Abstract
We have previously described the interactions of aquaporin-0 (AQP0) with dimyristoyl phosphatidylcholine (DMPC) lipids. We have now determined the 2.5 Å structure of AQP0 in two-dimensional (2D) crystals formed with Escherichia coli polar lipids (EPLs), which differ from DMPC both in headgroups and acyl chains. Comparison of the two structures shows that AQP0 does not adapt to the different length of the acyl chains in EPLs and that the distance between the phosphodiester groups in the two leaflets of the DMPC and EPL bilayers is almost identical. The EPL headgroups interact differently with AQP0 than do those of DMPC, but the acyl chains in the EPL and DMPC bilayers occupy similar positions. The interactions of annular lipids with membrane proteins seem to be driven by the propensity of the acyl chains to fill gaps in the protein surface. Interactions of the lipid headgroups may be responsible for the specific interactions found in tightly bound lipids but seem to have a negligible effect on interactions of generic annular lipids with membrane proteins.
Keywords: electron crystallography, lens, lipid–protein interactions, water channel
Introduction
How do membrane proteins interact with lipids? Spin-labelling and fluorescence-quenching studies have provided a thermodynamic understanding of lipid–protein interactions, but these methods do not allow a direct visualization of individual interactions between a protein and a lipid. Most of the available atomic resolution structural information on lipid–protein interactions comes from lipids in crystal structures of membrane proteins in detergent micelles. A careful analysis of all lipids bound to membrane proteins seen in crystal structures deposited in the Protein Data Bank established a lipid-binding motif. The motif consists of a positively charged residue and a polar residue that specifically interact with the negatively charged phosphodiester groups of the lipids (Palsdottir and Hunte, 2004). Most of the co-crystallized lipids originate from the donor membrane and must have remained associated with the protein during solubilization and purification to be incorporated in the three-dimensional (3D) crystal. Therefore, lipids in crystal structures must be strongly bound to the membrane proteins. These lipids are a special case of ‘annular' lipids, the lipids in direct contact with a membrane protein, because spin-labelling and fluorescence-quenching studies demonstrated that most annular lipids form only weak and non-specific interactions with membrane proteins (Lee, 2003). Generic annular lipids are thus typically lost during solubilization and/or purification of membrane proteins and are usually not observed in crystal structures.
The analysis of the lipids in 3D crystals also showed that the lipid headgroups, and in particular the phosphodiester groups, were tightly associated with the membrane proteins, and thus were the best-ordered atoms of the lipids in these structures (Palsdottir and Hunte, 2004). In contrast, density for the lipid headgroups was poor in electron and X-ray crystallographic density maps of bacteriorhodospsin (e.g., Grigorieff et al, 1996; Luecke et al, 1999), a light-driven proton pump that forms crystalline arrays in the membrane of Halobacterium salinarum, raising the question of what general principles govern the interactions of membrane proteins with their annular lipids.
Previously, the structure of the lens-specific water channel aquaporin-0 (AQP0) was determined by electron crystallography of double-layered, two-dimensional (2D) crystals, revealing seven annular dimyristoyl phosphatidylcholine (DMPC) molecules that surround each monomer and two bulk lipids not in direct contact with AQP0 (Gonen et al, 2005). As with bacteriorhodopsin, examination of the electron crystallographic structure of AQP0 in the DMPC bilayer revealed few favourable interactions of the lipid headgroups with the protein surface (Hite et al, 2008). Although the bacteriorhodopsin structures were obtained with the native purple membrane lipids, AQP0 was completely delipidated before reconstitution, and DMPC, a synthetic lipid, is not found in biological membranes. Although the interactions between bacteriorhodopsin and purple membrane lipids are structurally and functionally best characterized, the lipid–protein interactions seen in the AQP0 structure may thus be the most generic in nature. The situation of the lipids in AQP0 2D crystals is, however, special, because they are sandwiched in between two AQP0 tetramers, which is not typically the case for lipids surrounding membrane proteins in biological membranes.
