Abstract
Abasic sites represent the most frequent DNA lesions in the genome that have high mutagenic potential and lead to mutations commonly found in human cancers. Although these lesions are devoid of the genetic information, adenine is most efficiently inserted when abasic sites are bypassed by DNA polymerases, a phenomenon termed A-rule. In this study, we present X-ray structures of a DNA polymerase caught while incorporating a nucleotide opposite an abasic site. We found that a functionally important tyrosine side chain directs for nucleotide incorporation rather than DNA. It fills the vacant space of the absent template nucleobase and thereby mimics a pyrimidine nucleobase directing for preferential purine incorporation opposite abasic residues because of enhanced geometric fit to the active site. This amino acid templating mechanism was corroborated by switching to pyrimidine specificity because of mutation of the templating tyrosine into tryptophan. The tyrosine is located in motif B and highly conserved throughout evolution from bacteria to humans indicating a general amino acid templating mechanism for bypass of non-instructive lesions by DNA polymerases at least from this sequence family.
Keywords: abasic sites, DNA polymerases, DNA repair, DNA replication, translesion synthesis
Introduction
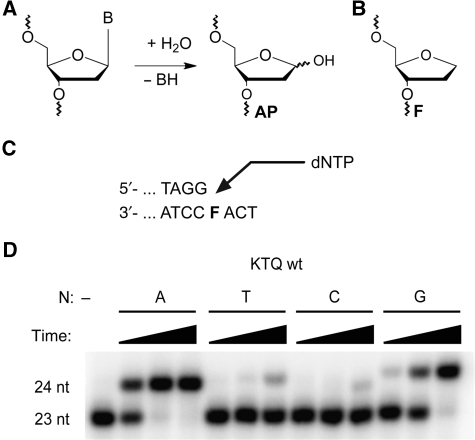
DNA is constantly damaged by endo- and exogenous agents. The most frequent DNA damage observed under physiological conditions are abasic sites resulting from spontaneous hydrolysis of the bond that connects the sugar and the nucleobase in DNA (Lindahl, 1993). It has been estimated that ∼10 000 abasic sites are formed in a human cell per day (Lindahl and Nyberg, 1972; Loeb and Preston, 1986; Lindahl, 1993). Guanine and adenine nucleobase residues are cleaved most efficiently resulting in the abasic sugar moiety (AP, Figure 1A) with the loss of the genetic information stored in the nucleobase (Loeb and Preston, 1986). As these lesions are devoid of the genetic information they are potentially mutagenic. The bulk of this damage is removed by DNA repair systems, which use the sister strand to guide incorporation of the right nucleotide in places of the lesion. However, undetected lesions or those which are formed during S phase pose a challenge to DNA polymerases. Indeed, abasic sites are strong blocks for bypass DNA synthesis catalysed by DNA polymerases (Goodman, 2002; Hübscher et al, 2002). Additionally, several studies indicated the mutagenic potential of these lesions in translesion synthesis, which is more pronounced in animal compared with bacterial cells presumably because of higher translesion synthesis in eukaryotes (Schaaper et al, 1983; Avkin et al, 2002; Pagès et al, 2008). Although abasic sites are considered being non-instructive, in vitro and in vivo studies of the abasic site or the stabilized tetrahydrofuran analogue F (Figure 1B) have shown that adenine, and to a lesser extent guanine, is most frequently incorporated opposite the lesion. DNA polymerases are categorized into seven families according to their sequence homology (Ito and Braithwaite, 1991; Braithwaite and Ito, 1993; Ohmori et al, 2001). The strong preference of DNA polymerase for adenine incorporation is mainly found for DNA polymerases from family A (including human DNA polymerases γ and θ) and B (including human DNA polymerases α, ɛ, and δ) and has been termed ‘A-rule' (Sagher and Strauss, 1983; Schaaper et al, 1983; Loeb and Preston, 1986; Lawrence et al, 1990; Goodman et al, 1994; Shibutani et al, 1997; Avkin et al, 2002; Strauss, 2002; Pagès et al, 2008). The apparent selectivity for incorporation of purines ultimately results in transversion mutations commonly found in human cancers (Hoeijmakers, 2001).
Figure 1.
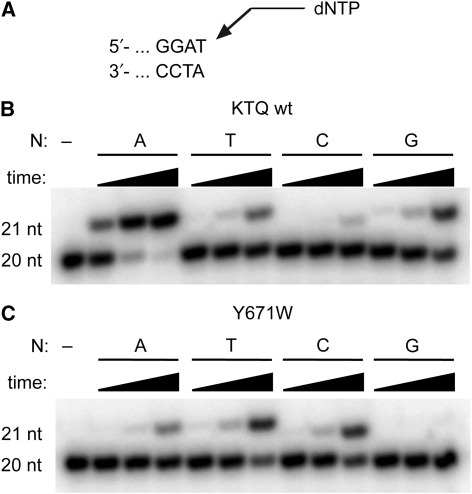
Nucleotide incorporation opposite an abasic site. (A) Hydrolysis of the glycosidic bond leading to nucleobase loss and formation of abasic site AP. B: nucleobase. (B) Structure of the abasic site analogue F. (C) Partial primer template sequence used in primer extension experiments. (D) Single nucleotide incorporation of KlenTaq opposite F for 2, 10, or 60 min, respectively. The respective dNTP is indicated.
What are the determinants of enzymatic specificity for purines? The mechanistic basis for the A-rule is still under debate (Randall et al, 1987; Matray and Kool, 1999; Kool, 2002; Taylor, 2002; Freisinger et al, 2004; Hogg et al, 2004; Seki et al, 2004; Reineks and Berdis, 2004; Ling et al, 2004a; Fiala et al, 2007; Zahn et al, 2007; Chen et al, 2008; Beard et al, 2009; Nair et al, 2009). Structural and functional studies have added significantly to our understanding of the basic mechanisms of translesion synthesis by DNA polymerases (Yang and Woodgate, 2007). Intrinsic properties of purines like superior base-stacking ability of the incoming purine nucleotide to the nucleobase π-system at the primer end were proposed as being the driving forces for preferential purine selection when incorporating a nucleotide opposite an abasic site. Interestingly, it was found that DNA polymerases from diverse sequence families also catalyse non-templated nucleotide addition to the 3′-termini of blunt-ended DNA with a strong preference for incorporation of adenine that parallels the incorporation tendency opposite non-instructive abasic sites (Clark et al, 1987; Clark, 1988; Peliska and Benkovic, 1992; Hwang and Taylor, 2004; Fiala et al, 2007b). Here again superior base-stacking ability of adenine was suggested as a driving force for the enzymes to select an adenine nucleotide. Several structures from DNA polymerases belonging to DNA polymerase families X and Y in complex with a primer template duplex containing the abasic site analogue F and incoming 2′-deoxynucleoside-5′-triphosphate (dNTP) were recently reported (Ling et al, 2004a; Beard et al, 2009; Nair et al, 2009). DNA polymerases from these families use different, sequence-depending mechanisms that might compete with the A-rule when bypassing abasic sites. However, DNA polymerases from family A and B, which are involved in the majority of DNA synthesis in DNA replication and repair, follow the A-rule when bypassing abasic sites. Up to now only structures of RB69 DNA polymerase from family B were reported including one structure in complex with an abasic site lesion and an incoming dGTP (Freisinger et al, 2004). This structure as well as another structure of RB69 DNA polymerase that captures an artificial 5-nitro-1-indoyl-nucleotide, which is unable to undergo hydrogen bonds like natural nucleotides but is more efficiently incorporated opposite abasic sites than dAMP because of increased stacking ability, suggest that base stacking is likely to have a paramount role in the selective incorporation of dAMP opposite abasic sites (Zahn et al, 2007).
