpoint response, revealing a new avenue to deactivate checkpoint signalling.
Results
Time-dependent proteolysis of Rad17 in response to UV in human primary cells
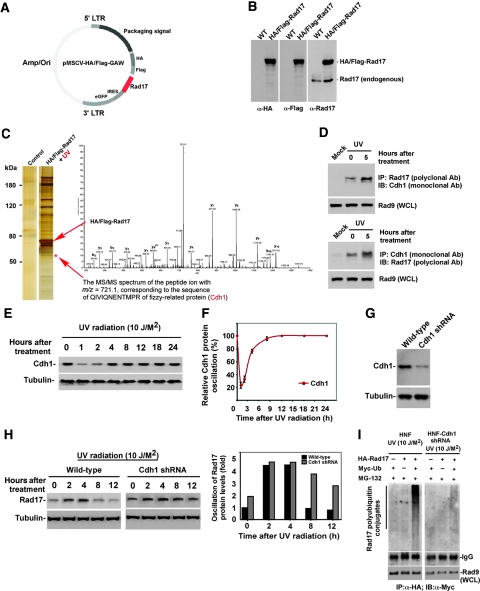
Recent studies have revealed the critical function of the ubiquitin-proteasome system (UPS) in the surveillance of DNA damage and repair of DNA lesions with many proteins that function as sensors, transducers or effectors are regulated by either mono-ubiquitylation or polyubiquitylation (Zhou and Elledge, 2000; Huang and D'Andrea, 2006). To understand the function of poly-ubiquitylation-mediated proteolysis in genomic integrity, we systematically identified genotoxic stress-induced turnover proteins in DNA damage signalling pathways by using a small-pool expression cloning and large-scale immunoblotting analysis (Zou et al, 1999; Wood et al, 2005). Our effort led to the identification of several genotoxic stress-induced fluctuating proteins, with Rad17 being one of the most interesting candidates. Its dramatic oscillating expression triggered by UV radiation had not been reported earlier.
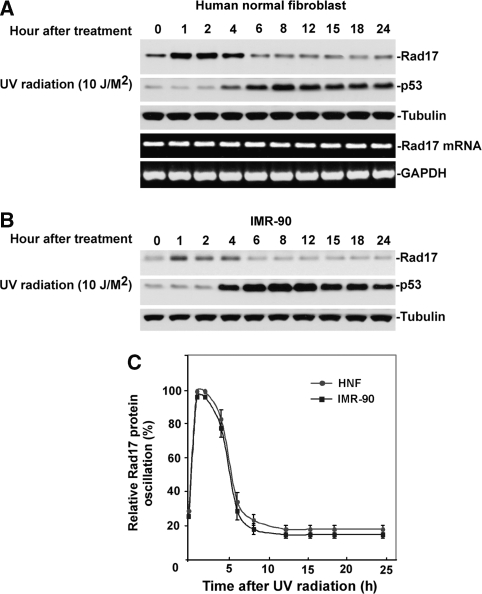
To confirm that this time-dependent variation of Rad17 after exposure to UV is a consequence of post-transcriptional modification, we examined the kinetics of Rad17 protein alteration in response to UV in both human normal skin fibroblast (HNF) cells as well as another human lung fibroblast cell, IMR-90. As shown in Figure 1A–C, on exposure to UV, Rad17 accumulates reaching a plateau in 1 h (in both HNF and IMR-90). Rad17 protein levels then gradually drops after 4 h and return to a basal level after 6 to 8 h. Examination of the kinetics of Rad17 mRNA in HNF suggests the drop in Rad17 levels after 4 h of exposure to UV is mediated by post-transcriptional events and may have important biological relevance (Figure 1A).
Figure 1.
Time-dependent Rad17 proteolysis in response to UV in human primary cells. (A) On DNA damage, Rad17 accumulates, reaching a plateau in 1 h in HNF cells. Rad17 protein levels then gradually decline after 4 h and reach a basal level after 6 to 8 h. The UV-activated damage response was indicated by the kinetics of p53. The kinetics of Rad17 mRNA after exposure of cells to UV was examined using RT–PCR, with no significant alteration observed in Rad17 mRNA levels. Equivalent amount of loaded protein was estimated by measuring tubulin. (B) A similar pattern of Rad17 alteration induced by UV was found in another human primary cell type, IMR-90. (C) Summary of UV-induced alterations of Rad17 protein levels in HNF and IMR-90 cells.
Rad17 undergoes UPS-mediated proteolysis after exposure to UV radiation
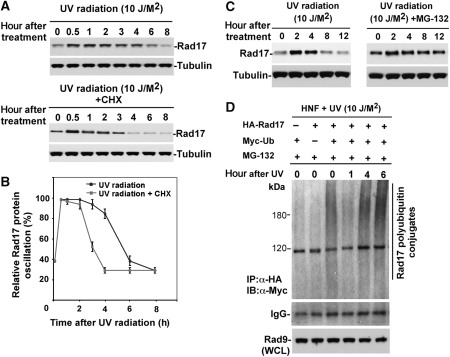
To determine whether UV exposure alters Rad17 protein stability, we treated HNF cells with cycloheximide (20 mM) to block protein synthesis, and then monitored the rate of decline of the endogenous Rad17 in control and UV-treated cells by immunoblotting (Wan et al, 2001; Liu et al, 2007). The result indicated that UV exposure decreased the apparent half-life of the Rad17 protein from ∼8 to 4 h (Figure 2A and B), where the Rad17 half-life in HNF cell without UV treatment is ∼8 h (Supplementary Figure S2). To determine whether proteasomal activity was involved in Rad17 downregulation, we treated HNF cells with UV in the absence or presence of proteasome inhibitor MG-132 (10 mM) (Liu et al, 2007). As shown in Figure 2C, treatment with MG-132 attenuated the decrease in Rad17 protein levels induced by UV radiation. Similar results were obtained in experiments with a second proteasome inhibitor, ALLN (data not shown), suggesting that the time-dependent drop in Rad17 protein levels induced by UV is mediated by the UPS. We next determined whether the UV-induced turnover of Rad17 protein levels occurs through a ubiquitylation cascade, a common prelude to proteasomal degradation. We co-transfected Myc-tagged ubiquitin and HA-tagged Rad17 into HNF cells and collected cells at different time points after exposure to UV (a special heat-shock method was used to enhance efficiency of transfection in the fibroblast cells) (Figure 2D). UV-induced ubiquitylation of Rad17 was measured by immunoprecipitation of Rad17 by anti-HA antibody (lysates were prepared with denaturing condition) coupled with immunoblotting using anti-Myc antibody (Liu et al, 2007). As indicated in Figure 2D, the levels of ubiquitin-conjugated Rad17 dropped initially but elevated after 1 h after exposure to UV radiation, particularly in the presence of MG-132. Taken together, the above results suggested that UV-induced Rad17 turnover is mediated by the UPS.