The tight positioning of the lipids in AQP0 crystals reduced their mobility and rendered them visible in the density maps, making it possible to describe the lipid–protein interactions. If AQP0 2D crystals could be grown with other lipids, such crystals would provide an opportunity to study how a membrane protein interacts with different lipids in a near-native environment. While formation of high-quality 2D crystals usually requires the use of a specific lipid, aquaporins can often form 2D crystals with a variety of lipids. AQP1, for example, formed large, well-ordered 2D crystals with three different lipids (Jap and Li, 1995; Murata et al, 2000; Ren et al, 2000). In the case of AQP0, 2D crystallization screens showed that, in addition to DMPC, AQP0 also forms 2D crystals with E. coli polar lipids (EPLs). E. coli polar lipids differ from DMPC in headgroup chemistry, as well as acyl chain length and saturation. The headgroups of EPLs are a mixture of phosphatidylethanolamine (PE), phosphatidylglycerol (PG) and cardiolipin (CL) rather than the pure phosphatidylcholine (PC) headgroup of DMPC. The acyl chains of DMPC are two saturated 14-carbon acyl chains, whereas the average length of the acyl chains of EPLs is 16 carbon atoms, and approximately 55% of the acyl chains are unsaturated (Lugtenberg and Peters, 1976). We have now optimized the quality of the AQP0 2D crystals formed with EPLs and produced a density map at 2.5 Å resolution. As with the previous density map produced with 2D crystals formed with DMPC, the current density map revealed the seven annular lipids, allowing us to compare the interactions of AQP0 with two very different lipids, DMPC and EPLs.
Results
Structure of AQP0 in DMPC and EPL bilayers
Well-ordered, double-layered 2D crystals of AQP0 formed with EPLs at a lipid-to-protein ratio of 0.25 (mg/mg; Figure 1A). These crystals were used to record electron diffraction patterns at liquid helium temperature (∼6 K). Diffraction patterns from untilted crystals showed reflections beyond 1.9 Å (Supplementary Figure S1), but the resolution was more limited in diffraction patterns of highly tilted crystals (Supplementary Figure S2). After merging 281 diffraction patterns and phasing by molecular replacement, we produced a density map at 2.5 Å resolution (Figure 1B) and modelled and refined the structure of AQP0 (residues 7–226) in the membrane junction (Figure 1C; Table 1). Residues 1–6 and 227–263, which include the C-terminal helix modelled in the previous structure of AQP0 in a DMPC bilayer (Gonen et al, 2005), did not show clear density in the map and were therefore excluded from the model of AQP0 in the EPL bilayer.
Figure 1.
Double-layered 2D crystals of AQP0 in Escherichia coli polar lipids (EPLs). (A) Representative AQP0 2D crystal formed with EPLs in negative stain. Scale bar: 1 μm. (B) Region of the final 2Fo–Fc map refined to 2.5 Å resolution showing pore-lining residues and a water molecule. (C) Atomic model of the AQP0 membrane junction. (D) Overlay of the AQP0EPL (gold) and AQP0DMPC (light blue) structures. (E) The water pore in AQP0EPL (gold) and AQP0DMPC (light blue). The three water molecules in the pore of AQP0EPL (red) are at similar positions as those previously seen in AQP0DMPC (blue). The 2Fo–Fc density map for the water molecules is shown at a contouring level of 1σ (blue wire mesh).
Table 1. Electron crystallographic data.