In this study, we present two structures of the large fragment of Thermus aquaticus DNA polymerase that follows the A-rule. In these structures, the enzyme is caught while incorporating either an adenine opposite an abasic site or at the 3′-terminus of a blunt-end primer template duplex. These structures provide insights into the mechanistic origin for purine selectivity of this DNA polymerase in absence of nucleobase information. Interestingly, in both structures we found the incoming triphosphate positioned in a way that obviates stacking interactions to the primer template duplex. Instead, a tyrosine protein side chain fills the space of the absent template nucleobase at the site of the nascent nucleobase pair and thereby supports hydrogen bond networks with the incoming adenine nucleotide. Furthermore, the shape and size of the adenine tyrosine pair mimics the geometries of canonical base pairs indicating that geometric complementarity to the active site of the enzyme adds to the selection of purines in the absence of templating nucleobase information. This tyrosine is highly conserved throughout evolution in DNA polymerase family A from bacteria to humans suggesting that this ‘amino acid side chain templating' is a general mechanism for bypass of non-instructive lesions by DNA polymerases from this sequence family. The mechanistic model of ‘amino acid side chain templating' is further corroborated by the results of mutagenesis studies in which the selectivity for purine incorporation was switched to pyrimidines on substituting the six-membered aromatic phenol of the templating tyrosine to the bicyclic indole of tryptophan.
Results
KlenTaq follows the A-rule
Significant mechanistic insights of nucleotide incorporation during DNA polymerization were derived from high-resolution structures of KlenTaq, an N-terminally truncated form of T. aquaticus (Taq) DNA polymerase (Korolev et al, 1995; Li et al, 1998; Li and Waksman, 2001; Rothwell et al, 2005). KlenTaq is a member of family A DNA polymerases that are involved in prokaryotic and eukaryotic repair of DNA lesions (Goodman, 2002; Hübscher et al, 2002; Seki et al, 2004). It has been shown that DNA polymerases from this family follow the A-rule (Sagher and Strauss, 1983; Shibutani et al, 1997; Seki et al, 2004), which we found for KlenTaq as well (Figure 1C and D). Single nucleotide incorporation opposite F was conducted with all four dNTPs and preferential incorporation of dAMP was observed (Figure 1D). Quantification of these findings by pre-steady-state kinetics confirmed that indeed dAMP is most efficiently incorporated opposite the abasic site F followed by dGMP (Table 1). Consistently with earlier findings of A family enzymes pyrimidines are significantly less efficiently processed opposite the lesion.
Table 1. Transient kinetic analysis of nucleotide incorporation opposite abasic site F by KlenTaq and mutants.
| Enzyme | Template | dNTP | kpol (s−1 × 10−2) | Kd (μM) | kpol/Kd (μM−1 s−1 × 10−4) |
|---|---|---|---|---|---|
| Wild type | dT | A | 516±44 | 15.3±4.7 | 3373 |
| Wild type | F | A | 2.73±0.23 | 149±35 | 1.83 |
| Wild type | F | G | 0.43±0.01 | 65.5±5.2 | 0.66 |
| Wild type | F | T | 0.03±0.006 | 299±122 | 0.01 |
| Wild type | F | C | 0.02±0.001 | 294±17 | 0.007 |
| Y671A | dT | A | 1.06±0.06 | 168±26 | 0.63 |
| Y671A | F | A | 0.02±0.002 | 304±68 | 0.007 |
| Y671A | F | G | 0.05±0.002 | 182±23 | 0.03 |
| Y671A | F | T | NA | NA | NA |
| Y671A | F | C | NA | NA | NA |
| Y671F | dT | A | 2265±161 | 54.1±12.0 | 4187 |
| Y671F | F | A | 1.16±0.06 | 233±30 | 0.50 |
| Y671F | F | G | 0.33±0.03 | 294±54 | 0.11 |
| Y671F | F | T | 0.02±0.002 | 245±64 | 0.008 |
| Y671F | F | C | 0.01±0.001 | 262±65 | 0.004 |
| Wild type | dT | d3A | 59.6±3.3 | 95.4±13.4 | 62.5 |
| Wild type | F | d3A | 0.12±0.01 | 572±67 | 0.02 |
| Y671W | dT | A | 91.6±15.2 | 363±104 | 25.2 |
| Y671W | F | A | 0.10±0.02 | 514±182 | 0.02 |
| Y671W | F | G | NA | NA | NA |
| Y671W | F | T | 0.35±0.03 | 382±76 | 0.09 |
| Y671W | F | C | 0.92±0.12 | 845±161 | 0.11 |
| NA, Not accessible because the turnover after 1 h using 600 μM dNTP was <20%. | |||||
Crystallization and structure determination of KlenTaq in complex with incoming nucleoside triphosphate opposite abasic site
To show the structural basis for the ability of DNA polymerases to promote abasic site bypass according to the A-rule we crystallized KlenTaq in complex with relevant substrates. We were able to obtain several crystals and could solve the structure of a ternary complex of KlenTaq bound to an abasic site, containing a primer, template, and an incoming 2′,3′-dideoxyadenosine-5′-triphosphate (ddATP) opposite the lesion (henceforth termed KlenTaqAP for simplicity). The crystals were obtained by a strategy used earlier for KlenTaq (Li and Waksman, 2001) bound to non-damaged substrates. The structure was solved by difference Fourier techniques and provides a snapshot of nucleotide incorporation opposite an abasic site F in the template strand at a resolution of 2.3 Å (Supplementary Table I).
