Abstract
Human metapneumovirus (HMPV) is a recently discovered paramyxovirus of the subfamily Pneumovirinae, which also includes avian pneumovirus and human respiratory syncytial virus (HRSV). HMPV is an important cause of respiratory disease worldwide. To understand early events in HMPV replication, cDNAs encoding the HMPV nucleoprotein (N), phosphoprotein (P), matrix protein (M), M2-1 protein and M2-2 protein were cloned from cells infected with the genotype A1 HMPV wild-type strain TN/96-12. HMPV N and P were shown to interact using a variety of techniques: yeast two-hybrid assays, co-immunoprecipitation and fluorescence resonance energy transfer (FRET). Confocal microscopy studies showed that, when expressed individually, fluorescently tagged HMPV N and P exhibited a diffuse expression pattern in the host-cell cytoplasm of uninfected cells but were recruited to cytoplasmic viral inclusion bodies in HMPV-infected cells. Furthermore, when HMPV N and P were expressed together, they also formed cytoplasmic inclusion-like complexes, even in the absence of viral infection. FRET microscopy revealed that HMPV N and P interacted directly within cytoplasmic inclusion-like complexes. Moreover, it was shown by yeast two-hybrid analysis that the N-terminal 28 aa are required for the recruitment to and formation of cytoplasmic inclusions, but are dispensable for binding to HMPV P. This work showed that HMPV N and P proteins provide the minimal viral requirements for HMPV inclusion body formation, which may be a distinguishing characteristic of members of the subfamily Pneumovirinae.
INTRODUCTION
The discovery of human metapneumovirus (HMPV) was first reported in 2001 following its isolation from 28 young children with symptoms ranging from upper respiratory tract disease to severe bronchiolitis and pneumonia (van den Hoogen et al., 2001). Human infection has since been reported in children worldwide (Crowe, 2004), and these epidemiological studies reveal several disease manifestations including severe lower respiratory tract disease (Bosis et al., 2005; Esper et al., 2004; Falsey et al., 2003; Mullins et al., 2004; Ulloa-Gutierrez et al., 2004; van den Hoogen et al., 2003; Williams et al., 2004). Initial studies of infected cells using electron microscopy showed paramyxovirus-like particles (van den Hoogen et al., 2001), and sequence analysis revealed a non-segmented, single-stranded RNA genome of negative polarity with a gene order characteristic of the genus Metapneumovirus (3′-N-P-M-F-M2-SH-G-L-5′) (Herfst et al., 2004; van den Hoogen et al., 2002; Yu et al., 1992). HMPV belongs to the family Paramyxoviridae, subfamily Pneumovirinae, which also includes human respiratory syncytial virus (HRSV). We have undertaken experiments to identify and characterize interactions between proteins of the HMPV nucleocapsid complex to understand HMPV replication and assembly in more detail.
Extensive experimental resources and a large body of knowledge about paramyxovirus biology have emerged from work in numerous well-developed experimental systems including HRSV (Lamb & Kolakofsky, 2001). Whilst compelling questions remain regarding regulation of paramyxovirus assembly, the detailed understanding of shared paramyxovirus gene regulation, structure and cell entry characteristics afford a substantial understanding of HMPV replication based on its genomic sequence. The HMPV genome sequence has been determined and analysed for all HMPV open reading frames and intergenic sequences (Herfst et al., 2004; van den Hoogen et al., 2002).
Our understanding of the replication of the pneumovirus HRSV provides a framework to generate hypotheses regarding early virus assembly events in HMPV replication. HRSV replication results in the formation of dense cytoplasmic inclusion bodies that appear to contain aggregated nucleocapsids. The inclusion bodies contain viral RNA, HRSV nucleoprotein (N) and phosphoprotein (P), and the viral polymerase complex (García et al., 1993; García-Barreno et al., 1996; Lamb & Kolakofsky, 2001). We use the term ‘inclusion body’ in reference to these cytoplasmic structures seen in viral infection. Co-expression of HRSV N and P, independent of other HRSV proteins or viral RNA, results in the formation of inclusion-like complexes that resemble inclusions found in RSV-infected cells (García et al., 1993; García-Barreno et al., 1996). We use the term ‘inclusion-like complex’ in reference to the cytoplasmic structures in uninfected cells expressing viral replication complex proteins. Similarly, rabies virus N and P proteins are both required for the formation of inclusion bodies (Chenik et al., 1994). In contrast, individual expression of the nucleocapsid protein is sufficient to induce inclusion-like complex formation in measles virus (MeV) (Spehner et al., 1991). HRSV N binds viral genomic RNA and HRSV P, and HRSV P participates in the function of the viral RNA-dependent RNA polymerase. The domains of HRSV N and P required for their interaction have been identified (Castagne et al., 2004; Slack & Easton, 1998; Stokes et al., 2003). Host-cell proteins required for the formation of HRSV inclusion bodies are not known, but cellular actin and associated proteins have been implicated in HRSV genome expression and assembly (Bitko et al., 2003; Burke et al., 1998, 2000; Kallewaard et al., 2005; Ulloa et al., 1998). Hypothesized similarities between HMPV and HRSV assembly require experimental confirmation, as the N proteins of HMPV and HRSV share just 41 % amino acid sequence identity, whilst the P proteins share just 24 % (van den Hoogen et al., 2002). Furthermore, HMPV provides an additional threat as a respiratory disease separate from that of HRSV, making work on this specific virus critical to human health.