| Two-dimensional crystals | |
| Layer group | p422 |
| Unit cell | a=b=65.5 Å |
| Thickness (assumed) | 200 Å |
| Electron diffraction | |
| Number of patterns merged | 281 (0°: 11; 20°: 22; 45°: 63; 60°: 108; 70°: 77) |
| Resolution limit for merging | 2.3 Å |
| RFriedel | 18.9% |
| RMerge | 22.6% |
| Observed amplitudes to 2.5 Å | 129 893 |
| Unique reflections | 14 417 |
| Maximum tilt angle | 74.2° |
| Fourier space sampled | 92.3% (83.5% at 2.6–2.5 Å) |
| Multiplicity | 8.1 (4.0 at 2.6–2.5 Å) |
| Crystallographic refinement (10.0–2.5 Å) | |
| Resolution limit for refinement | 2.5 Å |
| Crystallographic R factor | 24.98% |
| Free R factor | 28.43% |
| Reflections in working/test set | 12 801/1453 |
| Non-hydrogen protein atoms | 1668 |
| Non-hydrogen lipid atoms | 273 |
| Solvent molecules | 8 |
| Average protein B factor (Å2) | 42.3 |
| Average lipid B factor (Å2) | 88.0 |
| Ramachandran plot (%) | 98.4/1.6/0.0 (allowed; generous; disallowed) |
| Rfree is calculated from a randomly chosen 10% of reflections, and Rcryst is calculated over the remaining 90% of reflections. | |
Other than the C-terminal helix, which was disordered and therefore could not be modelled, the structure of AQP0 in the EPL bilayer (AQP0EPL) is virtually identical with its structure in the DMPC bilayer (AQP0DMPC; Figure 1D). The r.m.s.d. values between all modelled backbone atoms is 0.48 Å, and 0.4 Å if only the transmembrane domains are compared. The pore-lining residues in AQP0EPL and AQP0DMPC are also essentially the same, and both structures show three water molecules in the centre of the channel at the same positions (Figure 1E).
Modelling the EPL bilayer
In addition to the protein, the density map also allowed us to build all seven annular lipids surrounding each AQP0 monomer, but not the lipids in the central area between four adjacent tetramers (asterisks in Figure 2A). The acyl chains of the annular lipids were initially modelled as 10 carbon chains and then extended to occupy the maximal length that was clearly resolved in the density map. In the initial density map, densities representing the acyl chains were often branched, indicating that they can adopt multiple conformations. As the data were insufficient for the refinement of alternative conformations, we chose to build each acyl chain into the strongest density, representing the predominant lipid conformation. After several rounds of refinement, the branches disappeared, and all acyl chains were represented by a single, mostly continuous density.
Figure 2.
The EPL bilayer. (A) Top view of the AQP0 2D crystal showing AQP0 tetramers (gold) and the surrounding EPLs (red). (B, C) The seven (B) annular EPLs and (C) DMPC molecules surrounding an AQP0 monomer. As lipids are sandwiched between two adjacent AQP0 subunits, their positions relative to both AQP0 subunits are shown.
Approximately 55% of the acyl chains of EPLs contain an unsaturated bond, with the two most abundant species being 16c1:9 and 18c1:11 (Lugtenberg and Peters, 1976). Although some lipids showed kinks that could indicate the presence of a double bond, due to the heterogeneity of the acyl chains in EPLs, we modelled all acyl chains as being fully saturated.
Escherichia coli polar lipids are a mixture of three different headgroups (67% PE, 23% PG and 10% CL lipids; Oursel et al (2007)) with PE being by far the most abundant headgroup. We therefore initially modelled all headgroups as PE. After refinement, we found no evidence in the density map, which suggested that any of the lipid positions is preferentially occupied by a lipid with a particular headgroup. Our final model of the EPL bilayer thus contains seven PE molecules with saturated acyl chains ranging from 5 to 17 carbon atoms (Figure 2B).