Overall DNA polymerase structure in complex with abasic site template
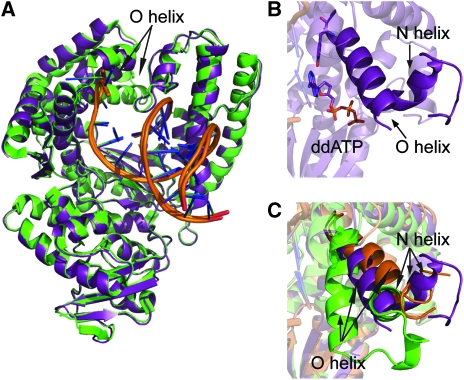
KlenTaq in complex with ddATP opposite an abasic site template adopted an overall conformation very similar to the one in a ternary complex of the enzyme bound to non-damaged DNA and ddATP (PDB 1QSY; Li and Waksman, 2001) described earlier resulting in a root mean square deviation for Cα of 0.45 Å (Figure 2A). However, remarkable structural changes are found in the finger domain of the enzyme especially in the orientation of the O helix near the primer terminus and the dNTP-binding site (Figure 2B and C). Previously described structures of the ternary complexes of KlenTaq bound to undamaged DNA and ddNTP in an open (PDB 2KTQ, Li et al, 1998) and a closed (PDB 1QSY) conformation show that switching from the open form to the closed conformation causes a structural change of 46° of the finger domain. In the open conformation the orientation of the O helix resembles the binary structure of KlenTaq in complex with the primer template (PDB 4KTQ). The positioning of the O helix allows binding of the substrates in the open conformation, whereas in the closed conformation the O helix packs against the templating nucleobase and incoming dNTP and thereby closes the active site. However, in the KlenTaqAP structure, the O helix is in a conformation that leaves the active site more open compared with the closed structure containing the undamaged primer template complex. The overall orientation of the O helix is somehow between the open and closed conformations, illustrated in the superimposition of the structures (Figure 2C; Supplementary Figure S2).
Figure 2.
Structures of KlenTaq in complex with substrates. (A) Structure of KlenTaqAP (purple) superimposed with KlenTaq (green, PDB 1QSY) bound to undamaged DNA. The location of the O helix is indicated. (B) Close-up view highlights the location of the O and N helices in KlenTaqAP. (C) Superimposition of KlenTaqAP, KlenTaq (green, PDB 1QSY, closed conformation) and KlenTaq (orange, PDB 2KTQ, open conformation).
Protein side chain and substrate conformations at the abasic site
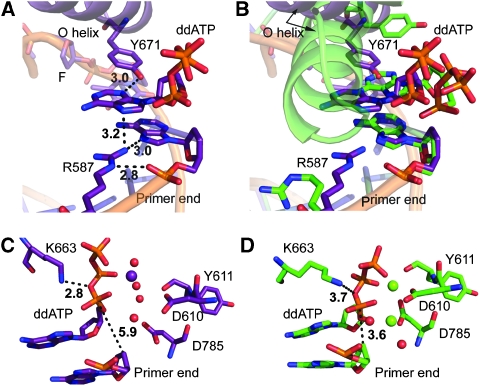
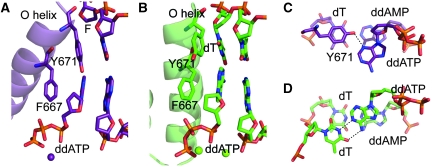
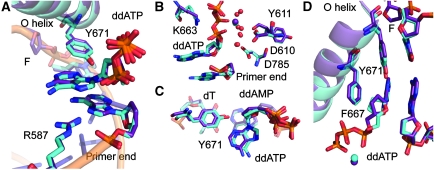
The abasic site is intrahelically placed within the DNA substrate and the tetrahydrofuran moiety rotated by about 90° in comparison to the respective sugar conformation found in canonical KlenTaq structures (Supplementary Figure S1). The incoming ddATP is stabilized by a network of hydrogen bond interactions. Arg587 adopts a conformation that allows the formation of hydrogen bonds between the primer strand and the incoming nucleotide (Figure 3A). The distances found indicate bonding of the Arg587 guanidinium group to the 3′-terminal nucleotide in the primer strand by binding to N7 of the adenine and the phosphodiester as well as to N7 of the adenine of the incoming ddATP. The conformation of Arg587 in the KlenTaqAP structure differs from the conformations described in the earlier structures where such interactions were not found (Figure 3A and B). Furthermore, the nucleobase of the incoming ddATP is positioned in a cleft formed by Phe667 and Tyr671 (Figures 3A and 4A). The distance between the hydroxyl group of Tyr671 and the N3 of adenine indicates a hydrogen bonding interaction between these residues (Supplementary Figure S3). Thereby, the aromatic side chain of Tyr671 fills the vacant space that has been left by the missing nucleobase and may act as a positioning device to substitute for the missing nucleobase (Figure 4A). These interactions place the incoming ddATP opposite the abasic site in a position where stacking interactions to the nucleobase π-system at the primer end, which are found in the non-damaged complexes, are lost (Figure 4C and D). The Tyr671, on the other hand, is positioned in a way that it nicely stacks to the template nucleobase positioned 3′ of the abasic site (Figure 4C and D).
Figure 3.
Interaction network of incoming ddATP opposite abasic site F. (A) Hydrogen bond network stabilizing ddATP. Labelled are the amino acid side chains R587 and Y671. (B) Superimposed structures of KlenTaqAP (purple) and KlenTaq (green, PDB 1QSY) show the difference in orientation of residues R587 and Y671. (C) Inner coordination spheres showing metal ion in KlenTaqAP. A Mg2+ ion (purple sphere) is coordinated by the triphosphate moiety of ddATP and two water molecules (red spheres). Two additional water molecules form hydrogen bonds to residues D610, Y611, and D785. Residue K663, recently, discussed to act as general acid in catalysis, is shown. (D) KlenTaq (PDB 1QSY) showing the same residues as in (C). All distances are in Å.
Figure 4.
Active site and nascent base pair assemblies. (A) Close-up view of KlenTaqAP active site processing ddATP opposite abasic site F. Shown are O helix, residues Y671, and F667, the respective template residues and incoming ddATP. (B) Structure of KlenTaq (PDB 1QSY) active site processing ddATP opposite template dT. (C) Top view to the nascent base pair opposite F. In the front the incoming ddATP opposite Y671 is depicted. The hydrogen bond between the hydroxyl group of Y671 and N3 of adenine is indicated as dashed line. In transparent the first nucleobase pair of the primer template terminus is shown. (D) Top view of the nascent base pair in KlenTaq (PDB 1QSY). The hydrogen bonding between the incoming ddATP and the templating dT is shown in dashed lines. The primer template terminus is illustrated in transparent.
Compared with the earlier KlenTaq structure bound to canonical substrates (PDB 1QSY) the incoming nucleotide aligns in a different manner in the KlenTaqAP structure. The distance from the primer 3′-terminus to the α-phosphate of the ddATP is larger by 2.3 Å (Figure 3C and D). However, the distance from the oxygen that bridges α- and β-phosphate groups of the triphosphate moiety and Lys663, which was previously discussed to act as a general acid in catalysis of nucleotide bond formation (Castro et al, 2009), is found to be shorter by 0.9 Å (Figure 3C and D). DNA polymerases use a general two-metal ion mechanism (Fothergill et al, 1995; Steitz, 1998; Yang et al, 2006) to catalyse the nucleophilic substitution of pyrophosphate by the 3′-hydroxyl group of the primer. However, in the KlenTaqAP structure only one Mg2+ is found coordinating to the triphosphate moiety and involved in interaction with two water molecules (Figure 3C and D). The two other water molecules form several hydrogen bonds to the amino acid residues known to be essential for catalysis. This altered arrangement may account for the observed 1840-fold decline in dAMP incorporation efficiency (kpol/Kd) opposite F compared with incorporation opposite dT (Table 1) and stalling of the DNA polymerase when encountering the lesion.