The precise role of cytoplasmic inclusion bodies in paramyxovirus replication is unknown. However, paramyxovirus inclusion bodies are observed in cells at the earliest times of detectable viral protein translation. This observation supports the hypothesis that paramyxovirus inclusion bodies form because of intrinsic properties of viral proteins rather than as a result of high intracellular concentrations of viral proteins found late in viral replication (García-Barreno et al., 1996). Furthermore, when HRSV N and P are co-translated in vitro, aggregate formation is not observed, suggesting that specific cellular factors are required for the formation of inclusion bodies (García et al., 1993).
Here, we performed experiments to identify interactions between HMPV proteins important in nucleocapsid formation and polymerase complex function, and observed interactions between HMPV N and P using a variety of techniques. We found that, like HRSV but unlike MeV (Spehner et al., 1991), HMPV N and P together are required for the formation of cytoplasmic inclusion bodies. We also performed mutational analysis and found that the N terminus of HMPV N is dispensable for binding to HMPV P but is required for the formation of viral inclusion-like complexes. These studies are an important step towards understanding early events in paramyxovirus replication, as well as distinguishing key differences between the emerging pathogen HMPV and other members of the paramyxovirus family including HRSV.
METHODS
Cells, viruses and transfections
Rhesus monkey kidney LLC-MK2 cells (ATCC CCL-7) were grown in Opti-MEM I medium (Invitrogen) supplemented with 2 % fetal bovine serum, 2 mM L-glutamine, 100 U penicillin G ml−1 and 100 μg streptomycin ml−1 (all from Atlanta Biologicals). The HMPV virus strain used (TN/96-12) was a clinical isolate, obtained as described previously (Williams et al., 2004), resulting in HMPV stocks of approximately 5×105 p.f.u. ml−1. Plasmid transfections were performed using Effectene lipofection (Qiagen). LLC-MK2 cells (1×105) were added to wells containing 12 mm glass coverslips and transfected with a total of 1 μg plasmid DNA per well in a 12-well plate and 7 μg per plate in 10 mm tissue culture dishes. Infection of cells with HMPV was performed by the addition of HMPV at an m.o.i. of 1–2 and cells were incubated for 24 h prior to immunofluorescence staining.
Antibodies
The production of HMPV antiserum has been described previously (Williams et al., 2005). HMPV N- and P-specific antisera were obtained using a synthesized peptide corresponding to aa 48–64 of HMPV N and aa 21–35 of HMPV P, respectively (Sigma-Genosys). Polyclonal antiserum was obtained by inoculating one rabbit with 200 μg synthetic peptide in complete Freund’s adjuvant (both from Sigma), followed by 100 μg booster doses in incomplete Freund’s adjuvant at 2, 4, 6, 8 and 10 weeks post-inoculation. Antiserum was obtained 7 days after the last boost.
Plasmid construction
Plasmid expression constructs encoding cDNA for the genes of HMPV strain TN/96-12 were obtained by RT-PCR of viral RNA extracted from HMPV stocks. This RNA was used as template for RT-PCR amplification of cDNA using random hexamer primers (ABI) and a Q Omniscript RT kit, following the protocol of the manufacturer (Qiagen). The resultant HMPV cDNA was used as template for PCR amplification of sequences encoding each viral protein studied. The sequences of the primers used are given in Supplementary Table S1, available in JGV Online.
The yeast two-hybrid (Y2H) cloning vector pGBKT7 was used as bait constructs, and pGADT7 was used as prey constructs (Clontech). pGEM-T Easy vectors encoding HMPV N, P, M, M2-1 or M2-2 protein were digested and ligated into pGBKT7 and pGADT7 to generate the viral protein bait and prey plasmids. Mammalian expression vectors encoding each HMPV protein were constructed by digesting the respective bait plasmid followed by ligation into pcDNA3.1(−) [pcDNA3.1(+) was used for HMPV P]. Each of these coding sequences was also used to construct mammalian expression plasmids encoding green fluorescent protein (GFP) fused to the N terminus of expressed HMPV N, M or P. Plasmids expressing HMPV N deletion mutants as Y2H bait fusion proteins were constructed as above; the specific primers and restriction enzymes used are given in the Supplementary Table S1.