Organization of the EPL and DMPC bilayers surrounding AQP0
The EPL and DMPC bilayers surrounding AQP0 are remarkably similar (Figure 2B and C). The two bilayers contain the same number of lipids at comparable positions (Figure 3C) and have almost the same thickness (the average distance between the phosphodiester groups in the two leaflets is 31.9 Å for the EPL bilayer and 33.6 Å for the DMPC bilayer; Figure 3A). As acyl chains of EPLs are on average longer than those of DMPC (16 versus 14 carbon atoms), this finding raises the question how the longer EPL acyl chains are accommodated. Unexpectedly, despite the longer acyl chains of EPLs, the average distance between the C2 atoms of glycerols in the two leaflets of the EPL bilayer, 27.0 Å, is smaller than the corresponding average distance in the DMPC bilayer, 31.2 Å (Figure 3B). Further comparison of the AQP0EPL and AQP0DMPC structures reveals that the DMPC molecules cover less surface area on AQP0 than EPLs (Figures 2B and C). Indeed, the DMPC bilayer leaves areas of the hydrophobic surface of AQP0 uncovered (Figure 2C, e.g. area in between PC3 and PC4 of the extracellular leaflet and PC7 of the cytoplasmic leaflet), suggesting that 14-carbon acyl chains are close to the minimum needed to saturate the hydrophobic belt of AQP0. Furthermore, unlike the lipids in the DMPC bilayer, several of the longer EPL acyl chains interdigitate in the middle of the bilayer, filling gaps between acyl chains in the opposite leaflet (Figure 2B; e.g. one acyl chain of PE7 inserts between the two acyl chains of PE3 and the other acyl chain of PE7 inserts between the two acyl chains of PE4). Other acyl chains bend sharply as they approach the midpoint of the bilayer and then extend parallel to the membrane plane (Figure 2B; e.g. acyl chains of PE4, PE6, and PE7). Such bending may be facilitated by unsaturated bonds present in acyl chains of EPLs, which have previously been proposed to explain the bent conformations seen in lipids associated with cytochrome b–c1 complexes (Palsdottir and Hunte, 2004). Indeed, the positions of the kinks seen in the acyl chains of lipids PE3, PE4, and PE6 occur close to the positions of the double bonds in the most abundant unsaturated acyl chains of EPLs, 16c1:9 and 18c1:11, whereas the kink in the acyl chain of PE7 is located between C6 and C7.
Figure 3.
Comparisons between the EPL and DMPC bilayers. (A) The average distance between the phosphodiester groups in the two leaflets of the bilayer formed by EPLs (31.9 Å) is very similar to that between the phosphodiester groups in the DMPC bilayers (33.6 Å). Phosphorous atoms are shown in orange. Red shading represents the region between the most distantly located phosphorous atoms in each leaflet. (B) Despite the longer acyl chains of EPLs, the average distance between the C2 atoms of the lipids' glycerol backbones in the two leaflets of the bilayer formed by EPLs (27.0 Å) is shorter than that between the C2 atoms in the DMPC bilayers (31.2 Å). The glycerol groups are shown in green. The green shading represents the region between the two most distantly located C2 atoms in each leaflet. (C) Overlay of the lipids seen in the AQP0EPL (red) and AQP0DMPC (blue) structures. (D) Same as in (C) with the lipids coloured according to their B-factors. Colour coding: intense blue, B-factors below 75 Å2; pale blue, B-factors between 75 and 94 Å2; pale red, B-factors between 95 and 114 Å2; intense red, B-factors above 115 Å2.
Interaction of the lipid headgroups with AQP0
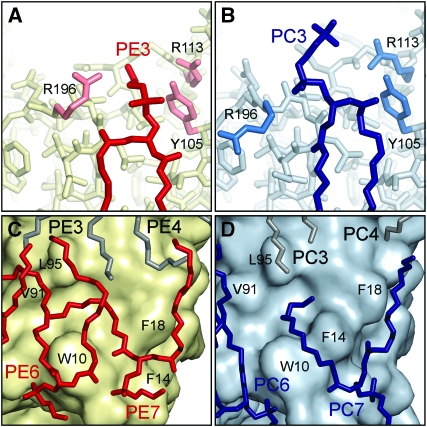
The AQP0EPL and AQP0DMPC structures make it possible to analyse the interactions of AQP0 with different lipids. Although the corresponding lipids in the two bilayers are at very similar positions (Figure 3C), all the headgroups of the corresponding lipids in the two bilayers adopt different conformations. Furthermore, none of the DMPC lipids in the AQP0DMPC structure interacted with AQP0 through a lipid-binding motif (Gonen et al, 2005), and only one lipid, PE3, in the AQP0EPL structure is in an environment consistent with a lipid-binding motif: the phosphodiester group of PE3 interacts with a positively charged residue, Arg 196, and a polar residue, Tyr 105 (Figure 4A). In PC3, the corresponding lipid in the AQP0DMPC structure (Figure 4B), the glycerol backbone is shifted by ∼3.4 Å closer to the centre of the lipid bilayer. When Hunte and coworkers identified the lipid-binding motif, they noted that it did not seem to apply to PC lipids and suggested that this may be due to the large choline headgroup that could cause steric clashes (Palsdottir and Hunte, 2004). We were therefore unsure whether the changes in the conformation of PE3 with respect to PC3 were the result of the presence of a lipid-binding motif. When we attempted to reposition PC3 to a location more similar to PE3, we found that there was sufficient space for a choline headgroup to occupy the same space as the ethanolamine headgroup of PE3. As the choline headgroup of PC3 does not occupy this position, even though it would be sterically possible, and therefore does not interact with AQP0 through Arg 196 and Tyr 105, we conclude that these two residues do not constitute a true lipid-binding motif. Together, these observations suggest that the exact chemical identities of the phospholipid headgroups have a negligible role in the interaction of annular lipids with membrane proteins, corroborating previous results obtained by spin-labelling studies (Lee, 2003).