Figure 2.
UV-induced Rad17 degradation is through a ubiquitin-proteasome pathway. (A) Alteration of Rad17 protein half-life in response to UV radiation. HNF cells were pretreated for 15 min with 20 mM cycloheximide followed by UV treatment. Rad17 protein levels drop to basal levels in 8 h after exposure to UV in the absence of cycloheximide, whereas UV exposure decreased the apparent half-life of the Rad17 protein from ∼8 to 4 h in the presence of cycloheximide. (B) Summary of UV-induced Rad17 degradation in the absence or presence of cycloheximide. (C) Pre-incubation of proteasome inhibitor MG-132 (10 uM) attenuates the UV-induced Rad17 downregulation in HNF cells. (D) Rad17 is ubiquitylated responding to UV radiation. Myc-tagged ubiquitin and HA-tagged Rad17 were co-transfected into HNF cells. Cells were exposed to UV and harvested at the indicated time points. HA-Rad17 was pulled down by antibody against HA. The ubiquitin-conjugated HA-Rad17 was subsequently detected by immunoblotting using anti-Myc antibody.
Association of UV-induced Rad17 degradation with localization on chromatin
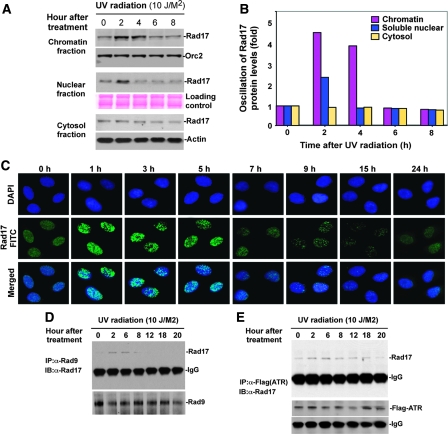
A requirement that damage-induced Rad17 to translocate onto chromatin has been controversial (Bao et al, 2001; Zou et al, 2002). A model for UV-induced association of Rad17 and chromatin was suggested for MCF7, a breast cancer cell line but was not observed for HEK293T, an immortalized human embryonic kidney cell line (Bao et al, 2001; Zou et al, 2002). Given the difference between Rad17 protein levels in cancer cells and normal cells (data not shown), we decided to examine the association of UV-induced Rad17 proteolysis with chromatin in human primary cultured cells. HNF cells were treated with UV and collected at intervals (Figure 3A). Cells were fractionated into cytoplasmic proteins, soluble nuclear proteins and chromatin-bound proteins (Zou et al, 2002). As shown in Figure 3A and B, a basal level of Rad17 was detected in the chromatin fraction. On DNA damage, chromatin-associated Rad17 protein increased sharply, reaching a peak in 2 h and then gradually declining to basal level after 6 to 8 h. Interestingly, a minor oscillation of Rad17 protein levels was observed in soluble nuclear fraction, whereas no significant Rad17 alteration was measured in the cytoplasmic fractions (Figure 3A). To determine whether Rad17 could form foci on UV-damaged DNA, we performed immunocytochemistry of Rad17 in HNF cells at indicated time points after exposure to UV (Figure 3C). The results showed that Rad17 formed transient nuclear foci in response to UV radiation. Collectively, the above results suggest that in human primary cells, proteolytic regulation of Rad17 in response to DNA damage occurs during its association with chromatin.
Figure 3.
Proteolytic regulation of Rad17 is associated with chromatin-based DNA damage response and dissociation of checkpoint complex. (A) UV-induced Rad17 translocation and its degradation on chromatin. HNF cells were treated with UV and harvested at indicated time points. Cell pellets were fractionated into cytosol, soluble nuclear and chromatin-enriched portions. UV-induced Rad17 degradation was examined for different cellular compartments. Normalized protein loading for chromatin, nuclear and cytosol fractions was measured by ORC2, Ponceau S staining and actin, respectively. (B) Summary of UV-induced Rad17 alterations in different cellular compartment. (C) Transient formation of Rad17 nuclear foci in response to UV radiation. HNF cells were treated with UV and fixed at the different time points followed by immunostaining with antibody against Rad17. (D) Kinetics of interaction of Rad17 and 9-1-1 induced by DNA damage is regulated by Rad17 proteolysis. HNF cells were exposed to UV and collected at the indicated time points. Endogenous Rad9 complexes were immunoprecipitated with anti-Rad9 antibody (polyclonal). Rad9 IP complex was then immunoblotted with antibody against Rad17. Co-IP Rad9 was determined by immunoblotting using antibody (monoclonal) against Rad9. (E) Kinetics of interaction of Rad17 and ATR in response to DNA damage. Flag-tagged ATR was transfected into HNF cells. Ectopically expressed Flag-ATR was immunoprecipitated with anti-Flag antibody. ATR IP complexes were immunoblotted with Rad17 antibody. Co-IP Flag-ATR was measured by immunoblotting using antibody against Flag.
To further test the protein abundance of Rad17 in association with the checkpoint complex after cellular exposure to UV radiation, we measured the specific interaction of Rad17 with the 9-1-1 complexes and the checkpoint kinase, ATR. We examined protein–protein interactions between endogenous Rad17 with Rad9 and endogenous Rad17 with ectopic Flag-tagged ATR. As indicated in Figure 3D, the protein abundance of Rad17 interacting with Rad9 increased after UV radiation and gradually dropped after 6 h. Similar observation was seen between Rad17 with ATR in response to UV (Figure 3E). Given the oscillation of Rad17 protein abundance after cellular exposure to UV (Figures 1A–C and 2A), any alteration of Rad17 abundance in association with the checkpoint complex is potentially due to its change in protein levels. Taken together, the above results suggest that time-dependent proteolysis of Rad17 is involved in checkpoint function.
