Abstract
The gene mutated in Bloom's syndrome, BLM, encodes a member of the RecQ family of DNA helicases that is needed to suppress genome instability and cancer predisposition. BLM is highly conserved and all BLM orthologs, including budding yeast Sgs1, have a large N-terminus that binds Top3–Rmi1 but has no known catalytic activity. In this study, we describe a sub-domain of the Sgs1 N-terminus that shows in vitro single-strand DNA (ssDNA) binding, ssDNA annealing and strand-exchange (SE) activities. These activities are conserved in the human and Drosophila orthologs. SE between duplex DNA and homologous ssDNA requires no cofactors and is inhibited by a single mismatched base pair. The SE domain of Sgs1 is required in vivo for the suppression of hyper-recombination, suppression of synthetic lethality and heteroduplex rejection. The top3Δ slow-growth phenotype is also SE dependent. Surprisingly, the highly divergent human SE domain functions in yeast. This work identifies SE as a new molecular function of BLM/Sgs1, and we propose that at least one role of SE is to mediate the strand-passage events catalysed by Top3–Rmi1.
Keywords: BLM, Bloom's syndrome, DNA helicase, Sgs1, synthetic lethality
Introduction
The RecQ family of DNA helicases comprises five eukaryotic members that participate in homologous recombination to repair double-stranded DNA breaks (DSBs) and stalled replication forks. Defects in all five helicases are associated with genome instability, and defects in three (BLM, WRN and RecQ4) are known to cause cancer predisposition syndromes in humans (Wang et al, 2003; Hu et al, 2005; Chu and Hickson, 2009). Structurally, these enzymes are well characterized. All members contain a highly conserved DNA helicase domain (Supplementary Figure S1A), and most contain an RQC domain that participates in DNA binding and protein–protein interactions (Bernstein et al, 2003; Bennett and Keck, 2004) (Figure 1A). Some RecQ members also contain a C-terminal HRDC domain that assists in DNA binding (Bernstein and Keck, 2005) and is required for in vitro activity (Wu et al, 2005). These helicases efficiently unwind a variety of model recombination intermediates such as Holliday Junctions (HJs), D-loops, replication forks and G-quadruplex DNA (Sun et al, 1998; Karow et al, 2000; LeRoy et al, 2005; Bachrati et al, 2006; Ralf et al, 2006; Capp et al, 2009). However, a clear understanding of how these enzymes suppress inappropriate recombination in vivo is lacking.
Figure 1.
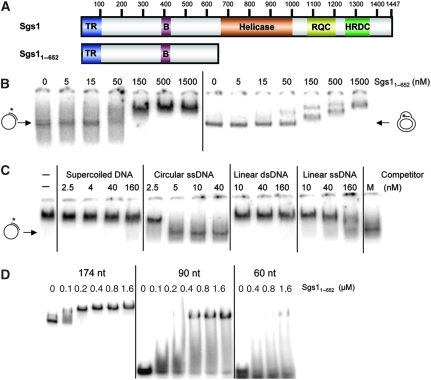
Identification of a ssDNA-binding activity in Sgs11−652 (A) Schematic representations of the full-length 1447 aa Sgs1 protein and Sgs11−652. Domains: TR, Top3–Rmi1 binding; B, C-terminal domain in Bloom's syndrome DEAD helicases (BDHCT) homology; RQC, RecQ C-terminal homology; HRDC, Human RecQ and RnaseD C-terminal homology. (B) EMSA assays contained the indicated concentrations of His6-tagged Sgs11−652 and 1 nM of either primed ssDNA (oligo #16 annealed to ∅X174 ssDNA) or a plasmid-based D-loop (oligo #17 annealed to pSK+ DNA by RecA). Asterisks represent positions of 32P-labelling. The reaction mixtures were incubated for 20 min at room temperature under standard conditions as described in ‘Materials and methods'. After incubation, the products were subjected to electrophoresis in composite 2.5% polyacrylamide/0.8% agarose gels followed by phosphorimager analysis. (C) Sgs11−652 (300 nM) was incubated together with 1 nM primed ssDNA and various concentrations of the four unlabelled versions of pSK+ DNA as indicated. Incubation and analysis was performed as in (B). M is a mock incubation in the absence of Sgs1 protein. (D) The indicated concentrations of Sgs11−652 were incubated with one of three 32P-labelled ssDNAs at a concentration of 1 nM: poly(dT)174, a 90 nt oligo of random sequence (oligo #18), or oligo(dT)60. Incubation and analysis was performed as in (B) except for the use of 10% PAGE.
The only RecQ DNA helicase conserved in lower eukaryotes is the ortholog of the gene defective in Bloom's syndrome (BS) BLM. BS is a rare autosomal disease associated with elevated levels of sister chromatid exchange and susceptibility to a wide variety of cancers (Chaganti et al, 1974; German et al, 2007). Similar to human BLM, budding yeast Sgs1 forms a tight complex with two other subunits, Top3 and Rmi1 (Gangloff et al, 1994; Bennett et al, 2000; Wu et al, 2000; Fricke et al, 2001; Ui et al, 2001; Mullen et al, 2005). The hyper-recombinational phenotypes of BS cells and yeast sgs1Δ mutants (Gangloff et al, 1994) indicate that the BLM–TOP3–RMI1 complex functions as an anti-recombinase that may be related to its ability to dissolve double HJs and/or unwind D-loops in vitro (Wu and Hickson, 2003; Bachrati et al, 2006). However, the complex is known to be multi-functional. It interacts with the Rad51 strand-exchange (SE) protein (Wu et al, 2001; Bugreev et al, 2009), it is required for recombination in model systems such as fission yeast and Drosophila (Adams et al, 2003; Cromie et al, 2008), and it promotes 5′-end resection (Mimitou and Symington, 2008; Nimonkar et al, 2008; Zhu et al, 2008). Furthermore, BLM promotes single-strand DNA (ssDNA) annealing (SA) in vitro. Considerable evidence indicates that this SA activity is intrinsic to the RecQ helicase domain: annealase activity has been identified in all five RecQ members, including the two smallest (RecQ1 and RecQ5), and although it is ATP independent, it is typically inhibited by non-hydrolyzable ATP analogues (Garcia et al, 2004; Cheok et al, 2005; Machwe et al, 2005; Sharma et al, 2005; Macris et al, 2006; Muftuoglu et al, 2008; Xu and Liu, 2009). For RecQ5 and WRN, SA activity has been localized to RecQ C-terminal domains (Garcia et al, 2004; Muftuoglu et al, 2008). Similarly, the N-terminal extension of BLM is dispensible for an SA activity that maps to residues 642–1350 (Cheok et al, 2005). Finally, BLM shows DNA SE activities that are both ATP dependent (Machwe et al, 2005; Weinert and Rio, 2007) and ATP independent (Machwe et al, 2005; Bugreev et al, 2009). For reasons that are unclear, the ATP-independent SE activities are sensitive to non-hydrolyzable ATP analogues and certain DNA helicase mutations (Machwe et al, 2005; Bugreev et al, 2009).
Apart from the DNA helicase domain, BLM/Sgs1orthologs contain a poorly characterized N-terminal domain of about 650 amino acids (aa) (Figure 1A). Functional analysis of the N-terminus has been hindered in part by the lack of sequence conservation between orthologs (Supplementary Figure S1B). In yeast, this domain (Sgs11−652) is known to be physiologically important (Mullen et al, 2000; Rockmill et al, 2003; Bernstein et al, 2009) and at least one essential role is to bind Top3 and Rmi1 through its N-terminal 100 aa. In searching for a biochemical function of Sgs11−652 we identified a sub-domain that shows ssDNA binding and SA activity. Despite the lack of sequence conservation, this domain is functionally conserved in multiple BLM orthologs. In addition, it shows an SE activity that is inactive on homologous templates containing a single mismatch. To determine the physiological role of this activity we characterized an SGS1 allele lacking the SE domain. This sgs1-ΔSE allele showed a null phenotype in most in vivo assays including suppression of hyper-recombination. These data indicate that DNA SE is an essential conserved function of BLM/Sgs1 orthologs, and we speculate that its in vivo role is to promote DNA SE in conjunction with Top3–Rmi1 at double HJs and D-loops.