Figure 4.
EPL headgroups and acyl chains. (A) Lipid PE3 interacts with AQP0 through a putative lipid-binding motif formed by Arg196 and Tyr105. (B) In AQP0DMPC the same residues do not interact with the corresponding lipid, PC3. (C) Lipid PE7 in AQP0EPL bends to follow the surface features of AQP0 and fills in a pocket formed by residues Trp10, Val91, and Leu95. (D) The corresponding lipid in AQP0DMPC, PC7, follows a different path but fills in the same pocket in the AQP0 surface.
Interaction of the lipid acyl chains with AQP0
In contrast to the headgroups, acyl chains seem to be the crucial element guiding the interaction of annular lipids with the protein. Comparison of the lipids in AQP0EPL and AQP0DMPC reveals that the acyl chains in the extracellular leaflet occupy quite similar positions. The acyl chains of PE3 and PE4 extend along the same paths as those of the corresponding lipids, PC3, and PC4. The two acyl chains of PE2 are in similar positions as those occupied by one of the acyl chains of PC1 and one of the acyl chains of PC2 (Figure 3C and Supplementary Video S1).
The positions of the acyl chains in the cytoplasmic leaflet vary to a greater degree between AQP0EPL and AQP0DMPC than those of the acyl chains in the extracellular leaflet (Figure 3C). The gap between two adjacent AQP0 tetramers is narrower on the extracellular side of the bilayer, which may force the lipids in this leaflet into more defined positions than those in the cytoplasmic leaflet. This idea is supported by the average B-factors of the lipids, which tend to be lower for the lipids in the extracellular leaflet, suggesting that their mobility is more restricted compared with those in the cytoplasmic leaflet (Figure 3D and Supplementary Figure S3).
Lipid PE7 in AQP0EPL illustrates how the protein surface defines the positions of the acyl chains of annular lipids (Figure 4C). One acyl chain of PE7 lies in a groove on the surface of AQP0 that guides it straight towards the centre of the bilayer. The other acyl chain runs in between the bulky side chains of Trp 10 and Phe 18 and then bends sharply to evade the side chain of Leu 95, thus filling in a cleft formed by Trp 10 on one side and Val 91 and Leu 95 on the other. The two acyl chains of PE6 extend over the surface of this cleft, completely covering it. Phosphatidylcholine 7, the lipid in AQP0DMPC that corresponds to PE7 in AQP0EPL, adapts differently to the protein surface (Figure 4D). Compared with PE7, the glycerol backbone of PC7 is shifted by about 3 Å away from the centre of the bilayer, which is probably caused by the bulky side chain of Phe 14 adopting a different conformation in AQP0DMPC. One of the acyl chains of PC7 follows the same groove in the AQP0 surface as the corresponding acyl chain of PE7, extending straight to the centre of the bilayer. The second acyl chain, however, follows a gap in between the side chains of Trp 10 and Phe 14 in its different position. It then immediately fills the cleft formed by residues Trp 10, Phe 14, Val 91, and Leu 95. Thus, both PE7 and PC7 adopt conformations that allow the acyl chains to fill in the large hydrophobic pocket in the AQP0 surface, but in different ways. To even out the protein surface is a crucial function of annular lipids, because it ensures that the bilayer forms a tight seal around the membrane protein and preserves the separation of the cellular interior from the external environment.