Sequences encoding HMPV N or P, which were codon-optimized for expression in mammalian cells, were synthesized commercially and cloned into the pPCR-Script backbone plasmid (GeneArt). Mammalian expression vectors encoding codon-optimized HMPV N or P were constructed by digesting PCR-ScriptMPVNopt or PCR-ScriptMPVPopt followed by ligation into pcDNA3.1(−) to create pCMVMPVNopt or pCMVMPVPopt. Each of these coding sequences was also used to construct mammalian expression plasmids encoding fluorescent proteins fused to the N terminus of expressed HMPV N or P. Fusion constructs were also created with the fluorescent proteins Cerulean (kindly provided by D. Piston; Rizzo et al., 2004) and Venus (kindly provided by A. Miyawaki courtesy of D. Piston; Nagai et al., 2002), and red fluorescent protein (RFP).
Y2H analysis
Budding Y2H strain AH109 (Clontech) with the HIS3, ADE2 and MEL1 reporter genes downstream of heterologous GAL4-responsive promoter elements was transformed with pBMPVN, pBMPVP, pBMPVM, pBMPVM2-1 and pBMPV2-2 in pairwise combinations using lithium acetate. Cells were plated onto His−/Leu−/Ade−/Trp− medium and supplemented with X-Gal to select for interacting proteins. The level of β-galactosidase enzyme activity produced was measured using a solution of chlorophenol red-β-D-galactopyranoside as substrate (Stratagene) with the absorbance of the coloured product measured at a wavelength of 580 nm. β-Galactosidase reporter gene activation was scored as positive only if the absorbance exceeded 300 % of the level of the control cells for each experiment performed in triplicate.
In vitro co-immunoprecipitation of the N and P proteins
In vitro translation of HMPV proteins was performed using the TNT T7 Quick-Coupled Transcription/Translation System (Promega). The bait, prey and pcDNA3.1 mammalian expression plasmids described above, encoding HMPV N or P, were used as templates to generate native proteins and fusion proteins with an N-terminal c-Myc epitope tag in the absence or presence of [35S]methionine and [35S]cysteine (Easy Tag Express 35S Protein Labelling Mix; Perkin Elmer Life Sciences). Plasmids pGBKT7-53 and pGBKT7-LAM (encoding p53 and lamin C, respectively) were used as controls. Approximately equal amounts of translated proteins were mixed in PBS with protease inhibitor cocktail and incubated at 4 °C for 120 min with gentle agitation. Mouse monoclonal anti-c-Myc or rabbit polyclonal anti-haemagglutinin antibodies (Clontech) bound to protein G–Sepharose (Amersham Pharmacia Biotech) were added, and the protein–antibody mixtures were incubated at 4 °C for an additional 90 min. The Sepharose beads were washed with 1 % Triton X-100 in PBS and proteins were resolved on a 12 % polyacrylamide gel.
Immunofluorescence staining and confocal microscopy
LLC-MK2 cells were washed with PBS, fixed in 3.7 % formaldehyde for 10 min and permeabilized with 1 % Triton X-100. Cells were blocked with PBS containing 5 % γ-globulin-free BSA (Sigma), 0.1 M glycine and 0.05 % Tween 20 (PBS-BG), incubated with a rabbit HMPV P-specific antiserum (diluted 1: 1000) or a rabbit HMPV N-specific antiserum (diluted 1: 500) in PBS-BG for 1 h, and washed and incubated with either Alexa Fluor 488-labelled or Alexa Fluor 546-labelled goat anti-rabbit secondary antibodies (diluted 1: 3000 in PBS-BG) for 1 h (Invitrogen). Nuclear staining was performed using TO-PRO-3 iodide (Invitrogen). Cells were washed and stained coverslips were mounted on glass slides using Aqua Poly/Mount (Polysciences). Images were obtained with an LSM 510 META confocal microscope (Carl Zeiss). Image reconstruction and analysis were performed with MetaMorph image analysis software (Universal Imaging Corporation). For fluorescence resonance energy transfer (FRET) microscopy experiments, a 514 nm laser was used to provide photobleaching of Venus in specified regions of interest.