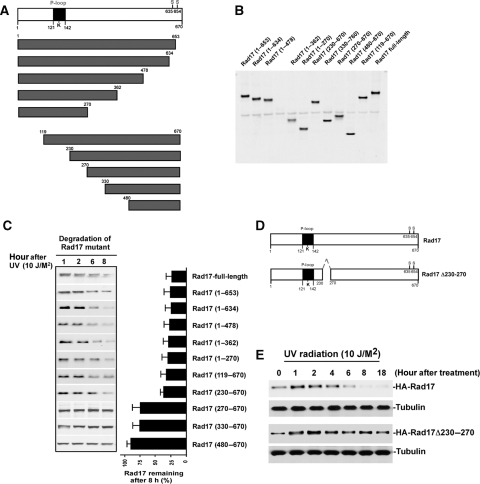
Molecular mapping of the region on Rad17 that mediates UV-induced Rad17 degradation
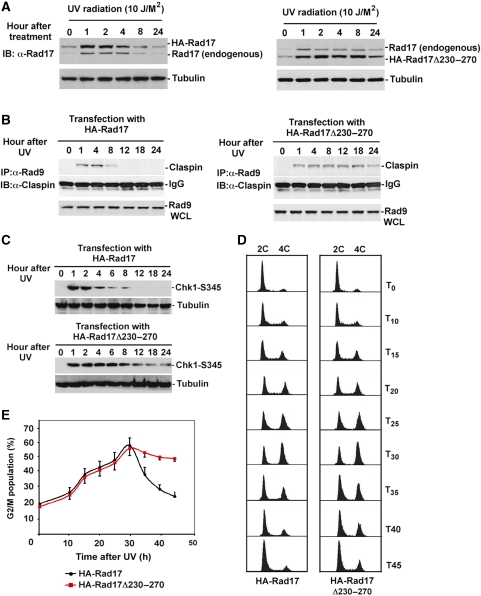
Determination of molecular sites in Rad17 that facilitate its ubiquitylation for subsequent destruction could provide a mean of engineering a stable mutant to dissect the biological consequences of UV-induced Rad17 degradation. To map such a molecular degron, we engineered a series of HA-tagged deletion mutants for Rad17 as indicated in Figure 4A. To evaluate the stability of the mutants in response to UV, we transfected 10 Rad17 mutants into HNF cells, respectively, and measured their alteration after exposure to UV by immunoblotting. The results in Figure 4C showed that the amino-acid stretch from 230–270 on Rad17 is vital in determining its stability in response to DNA damage. The absence of this region significantly stabilized Rad17 protein in response to UV. To engineer a Rad17 functionally stable mutant to address the consequence of its degradation by functional interference, we designed a Rad17 mutant, where the degron region (amino acid 230–270) was eliminated (Figure 4D). As indicated in Figure 4E, the absence of this degron region on Rad17 significantly attenuates UV-induced Rad17 degradation.
Figure 4.
Mapping of the functional domain that mediates UV-induced Rad17 degradation. (A) Schematic presentation of Rad17 structure and strategy for mutagenesis. (B) Expression of a set of Rad17 mutants. (C) Stability of Rad17 mutants in response to UV radiation. A series of HA-tagged Rad17 mutants were transfected into HNF cells. Stability for each mutant was measured by immunoblotting. (D) Design of Rad17 stable mutant based on information of protein degradation summarized in (C). Amino acids from 230 to 270 were deleted for a stable Rad17 mutant. (E) Both HA-tagged wild type and stable Rad17 were transfected into HNF cells and their stability in response to UV radiation was evaluated by immunoblotting.
Identification of Cdh1/APC as a putative E3 ligase that mediates UV-induced Rad17 degradation
To search for the potential ubiquitin-protein ligase that governs UV-induced Rad17 degradation, we chose to identify Rad17 interacting proteins by purifying Rad17 complexes after exposure to UV. Exploiting a well-established mammalian TAP purification system (a gift from Dr Wade J Harper), we created a TAP-Rad17 construct (Figure 5A) and further established a TAP-Rad17 stable clone in HeLa S3 cells due to its convenient feature for protein purification (Figure 5B) (Knuesel et al, 2003; Sowa et al, 2009). We purified Rad17 complexes from the cell pellets collected from cells exposed over 5 h to UV radiation (the time point of 5 h was decided according to the Rad17 alteration profile of total protein and ubiquitin conjugates in Figures 1A–C and 2D) following the method described earlier (Knuesel et al, 2003; Sowa et al, 2009). The IgG-agroase beads were used as a control resin for the purification. The purified Rad17 complex was then resolved by SDS–PAGE following by silver staining (Figure 5C). The differential protein bands between the control and treatment were isolated and subjected to mass spectrometry (Candiello et al, 2007). As shown in Figure 5C, although we found the UV-induced interaction between Rad17 with Rad9, RFC and some novel associating proteins, we also observed the binding between Rad17 and Cdh1, a substrate recruiting factor for an E3 ligase APC. We subsequently examined the interaction between Rad17 and Cdh1 in human primary cells used in most of our experiments to confirm the specificity of this observed protein–protein interaction. As shown in Figure 5D, the specific interaction between Rad17 and Cdh1 was observed in cells after exposure to UV. To our surprise, we also observed a moderate interaction between Rad17 and Cdh1 from the human primary cells not treating with UV. Our earlier studies implicated Cdh1/APC function in cellular response to UV radiation, in which high-dose UV treatment (>50 J/M2) induces destruction of Cdh1 (Liu et al, 2008). To test whether Cdh1 protein levels are regulated after exposure to low-dose UV treatment (<10 J/M2), we monitored the oscillation of Cdh1 after exposure to UV. As shown in Figure 5E and F, Cdh1 is degraded after UV treatment at dose of 10 J/M2 and the UV-induced decline of Cdh1 levels gradually recovered 2 h after exposure to UV. The complemented overlap of pattern of protein oscillation between Cdh1 and Rad17 after exposure to UV radiation suggests a connection between Cdh1 and Rad17. To further verify whether Cdh1/APC is the E3 ligase that mediates UV-induced Rad17 proteolysis in vivo, we generated human primary cells depleted of Cdh1 (Figure 5G) (Open Biosystem, Inc) following protocol described earlier (Liu et al, 2007) and further evaluated the effect of Cdh1 depletion on UV-mediated Rad17 proteolysis. As shown in Figure 5H and I, depletion of Cdh1 led to slight elevation of Rad17 in HNF cells. Moreover, knockdown of Cdh1 significantly changed the UV-induced pattern of Rad17 protein levels with drastic drop and ubiquitylation over 5 to 6 h after UV treatment in the wild-type cells. All together, the above results suggest that Cdh1/APC is the putative ubiquitin-protein ligase that regulates UV-induced Rad17 proteolysis.