Discussion
Comparison of our new AQP0EPL structure with the previously determined AQP0DMPC structure shows that the annular lipids studied here have very little influence on the structure of AQP0. With only a few exceptions, such as Phe 14, the amino-acid residues in the AQP0EPL and AQP0DMPC structures, in particular the ones lining the water channel, have nearly identical conformations. The virtually identical channel structure in AQP0EPL and AQP0DMPC is consistent with a previous study that showed that the lipid environment did not affect water conduction by AQP1 (Zeidel et al, 1994), although only a very limited range of lipid compositions were tested in this study.
Electron crystallography of AQP0 2D crystals makes it possible to visualize the lipids surrounding the membrane protein. In our studies, only a single layer of lipid molecules separates the individual tetramers in the 2D crystals (Figure 2A). This tight packing of the lipids in the 2D crystals restricts their mobility and makes it possible to see the lipids in the density maps, providing us with the opportunity to study the interactions of annular lipids with AQP0. However, this situation is not typical for a biological membrane, in which membrane proteins tend to be separated by a larger number of lipids. Nevertheless, the 2D crystals formed in vitro display the same lattice parameters as the 2D arrays formed by AQP0 in native lens membranes (Buzhynskyy et al, 2007). The 2D crystals are therefore a good representation of the AQP0 arrays in the lens membrane. Furthermore, AQP0 forms 2D crystals with lipids as different as DMPC and EPLs. The sandwiching between two adjacent tetramers may thus limit the mobility of the lipids but may not impose further restrictions on the behaviour of the lipids surrounding AQP0. Our findings concerning the interaction of annular lipids with AQP0 in the 2D crystals should therefore bear relevance for membrane proteins surrounded by a larger number of lipids. Nevertheless, further studies will be required to confirm this notion.
The structures of AQP0 in two different lipid environments allow us to see how the protein and the lipid bilayer adapt to each other. To accommodate proteins in a lipid bilayer with a different hydrophobic thickness, a situation known as hydrophobic mismatch, it has been proposed that either the lipids adjust their length to match the protein or that α-helical membrane proteins may stretch or contract axially to adapt to the thickness of the surrounding lipid bilayer (Lee, 2004). It has been shown for several membrane proteins that hydrophobic mismatch affects their activity (Lee, 2004), suggesting changes in protein structure. It is not clear, however, whether all membrane proteins have the flexibility to adjust to the hydrophobic thickness of the surrounding lipid bilayer. Aquaporin-0, which is a very stable and presumably a very rigid membrane protein, has the same structure in the EPL and DMPC bilayers (Figure 1D). In the two structures, it is exclusively the lipids that adapt to the protein, which bend and interdigitate to accommodate the longer acyl chains of the EPLs. A caveat of this observation is that the acyl chains of EPLs are not only longer than those of DMPC, but 55% of the acyl chains of EPLs are unsaturated (Lugtenberg and Peters, 1976), and double bonds reduce the hydrophobic thickness of the bilayer formed by unsaturated lipids (Marsh, 2008). Thus, although the AQP0DMPC and AQP0EPL structures suggest that it is exclusively the lipids that adapt to the protein, the hydrophobic thickness of the DMPC and EPL bilayers may be too similar to induce observable changes in the AQP0 structure. To conclusively rule out structural changes in AQP0 due to hydrophobic mismatch, it will be necessary to determine structures of AQP0, in which the protein is surrounded by lipids that form bilayers with a hydrophobic thickness that is significantly different from that of AQP0.
After modelling all the headgroups as ethanolamine and refining the lipids in the AQP0EPL structure, we did not observe any density that indicated PG or CL headgroups. This result suggests that there are no lipid positions on the surface of AQP0 that are preferentially occupied by PG or CL lipids. In the absence of preferential binding sites, averaging of the three headgroups would probably result in all headgroups appearing as the predominant PE headgroup. Furthermore, inspection of the protein surface did not reveal specific CL-binding sites as defined by Palsdottir and Hunte (2004). As CL, a lipid composed of four acyl chains and two phosphodiester groups linked by a glycerol, constitutes only 10% of EPLs, it may randomly occupy two adjacent lipid positions and thus be obscured as a result of averaging. Alternatively, due to its larger size, CL may preferentially localize to the larger lipid area in the middle of four neighbouring AQP0 tetramers in which no ordered lipid molecules were found (asterisks in Figure 2A). As PG and CL lipids do not occur in lens membranes, it is not surprising that AQP0 does not select for these lipids at particular positions.