Results
Identification of a novel ssDNA-binding activity in BLM orthologs
Structure–function analysis of Sgs1 showed earlier that deletion of the Top3–Rmi1-binding domain (TR; Figure 1A) creates a hypermorphic phenotype in yeast (Mullen et al, 2000; Bennett and Wang, 2001; Weinstein and Rothstein, 2008); that is, removal of the first 80–150 aa results in slow-growth and hyper-recombinational phenotypes that are more extreme than the sgs1Δ null. These hypermorphic phenotypes are suppressed either by a point mutation that eliminates Sgs1 DNA helicase activity or by deleting more of the N-terminus to aa 323 (Mullen et al, 2000; Weinstein and Rothstein, 2008). The simplest interpretation of this result is that the first 323 aa of Sgs1 contain a helicase-dependent activity that is toxic when untethered to Top3–Rmi1. As prior studies had failed to identify deoxyoligonucleotide (oligo)-binding activity in the N-terminus of Sgs1 or BLM (Cheok et al, 2005; Chen and Brill, 2007), we assayed it for structure-specific DNA-binding activity. To this end, we incubated Sgs11−652 protein with two large radiolablled probes consisting of either primed ∅X174 ssDNA or a plasmid-based D-loop. Analysis by electrophoretic mobility shift assay (EMSA) indicated that the migration of both the probes was retarded by Sgs11−652 (Figure 1B). Surprisingly, further characterization showed that the binding of Sgs11−652 to primed ∅X174 ssDNA was sensitive to unprimed plasmid-length ssDNA competitor (Figure 1C). This suggested that Sgs11−652 does bind ssDNA and that it might bind oligonucleotide substrates if they were unusually long. To test this hypothesis, we incubated Sgs11−652 with oligonucleotides of 60, 90 or 174 nt. As predicted, Sgs11−652 efficiently bound d(T)174, but bound the 90- and 60-mer oligos progressively less well (Figure 1D). We note that Sgs11−652 binds d(T)174 with high affinity because it could be detected at nM concentrations of both protein and substrate. As the D-loop is unlikely to contain significant ssDNA character, its binding by Sgs11−652 may involve different structural determinants.
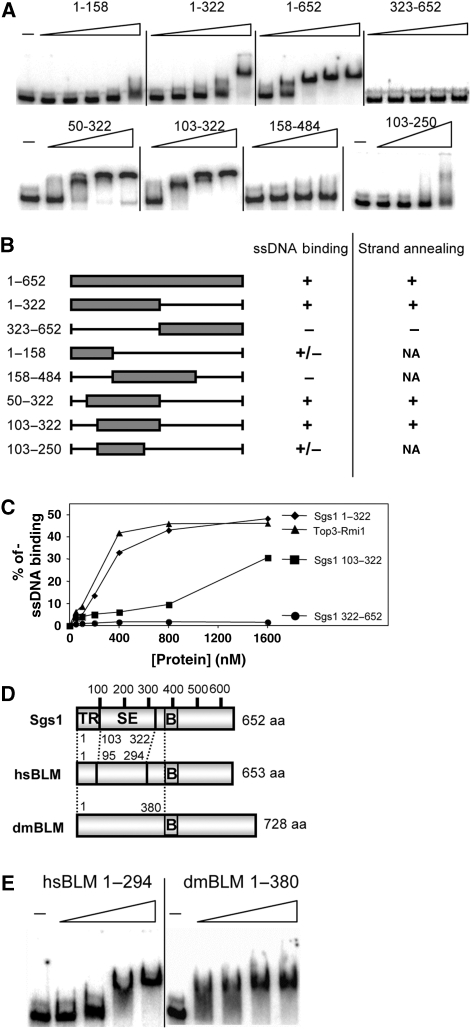
To further localize this activity, we expressed and purified sub-domains of Sgs11−652 as GST-fusion proteins and assayed them for ssDNA binding (Figure 2A). On the basis of these assays we determined that the C-terminal half of Sgs11−652 was dispensible for binding, and that Sgs1103−322 was the minimal region required for activity (Figure 2B). To confirm this result, and show that ssDNA binding could be detected by methods other than EMSA, we performed nitrocellulose filter-binding assays with His6-tagged proteins. These assays confirmed that ssDNA-binding activity could be detected in both Sgs11−322 and Sgs1103−322 (Figure 2C).
Figure 2.
The ssDNA-binding activity of Sgs11−652 maps to a sub-domain and is conserved in human and Drosophila BLM orthologs. (A) The following GST–Sgs1 fusion proteins were subjected to EMSA assay as in Figure 1D using 32P-labelled poly(dT)174 as probe: Sgs11−158, Sgs11−322, Sgs11−652 and Sgs1322−652 at 18, 37, 75, 150 and 300 nM; Sgs150−322, Sgs1103−322 and Sgs1158−484 at 100, 200, 400 and 800 nM; and Sgs1103−250 at 150, 300, 600 and 1200 nM. Dashes (−) indicate no-protein control lanes. (B) Summary of ssDNA binding and SA results. Symbols in the ssDNA-binding column represent the following results: +, strong; +/−, weak; and −, no ssDNA-binding activity. Symbols in the SA column represent the following results: +, strong; +/−, weak; and −, no SA activity. (C) The indicated His6-tagged proteins were assayed for ssDNA-binding using a nitrocellulose filter-binding assay. Reactions were performed as in Figure 1D, but were analysed by filtering through alkalai-treated nitrocellulose and quantifying the bound products by scintillation counting. The data are presented as a percentage of input CPM. (D) The N-termini of Sgs1, hsBLM and dmBlm are presented schematically with putative domain boundaries indicated by dotted lines. SE, strand-exchange domain. (E) EMSA assays were performed as above using GST–hsBLM1−294 at 0, 100, 200, 400 and 800 nM, and dmBlm1−380 at 0, 140, 280, 560 and 840 nM.
To determine whether these results generalized to other BLM orthologs, we assayed comparable regions of human and Drosophila BLM for ssDNA binding. Pairwise amino acid sequence alignments were of limited usefulness in identifying homologous regions in these orthologs. However, vertebrate BLM orthologs contain a conserved 40 aa region of unknown function, BDHCT (InterPro: IPR012532; hsBLM372−411) that showed weak similarity to the fly and yeast orthologs in multiple sequence alignments (Supplementary Figure S1B). Using this alignment we chose to express hsBLM1−294 and dmBLM1−380 as approximations of Sgs11−322 and Sgs11−386, respectively (Figure 2D). These domains were then purified as GST-fusion proteins for use in an EMSA assay. As shown in Figure 2E, titrations of both metazoan proteins resulted in a mobility shift of the d(T)174 probe. When the reaction products were incubated with an antibody to GST before electrophoresis, the resulting signals were further retarded indicating that the GST–Sgs1 and GST–hsBLM fusion proteins are responsible for this activity (Supplementary Figure S2). The GST portions of the proteins did not contribute to this activity as hexahistidine-tagged versions of all three proteins bound ssDNA (Supplementary Figure S3).
Characterization of a strand annealing activity
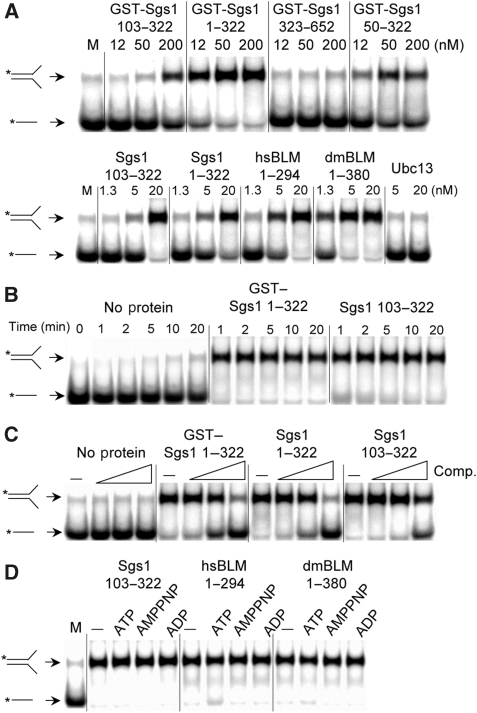
We assayed GST-tagged Sgs1 proteins for SA activity by incubating them with two partially homologous oligos one of which was 32P-labelled. Compared with mock treatment, Sgs1 proteins that included residues 103–322 accelerated the rate of strand pairing (Figure 3A, upper). By assaying a variety of sub-domains we observed a correlation between ssDNA binding and SA activity (Figure 2B). Sgs1103−322 is the minimal domain required for this activity and, for reasons described below, we hereafter refer to it as the SE domain. SA activity was conserved in the human and fly domains as His6-tagged versions of all three proteins accelerated strand pairing (Figure 3A, lower). The His6-tagged proteins, which showed SA activity at concentrations as low as 5 nM, were judged to be superior to the GST-tagged versions presumably because of the smaller size of the epitope tag.