Tyrosine 671 mimics the absent nucleobase in the template strand
The positioning of Tyr671 opposite the incoming ddATP at the place that is usually occupied by the templating nucleobase suggests that the protein side chain is a templating device and mimics the shape and size of a six-membered pyrimidine nucleobase in the template strand. These active site constraints might determine the preferential incorporation of adenine by a close geometric fit to the enzyme active site (Goodman, 1997; Kool, 2002). To study the functional role of Tyr671 in abasic site lesion bypass we constructed mutants at this position. The Tyr671Ala mutant was constructed to test the requirement of an aromatic residue at this position. Indeed, this mutation led to an enzyme that has significantly reduced activity in general. The efficiency of dAMP incorporation opposite canonical template dT is more than 5350-fold reduced (Table 1). Incorporation efficiency opposite an abasic site is further reduced and we were able to measure the incorporation of dAMP and dGMP only. Interestingly, the enzyme lost its preference of incorporation of dAMP opposite an abasic site and instead incorporates dGMP four-fold more efficient. To probe the effects of the hydroxyl group in Tyr671 on enzyme activity we generated the Tyr671Phe mutant next. We found that this enzyme is highly active when non-damaged substrates are processed. The Tyr671Phe mutant follows the A-rule; however, in comparison to the wild type, this enzyme incorporates dAMP opposite an abasic site with four-fold reduced efficiency resulting from both, an increased Kd and a reduced kpol, indicating the requirement of the hydroxyl group for efficient catalysis.
To further analyse the interaction between the hydroxyl group of Tyr671 and N3 of adenine in the incoming nucleotide an adenine analogue in which the N3 is substituted by a non-polar CH (namely 3-deaza-2′-deoxyadenosine-5′-triphosphate (d3DATP)) was used in primer extension studies (Supplementary Figure S4). These kinds of purine analogues were intensively studied to access the role of N3–hydrogen bond acceptors in DNA polymerase function (Spratt, 2001; Washington et al, 2003; Meyer et al, 2004; Moore et al, 2004; McCain et al, 2005; Wolfle et al, 2005; Beckman et al, 2007; Cavanaugh et al, 2009; Trostler et al, 2009). In accordance with earlier reports (Trostler et al, 2009) we found that incorporation efficiency of the analogue was decreased by 54-fold compared with the natural substrate (Table 1). Interestingly, incorporation efficiency of d3DAMP opposite the abasic site was decreased by more than 90-fold compared with dAMP incorporation opposite the lesion. These studies further highlight the importance of the interaction of the hydroxyl group of Tyr671 and N3-adenine in the incoming nucleotide.
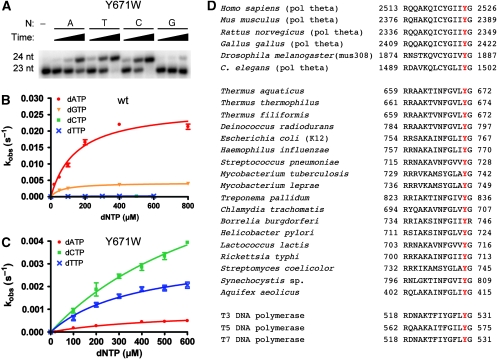
Next, the mutation Tyr671Trp was constructed. As this mutation will transform the six-membered phenol ring of Tyr671 into a bicyclic indole consisting of a six-membered ring fused to a five-membered ring, the amino acid side adopts an approximate size of a purine. We assumed that, if the amino acid side chain at this position indeed inherited the templating role of the absent nucleobase, the smaller pyrimidine nucleotides thymidine and cytidine should be more preferentially incorporated opposite an abasic site because of the closer geometric fit of the tryptophane side chain to purines. Indeed, this was observed in primer extension studies and quantified by measurement of the kinetics (Figure 5A–C; Table 1). Although the wild type enzyme incorporates dTMP and dCMP with 183- and 261-fold reduced efficiency opposite the abasic site compared to dAMP, respectively, the contrary is observed for Tyr671Trp. In the Tyr671Trp mutant enzyme the preference for dATP vanished and instead the pyrimidines dTMP and dCMP are about five-fold more efficiently incorporated than dAMP (Figure 5A–C; Table 1).
Figure 5.
Nucleotide incorporation opposite abasic site F. (A) Single nucleotide incorporation of KlenTaq Tyr671Trp mutant opposite F for 2, 10, or 60 min, respectively. The respective dNTP is indicated. (B, C) Pre-steady-state kinetics of single nucleotide incorporation opposite F catalysed by KlenTaq wild type (B) and Tyr671Trp mutant (C), respectively. The curves show dependence of the observed pre-steady-state rates (kobs) on dNTP concentration. The kobs values were plotted versus the concentration of the used dNTP and fitted to a hyperbolic equation. (D) Amino acid sequence alignment of DNA polymerases highlighting the conserved position equivalent to Y671 in KlenTaq.
Template-independent extension of blunt-end primer templates by KlenTaq
Many DNA polymerases catalyse non-templated nucleotide addition to the 3′-termini of blunt-ended DNA with a strong preference for incorporation of adenine (Clark et al, 1987; Clark, 1988; Peliska and Benkovic, 1992; Hwang and Taylor, 2004; Fiala et al, 2007b). The same holds true for KlenTaq as our analyses show (Figure 6; Table 2). KlenTaq extends the blunt-end primer terminus by incorporation of dAMP about 19-fold more efficient than of dGMP. The pyrimidines are even less efficiently incorporated compared with dAMP (38-fold and 280-fold reduced efficiency for dTMP and dCMP, respectively). To study a potential templating effect of Tyr671 we analysed the Tyr671Trp mutant in blunt-end extension, next. Again, the mutation caused a shift of the incorporation specificity to preferential incorporation of pyrimidines (Figure 6; Table 2). Thus, dTMP and dCMP are about two-fold more efficiently incorporated than dAMP by the mutated enzyme. We could not observe any significant incorporation of dGMP even after prolonged incubation times.
Figure 6.
Nucleotide incorporation at a blunt-end primer template duplex. (A) Partial sequence used in primer extension experiments. (B) Single nucleotide incorporation of KlenTaq by extension of blunt-end primer template duplex for 2, 10, or 60 min, respectively. The respective dNTP is indicated. (C) As in (B) for KlenTaq mutant Tyr671Trp in presence of a blunt-end primer template duplex.