Figure 5.
Identification of Cdh1/APC as a putative E3 ligase that mediates UV-induced Rad17 degradation. (A) Engineered Rad17-TAP construct. (B) Verification of engineered Rad17-TAP expression. Stably expressed and HA and Flag-tagged Rad17 was examined by antibodies against HA and Flag. (C) A silver-stained protein profile of purified Rad17 complexes in response to UV. Analyses of mass spectrometry indicate the presence of Cdh1 in the Rad17 complex. The IgG-agroase was used as control resin for the purification. Analyses of mass spectrometry indicate the presence of Cdh1 in the Rad17 complex. (D) Verification of interaction between Rad17 and Cdh1 by co-immunoprecipitation. Reversed immunoprecipitation coupled western blotting were carried out by using polyclonal anti-Rad17 and monoclonal anti-Cdh1. (E) Measurement of alteration in Cdh1 protein levels at various time points after exposure of cell to UV radiation. (F) Summary of (E). (G) Depletion of Cdh1 in HNF cells by RNA interference. (H) Effect of Cdh1 depletion on UV-induced Rad17 proteolysis. Knockdown of Cdh1 attenuates the UV-mediated Rad17 proteolysis. (I) Depletion of Cdh1 leads to attenuation of UV-induced Rad17 ubiquitylation. Cells were transfected with plasmid as indicated and were harvested 5 h after exposure to UV. HA-tagged Rad17 was pulled down by antibody against HA. The ubiquitin-conjugated HA-Rad17 was then detected by immunoblotting using anti-Myc antibody.
Stabilization of Rad17 results in a prolonged interaction between Rad17 and checkpoint complex
To determine the biological significance of UV-induced Rad17 proteolytic regulation, we attempted to carry out a functional interference analysis by using a non-degradable Rad17 mutant. To determine whether the absence of the degron region in Rad17 impairs the substrate recognition needed for ubiquitylation of Rad17 without impairing its function in the checkpoint response, we initially analysed its association with chromatin and with the checkpoint network in response to DNA damage. As indicated in Figure 6A and B, although protein abundance of wild-type Rad17 associated with checkpoint complex increased at the beginning and dropped over 4 h after exposure to UV, the stabilized Rad17 protein showed a significantly extended period of interaction with Rad9. We next tested the kinetics of protein alteration for both wild type and Rad17Δ230–270 on chromatin after exposure to UV. As shown in Figure 6C and D, the loss of the region 230–270 on Rad17 largely stabilized Rad17 protein levels on chromatin after DNA damage, whereas wild-type Rad17 protein levels dropped over 4 h after exposure to UV. The above results suggest that Rad17Δ230–270 fits the criteria of a stable mutant after cellular treatment with UV, and can be used for functional interference studies.
Figure 6.
Stabilization of Rad17 results in a prolonged interaction between Rad17 and checkpoint complex. (A, B) Measurement of kinetics of interacted wild type and non-degradable Rad17 with checkpoint complex after exposure of cell to UV radiation. A HA-tagged Rad17 stable mutant as well as a control (HA-Rad17) were transfected into HNF cells. Association of the Rad17 stable mutant as well as HA-Rad17 with 9-1-1 complexes in response to UV was detected by immunoprecipitation using antibody against Rad9 followed by immunoblotting with anti-HA antibody. Total expression of Rad9 was determined with 1/12th of the volume of the total cellular extract used for immunoprecipitation. (C) Both wild type and stable Rad17 were transfected into HNF cells. Transfected cells were subsequently exposed to UV and harvested at indicated time points. Protein stability for wild type and stable Rad17 were measured by immunoblotting using anti-HA antibody based on chromatin fraction. Equal protein loading was estimated by measuring Orc2. (D) Summary of UV-induced protein turnover for wild type and stable Rad17.
Stabilized Rad17 disrupts termination of checkpoint signalling, which in turn impairs re-entry into cell-cycle progression
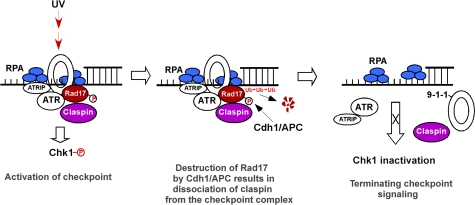
The function of Rad17 in the activation of checkpoint response is thought to be through loading the checkpoint complexes 9-1-1 to chromatin and also presenting Claspin, a component that recruits Chk1, to the checkpoint complex. Afterwards, Claspin subsequently brings Chk1 to ATR for activation using Rad17 (Wang et al, 2006). Our hypothesis is that UV-induced acute accumulation of Rad17 in the first 1 h is important for enhancing the ATR/9-1-1/Rad17/Claspin complex or amplifying checkpoint signal, which permits the activation of Chk1. On completion of the checkpoint response, Rad17 undergoes ubiquitin-dependent proteasomal degradation ∼4 h after exposure to UV radiation. The degradation of Rad17 would dissociate Claspin from the checkpoint complex disconnecting Chk1 and ATR, thereby terminating the upstream activating signal to Chk1 (Figure 8). To test the effect of the stabilized Rad17 on regulating checkpoint response, we have examined the effects of stabilized Rad17 on the association of Claspin with the checkpoint complexes as represented by its association with Rad9 by co-IP analysis. Finally, we have measured the effect of stabilized Rad17 on the phosphorylation of Chk1 by immnoblotting. As shown in Figure 7A, expression of stable Rad17 reduces turnover rate of endogenous Rad17 after exposure to UV. We have also demonstrated that ectopically expressed Rad17 interacts with endogenous Rad17 and probably forms a dimmer or oligomer (Supplementary Figure S3). Accordingly, the altered turnover rate for the endogenous Rad17 could be due to its interaction with the expressed stabilized Rad17. Co-IP experiment has demonstrated that expression of stabilized Rad17 results in a prolonged presence of Claspin with the checkpoint complexes as reflected by the duration of association between Claspin and Rad9 after UV exposure (Figure 7B). In addition, expression of the stabilized Rad17 leads to Chk1 activated for a significantly extended duration, whereas Chk1 phosphorylation diminished 8 h after exposure to UV when cells were transfected with wild-type Rad17 (Figure 7C). The above examinations indicate that proteolysis of Rad17 could be critical for terminating checkpoint signalling by facilitating the dissociation of Claspin from the checkpoint complex.