Figure 3.
The SE domains from BLM/Sgs1 orthologs show strand annealing activity. (A) SA assays contained the indicated concentrations of GST- (upper) or His6-tagged (lower) proteins plus 1 nM each of a 32P-labelled 50 nt oligo (#1) and an unlabelled 50 nt oligo (#2) that share 25 nt of perfect complementarity. The reactions were incubated at 37°C for 5 min under standard conditions as described in ‘Materials and methods'. Reactions were stopped and the products were resolved by 10% PAGE followed by phosphorimager analysis. M is a mock reaction lacking protein. (B) GST–Sgs11−322 (50 nM) or Sgs1103−322 (20 nM) were assayed as in (A) except that the reactions were stopped at the indicated times before analysis. (C) The indicated SE domain proteins (50 nM) were assayed as in (A) except that the reactions contained either no competitor (−) or a 10-, 100- or 1000-fold excess of oligo #16 before the addition of proteins. (D) The indicated SE domain proteins (50 nM) were assayed as in (A) except that the reaction contained 1 mM Mg2+ and either no additions (−) or 1 mM of the indicated cofactor. Throughout, all proteins are His6-tagged unless indicated as GST-tagged. Asterisks represent positions of 32P-labelling.
Further characterization of the SA activity indicated that it is rapid. In contrast to spontaneous annealing, which was just detectable at 20 min, the enzyme-catalysed reaction was complete within 1 min (Figure 3B). The effect of non-homologous competitor ssDNA was tested by including high concentrations of an unrelated oligo in the reaction. As shown in Figure 3C, SA was resistant to a 100-fold excess of competitor, whereas a 1000-fold excess resulted in inhibition. Thus, high levels of non-homologous ssDNA are required to inhibit SA activity. We next examined the role that cofactors may have in SA. As shown in Figure 3D, the SA activities of all three orthologous SE domains were unaffected by ATP, ADP or the non-hydrolyzable analogue AMPPNP. Thus, the BLM/Sgs1 SA activity identified here behaves differently than activities identified in the full-length protein, which are presumably dependent on the RecQ helicase domain.
The SE domain shows DNA SE activity
The SE domain was tested for the ability to catalyse SE between a duplex DNA substrate containing one labelled strand and a complementary ssDNA oligo. A similar ATP-dependent or ATP-stimulated reaction has been observed with multiple RecQ homologs (Machwe et al, 2005; Weinert and Rio, 2007; Xu and Liu, 2009). One version of this reaction uses an excess of recipient ssDNA that has the same sense as the duplex's labelled strand. Denaturation of the fork is expected to result in annealing of the unlabelled complementary strands and release of the free 32P-labelled ssDNA oligo. This reaction is essentially unidirectional as there is little chance of the duplex's unlabelled strand exchanging back to the less abundant labelled strand. Therefore, our substrates consisted of a synthetic forked donor DNA with a radiolabelled top strand plus a five-fold molar excess of unlabelled top strand as recipient. As shown in Figure 4A, SE domains from all three species promoted the exchange reaction. SE was catalysed by SE protein, and did not result from spontaneous denaturation of the duplex, because neither incubation with non-specific protein (GST), nor excess complementary DNA alone, resulted in SE (Figure 4A). To eliminate the possibility that the donor DNA was simply melted by SE after which it passively annealed to the recipient oligo during the protease step before electrophoresis, we included a high concentration of a second recipient oligo during the protease incubation. The failure to detect annealing to this larger 94 nt oligo during the protease reaction indicates that ssDNA was not present after the assay or during protease treatment (Supplementary Figure S4A). Moreover, the SE protein lacked detectable nuclease activity that might result in an artefactual DNA SE activity (Supplementary Figure S4B). Thus, the simplest explanation for SE is that the SE domain melts double-stranded DNA while it simultaneously anneals complementary DNA strands. Such a coordinated reaction might explain why higher protein concentrations were required for SE (200–400 nM) than for SA (5–20 nM).
Figure 4.
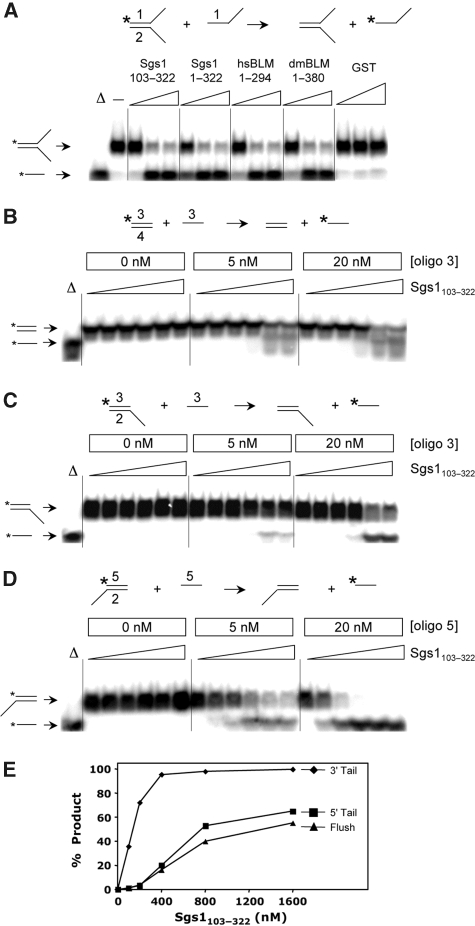
The SE domains from BLM/Sgs1 orthologs show DNA SE activity. (A) The SE assay is illustrated at the top of the panel. Reactions contained the indicated SE domain proteins at 0 (−), 50, 200 or 400 nM plus 2 nM forked DNA (where oligo #1 is 32P-labelled) plus 10 nM oligo #1. Substrate DNAs were incubated with GST at 50, 200 or 800 nM as negative control. The reactions were incubated at 37°C for 30 min under standard conditions and the products were analysed by 8% PAGE and phosphorimaging. The first lane (Δ) contains 32P-labelled oligo #1 as marker. (B) Sgs1103−322 (0, 100, 200, 400, 800 and 1600 nM) was used in an SE assay using blunt-ended substrate as indicated in the reaction at the top of the panel. Reactions contained 1 nM duplex DNA plus the indicated amounts of oligo 3. Assays were performed as in (A) except for use of 10% PAGE. (C) Sgs1103−322 was assayed using 5′-tail duplex DNA as substrate as in (B). (D) Sgs1103−322 was assayed using 3′-tail duplex DNA as substrate as in (B). (E) The SE reactions shown in (B–D) (20 nM unlabelled oligo) were quantified and are presented as a function of protein concentration. Sequences of the indicated oligos are presented in Supplementary Table 1. Throughout, all proteins are His6-tagged. Asterisks represent positions of 32P-labelling.
The substrate requirements for SE were examined by preparing radiolabelled duplex substrates whose ends were either flushed or contained a free 5′-tail or 3′-tail. Sgs1103−322 was then titrated into the reactions that contained different concentrations of unlabelled recipient DNA. In all cases, SE required a five-fold excess of recipient DNA (Figure 4B–D). SE also took place using the blunt donor; however, lower levels of protein were required when the donor duplex contained a 3′-tail (Figure 4E). The stimulation by a 3′-tail suggests that unwinding and annealing has a specific polarity.
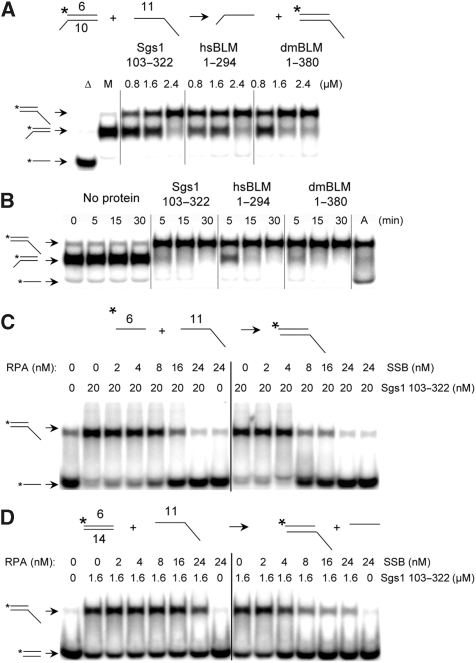
To further characterize the Sgs1103−322 protein, we used an SE reaction in which the recipient DNA was complementary to the labelled strand of the duplex but larger in size. Thus, the appearance of a retarded signal in native gel electrophoresis is diagnostic for SE. Using these substrates, the SE domains from three BLM orthologs efficiently converted the donor DNA signal into a slower-migrating form (Figure 5A). Time-course experiments confirmed that the three orthologs had similar kinetics and that the reactions were essentially complete within 5 min (Figure 5B).