Table 2. Transient kinetic analysis blunt-end DNA duplex extension by KlenTaq and mutants.
| Enzyme | dNTP | kpol (s−1 × 10−2) | Kd (μM) | kpol/Kd (μM−1 s−1 × 10−4) |
|---|---|---|---|---|
| Wild type | A | 3.11±0.08 | 275±13 | 1.13 |
| Wild type | G | 0.21±0.01 | 371±39 | 0.06 |
| Wild type | T | 0.13±0.01 | 503±98 | 0.03 |
| Wild type | C | 0.07±0.02 | 1729±620 | 0.004 |
| Y671W | A | 0.12±0.01 | 450±42 | 0.03 |
| Y671W | G | NA | NA | NA |
| Y671W | T | 0.52±0.03 | 747±69 | 0.07 |
| Y671W | C | 0.44±0.04 | 746±108 | 0.06 |
| NA, Not accessible because the turnover after 1 h using 600 μM dNTP was <20%. | ||||
KlenTaq structure in complex with blunt-end primer template duplex and incoming nucleoside triphosphate
To generate structural insights into template-independent nucleotidyl transferase activity of KlenTaq we crystallized the enzyme in presence of the same primer used before, a template and ddATP. By the incorporation of ddAMP at the primer terminus a blunt-end primer template duplex lacking a 3′-terminal hydroxyl group is formed and a ddATP captured at the active site for template-independent elongation of the primer end. This structure (henceforth termed KlenTaqBE) was determined with a resolution of 2.2 Å (Supplementary Table I).
Comparison of the KlenTaqBE and KlenTaqAP structures shows high similarity resulting in a very low root mean square deviation for Cα of about 0.26 Å (Supplementary Figures S3 and S5). The close-up view of the O helix in KlenTaqBE and its superimposition to KlenTaqAP highlight the similarity (Figure 7). Both structures show the same orientation of the O helix and the interaction patterns of the amino acids Arg587 and Tyr671 are very much alike. The same holds true for the position of the incoming ddATP that is again placed in a way obviating significant stacking interactions to the nucleobase π-system at the primer end (Figure 7).
Figure 7.
Comparison of KlenTaqBE (cyan) and KlenTaqAP (purple). (A) Superimposed structures of KlenTaqBE and KlenTaqAP. (B) Comparison of the inner coordination sphere of metal ion in KlenTaqBE and KlenTaqAP. (C) Superimposition of the top view of the nascent nucleobase pair of KlenTaqBE and KlenTaqAP. (D) Close-up views of the active sites of KlenTaqBE and KlenTaqAP. Labelled are O helix, residues F667, and Y671.
Discussion
Several studies show that adenine is most frequently incorporated opposite abasic site lesions in cells resulting in transversion mutations. Indeed, it has been shown that DNA polymerases from the sequence families A and B, which are involved in the majority of DNA synthesis in a cell, follow the A-rule (Sagher and Strauss, 1983; Randall et al, 1987; Shibutani et al, 1997; Matray and Kool, 1999; Taylor, 2002; Freisinger et al, 2004; Hogg et al, 2004; Seki et al, 2004). On the other hand, DNA polymerases from other sequence families like X and Y use different, sequence-depending mechanisms that might compete with the A-rule. Structures of these DNA polymerases complexed to abasic site containing DNA duplexes were reported for human DNA polymerases ι and β, and DNA polymerase IV (Dpo4) of Sulfolobus Solfataricus (Ling et al, 2004a; Beard et al, 2009; Nair et al, 2009). Human DNA polymerase ι incorporates a nucleotide opposite an abasic site with relatively small preference for G, T, and A over C. Structures of this enzyme in complex with an abasic site lesion and several dNTPs suggest that the enzyme forms an active site that stabilizes the incoming dNTP by distinct networks of hydrogen bonds (Nair et al, 2009). In contrast, DNA polymerase β and Dpo4 show obviation of the A-rule by predominantly inserting a nucleotide that is complementary to the first downstream templating nucleobase (Efrati et al, 1997; Ling et al, 2004a; Fiala et al, 2007; Beard et al, 2009). The structural data of Dpo4 show that the tendency for using the nucleobase downstream to the lesion stems from the ability of the enzyme to bulge out the lesion in an extrahelical position. In contrast, structures of RB69 DNA polymerase from family B that obeys the A-rule suggest that intrinsic properties of the purine nucleobase of the incoming dNTP like base stacking and solvation properties are likely crucial for selection of dATP opposite the abasic site by this DNA polymerase sequence family (Freisinger et al, 2004; Hogg et al, 2004; Zahn et al, 2007).
Sequence family A members Taq DNA polymerase and Escherichia coli DNA polymerase I have been studied extensively in the past in their structure–function relationship (Patel et al 2001; Loh and Loeb 2005, Loh et al, 2007). We present here the first structure of a DNA polymerase that follows the A-rule and is complexed with an abasic site lesion and an incoming adenine opposite the lesion. The structure and mutagenesis studies show that the amino acid side chain of Tyr671, which is placed opposite the incoming ddATP at the location that is usually occupied by the templating nucleobase, confers the templating ability of the missing nucleobase at least to some extent. Previous studies on Tyr671 of Taq DNA polymerase and the corresponding Tyr766 residue in DNA polymerase I from E. coli have shown the involvement of this tyrosine residue in discriminating ribonucleotides and non-canonical nucleotides (Suzuki et al, 1996; Bell et al, 1997; Minnick et al, 1999). However, the role in translesion synthesis of this tyrosine has not been shown before. That amino acid templating of Tyr761 is involved in selecting a purine nucleotide is further corroborated by the finding that the respective tryptophan mutant lost its purine specificity and instead incorporates pyrimidines more efficiently. The result can be rationalized by assuming that the bicyclic indole consisting of a six-membered ring fused to a five-membered ring will mimic the approximate size and shape of a purine, and as a consequence, will direct for pyrimidine incorporation because of enhanced geometric fit to the enzyme active site. These findings corroborate the model of amino acid templating for abasic site bypass and highlight the importance of geometric fit of the substrates to the active site of the enzyme for DNA polymerase activity. In agreement with these observations, it has been shown that DNA polymerases are able to process non-natural nucleobase surrogates placed in template strands that mimic the shape and size of the natural nucleobase but have decreased hydrogen bonding capabilities (Kool, 2002).
Amino acid sequence alignment (Figure 5D) of A-family DNA polymerases discloses that the respective tyrosine is within motif B (Delarue et al, 1990) and is highly conserved throughout evolution from bacteria to humans. Owing to the high conservation of amino acids structure and sequence at these positions in DNA polymerases from prokaryotes to eukaryotes it is likely that the depicted mechanism of abasic site bypass is general and applies to other DNA polymerases in this sequence family as well. The mechanism of using protein residues to direct nucleotide incorporation is reminiscent of a transfer RNA CCA-adding enzyme (Xiong and Steitz, 2004) and the specialized DNA polymerase Rev1 that exclusively incorporates cytosine nucleotides (Nair et al, 2005). Unlike other DNA lesions abasic sites are devoid of any structural information that is at least residual in damaged nucleobases. Residual information is used by DNA polymerases to catalyse translesion synthesis through aberrant hydrogen bonding patterns of the incoming dNTP to the damaged template nucleobase. Bypass of DNA lesions such as thymidine dimers (Ling et al, 2003), 8-oxidized guanosine residues (Brieba et al, 2004; Hsu et al, 2004; Rechkoblit et al, 2006), or nucleobase adducts (Ling et al, 2004b; Nair et al, 2006) is promoted in this fashion.