Figure 7.
Stabilized Rad17 disrupts termination of checkpoint signalling, which in turn impairs re-entry into cell-cycle progression. (A) Expression of stabilized Rad17 interferes the UV-induced proteolysis of endogenous Rad17, which in turn reduces the turnover rate of endogenous Rad17 after exposure to UV. (B) Stabilization of Rad17 enhances a prolonged association between Claspin and checkpoint complex as demonstrated by co-IP assay. (C) Stabilized Rad17 results in Chk1 being phosphorylated for an expanded time. (D) Stabilization of Rad17 is associated with delayed re-entry into cell-cycle progression after exposure to UV radiation. U2OS cells were transfected with wild type and stable Rad17 and then were treated with UV radiation (10 J/M2). Cells were collected at the indicated time points and stained with propidium iodide for FACS analyses. UV radiation causes a delay at G2/M. UV-induced G2/M delay in cells transfected with wild-type Rad17 gradually recovered after 30 h of exposure to UV, whereas transfection of Rad17 stable mutant significantly attenuated the time-dependent recovery from G2/M delay. (E) Summary for experiment in (D).
The failure to orchestrate termination of the checkpoint signalling and the subsequent operation of DNA repair machinery could disturb the cellular re-entry into the cell cycle (Calonge and O'Connell, 2008). To examine whether stabilized Rad17 could influence the re-entry into cell-cycle progression, we measured the kinetics of the cell-cycle profile using a FACS analysis (Liu et al, 2008). As indicated in Figure 7D and E, expression of Rad17 stable mutant in U2OS significantly delayed recovery of G2/M population after 30 h exposure to UV compared with expression of wild-type Rad17. Taken together, the above results suggest that UV-induced Rad17 proteolysis is critical in regulating DNA damage checkpoint. Failure in Rad17 proteolysis disrupts termination of checkpoint signalling, which in turn impairs the recovery from the genotoxic stress (Figure 8).
Figure 8.
A proposed model for the function of Rad17 proteolytic regulation in regulating UV-induced checkpoint response.
Discussion
Proteolysis of Rad17 has critical function in regulating checkpoint termination
Discovering that UV induces Rad17 proteolysis using our non-biased screening has further expanded our understanding of its importance in regulating DNA damage checkpoint response. Although tremendous efforts have been made contributing to understanding the function of Rad17 in the activation of checkpoint, the present findings have significant implication for a novel regulatory function of Rad17 in the termination of checkpoint through its degradation. Activation of Chk1 by ATR in response to genotoxic stress generates a signal to delay cell-cycle progression. Earlier works have shown the crucial function for Rad17 in activating Chk1 by loading checkpoint complex 9-1-1 onto the damaged DNA and uploading Claspin to checkpoint complexes (Bao et al, 2001; Kastan and Bartek, 2004; Harper and Elledge, 2007). The present results have revealed a new function for Rad17 in orchestrating checkpoint control. Time-dependent Rad17 proteolysis results in turning off checkpoint signalling by disconnecting Chk1 from ATR through dissociating Claspin from the checkpoint complexes. Failure on Rad17 proteolysis leads to a disrupted recovery from genotoxic stress. Results from these works advance the current paradigm in regulation of the checkpoint response.
Time-dependent proteolysis of Rad17 regulates checkpoint response
Previous works have concentrated on the importance of Rad17 in first 2h after cellular exposure to genotoxic stress (Dean et al, 1998; Li et al, 1999; Bao et al, 2001). Our monitoring of Rad17 kinetics for a 24-h span in response to DNA damage signalling uncovers a significant Rad17 oscillation after exposure to UV. Result from the ATR mutant cell line further indicated the possible involvement of ATR in this UV-induced alteration of Rad17 (Supplementary Figure S1). The fluctuation of Rad17 in response to UV suggests the presence of a basal level of turnover for Rad17, in which DNA damage would downregulate Rad17 turnover resulting in a transient Rad17 accumulation that is required for the activation of the checkpoint signalling. The result showed that Cdh1 also could interact with Rad17 in the absence of UV radiation imply a basal turnover of Rad17 is mediated by the Cdh1/APC pathway, although more solid work needs to be done to confirm this point. The observation of UV-induced drop of Cdh1 in the first 2 h further indicates a possible connection between the time-dependent downregulation of Cdh1/APC with the upregulation of Rad17 at the beginning after exposure to UV radiation. In 4 to 6 h following the completion of cellular checkpoint activation after exposure to UV radiation, Rad17 undergoes proteolysis, where on the destruction of Rad17 results in loss of the platform, which is required to bridging Claspin to the checkpoint complexes. Recent reports indicate that Chk1 activation requires phosphorylation of its residues Ser317 and Ser345 (Cimprich and Cortez, 2008). Claspin is critical for recruiting Chk1 to ATR, whereas Rad17 functions as a platform for Claspin, thereby facilitating a special interaction between Claspin and ATR (Chini and Chen, 2004; Liu et al, 2006; Wang et al, 2006). We therefore speculate that the dissociation of Claspin from the checkpoint complexes due to Rad17 degradation destroys the interaction between ATR and Chk1, which could be a mechanism that terminates checkpoint signalling.