Figure 5.
Characterization of SA and SE reactions. (A) The indicated SE reaction was performed by titrating Sgs1103−322, hsBLM1−294 or dmBLM1−380 into the standard SE assay. After incubation at 37°C for 30 min, the products were analysed as in Figure 4. Δ, boiled substrate; M, mock reaction without protein. (B) Time courses of the SE reaction illustrated in (A) were carried out with Sgs1103−322 (2.4 μM), hsBLM1−294 (1.2 μM) or dmBLM1−380 (1.2 μM). A, annealed oligos were obtained by slow cooling and used as marker. (C) The indicated SA reaction was performed by incubating 2 nM each of oligo #6 (32 nt) and oligo 11 (94 nt) together with various concentrations of either E. coli SSB or yeast RPA. Reactions were assembled on ice before incubation at 37°C for 5 min. (D) An SE reaction was performed using the indicated duplex DNA (0.5 nM) as donor and a 94-nt ssDNA (2.5 nM) as recipient. Reactions were assembled on ice before incubation at 37°C for 30 min. Throughout, all proteins are His6-tagged. Asterisks represent positions of 32P-labelling.
The impact of ssDNA-binding proteins on SA and SE activities was then assessed. The annealing of complementary 32- and 94-nt oligos (2 nM each) was inhibited by 8 nM Escherichia coli SSB and 16 nM yeast RPA (Figure 5C). Under these conditions, we estimate that both ssDNA-binding proteins occlude 30 nt of ssDNA, so that there are 8 nM of binding sites in the substrate. Thus, access of Sgs1103−322 to ssDNA appears to be blocked by excess concentrations of RPA and SSB. The corresponding substrates were then assayed in an analogous SE reaction: a 32 bp duplex (0.5 nM) and 94-nt recipient oligo (2.5 nM). E. coli SSB again inhibited the reaction at 8 nM, which is expected to saturate the 7.5 nM binding sites. Higher levels of RPA partially inhibited the SE reaction (Figure 5D). Thus, although there are quantitative differences, Sgs1103−322-promoted SE is inhibited by high levels of both ssDNA-binding proteins.
Two additional experiments were performed to characterize the SE reaction. First, because the above experiments were performed in the presence of EDTA, we tested whether it was influenced by divalent cations. The results (Supplementary Figure S5A and B) indicated that the SE reaction was unaltered by physiological levels of Mg2+. Second, to examine its stoichiometry we titrated Sgs1103−322 into an SE reaction and quantified the products. The results indicated that 40–54% of the flush-end DNA duplex was exchanged onto the recipient ssDNA over a range of 0.4–1.6 μM Sgs1103−322 (Supplementary Figure S5C). On the basis of these values, SE requires a minimum of one molecule of protein for each 7 nts of ssDNA.
The SE domain is required for Sgs1 function in vivo
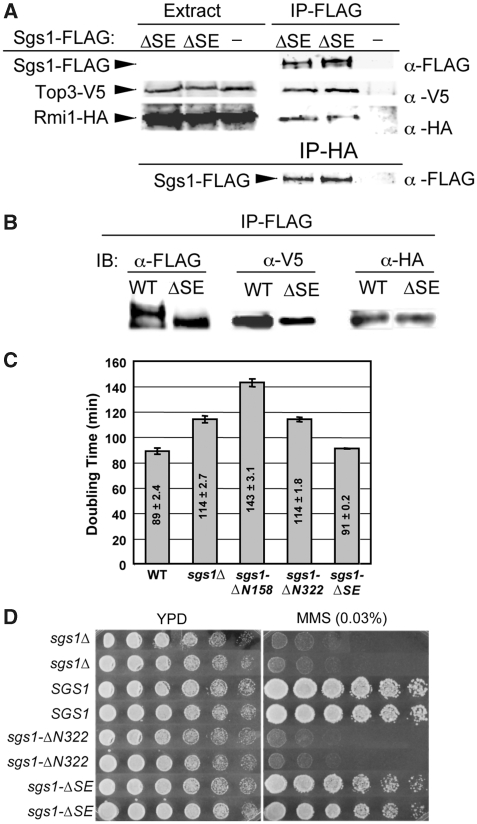
To determine the in vivo function of the SE domain, we constructed the sgs1-ΔSE allele that lacks the SE coding region. This allele, which expresses Sgs1Δ103−322 from its own promoter, was tested for its ability to complement sgs1Δ phenotypes. To eliminate the possibility that sgs1-ΔSE phenotypes were due to a defect in Top3–Rmi1 binding, we first assayed the interaction by immunoprecipitating (IP'ing) Sgs1Δ103−322 and immunoblotting for Top3 and Rmi1. As demonstrated earlier for WT Sgs1 (Mullen et al, 2005), epitope-tagged versions of Top3 and Rmi1 were present in precipitates of FLAG-tagged Sgs1Δ103−322, and Sgs1Δ103−322 was found in Rmi1 precipitates (Figure 6A). In a side-by-side comparison, approximately equal amounts of the Top3 and Rmi1 subunits were co-IP'd with Sgs1-WT and Sgs1Δ103−322, respectively (Figure 6B). On the basis of these results and the data presented below, we conclude that Sgs1Δ103−322 interacts properly with Top3 and Rmi1.
Figure 6.
Sgs1Δ103−322 physically interacts with Top3–Rmi1. (A) Yeast strains were constructed to express integrated versions of Top3-V5, Rmi1-HA, and either Sgs1Δ103−322-FLAG (ΔSE) or no Sgs1 (−) as the sole copies of these subunits. Cell extracts were prepared and either immunoblotted directly (extract) or subjected to IP with α-FLAG beads (IP-FLAG) before immunoblotting with α-FLAG, α-V5 and α-HA antibodies. Similar to WT Sgs1-FLAG, Sgs1Δ103−322-FLAG is insufficiently abundant to be detected in crude cell extracts (Mullen et al, 2005). In the lower panel, extracts were IP'd with α-HA and immunoblotted with α-FLAG to detect Sgs1Δ103−322-FLAG. (B) Cell extracts were prepared from strains expressing Top3-V5, Rmi1-HA and either Sgs1-FLAG (WT) or Sgs1Δ103−322-FLAG (ΔSE) as above. Extracts containing 2 mg of total protein were subjected to IP with α-FLAG beads (IP-FLAG) and immunoblotted with α-FLAG, α-V5 and α-HA antibodies. (C) Cells of the indicated genotype were grown in liquid YPD at 30°C and doubling times were determined. Shown are the average values±s.d. (D) Cells of the indicated genotype were resuspended at OD=3, serially diluted in three-fold increments and approximately 5 μl were spotted onto YPD plates with or without 0.03% MMS. Plates were photographed after 2 (YPD) or 3 days (MMS) growth at 30°C.
Different alleles of SGS1 have been shown to confer distinct slow-growth phenotypes (Weinstein and Rothstein, 2008). For example, the sgs1Δ null strain grows more slowly than WT, but a strain lacking the Top3–Rmi1-binding domain of Sgs1 (e.g. sgs1-ΔN158 encoding Sgs1159−1447) grows even more slowly than sgs1Δ (Figure 6C). As previously observed (Mullen et al, 2000), this hypermorphic phenotype was eliminated by a larger N-terminal truncation (e.g. sgs1-ΔN322 encoding Sgs1323−1447). In contrast to these mutants, the doubling time of the sgs1-ΔSE strain was identical to WT (Figure 6C). Furthermore, when we examined its ability to complement the MMS sensitivity of sgs1Δ, the sgs1-ΔSE allele conferred a WT level of resistance (Figure 6D). This result is consistent with other internal deletions made within the Sgs1 N-terminus (Ui et al, 2001) and indicates that the sgs1-ΔSE allele functions similar to WT in these assays.