Interestingly, all studied DNA polymerases that obviate the A-rule when encountering an abasic site show preference for incorporation of dAMP when catalysing the template-independent extension of blunt-end 3′ termini of DNA duplexes (Clark, 1988; Fiala et al, 2007b). These enzymes include DNA polymerases β, η, and Dpo4. A structure of Dpo4 in complex with a blunt-end DNA duplex and ddATP shows that the nucleobase and the sugar of the incoming ddATP stack to the nucleobase π-system of the blunt-end DNA duplex (Fiala et al, 2007b). This result corroborates earlier conclusions drawn from functional studies that intrinsic stacking interactions and solvation properties are driving forces for selection of adenine. However, our structural data presented here do not provide any evidence for stacking interactions of the incoming ddATP with the primer template terminus and rather indicate that amino acid templating applies for selection of adenine for DNA polymerases from sequence family A. This conclusion is corroborated by the findings that (a) no significant stacking of the incoming ddATP to the π-system of the DNA duplexes is observed in the structures and (b) mutation of the templating tyrosine to tryptophan switches the selectivity for purines to pyrimidines. Nevertheless, it cannot be excluded that stacking interactions might have a role in processes that are unresolved by the structural data, for example, at states prior or later on the reaction coordinate than the one resolved here.
Both structures presented here show different protein and substrate arrangements in the active site in the absence of any templating nucleobase in comparison to the canonical complexes. They differ from the corresponding structure of ddATP in the KlenTaq structure with undamaged template DNA, which exhibits more favourable distances for formation of the new bond from the α-phosphate of the incoming nucleotide to the 3′-hydroxyl end of the primer (Korolev et al, 1995; Li et al, 1998; Li and Waksman, 2001). Moreover, the location of the α-phosphate of the incoming ddATP in KlenTaqAP obviates tight complexation of a Mg2+ ion by coordination together with D785 as found in the KlenTaq structure (Figure 3C and D) and, as a consequence, only one Mg2+ ion was found in the active site. This indicates that in the absence of the nucleobase larger conformational fluctuations as well as binding of a second Mg2+ ion would be required for phosphodiester bond formation, in accord with the observed decline in nucleotide incorporation efficiency and the strong block of the abasic site lesion. Bypass of the non-instructive abasic site by DNA polymerases is intrinsically error-prone. Thus, pausing resulting from the lesion might allow the DNA polymerase to be replaced by repair systems that use the sister strand for error-free repair. However, recently it was reported that the A-family member human DNA polymerase θ is able to bypass abasic sites with high activity and thereby incorporates dAMP opposite the lesion preferentially (Seki et al, 2004). DNA polymerase θ is involved in the diversification of immunoglobulin (Ig) genes during somatic hypermutation (Masuda et al, 2007). Hence, the efficient but intrinsically error-prone bypass of abasic sites by DNA polymerase θ following the A-rule might be of advantage for physiological mutation in somatic hypermutation of Ig genes.
Although our structural data show interaction patterns that stabilize purines in favour of pyrimidines as incoming nucleoside triphosphates, they do not directly allow drawing conclusions on the preference of adenine over guanine opposite an abasic site as well as at the blunt-end duplex. Intrinsic properties of the respective nucleobase like stacking and solvation properties may likely account for the preference of adenine. Modelling of a guanine in the position that is occupied by adenine in the structure indicates that the exocyclic C2–NH2 group of guanine shortens the distance to Tyr671 in comparison to the C2–H in adenine (Supplementary Figure S6). This restriction of the active site might perturb conformational changes required for catalysis of nucleotide incorporation, and thereby adds to lowering the efficiency of dGMP incorporation by the enzyme.
Materials and methods
Proteins and oligonucleotides
Protein mutation and purification were conducted as described (Li et al, 1998). All oligonucleotides were purchased from Purimex, Germany and double HPLC purified. The nucleotide d3DATP was synthesized starting from d3DA purchased from Berry & Associates Inc., USA, according to published procedures (Di Pasquale et al, 2008).
Primer extension assays
Incorporation opposite dT or F: 20 μl of the KlenTaq reactions contained 100 nM primer (5′-d(CGT TGG TCC TGA AGG AGG ATA GG)-3′), 130 nM template (5′-d(AAA TCA TCC TAT CCT CCT TCA GGA CCA ACG TAC)-3′), or F-containing template (5′-d(AAA TCA FCC TAT CCT CCT TCA GGA CCA ACG TAC)-3′), respectively, 100 μM dNTPs in buffer (20 mM Tris–HCl pH 7.5, 50 mM NaCl, and 2 mM MgCl2) and 500 nM of the respective KlenTaq polymerase. Reaction mixtures were incubated at 37°C. Incubation times are provided in the respective figure legends. Primer was labelled using [γ32P]-ATP according to standard techniques. Reactions were stopped by addition of 45 μl stop solution (80% [v/v] formamide, 20 mM EDTA, 0.25% [w/v] bromophenol blue, 0.25% [w/v] xylene cyanol) and analysed by 12% denaturing PAGE. Visualization was performed by phosphoimaging. Blunt-end extension experiments were performed as described above using DNA primer (5′-d(CGT TGG TCC TGA AGG AGG AT)-3′) template (5′-d(ATC CTC CTT CAG GAC CAA CGA AA)-3′) strands that form a blunt-end only at one terminus.
Enzyme kinetics
The rate of single turnover, single nucleotide incorporation was determined by rapid quench flow kinetics using a chemical quench flow apparatus (RQF-3, KinTek Corp., University Park, PA) as described earlier (Di Pasquale et al, 2008). For reaction times longer than 5 s, a manual quench was performed. In brief, 15 μl of radiolabelled primer/template complex (200 nM) and DNA polymerase (2 μM) in reaction buffer were rapidly mixed with 15 μl of a dNTP solution in buffer at 37°C. Quenching was achieved by adding 0.3 M EDTA solution at defined time intervals before mixing with the previously described stop solution. For the analysis of dNTP incorporation opposite to the abasic site primer (sequences see above) and templates (sequences see above) were applied. Quenched samples were analysed on a 12% denaturing PAGE followed by phosphoimaging. For kinetic analysis experimental data were fit by nonlinear regression using the program GraphPad Prism 4. The data were fit to a single exponential equation: [conversion]=A*(1−exp(−kobs t)). The observed catalytic rates (kobs) were then plotted against the dNTP concentrations used and the data were fitted to a hyperbolic equation to determine the Kd of the incoming nucleotide. The incorporation efficiency is given by kpol/Kd. Kinetic experiments for blunt-end measurements were performed under the same conditions. The identical primer/template duplex was used as described above.