Despite the different acyl chain length and the different orientations of the headgroups, the average distance between the phosphodiester groups in the two leaflets is almost identical in the bilayers formed by EPLs, 31.9 Å, and DMPC, 33.6 Å (Figure 3A). More surprisingly, the average distance between the C2 atoms of the glycerol of the lipids in the two leaflets is larger for the shorter DMPC lipids, 31.2 Å, than for the longer EPLs, 27.0 Å (Figure 3B). This finding suggests that the vertical position of the lipids is defined by the strong charges of the phosphodiester groups that have to be positioned outside the hydrophobic belt of the membrane protein. The glycerol backbone, however, which is largely hydrophobic in nature, is allowed more flexibility in its positioning within the hydrophobic region of the membrane protein and can occupy the same region as the acyl chains.
In conclusion, we propose that the vertical position of the annular lipids surrounding a membrane protein is defined by the thickness of the hydrophobic surface of the membrane protein, with the charged phosphodiester groups of the lipids in the two leaflets being positioned immediately outside of the hydrophobic belt of the protein. Although some membrane proteins may change their structure to adjust to bilayers with a significantly different hydrophobic thickness, it remains to be determined whether rigid membrane proteins, such as AQP0, can adapt in a similar manner. The requirement that acyl chains fill in the gaps on the protein surface constitutes the guiding principle for the lateral interactions of annular lipids with membrane proteins. Additional interactions of the protein with headgroups of annular lipids, that is, lipid-binding sites, may allow membrane proteins to select for specific lipids and/or to form tight interactions with lipids important for their function or structural integrity. Furthermore, the fact that AQP0 forms well-ordered 2D crystals with lipids as different as DMPC and EPLs suggests that it will be possible to grow 2D crystals of AQP0 with many other lipids, opening an avenue for the systematic study of lipid-protein interactions.
Materials and methods
Protein purification and crystallization
The core tissue of sheep lenses (Wolverine Packing Company, Detroit, MI) was dissected away from the soft cortical tissue and membranes were prepared as previously described (Gonen et al, 2004). Purified membranes were solubilized in 4% (w/v) octyl glucoside in 10 mM Tris (pH 8.0) for 30 min at 22°C. Insoluble material was separated by centrifugation at 300 000 × g. Solubilized proteins were bound to a MonoQ column (GE Healthcare) and eluted with 150 mM NaCl. Pooled fractions were run over a Superose 12 (GE Healthcare) and eluted with 1.2% octyl glucoside in 10 mM Tris (pH 8.0) and 150 mM NaCl. Purified AQP0 was reconstituted into 2D crystals using E. coli polar lipids (Avanti Polar Lipids) at a lipid-to-protein ratio of 0.25 (mg/mg) by dialysis against 10 mM MES (pH 6), 100 mM NaCl, and 50 mM MgCl2 at 37°C for 1 week with daily buffer exchanges.
Data collection, data processing, model building, and refinement
Specimens for cryo-electron microscopy were prepared using the carbon sandwich technique (Gyobu et al, 2004). Grids were transferred into an FEI Tecnai Polara electron microscope operated at an acceleration voltage of 300 kV. Low-dose electron diffraction patterns were recorded at liquid helium temperature (∼6 K) with a 4 × 4 K CCD camera (Gatan) and a camera length of 1.9 m. Electron diffraction patterns were analysed and merged as described previously (Gonen et al, 2004). The structure of AQP0 was determined by molecular replacement in Phaser 2.1 (McCoy et al, 2007) using as search model the 1.9-Å electron crystallographic model of AQP0 without the C-terminal helix ((Gonen et al, 2005); PDB ID: 2B6O).
The protein was rebuilt in Coot (Emsley and Cowtan, 2004),and the model was refined in CNS version 1.2 (Brunger et al, 1998) using 300 kV electron scattering factors. Protein residues 7–226 were visible and were modelled and refined.