Deregulated Rad17 proteolysis disrupts checkpoint response that impairs recovery of cell-cycle progression
On exposure to genotoxic stress, a cell would activate the checkpoint to delay cell-cycle progression and acquire time to fix the damaged DNA before a commitment to the next genome duplication. Once the repair is completed, cell survival relies on CDK1 being activated to permit mitosis to occur. Prior studies have shown that phosphorylation of Chk1 at its residues Ser317 and Ser345 induces cell-cycle arrest in G2/M or S phase (Cimprich and Cortez, 2008). An important Chk1 substrate for controlling cell-cycle progression is Cdc25 phosphatase (Abraham, 2001). Phosphorylatation of Ser216 on Cdc25C by Chk1 prevents its activation at G2, at least partially, due to its 14-3-3-mediated translocation to the cytoplasm (Abraham, 2001; Kastan and Bartek, 2004). The consequence of Cdc25A phosphorylation at Ser123 is its degradation (Abraham, 2001; Sancar et al, 2004). Both phosphorylation of Cdc25C and Cdc25A result in cellular arrest at G2/M through inhibition of CDK1 (Abraham, 2001; Sancar et al, 2004). Present results have shown that stabilization of Rad17 disrupts the cell-cycle recovery after completion of checkpoint activation and DNA repair. Stabilized Rad17 probably enables an extended Claspin-mediated communication between Chk1 and ATR, which results in prolonged activation of Chk1. Failure to terminate Chk1 activation leads to uncontrolled phosphorylation of Cdc25 that in turn results in a prolonged suppression of CDK1.
UV-mediated proteolysis of Rad17 is regulated by Cdh1/APC
Recent studies have unveiled a function for Cdh1/APC in genomic integrity and carcinogenesis. Cdh1/APC was believed to target ribonucleotide reductase R2, which is critical for DNA repair (Chabes et al, 2003). Regulation of Cdh1 is implicated in UV-induced response (Liu et al, 2008). Both conditional targeted disruption of Cdh1 in mice and Cdh1-depleted primary cultured cells showed severe aberrant genomic disruption and various types of cancer (Engelbert et al, 2008; Garcia-Higuera et al, 2008). Very recent work indicated that Cdh1 could target Claspin during G1 stage of the cell cycle, whereas inhibiting Cdh1/APC-mediated destruction through deubiquitylation could be important for G2 checkpoint (Bassermann et al, 2008; Gao et al, 2009). Identification of Cdh1/APC as the E3 ligase that mediates UV-induced Rad17 proteolysis implicated in checkpoint control advances the knowledge of Cdh1/APC in genomic integrity. Demonstration of the interplay between Cdh1 and Rad17 opens a new avenue to understand the mechanism of checkpoint regulation.
Integration of the present findings with the current paradigm
Regulation of checkpoint termination has attracted substantial interest for its critical function in orchestrating genomic integrity. Until recently, the mechanism by which checkpoint is turned off after completion of checkpoint activation remains largely unclear. Earlier work from yeast to mammalian cells attempted to uncover the mystery behind checkpoint termination (Calonge and O'Connell, 2008). Dephosphorylation of Chk1 has been thought as one important way to attenuate Chk1 activity with phosphtases Dis2 (PP1 isoform) and PPMID/Wip1 implicated in Chk1 dephosphorylation (den Elzen and O'Connell, 2004; Lu et al, 2005). Dephosphorylation of other components, including TopBP1, Rad3 and H2A, in the checkpoint pathway has been linked to ending the checkpoint signalling (Calonge and O'Connell, 2008). The function of the ubiquitin-proteosome system is connected to the mechanism of checkpoint termination, in which phosphorylated Chk1 could be degraded by SCF, whereas Claspin is destroyed through SCF/β-TRCP (Zhang et al, 2005; Francia et al, 2006; Mailand et al, 2006; Peschiaroli et al, 2006). In addition, polo-like kinase-mediated degradation of Wee1 is thought to be involved in re-entry into cell-cycle progression (van Vugt et al, 2004). The present work discovers a novel regulating mechanism to control checkpoint termination through regulated Rad17 protein degradation, which enhances our current knowledge of checkpoint termination and complements the notions concluded by other groups (Calonge and O'Connell, 2008).
Materials and methods
Plasmids and constructs
Rad17 mutants, including Rad17 (1–653), Rad17 (1–634), Rad17 (478), Rad17 (1–362), Rad17 (1–270), Rad17 (119–670), Rad17 (230–670), Rad17 (270–670), Rad17 (330–670), Rad17 (480–670) and Rad17 Δ230–270, were engineered by PCR using the following primers and then cloned into pCS2-HA, a mammalian expression vector.
5′-AAAAATCGATACC ATGTCAAAAACTTTTCTTAGA-3′
5′-AAAAATCGATACCATGGGATCTATTTTATTAATAACA-3′
5′-AAAAATCGATACCATGATCTCAAATATTAGTTTCAAC-3′
5′-AAAAATCGATACCATGCAGTTTTCTTCTTCAAAAGGA-3′
5′-AAAAATCGAT ACCATGAGGGAATATAGCACATCTATA-3′
5′-AAAAATCGATACCATGATCCAAGATATTGGAAGGCTC-3′
5′-TTGGCGCGCCTGTCCCATCACTCTCGTAGTC-3′
5′-TTGGCGCGCCTCCATTAAGCTGGGCTTCATC-3′
5′-TTGGCGCGCCTTCCGGCACAGTGGCTTGAGT-3′
5′-TTGGCGCGCCTAAAGAGCGTGTATTCCAGTC-3′
5′-TTGGCGCGCCTTTTCTTCGTTTTGATTTTGA-3′
5′-TTGGCGCGCCAGAACACTCTTCCTGAATTTC-3′
5′-AAAATCGATACCATGTCAAAAACTTTTCTTAGACCAAAGGTATCT-3′
5′-AAAAATCGATACCATGAATCAGGTAACAGACTGGG-3′
Antibodies and tissue specimens
Antibodies against Rad17 (H-300), Rad9 (M-389), Rad 9 (B-8), HA (F-7), Myc, Flag, p53 (DO-1), Chk1(S345) were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Antibodies β-actin and α-tubulin were purchased from Sigma-Aldrich (St Louis, MO). Rabbit polyclonal antibody against Claspin was purchased from Bethyl Laboratories (Montgomery, TX). Mouse monoclonal antibody ORC-2 was purchased from Abcam (Cambridge, MA). Goat-anti-rabbit and goat-anti-mouse HRP-conjugated secondary antibodies were purchased from Promega (Madison, WI).