Crystallization and structure determination
KlenTaqAP. The crystallization was set up using a solution containing KlenTaq (102 μM) in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, DNA template (5′-d(AAA FTG CGC CGT GGT C)-3′ (300 μM), DNA primer (5′-d(GAC CAC GGC GC)-3′ (300 μM), ddATP (4 mM), 20 mM MgCl2 and mixed in 1:1 ratio with the reservoir solution (0.05 M sodium cacodylate, pH 7, 0.2 M ammonium acetate, 0.01 M magnesium acetate, and 30% PEG 8000). Crystals were produced by the hanging drop vapour diffusion method by equilibrating against 1 ml of the reservoir solution for 5 days at 18°C.
KlenTaqBE. The crystallization was set up with KlenTaq (113 μM) in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, template (5′-d(TG CGC CGT GGT C)-3′ (340 μM), primer (5′-d(GAC CAC GGC GC)-3′ (340 μM), ddATP (2.3 mM), 20 mM MgCl2 and mixed in 1:1 ratio with the reservoir solution containing 0.05 M Tris–HCl, pH 8, 0.2 M ammonium chloride, 0.01 M calcium chloride, and 35% PEG 4000. Crystals were produced by the sitting drop vapour diffusion method by equilibrating against 1 ml of the reservoir solution for 5 days at 18°C. The crystals were frozen and analysed. Datasets were collected at the beamline PXIII (X06DA) at the Swiss Light Source of the Paul Scherrer Institute (PSI) in Villigen, Switzerland, at a wavelength 1.0 Å and using a Mar225 CCD detector. Data reduction was performed with the XDS package (Kabsch, 1993). The structures were solved by difference Fourier techniques using KlenTaq wild type (PDB 1QSY) as model. Refinement was performed with PHENIX (Adams et al, 2002) and model rebuilding was done with COOT (Emsley and Cowtan, 2004). The structures were solved and refined to a resolution of 2.3 Å in KlenTaqAP and 2.2 Å in KlenTaqBE. Both structures were in the same space group P3121, with cell dimensions a, b=110.2 Å, c=91.3 Å for KlenTaqAP, and a ,b=109.6 Å, c=91.0 Å for KlenTaqBE, respectively. The Mg2+ ion in the enzyme active sites of both structures was coordinated characteristic for a bivalent metal ion. Figures were made with PyMOL (DeLano, 2002).
Accession numbers
The coordinates and structure factors have been deposited in the Protein Data Bank with the accession numbers 3LWL and 3LWM.
Supplementary Material
Acknowledgments
We thank the DFG for funding within SPP 1170 and the beamline staff of the Swiss Light Source (SLS) for support.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Avkin S, Adar S, Blander G, Livneh Z (2002) Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci USA 99: 3764–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard WA, Shock DD, Batra VK, Pedersen LC, Wilson SH (2009) DNA polymerase β substrate specificity: side chain modulation of the ‘A-rule'. J Biol Chem 284: 31680–31689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J, Kincaid K, Hocek M, Spratt T, Engels J, Cosstick R, Kuchta RD (2007) Human DNA polymerase alpha uses a combination of positive and negative selectivity to polymerize purine dNTPs with high fidelity. Biochemistry 46: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JB, Eckert KA, Joyce CM, Kunkel TA (1997) Base miscoding and strand misalignment errors by mutator Klenow polymerases with amino acid substitutions at tyrosine 766 in the O helix of the fingers subdomain. J Biol Chem 272: 7345–7351 [DOI] [PubMed] [Google Scholar]
- Braithwaite DK, Ito J (1993) Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res 21: 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieba LG, Eichman BF, Kokoska RJ, Doublié S, Kunkel TA, Ellenberger T (2004) Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J 23: 3452–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Götte M, Konigsberg W, Cameron CE (2009) Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol 16: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh NA, Urban M, Beckman J, Spratt TE, Kuchta RD (2009) Identifying the features of purine dNTPs that allow accurate and efficient DNA replication by herpes simplex virus I DNA polymerase. Biochemistry 48: 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Dupradeau FY, Case DA, Turner CJ, Stubbe J (2008) DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics. Nucleic Acids Res 36: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JM (1988) Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res 16: 9677–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JM, Joyce CM, Beardsley GP (1987) Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J Mol Biol 198: 123–127 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL User's Manual. Palo Alto, CA, USA: DeLano Scientific [Google Scholar]
- Delarue M, Poch O, Tordo N, Moras D, Argos P (1990) An attempt to unify the structure of polymerases. Protein Eng 3: 461–467 [DOI] [PubMed] [Google Scholar]
- Di Pasquale F, Fischer D, Grohmann D, Restle T, Geyer A, Marx A (2008) Opposed steric constraints in human DNA polymerase beta and E. coli DNA polymerase I. J Am Chem Soc 130: 10748–10757 [DOI] [PubMed] [Google Scholar]
- Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF (1997) Abasic translesion synthesis by DNA polymerase beta violates the ‘A-rule'. Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J Biol Chem 272: 2559–2569 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fiala KA, Brown JA, Ling H, Kshetry AK, Zhang J, Taylor J-S, Yang W, Suo Z (2007b) Mechanism of template-independent nucleotide incorporation catalyzed by a template-dependent DNA polymerase. J Mol Biol 365: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala KA, Hypes CD, Suo Z (2007) Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J Biol Chem 282: 8188–8198 [DOI] [PubMed] [Google Scholar]
- Fothergill M, Goodman MF, Petruska J, Warshel A (1995) Structure-energy analysis of the role of metal ions in phosphodiester bond hydrolysis by DNA polymerase I. J Am Chem Soc 117: 11619–11627 [Google Scholar]
- Freisinger E, Grollman AP, Miller H, Kisker C (2004) Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J 23: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF (1997) Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc Natl Acad Sci USA 94: 10493–10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem 71: 17–50 [DOI] [PubMed] [Google Scholar]
- Goodman MF, Cai H, Bloom LB, Eritja R (1994) Nucleotide insertion and primer extension at abasic template sites in different sequence contexts. Ann N Y Acad Sci 726: 132–142 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Hogg M, Wallace SS, Doublié S (2004) Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J 23: 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu GW, Ober M, Carell T, Beese LS (2004) Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature 431: 217–221 [DOI] [PubMed] [Google Scholar]
- Hübscher U, Maga G, Spadari S (2002) Eukaryotic DNA polymerases. Annu Rev Biochem 71: 133–163 [DOI] [PubMed] [Google Scholar]
- Hwang H, Taylor JS (2004) Role of base stacking and sequence context in the inhibition of yeast DNA polymerase eta by pyrene nucleotide. Biochemistry 43: 14612–14623 [DOI] [PubMed] [Google Scholar]
- Ito J, Braithwaite DK (1991) Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res 19: 4045–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kool ET (2002) Active site tightness and substrate fit in DNA replication. Annu Rev Biochem 71: 191–219 [DOI] [PubMed] [Google Scholar]