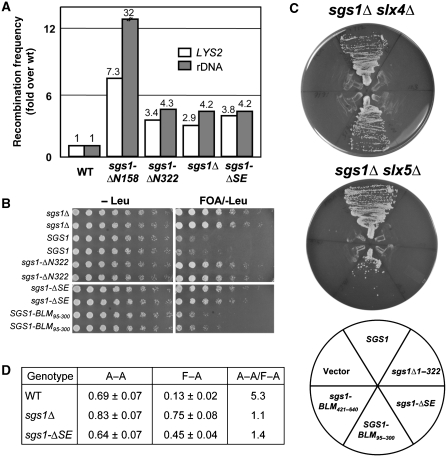
The sgs1-ΔSE allele was then used to test whether it would complement the sgs1Δ hyper-recombination phenotype. Intrachromosomal recombination was measured using a marker-excision assay in which CAN1 and URA3 are inserted between direct-repeat (DR) sequences at LYS2 and the rDNA, respectively (Mullen et al, 2000). Compared with WT, the sgsΔ null strain showed a 2.9-fold increase in recombination frequency at LYS2 and a 4.2-fold increase at the rDNA (Figure 7A), and consistent with its hypermorphic phenotype, sgs1-ΔN158 showed 7- and 32-fold increases at these loci. The sgs1-ΔSE allele generated recombination rates that were elevated by 3.8- and 4.2-fold, respectively. These levels closely match those obtained with sgs1Δ and sgs1-ΔN322. Thus, the SE domain is required to suppress hyper-recombination.
Figure 7.
The SE domain is required for multiple SGS1 functions. (A) Yeast strains of the indicated genotype were assayed for excision recombination at the LYS2 and rDNA loci as described earlier (Mullen et al, 2000). Recombination frequencies were determined and are presented as fold increase over WT. (B) Strain NJY728 (sgs1Δ top3Δ plus pJM555 (TOP3/URA3/ADE3/CEN)) was transformed with the indicated SGS1 alleles in pRS415 (LEU2/CEN). Transformants were streak purified on SD-leu plates, resuspended to OD600=3.0 and serially diluted in five-fold increments. Approximately 5 μl were spotted onto SD plates lacking leucine but with or without 5-FOA. Plates were photographed after 2 (−Leu) or 5 (5-FOA/−Leu) days growth at 30°C. (C) Strains NJY2083 (sgs1–11∷loxP slx4–11∷loxP plus pJM500 (SGS1/URA3/ADE3/CEN)) and NJY602 (sgs1–11∷KAN slx5-10∷TRP1 plus pJM500) were transformed with various SGS1 alleles in pRS415 as indicated in the key. Transformants were streaked onto plates containing 5-FOA and the plates were photographed after 2 (sgs1Δ slx4Δ) or 3 (sgs1Δ slx5Δ) days growth at 30°C. (D) Cells of the indicated genotype were assayed for viability after inducing HO endonuclease in SSA reporter strains that contained either homologous (A-A) or homeologous (F-A) direct repeats flanking the HO cut site (Sugawara et al, 2004). Results shown are the average viability±s.d.
The prototypical phenotype of SGS1 loss-of-function alleles is the suppression of top3Δ slow growth (Gangloff et al, 1994). However, some separation of function alleles, such as sgs1-D664Δ, confers this phenotype as well (Bernstein et al, 2009). To test the effect of sgs1-ΔSE in the top3Δ background, we introduced a variety of plasmid-borne SGS1 alleles into an sgs1Δ top3Δ double mutant that contained plasmid pJM555 (TOP3/URA3/CEN). These strains were then serially diluted and spotted into medium containing 5-FOA, which selects against pJM555. As expected, the sgs1Δ allele allowed good growth on this medium whereas SGS1 promoted slow growth (Figure 7B). The sgs1-ΔSE allele behaved like sgs1Δ (and sgs1-ΔN322) as indicated by the good growth of this strain on 5-FOA. On the basis of these data, sgs1-ΔSE resembles sgs1-D664Δ in that both alleles suppress top3Δ slow growth but confer MMS resistance. To test whether this phenotype was due to the loss of the SE activity, we replaced residues 103–322 of Sgs1 with residues 95–300 of hsBLM. Despite the fact that these domains are only 13% identical in aa sequence, this chimaeric allele SGS1-BLM95−300, showed WT function by promoting the slow growth of top3Δ cells (Figure 7B). This result supports the notion that SE activity is required for SGS1 to promote top3Δ slow growth.
One of the most sensitive assays for SGS1 function is its ability to complement sgs1Δ slxΔ synthetic lethality (Mullen et al, 2001). This was tested in two strains, sgs1Δ slx4Δ and sgs1Δ slx5Δ, that are kept alive by plasmid pJM500 (SGS1/URA3/CEN). Both strains were transformed with plasmid-borne SGS1 alleles, and the transformants were then streaked onto media that selects against pJM500. In contrast to WT SGS1, which promoted growth on 5-FOA, sgs1-ΔSE failed to complement either strain (Figure 7C). In this regard, sgs1-ΔSE resembled the null allele and sgs1-ΔN322. To test whether the loss of SE activity was responsible for this phenotype, we also transformed these tester strains with SGS1-BLM95−300. Again the human SE domain restored activity to the chimaeric yeast protein as indicated by the growth of yeast in the absence of SLX4 or SLX5. As control, we showed that another domain from the N-terminus of BLM could not provide SE function as the chimaeric allele SGS1-BLM421−640 failed to complement synthetic lethality (Figure 7C). It should be noted that the ability of SGS1-BLM95−300 to function in this assay most likely depended on productive interactions between the Sgs1 TR domain and Top3–Rmi1 because SGS1 alleles that replace Sgs11−322 with BLM1−300 fail to complement sgs1Δ slx4Δ or sgs1Δ slx5Δ synthetic lethality (Mullen and SJB, unpublished data). We conclude that the SE domain is essential for SGS1 function in these genetic backgrounds.
SGS1 has been shown to prevent recombination between homeologous DNA sequences in a single-strand annealing (SSA) assay (Sugawara et al, 2004). SSA is a DSB repair pathway in which a DSB between two DR sequences can be repaired by the annealing of the homologous 3′-ssDNA sequences that arise following 5′-end resection. We used strains engineered to contain an HO endonuclease site between two 205 bp DRs that have either 100% sequence homology (A-A strains) or 97% homeology (F-A strains) (Sugawara et al, 2004). Failure to repair the HO-induced break results in the loss of cell viability, which is exacerbated in F-A strains because WT cells prevent homeologous repair. For example, the viability of A-A strains after HO induction is five-fold better than that of F-A strains (Figure 7D). As demonstrated earlier, sgs1Δ cells show efficient repair using homeologous sequences such that the A-A/F-A ratio is 1.1 (Sugawara et al, 2004; Goldfarb and Alani, 2005). We found that sgs1-ΔSE strains show an A-A/F-A cell-viability ratio of 1.4, which is indicative of efficient homeologous repair. Thus, the SE domain is required for heteroduplex rejection. This result is consistent with the finding that Sgs11−652, as well as the Sgs1 DNA helicase domain, are required for heteroduplex rejection in this assay (Goldfarb and Alani, 2005).
Role of DNA homology in DNA SE in vitro
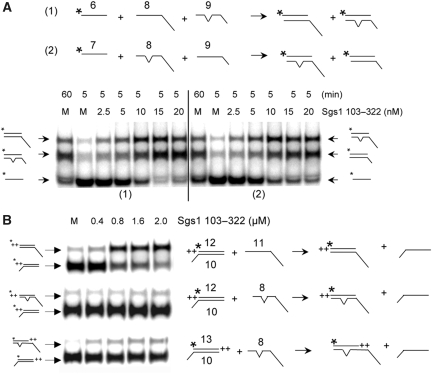
The results from the above experiment predicted that the SE domain might discriminate between homologous and homeologous DNA in in vitro assays. To test this hypothesis, we designed SA and SE assays using synthetic substrates that contained a single mismatched base pair within a 32 bp region of homology. The resulting 97% homeology would then approximate the in vivo conditions used in the experiment of Figure 7D. For the SA assay, we incubated a radiolabelled oligo (#6 in Figure 8A) with two versions of its complement: a perfectly homologous oligo that contained a long tail (#8, 94 nt), and one with a shorter tail that contained a single mismatched base at the centre of its 32 nt homologous region (#9; 57 nt). After SA, homologous and homeologous products were distinguishable by their differential migration in gel electrophoresis. As shown in Figure 8A, Sgs1103−322 promoted strand annealing on both complementary oligos regardless of the mismatch. Note that the ratio of homologous to homeologous products was identical to mock incubation controls used to measure spontaneous annealing after 5 or 60 min incubations (Figure 8A, left). This result was independent of whether the homologous strand or the homeologous strand contained the longer tail (Figure 8A, right). Homeologous DNA was also annealed as efficiently as homologous DNA in standard two-oligo reactions (data not shown). Thus, SE-stimulated annealing does not distinguish between homologous and homeologous DNA, at least at this level of homeology. It should also be noted that the Tms of the substrates used here must be sufficiently high that there is no impediment to the annealing of homeologous DNA in either spontaneous or SE-promoted reactions.