Cell culture and transfection
The HNF cell GM05757B (from ATCC) were maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Larenceville, GA), 1 × concentration of 2 mM L-glutamine, essential and non-essential amino acids and vitamins, 100 μg/ml streptomycin and 100 μg/ml penicillin in 5% CO2 at 37°C. IMR-90 and U2OS were grown in DMEM medium supplemented with 10% FBS, 100 μg/ml streptomycin and 100 μg/ml penicillin in 5% CO2 at 37°C.
Transfection of plasmids into HNF, IMR-90 was carried out using lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A gentle heat-shock strategy (submerge the saran-wrap sealed culture dish containing cells and transfection cocktail in 42°C water bath for 40 s) was used to enhance the transfection efficiency in HNF cells.
UV irradiation
Cells were irradiated with Ultraviolet C (245 nm) at a dose rate of 0.5 J/M2 per second. During irradiation, the cells were covered by a thin layer of phosphate-buffered saline (PBS). Afterwards, PBS was removed and fresh medium was added and the culture dish was returned to the incubator for the indicated times (Liu et al, 2008). Control cells were treated with PBS but were not irradiated.
Semi-quantitative RT–PCR
Total RNAs were extracted using TRIzol reagent (Invitrogen). The quanlity of the RNAs were assessed by 1% denaturing agarose gel electrophoresis and spectrophotometry. Five micrograms of total RNAs of each sample were reverse transcribed to the first strand of cDNA primed with oligo-(dT)12−18 using Transcriptase SuperScript II Preamplification System for First Strand cDNA kit (Invitrogen). Then 0.5–1 μl aliquots of the cDNA was used as template to amplify SLP-2 fragment with Rad17 primers: sense 5′-TTAGTTTCAACCCTGTGGCAC-3′; antisense: 5′-TGGCCGTAAGTTGTTTTCTCC-3′ at annealing temperature of 57°C for 28 cycles. The expression of the housekeeping gene GAPDH was used as an internal control (Liu et al, 2007).
Co-immunoprecipitation
Cell pellets collected at designated time points were lysed in buffer (25 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.5% Nodidet P-40, 1 mM EDTA, 5 mM MaF and 1 × protein inhibitor cocktail) on ice for 30 min. Then, 27G 1/2 syringes were used to shred the DNA. The supernatants were collected after centrifugation at 12 000 g for 30 min. Equal amount of protein lysates at designated time points were aliquoted, and equal amount of primary antibody was added to the above lysates. After rotation at 4°C overnight, equal amount of immobilized protein A/G beads (Pierce, Rockford, IL) were added to the tubes. After rotation again at 4°C for 4 h, the beads were collected by centrifugation at 2500 g for 3 min. Electrophoresis-loading buffer was added to the beads after washing with IP wash buffer (25 mM Tris–HCl, pH 7.5, 150 mM NaCl and 1 × protein inhibitor cocktail) five times. After denaturing at 95°C for 5 min, the supernatants were subject to western blot.
In vivo ubiquitylation assay for Rad17
Cells were co-transfected with Myc-Ub and HA-Rad17. Twenty-four hours later, cells were treated with UV. The in vivo ubiquitylation assay was performed under denaturing condition as described earlier (Liu et al, 2007). Immunoprecipitated complex with anti-HA antibody was resolved by SDS–PAGE. The immunoblotting was performed with anti-Myc antibody.
Chromatin fractionation
Chromatin fractionations were performed as described earlier (Mendez and Stillman, 2000). Cell pellets collected at designated time points were washed with PBS and resuspended in 200 μl solution A (10 mM Hepes, pH 7.9, 10 mM KCl, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 10 mM NaF, 1 mM Na3VO4 and 1 × protein inhibitor cocktail), and the cells were left on ice for 5 min. Cytosolic proteins were separated from nuclei by low-speed centrifugation at 1300 g for 4 min. Isolated nuclei were washed once with solution A and lysed in 200 μl solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT). After a 10-min incubation on ice, soluble nuclear proteins were separated from chromatin by centrifugation at 1700 g for 4 min. Isolated chromatin was washed once with solution B and spun down at high speed of 10 000 g for 1 min. Finally, Chromatin was resuspended in 200 μl of SDS sample buffer and sheared by sonication. The proteins were quantitated.
DNA damage detection by immunofluorescent staining
Cells grown on coverslips were briefly washed in PBS and fixed in −20°C absolute methonal at −20°C for 20 min. Cells were then washed in PBS and permeabilized with 1% Triton X-100 in PBS for 15 min at room temperature followed by washing in PBS and blocking with 3% BSA containing 0.1% Triton X-100 for 15 min at room temperature. Cells were then incubated with primary antibody Rad17 (1:200) overnight at 4°C in a humid chamber and incubated for another 3 h at 37°C the next morning. After a brief wash in PBS, cells were incubated with goat-anti-rabbit FITC-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) for at least 2 h at 37°C, washed with PBS and counterstained with 4′,6-diamidino-2-phenylindole (Vector Laboratories). Cells were analysed using an Olympus (Center Valley) IX81 epifluorescence microscope equipped with digital camera.
FACS analysis
Flow cytometry assay was performed by propidium iodide staining. Cells were digested with trypsin, washed twice with PBS and fixed overnight at 4°C in 70% ethanol. After washing twice with PBS, cells were incubated with 5 μg/ml propidium iodide and 50 μg/ml RNase A in PBS for 1 h at room temperature. Flow-activated cell sorter analysis was carried out using a FACSCalibur flow cytometer (Becton Dickson, Mountain View, CA) with CELLQUEST software.