Building the lipids into the density map was complicated by the structural heterogeneity of EPLs. All lipids were initially modelled into the Fo–Fc difference map with PE headgroups and short (∼8–10 carbon) acyl chains. Short acyl chains were used because some of the chains were poorly defined in the initial difference map and because several of the densities representing acyl chains were branched. After a round of simulated annealing refinement, further carbons were added to the acyl chains if additional density was clearly visible in the composite omit map. For branched densities, the strongest arm was chosen for model building. This process was repeated iteratively until up to a maximum of 18 carbon atoms were built (the longest abundant acyl chain length in EPLs) or until no additional density appeared upon further cycles. Figures were prepared with PyMol (www.pymol.org).
Accession codes
Atomic coordinates and structure factor files have been deposited with the Protein Data Bank under the accession code 3M9I.
Supplementary Material
Acknowledgments
This study was supported by NIH grant R01 EY015107 (to TW). TW is an investigator of the Howard Hughes Medical Institute. We thank S Harrison and Y Fujiyoshi for continuous support and advice; K Abe for advice regarding specimen preparation and T Rapoport for insightful discussions. RKH and TW conceived and designed the project. RKH performed the experiments. ZL assisted with data collection. RKH and TW wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Buzhynskyy N, Hite RK, Walz T, Scheuring S (2007) The supramolecular architecture of junctional microdomains in native lens membranes. EMBO Rep 8: 51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T (2005) Lipid–protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438: 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen T, Sliz P, Kistler J, Cheng Y, Walz T (2004) Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature 429: 193–197 [DOI] [PubMed] [Google Scholar]
- Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R (1996) Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol 259: 393–421 [DOI] [PubMed] [Google Scholar]
- Gyobu N, Tani K, Hiroaki Y, Kamegawa A, Mitsuoka K, Fujiyoshi Y (2004) Improved specimen preparation for cryo-electron microscopy using a symmetric carbon sandwich technique. J Struct Biol 146: 325–333 [DOI] [PubMed] [Google Scholar]
- Hite RK, Gonen T, Harrison SC, Walz T (2008) Interactions of lipids with aquaporin-0 and other membrane proteins. Pflugers Arch 456: 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jap BK, Li H (1995) Structure of the osmo-regulated H2O-channel, AQP-CHIP, in projection at 3.5 Å resolution. J Mol Biol 251: 413–420 [DOI] [PubMed] [Google Scholar]
- Lee AG (2003) Lipid–protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta 1612: 1–40 [DOI] [PubMed] [Google Scholar]
- Lee AG (2004) How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta 1666: 62–87 [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK (1999) Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol 291: 899–911 [DOI] [PubMed] [Google Scholar]
- Lugtenberg EJ, Peters R (1976) Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochim Biophys Acta 441: 38–47 [DOI] [PubMed] [Google Scholar]
- Marsh D (2008) Energetics of hydrophobic matching in lipid–protein interactions. Biophys J 94: 3996–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y (2000) Structural determinants of water permeation through aquaporin-1. Nature 407: 599–605 [DOI] [PubMed] [Google Scholar]
- Oursel D, Loutelier-Bourhis C, Orange N, Chevalier S, Norris V, Lange CM (2007) Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapid Commun Mass Spectrom 21: 1721–1728 [DOI] [PubMed] [Google Scholar]
- Palsdottir H, Hunte C (2004) Lipids in membrane protein structures. Biochim Biophys Acta 1666: 2–18 [DOI] [PubMed] [Google Scholar]
- Ren G, Cheng A, Reddy V, Melnyk P, Mitra AK (2000) Three-dimensional fold of the human AQP1 water channel determined at 4 Å resolution by electron crystallography of two-dimensional crystals embedded in ice. J Mol Biol 301: 369–387 [DOI] [PubMed] [Google Scholar]
- Zeidel ML, Nielsen S, Smith BL, Ambudkar SV, Maunsbach AB, Agre P (1994) Ultrastructure, pharmacologic inhibition, and transport selectivity of aquaporin channel-forming integral protein in proteoliposomes. Biochemistry 33: 1606–1615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.