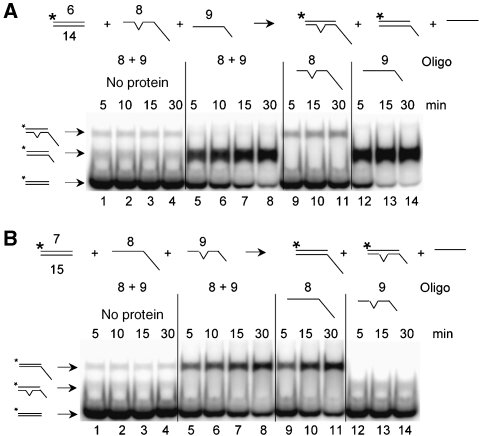
Figure 8.
Differential response of SA and SE to non-homology. (A) Two SA reactions are illustrated schematically in which a single non-complementary nucleotide at the centre of the 32 nt homologous region is indicated by a bulge. Reactions (1) and (2) differ by the lengths of the non-homologous tails on the recipient DNAs as indicated. Sgs1103−322 was titrated into a reaction containing the indicated 32P-labelled oligo (1 nM) plus recipient oligos (1 nM) and incubated for at 37°C for 5 min. Mock (M) reactions lack protein and were incubated for 5 or 60 min. (B) Three SE reactions were carried out at 37°C for 5 min using the indicated concentrations of Sgs1103−322, 1 nM of 32P-labelled donor dsDNA and 20 nM of recipient oligo. +, single nucleotide. Asterisks represent positions of 32P-labelling.
We next examined the effect of mismatches in SE reactions using standard methodology. Compared with substrates with perfect homology (Figure 8B, upper), Sgs1103−322 was unable to stimulate SE using an oligo with a single mismatch in the centre of the 32 nt region of homology (Figure 8B, middle). To compensate for the lower Tm of the mismatched products, substrates were designed such that the mismatched product would contain two additional bases of complementarity not found in the substrate (Figure 8B, lower). However, SE by Sgs1103−322 remained negligible.
As the above experment compared individual reactions, we designed a competitive SE reaction in which the labelled strand of the duplex would be allowed to exchange onto either homologous or homeologous ssDNA analogous to the experiment in Figure 8A. The diagram of Figure 9A illustrates the flush 32 bp duplex donor with a labelled top strand, and recipient DNAs consisting of either a 57 nt oligo with perfect homology or a 94 nt homeologous oligo containing a single mismatch. When incubated with the recipient DNAs individually, Sgs1103−322 efficiently exchanged the labelled donor strand onto the homologous substrate (#9) but not the homeologous substrate (#8). Note that the level of exchange onto oligo #8 (Figure 9A, lanes 9–11) approximated that obtained in the absence of protein (Figure 9A, lanes 1–4). Importantly, incubation of all three substrates together resulted in exchange exclusively onto the homologous substrate (Figure 9A, lanes 5–8).
Figure 9.
Sgs1103−322-catalysed SE is inhibited by a single mismatched base pair. (A) The indicated SE reaction was carried out using 1 nM of a 32P-labelled donor DNA with flush ends and 20 nM of either oligo 8 alone (lanes 9–11), oligo 9 alone (lanes 12–14) or both oligos (lanes 1–8) as recipient. Sgs1103−322 was either absent (lanes 1–4) or present at 1.6 μM (lanes 5–14). (B) The indicated SE reaction was performed as in (A). Note that reactions using flush donor substrates are slower than those with 3′ ssDNA extensions. Asterisks represent positions of 32P-labelling.
To test whether this result was biased by the fact that the homologous oligo contained a smaller non-homologous tail, we performed the control reaction in Figure 9B. Again exchange occurred exclusively onto the homologous oligo even when it contained the larger non-homologous tail. Thus, in contrast to strand annealing, SE catalysed by the SE domain in vitro is inhibited by as little as 3% non-homology. The ability of the SE domain to discriminate between homologous and homeologous substrates may be related to SGS1's role in heteroduplex rejection in vivo.
Discussion
BLM/Sgs1 forms part of a multi-functional complex with both DNA helicase and DNA topoisomerase activities. The SE activity we have characterized localizes to the N-terminus of Sgs1 and is a previously unidentified activity. Several results suggest that this SE activity is biologically relevant. First, SE is unusually stringent as it is inhibited on substrates containing a single mismatch. On the basis of this stringency, we are inclined to favour SE over SA as the bona fide in vivo activity. Second, SE is conserved in at least two other BLM orthologs. Third, the SE domain is required for most in vivo activities of SGS1, and in both cases that we tested the corresponding human domain was able to provide those activities. Fourth, control experiments rule out artefactual ‘meltase' or nuclease activities as explanations for SE in vitro.
The SA and SE activities reported here appear to be distinct from similar activities associated with the helicase domains of BLM and other RecQ family members (Cheok et al, 2005; Muftuoglu et al, 2008). However, their relationship to similar activities identified in full-length RecQ homologs is not clear. Athough the SA activity observed here is distinguished by its insensitivity to nucleotide analogues, the inhibition of SA by non-hydrolyzable analogues (Cheok et al, 2005; Machwe et al, 2005; Sharma et al, 2005; Muzzolini et al, 2007; Capp et al, 2009) seems to be explained in part by the fixation of RecQ helicase on ssDNA in the presence of AMPPNP (Capp et al, 2009). As this would affect both helicase- and non-helicase domains in the full-length protein, it is unclear whether the SA activities of all five RecQ homologs are limited to their helicase domains. In the case of SE, most RecQ family members require ATP. This includes the ability of WRN, BLM and RecQ5β to catalyse coordinated ATP-dependent DNA SE reactions (Machwe et al, 2005, 2006; Weinert and Rio, 2007) in addition to the reported second helicase domain of the RecQ4 N-terminus (Xu and Liu, 2009). However, SE by both BLM and WRN has been observed in the absence of ATP (Machwe et al, 2005; Bugreev et al, 2009). Therefore, an important next step is to determine whether the SA and SE activities we have described can be assayed when tethered to the RecQ helicase domain and whether they are regulated by the helicase.
Several details of this work suggest that the BLM/Sgs1 SE activity is linked to Top3–Rmi1 function. First, the location of the SE domain adjacent to the Top3–Rmi1-binding domain (TR) suggests that SE and Top3–Rmi1 act on the same substrate based simply on their proximity. Second, SE would be expected to generate topological problems in a chromosomal context that would require topoisomerase activity. Third, removal of the TR domain uncovers an SE-dependent toxicity (Mullen et al, 2000) that appears to phenocopy top3Δ (Wallis et al, 1989; Bennett and Wang, 2001; Weinstein and Rothstein, 2008). Although the hypermorphic phenotype of such sgs1-ΔTR alleles is suppressed by helicase-defective SGS1 alleles, it is not known whether translocation by the helicase is actually driving this genetic instability. As stable ssDNA binding by both BLM and RecQ4 is dependent on ATP (Weinert and Rio, 2007; Capp et al, 2009), mutations that reduce the ability of Sgs1 to bind and/or hydrolyse ATP may have multiple effects. Thus, it is possible that helicase-defective SGS1 alleles suppress the hypermorphic phenotype indirectly by inhibiting its ability to bind DNA. We suggest that the above results are most simply explained by a model in which SE generates a substrate for Top3–Rmi1. More specifically, SE may provide the access to ssDNA that is required by DNA topoisomerase III or the necessary directionality for strand passage.