Supplementary Material
Acknowledgments
We are grateful to the members of the Wan laboratory for their discussion and technical assistance. We thank Drs Vesna Rapic-Otrin and Christopher J Bakkenist for providing cell lines. We thank Drs Wade Harper and Jianping Jin for kindly providing the TAP purification vector and their advice on plasmid engineer, establishment of TAP expression stable cell line and protein purification. We also appreciate discussion with Drs Rick Wood, Patricia Opresko, Bennett Van Houten, Ze've Ronai and Gan Wang. This work is supported by National Institute of Health Grant CA115943. YW is a scholar of the American Cancer Society.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, Chen SM, Abraham RT, Wang XF (2001) ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature 411: 969–974 [DOI] [PubMed] [Google Scholar]
- Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M (2008) The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 134: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzowska M, Jaspers I, Essers J, de Waard H, van Drunen E, Hanada K, Beverloo B, Hendriks RW, de Klein A, Kanaar R, Hoeijmakers JH, Maas A (2004) Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J 23: 3548–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonge TM, O'Connell MJ (2008) Turning off the G2 DNA damage checkpoint. DNA Repair (Amst) 7: 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiello J, Balasubramani M, Schreiber EM, Cole GJ, Mayer U, Halfter W, Lin H (2007) Biomechanical properties of native basement membranes. FEBS J 274: 2897–2908 [DOI] [PubMed] [Google Scholar]
- Chabes AL, Pfleger CM, Kirschner MW, Thelander L (2003) Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci USA 100: 3925–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini CC, Chen J (2004) Claspin, a regulator of Chk1 in DNA replication stress pathway. DNA Repair (Amst) 3: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Lian L, O'Donnell M (1998) cDNA cloning and gene mapping of human homologs for Schizosaccharomyces pombe rad17, rad1, and hus1 and cloning of homologs from mouse, Caenorhabditis elegans, and Drosophila melanogaster. Genomics 54: 424–436 [DOI] [PubMed] [Google Scholar]
- den Elzen NR, O'Connell MJ (2004) Recovery from DNA damage checkpoint arrest by PP1-mediated inhibition of Chk1. EMBO J 23: 908–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbert D, Schnerch D, Baumgarten A, Wasch R (2008) The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene 27: 907–917 [DOI] [PubMed] [Google Scholar]
- Francia S, Weiss RS, Hande MP, Freire R, d'Adda di Fagagna F (2006) Telomere and telomerase modulation by the mammalian Rad9/Rad1/Hus1 DNA-damage-checkpoint complex. Curr Biol 16: 1551–1558 [DOI] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Korenjak M, Tseng A, Wu T, Wan L, Kirschner M, Dyson N, Wei W (2009) Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol Biol Cell 20: 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M (2008) Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol 10: 802–811 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Hannah J, Zhou P (2009) Regulation of DNA damage response pathways by the cullin-RING ubiquitin ligases. DNA Repair (Amst) 8: 536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA (2008) DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8: 193–204 [DOI] [PubMed] [Google Scholar]
- Huang TT, D'Andrea AD (2006) Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 7: 323–334 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Knuesel M, Wan Y, Xiao Z, Holinger E, Lowe N, Wang W, Liu X (2003) Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol Cell Proteomics 2: 1225–1233 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Hirano A, Kumano T, Xiang SL, Mihara K, Haseda Y, Matsui O, Shimizu H, Yamamoto K (2004) Critical role for chicken Rad17 and Rad9 in the cellular response to DNA damage and stalled DNA replication. Genes Cells 9: 291–303 [DOI] [PubMed] [Google Scholar]
- Li L, Peterson CA, Kanter-Smoler G, Wei YF, Ramagli LS, Sunnerhagen P, Siciliano MJ, Legerski RJ (1999) hRAD17, a structural homolog of the Schizosaccharomyces pombe RAD17 cell cycle checkpoint gene, stimulates p53 accumulation. Oncogene 18: 1689–1699 [DOI] [PubMed] [Google Scholar]
- Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J (2006) Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 26: 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li W, Fujita T, Yang Q, Wan Y (2008) Proteolysis of CDH1 enhances susceptibility to UV radiation-induced apoptosis. Carcinogenesis 29: 263–272 [DOI] [PubMed] [Google Scholar]
- Liu W, Wu G, Li W, Lobur D, Wan Y (2007) Cdh1-anaphase-promoting complex targets Skp2 for destruction in transforming growth factor beta-induced growth inhibition. Mol Cell Biol 27: 2967–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Nguyen TA, Donehower LA (2005) Reversal of the ATM/ATR-mediated DNA damage response by the oncogenic phosphatase PPM1D. Cell Cycle 4: 1060–1064 [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Bartek J, Lukas J (2006) Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell 23: 307–318 [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouspikel T (2008) Nucleotide excision repair and neurological diseases. DNA Repair (Amst) 7: 1155–1167 [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Margulies RU, Garvik BM, Hartwell LH (1997) RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 145: 45–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M (2006) SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell 23: 319–329 [DOI] [PubMed] [Google Scholar]
- Post S, Weng YC, Cimprich K, Chen LB, Xu Y, Lee EY (2001) Phosphorylation of serines 635 and 645 of human Rad17 is cell cycle regulated and is required for G(1)/S checkpoint activation in response to DNA damage. Proc Natl Acad Sci USA 98: 13102–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73: 39–85 [DOI] [PubMed] [Google Scholar]
- Siede W, Nusspaumer G, Portillo V, Rodriguez R, Friedberg EC (1996) Cloning and characterization of RAD17, a gene controlling cell cycle responses to DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res 24: 1669–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt MA, Bras A, Medema RH (2004) Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell 15: 799–811 [DOI] [PubMed] [Google Scholar]
- Venclovas C, Thelen MP (2000) Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res 28: 2481–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Liu X, Kirschner MW (2001) The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol Cell 8: 1027–1039 [DOI] [PubMed] [Google Scholar]
- Wang X, Zou L, Lu T, Bao S, Hurov KE, Hittelman WN, Elledge SJ, Li L (2006) Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell 23: 331–341 [DOI] [PubMed] [Google Scholar]
- Wang X, Zou L, Zheng H, Wei Q, Elledge SJ, Li L (2003) Genomic instability and endoreduplication triggered by RAD17 deletion. Genes Dev 17: 965–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Lindahl T (2005) Human DNA repair genes, 2005. Mutat Res 577: 275–283 [DOI] [PubMed] [Google Scholar]
- Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT (2005) Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell 19: 607–618 [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ (2000) The DNA damage response: putting checkpoints in perspective. Nature 408: 433–439 [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW (1999) Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285: 418–422 [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev 16: 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.