- Korolev S, Nayal M, Barnes WM, Di Cera E, Waksman G (1995) Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-A resolution: structural basis for thermostability. Proc Natl Acad Sci USA 92: 9264–9268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW, Borden A, Banerjee S K, LeClerc JE (1990) Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res 18: 2153–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Korolev S, Waksman G (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J 17: 7514–7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Waksman G (2001) Crystal structures of a ddATP-, ddTTP-, ddCTP, and ddGTP- trapped ternary complex of Klentaq1: insights into nucleotide incorporation and selectivity. Protein Sci 10: 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B (1972) Rate of depurination of native deoxyribonucleic acid. Biochemistry 11: 3610–3618 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W (2003) Replication of a cis–syn thymine dimer at atomic resolution. Nature 424: 1083–1087 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W (2004a) Snapshots of replication through an abasic lesion: structural basis for base substitutions and frameshifts. Mol Cell 13: 751–762 [DOI] [PubMed] [Google Scholar]
- Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, Jerina DM, Yang W (2004b) Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc Natl Acad Sci USA 101: 2265–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Preston BD (1986) Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet 20: 201–230 [DOI] [PubMed] [Google Scholar]
- Loh E, Choe J, Loeb LA (2007) Highly tolerated amino acid substitutions increase the fidelity of Escherichia coli DNA polymerase I. J Biol Chem 282: 12201–12209 [DOI] [PubMed] [Google Scholar]
- Loh E, Loeb LA (2005) Mutability of DNA polymerase I: implications for the creation of mutant DNA polymerases. DNA Repair 4: 1390–1398 [DOI] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, O-Wang J (2007) DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J Biol Chem 282: 17387–17394 [DOI] [PubMed] [Google Scholar]
- Matray TJ, Kool ET (1999) A specific partner for abasic damage in DNA. Nature 399: 704–708 [DOI] [PubMed] [Google Scholar]
- McCain MD, Meyer AS, Schultz SS, Glekas A, Spratt TE (2005) Fidelity of mispair formation and mispair extension is dependent on the interaction between the minor groove of the primer terminus and Arg668 of DNA polymerase I of Escherichia coli. Biochemistry 44: 5647–5659 [DOI] [PubMed] [Google Scholar]
- Meyer AS, Blandino M, Spratt TE (2004) Escherichia coli DNA polymerase I (Klenow fragment) uses a hydrogen-bonding fork from Arg668 to the primer terminus and incoming deoxynucleotide triphosphate to catalyze DNA replication. J Biol Chem 279: 33043–33046 [DOI] [PubMed] [Google Scholar]
- Minnick DT, Bebenek K, Osheroff WP, Turner RM Jr, Astatke M, Liu L, Kunkel TA, Joyce CM (1999) Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment). J Biol Chem 274: 3067–3075 [DOI] [PubMed] [Google Scholar]
- Moore CL, Zivkovic A, Engels JW, Kuchta RD (2004) Human DNA primase uses Watson-Crick hydrogen bonds to distinguish between correct and incorrect nucleoside triphosphates. Biochemistry 43: 12367–12374 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2005) Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309: 2219–2222 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2006) Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat Struct Mol Biol 13: 619–625 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2009) DNA synthesis across an abasic lesion by human DNA polymerase iota. Structure 17: 530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R (2001) The Y-family of DNA polymerases. Mol Cell 8: 486–487 [DOI] [PubMed] [Google Scholar]
- Pagès V, Johnson RE, Prakash L, Prakash S (2008) Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci USA 105: 1170–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA (2001) Prokaryotic DNA polymerase I: evolution, structure, and ‘base flipping' mechanism for nucleotide selection. J Mol Biol 308: 823–837 [DOI] [PubMed] [Google Scholar]
- Peliska JA, Benkovic SJ (1992) Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258: 1112–1118 [DOI] [PubMed] [Google Scholar]
- Randall SK, Eritja R, Kaplan BE, Petruska J, Goodman MF (1987) Nucleotide insertion kinetics opposite abasic lesions in DNA. J Biol Chem 262: 6864–6870 [PubMed] [Google Scholar]
- Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov N, Patel DJ (2006) Stepwise translocation of Dpo4 polymerase during error-free bypass of oxoG lesion. PLoS Biol 4: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineks EZ, Berdis AJ (2004) Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry 43: 393–404 [DOI] [PubMed] [Google Scholar]
- Rothwell PJ, Mitaksov V, Waksman G (2005) Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol Cell 19: 345–355 [DOI] [PubMed] [Google Scholar]
- Sagher D, Strauss B (1983) Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry 22: 4518–4526 [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Kunkel TA, Loeb LA (1983) Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci USA 80: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD (2004) High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J 23: 4484–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP (1997) Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the A rule. J Biol Chem 272: 13916–13922 [DOI] [PubMed] [Google Scholar]
- Spratt TE (2001) Identification of hydrogen bonds between Escherichia coli DNA polymerase I (Klenow fragment) and the minor groove of DNA by amino acid substitution of the polymerase and atomic substitution of the DNA. Biochemistry 40: 2647–2652 [DOI] [PubMed] [Google Scholar]
- Steitz TA (1998) A mechanism for all polymerases. Nature 391: 231–232 [DOI] [PubMed] [Google Scholar]
- Strauss BS (2002) The A rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst) 1: 125–135 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Baskin D, Hood L, Loeb LA (1996) Random mutagenesis of Thermus aquaticus DNA polymerase I: concordance of immutable sites in vivo with the crystal structure. Proc Natl Acad Sci USA 93: 9670–9675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS (2002) New structural and mechanistic insight into the A-rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions. Mutat Res 510: 55–70 [DOI] [PubMed] [Google Scholar]
- Trostler M, Delier A, Beckman J, Urban M, Patro JN, Spratt TE, Beese LS, Kuchta RD (2009) Discrimination between right and wrong purine dNTPs by DNA polymerase I from Bacillus stearothermophilus. Biochemistry 48: 4633–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington MT, Wolfle WT, Spratt TE, Prakash L, Prakash S (2003) Yeast DNA polymerase eta makes functional contacts with the DNA minor groove only at the incoming nucleoside triphosphate. Proc Natl Acad Sci USA 100: 5113–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfle WT, Washington MT, Kool ET, Spratt TE, Helquist SA, Prakash L, Prakash S (2005) Evidence for a Watson-Crick hydrogen bonding requirement in DNA synthesis by human DNA polymerase kappa. Mol Cell Biol 25: 7137–7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA (2004) Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 430: 640–645 [DOI] [PubMed] [Google Scholar]
- Yang W, Lee JY, Nowotny M (2006) Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell 22: 5–13 [DOI] [PubMed] [Google Scholar]
- Yang W, Woodgate R (2007) What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci USA 104: 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn KE, Belrhali H, Wallace SS, Doublié S (2007) Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry 46: 10551–10561 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.