In Figure 10, we relate these facts to two of the most likely pathways for BLM/Sgs1 function. BLM/Sgs1 is thought to have a role in dismantling D-loops given that D-loops are optimal substrates for BLM unwinding (Bachrati et al, 2006), that D-loop unwinding is an essential step in the synthesis-dependent strand annealing (SDSA) pathway for which BLM is required in Drosophila (Adams et al, 2003), and that the SE domain binds D-loops on its own (Figure 1B). As shown in Figure 10A, we suggest that an essential role of BLM/Sgs1 is not simply to displace the invading strand with its helicase activity but to properly restore the D-loop to its duplex state. D-loop formation should involve unwinding of the parental duplex especially in cases in which the invading strand is extended significantly by the DNA polymerase. Thus, in WT cells, BLM/Sgs1 could bind at the proximal end of the D-loop where the SE domain drives re-annealing of the donor duplex (Figure 10A, i) and displacement of the invading strand in a simplified SE reaction (ii). Importantly, the re-annealing step requires the catenating activity of Top3–Rmi1 to re-wind the donor duplex (iii). The BLM/Sgs1 3′–5′ DNA helicase activity is expected to assist in the displacement of the invading strand. In this model, the genomic instability of top3Δ strains may arise because of displacement of the invading strand in the absence of re-winding the parental duplex. The formation of such unwound DNA, which is expected to be recombinogenic, may be exacerbated in sgs1-ΔTR strains if Top3–Rmi1 regulates SE function.
Figure 10.
Proposed roles of SE in Sgs1–Top3–Rmi1 function. (A) SE-dependent D-loop unwinding in the SDSA recombination pathway. After strand invasion and elongation, the nascent strand (red) is displaced by the combined activities of Top3, Rmi1, Sgs-SE and Sgs1 helicase (T, R, SE and H, respectively). Shown in the bracket are intermediate steps: the displaced parental strand is rewound by SE, which anneals the blue parental strands (i) and displaces the red nascent strand (ii) whereas Top3–Rmi1 interlinks the parental strands (iii). Repeating this cycle ‘n' times leads to strand displacement. (B) SE mediates double HJ branch migration. The circular inset presents an example where the red DNA strand of the right-hand HJ is annealed back to its parental complement (in the direction of the dotted arrow). As in branch migration, this displaces the blue strand in the upper heteroduplex so that it can be re-annealed to the lower duplex (in this case by SE), however the DNA between two HJs is topologically constrained. Shown in the bracket is an example of the topological stress encountered on the bottom duplex (i) where the red strand must pass through the blue strand through Top3–Rmi1 activity (ii) yielding one unlinking of the bottom duplex (iii). Not shown is the rewinding of the lower blue strands or reciprocal reactions on the upper duplex. Repeating these cycles of SE- and Top3–Rmi1-mediated strand passage ‘n' times leads to displacement of the heteroduplex between the HJs and formation of a double hemi-catenane. Resolution of the hemi-catenane is promoted by the strand annealing activity of SE and catalysed by Top3–Rmi1 to yield non-crossover products.
Recent models of BLM–Top3α function have noted the need for directional SE (Plank and Hsieh, 2009) in double HJ dissolution (Wu and Hickson, 2003; Plank et al, 2006). Figure 10B illustrates how the ‘HJ migration' model (Plank and Hsieh, 2009) of dHJ dissolution could be aided by the SE domain by promoting the exchange of a HJ strand from one (lower) duplex to its complement in the other (upper) duplex (Figure 10B, dotted arrow). For example, such an exchange generates multiple topological constraints such as the intertwining on the bottom duplex (i). The SE domain is expected to cooperate with Top3–Rmi1 to catalyse strand passage (ii) to remove this intertwining (iii). This unwinding event on the lower duplex must be coordinated with re-winding on the top duplex, which is formally the same mechanism described in Figure 10A. The final decatenation step in this pathway provides the best example of how the SE domain provides directionality to Top3–Rmi1strand passage (Figure 10B). Active annealing of complementary strands by SE generates constraints to be relieved by Top3–Rmi1 and the directionality needed to separate the two hemicatenanes. Such forced annealing might also contribute to the sickness of top3Δ single mutants. Finally, the SE domain may be involved in sensing mismatches in heteroduplex, along with mismatch repair proteins, to promote gene conversion (Sugawara et al, 2004; Lo et al, 2006) or inhibit divergent dHJ branch migration.
It was reasoned earlier that Sgs11−652 contained an important functional domain as it supressed certain sgs1Δ phenotypes alone, and because SGS1 alleles lacking the TR domain showed a hypermorphic phenotype (Mullen et al, 2000). Although an intact Sgs1 DNA helicase domain is necessary to observe the hypermorphic phenotype (Mullen et al, 2000; Weinstein and Rothstein, 2008), it cannot be the sole cause of the instability because the SE domain is also required. Similarly, although Sgs1 DNA helicase activity has a role in the top3Δ slow-growth phenotype (Gangloff et al, 1994), the SE domain does as well (Figure 7B). Although further experiments are needed to determine exact roles of these two activities, it remains possible that the SE domain is the ultimate source of top3Δ instability. The ability of the SE domain to unwind and rewind DNA strands may explain why SGS1 alleles lacking DNA helicase activity retain the capacity to promote gene conversion, suppress MMS sensitivity, suppress hyper-recombination, suppress meiotic sporulation defects and induce slow growth in top3Δ cells (Lu et al, 1996; Miyajima et al, 2000; Mullen et al, 2000; Rockmill et al, 2003; Lo et al, 2006) or inhibit divergent dHJ branch migration.
Materials and methods
Proteins and DNA substrates
All GST-fusion proteins including Sgs11−158, Sgs11−322, Sgs11−652, Sgs1323−652, Sgs151−322, Sgs1103−322, Sgs1159−484, Sgs1103−250, hsBLM1−294 and dmBLM1−380 were expressed and purified from E. coli BL21(DE3)-RIL cells as described for GST–Sgs11−158−HA (Fricke et al, 2001). All His6-tagged proteins including Sgs11−652, Sgs11−322, Sgs1103−322, hsBLM1−294 and dmBLM1−380 were expressed and purified essentially as described for Sgs11−652-V5(His6) (Chen and Brill, 2007). E. coli BL21(DE3)-RIL cells were transformed with T7 expression plasmids and colonies were pooled and grown in 1 l LB media containing 0.1 mg/ml ampicillin at 37°C until OD600=0.4. The recombinant protein was induced by addition of 0.4 mM isopropyl-1-thio-D-galactopyranoside and the cells were grown at 16°C for 16 h. Induced cells were pelleted and resuspended in 40 ml Buffer N (25 mM Tris–HCl (pH7.5), 0.1 mM phenylmethylsulfonyl fluoride, 0.01% Nonidet P-40, 1 mM dithiothreitol, 10% glycerol and 500 mM NaCl) containing 10 mM imidazole and protease inhibitors (PIs) as above. The cells were sonicated for 2 min with a Branson sonifier 450 microtip at setting 2 and 25% duty cycle. The lysate was centrifuged at 13 500 r.p.m. in an SS34 rotor at 4°C for 15 min and the supernatant was filtered before loading onto a 5 ml Ni column. The column was washed with 10 CVs of Buffer N plus 10 mM imidazole and eluted with a 8 CV gradient from 10 to 500 mM imidazole in Buffer N. Peak fractions were pooled and dialysed against buffer A (25 mM Tris–HCl (pH7.5), 0.1 mM phenylmethylsulfonyl fluoride, 0.01% Nonidet P-40, 1 mM dithiothreitol, 10% glycerol and 1 mM EDTA) plus 200 mM NaCl and stored at −80°C. The oligos (IDT) used in this study are shown in Supplementary Table S1. Plasmid-based D-loop used for DNA-binding assay was prepared and purified as described earlier (McIlwraith et al, 2005).
EMSA DNA-binding assay
32P-labelled DNA substrates were prepared and assayed by EMSA essentially as described (Mullen et al, 2005). Proteins were added to 32P-labelled DNA substrate in a final volume of 20 μl containing 25 mM Tris (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 1.0 mg/ml BSA and incubated at 25°C for 20 min. Loading dye was added to a final concentration of 8% glycerol and 0.25% bromophenol blue. Oligonucleotide binding was tested by electrophoresis at 10 V/cm through a 10% polyacrylamide gel (29:1 acrylamide:bis) in 1 × TBE at room temperature. Binding to plasmid-based DNA probes was detected on a 2.5% polyacrylamide mixed with 0.8% agarose at room temperature. Gels were fixed in 50% EtOH/10% acetic acid for 15 min, dried and visualized by a Molecular Dynamics phosphorimager.
DNA filter-binding assay
Reactions were performed as described in the EMSA but analysed by alkali-treated nitrocellulose paper. Reactions were filtered under vacuum onto a 0.45 μm Protran nitrocellulose membrane (Whatman) and washed with 0.5 ml of reaction buffer twice at room temperature. Filters were dried and assayed for radioactivity by scintillation counting.
Strand annealing assay and SE assay
The standard strand annealing and SE reactions contained 25 mM Tris–HCl (pH 7.5), 50 mM sodium chloride, 1 mM EDTA, 10 μg/ml bovine serum albumin and 1 mM DDT in a final volume of 20 μl. The reactions were assembled on ice and initiated by shifting to 37°C for 5 min. Reactions were stopped by treating with a final concentration of 50 mM EDTA, 1% sodium dodecylsulfate (SDS) and 1 mg/ml proteinase K at 37°C for 15 min. The STOP buffer for all strand annealing reactions included the unlabelled version of the labelled oligo at a final concentration of 100 nM (oligo 1 or 6) (Machwe et al, 2006). After addition of loading dye the samples were resolved on a 10% polyacrylamide at room temperature.
Genetic assays
Synthetic lethality, MMS sensitivity and genetic recombination were assayed as described (Mullen et al, 2000). The heteroduplex rejection assay based on cell survival was performed as described earlier (Goldfarb and Alani, 2005).
Yeast extract preparation and immunoprecipitation
The cell pellet from 2 l of culture was resuspended in 2 × CE buffer (50 mM Tris–HCl (pH 7.5), 4 mM magnesium chloride, 1 mM EDTA, 20% glycerol, 5 mM DTT, 0.5 M sodium chloride) plus the following PIs: pepstatin, 10 mg/ml; leupeptin, 5 mg/ml; benzamidine, 10 mM; bacitracin, 100 mg/ml; aprotinin, 20 mg/ml; phenylmethylsulfonyl fluoride, 0.1 mM; and sodium metabisulfite, 10 mM. This cell suspension was frozen in liquid nitrogen and pulverized under liquid nitrogen using a SPEX 6850 freezer mill (Spex Inc., Metuchen, NJ). The sample was thawed and centrifuged in a Ti45 rotor at 44 000 r.p.m. for 30 min. Typical extracts contained 10 mg/ml protein, which were aliquoted and stored at −80°C.
IPs were performed at 4°C as follows. In all, 2 mg of total protein were incubated for 1 h with 40 μl protein G-Sepharose beads conjugated to anti-FLAG mouse monoclonal antibodies (Sigma), or with 1 μl of anti-HA (Roche, 5 μg/μl) or anti-V5 (Invitrogen, 1 μg/μl) monoclonal antibodies. Thirty microliters of Protein-A sepharose beads (Amersham-Pharmacia) were added to the HA and V5 samples, followed by rocking for 1 h. The immune complexes were then washed three times with 1 ml of RIPA buffer (150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 1% (v/v) NP40, 0.5% (w/v) deoxycholate, 0.1% (w/v) SDS). Bound proteins were resuspended in Laemmli buffer and resolved by SDS polyacrylamide gel electrophoresis (SDS–PAGE). After SDS–PAGE the gels were transferred to nitrocellulose membranes and treated with either anti-FLAG (Sigma), anti-V5 or anti-HA as the primary antibody at a 1:10 000 dilution. Blots were then treated with anti-mouse HRP-conjugated secondary antibody (1:10 000; Gibco-BRL) and developed with chemiluminescence reagents (Pierce) before capturing the image on a chemiluminescence camera (Fujifilm).
Supplementary Material
Acknowledgments
We thank Eric Alani and Jeff Sekelsky for strains and plasmids, Steve Kowalczykowski for advice on strand-exchange reactions and Jan Mullen for unpublished results. We are also grateful to Jac Nickoloff and Jan Mullen for valuable comments on the paper. This work was supported by NIH grant GM071268.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams MD, McVey M, Sekelsky JJ (2003) Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267 [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID (2006) Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res 34: 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Keck JL (2004) Structure and function of RecQ DNA helicases. Crit Rev Biochem Mol Biol 39: 79–97 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Noirot-Gros MF, Wang JC (2000) Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J Biol Chem 275: 26898–26905 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Wang JC (2001) Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: studies of fusion proteins. Proc Natl Acad Sci USA 98: 11108–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DA, Keck JL (2005) Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure 13: 1173–1182 [DOI] [PubMed] [Google Scholar]
- Bernstein DA, Zittel MC, Keck JL (2003) High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J 22: 4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R (2009) Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J 28: 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin AV (2009) Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J Biol Chem 284: 26349–26359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capp C, Wu J, Hsieh TS (2009) Drosophila RecQ4 has a 3′-5′DNA helicase activity that is essential for viability. J Biol Chem 284: 30845–30852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti RS, Schonberg S, German J (1974) A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci USA 71: 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Brill SJ (2007) Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem 282: 28971–28979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID (2005) The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res 33: 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID (2009) RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer 9: 644–654 [DOI] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Smith GR (2008) The fission yeast BLM homolog Rqh1 promotes meiotic recombination. Genetics 179: 1157–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Kaliraman V, Brill SJ (2001) Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J Biol Chem 276: 8848–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol 14: 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PL, Liu Y, Jiricny J, West SC, Janscak P (2004) Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J 23: 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA (2007) Syndrome-causing mutations of the BLM gene in persons in the Bloom's syndrome Registry. Hum Mutat 28: 743–753 [DOI] [PubMed] [Google Scholar]
- Goldfarb T, Alani E (2005) Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair and nonhomologous tail removal. Genetics 169: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G (2005) Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol Cell Biol 25: 3431–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID (2000) The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA 97: 6504–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Carroll R, Kyin S, Seki M, Cole MD (2005) Identification of RecQL1 as a Holliday junction processing enzyme in human cell lines. Nucleic Acids Res 33: 6251–6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Paffett KS, Amit O, Clikeman JA, Sterk R, Brenneman MA, Nickoloff JA (2006) Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol Cell Biol 26: 4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Mullen JR, Brill SJ, Kleff S, Romeo A, Sternglanz R (1996) Human homologs of yeast DNA helicase. Nature 383: 678–679 [DOI] [PubMed] [Google Scholar]
- Machwe A, Lozada EM, Xiao L, Orren DK (2006) Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol Biol 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Matson SW, Orren DK (2005) RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem 280: 23397–23407 [DOI] [PubMed] [Google Scholar]
- Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P (2006) Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 5: 172–180 [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC (2005) Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell 20: 783–792 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima A, Seki M, Onoda F, Ui A, Satoh Y, Ohno Y, Enomoto T (2000) Different domains of Sgs1 are required for mitotic and meiotic functions. Genes Genet Syst 75: 319–326 [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, Kulikowicz T, Beck G, Lee JW, Piotrowski J, Bohr VA (2008) Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry 47: 10247–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Brill SJ (2000) Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ (2005) Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol 25: 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, Rappas M, Freemont PS, Vindigni A (2007) Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol 5: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC (2008) Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA 105: 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank J, Hsieh TS (2009) Helicase-appended topoisomerases: new insight into the mechanism of directional strand exchange. J Biol Chem 284: 30737–30741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank JL, Wu J, Hsieh TS (2006) Topoisomerase IIIalpha and Bloom's helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci USA 103: 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L (2006) The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem 281: 22839–22846 [DOI] [PubMed] [Google Scholar]
- Rockmill B, Fung JC, Branda SS, Roeder GS (2003) The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr Biol 13: 1954–1962 [DOI] [PubMed] [Google Scholar]
- Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM Jr (2005) Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem 280: 28072–28084 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE (2004) Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci USA 101: 9315–9320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Karow JK, Hickson ID, Maizels N (1998) The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem 273: 27587–27592 [DOI] [PubMed] [Google Scholar]
- Ui A, Satoh Y, Onoda F, Miyajima A, Seki M, Enomoto T (2001) The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol Genet Genomics 265: 837–850 [DOI] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R (1989) A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58: 409–419 [DOI] [PubMed] [Google Scholar]
- Wang W, Seki M, Narita Y, Nakagawa T, Yoshimura A, Otsuki M, Kawabe Y, Tada S, Yagi H, Ishii Y, Enomoto T (2003) Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol Cell Biol 23: 3527–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Rio DC (2007) DNA strand displacement, strand annealing and strand swapping by the Drosophila Bloom's syndrome helicase. Nucleic Acids Res 35: 1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J, Rothstein R (2008) The genetic consequences of ablating helicase activity and the Top3 interaction domain of Sgs1. DNA Repair (Amst) 7: 558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID (2005) The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J 24: 2679–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SL, Levitt NC, Hickson ID (2001) Potential role for the BLM Helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem 276: 19375–19381 [DOI] [PubMed] [Google Scholar]
- Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID (2000) The Bloom's syndrome gene product interacts with topoisomerase III. J Biol Chem 275: 9636–9644 [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Xu X, Liu Y (2009) Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J 28: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.