FRET fluorometry analysis
One 10 cm2 dish of 293T cells was transfected for each experimental condition. Cells were harvested for analysis 24 h after transfection. Cells were then analysed by fluorometry in a PTI T-format scanning cuvette spectrofluorometer (Photon Technology International). Samples were excited at 433 nm, and the resulting emission scan ranging from 450 to 550 nm was obtained. For analysis of Venus, emission samples were exited at 514 nm, with a resulting emission scan of 526–600 nm.
RESULTS
HMPV N and P interact
To examine the capacity of HMPV proteins to interact with each other, we used Y2H analysis with bait and prey vectors encoding full-length HMPV proteins (Table 1). When bait and prey plasmids encoding the HMPV N or P protein were used in pairwise Y2H transformations, these proteins interacted. No interactions between other HMPV proteins were identified using this technique (Table 1).
Table 1.
Y2H analysis of HMPV N, P, M, M2-1 and M2-2 protein interactions
| GAL4-AD (prey) |
|||||
|---|---|---|---|---|---|
| GAL4-BD (bait) | N | P | M | M2-1 | M2-2 |
| N | − | + | − | − | − |
| P | + | − | − | − | − |
| M | − | − | − | − | − |
| M2-1 | − | − | − | − | − |
| M2-2 | − | − | − | − | − |
Y2H analysis was performed using bait and prey vectors encoding full-length HMPV proteins fused at the N terminus to the GAL4 DNA-binding domain (BD) as bait or activating domain (AD) as prey. Interactions were scored as positive only if transformed yeast grew on medium with highest stringency nutritional selection (His−/Leu−/Ade−/Trp−) and negative if there was no growth on this medium.
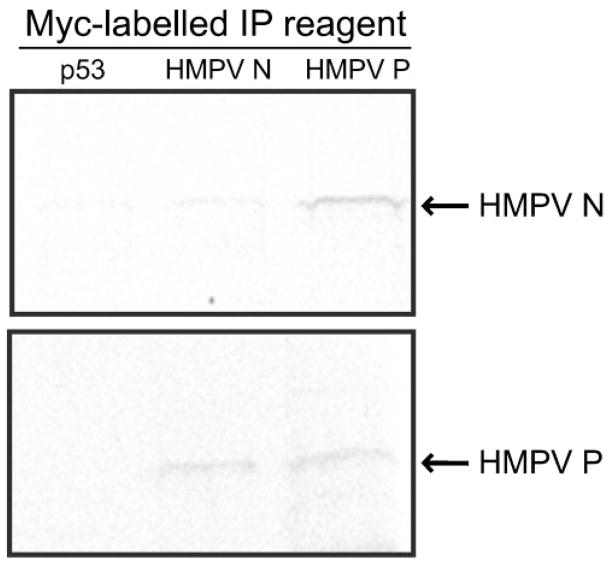
To confirm this genetic interaction between HMPV N and P, co-immunoprecipitation experiments were performed using in vitro-translated viral proteins, expressed as native amino acid sequence and appended with the c-Myc epitope tag at the N terminus. Radiolabelled native HMPV N co-immunoprecipitated specifically with c-Myc–HMPV P (Fig. 1, upper panel). Radiolabelled native HMPV P co-immunoprecipitated specifically with cold c-Myc–HMPV N (Fig. 1, lower panel). Immunoprecipitation was not detected when radiolabelled HMPV N or P was expressed with c-Myc–p53 or c-Myc–HMPV M (Fig. 1 and data not shown). We found in repeated experiments using this technique that HMPV N and P co-immunoprecipitation was consistent.
Fig. 1.
Co-immunoprecipitation of HMPV N and P. Upper panel: radiolabelled, native HMPV N was incubated with unlabelled c-Myc-tagged p53 (control), c-Myc–HMPV N and c-Myc–HMPV P. Lower panel: radiolabelled, native HMPV P was incubated with unlabelled c-Myc-tagged p53 (control), c-Myc–HMPV N and c-Myc–HMPV P. Anti-c-Myc antibody was used for immunoprecipitation (IP) and binding was analysed by SDS-PAGE and autoradiography.
Using in vitro co-immunoprecipitation, radiolabelled native HMPV P co-immunoprecipitated specifically with cold c-Myc–HMPV P (Fig. 1, lower panel), suggesting HMPV P homodimerization. The lack of HMPV P–P interaction in the Y2H experiments described above does not necessarily contradict these findings, but rather may suggest that the Y2H technique requiring large N-terminal protein fusions and using high-stringency nutritional selection may not be useful for studying HMPV P–P complex formation. We performed experiments to estimate the efficiency of HMPV N and P co-immunoprecipitation using this technique. Quantification was performed by immunoprecipitating radiolabelled HMPV N or P using anti-HMPV N or anti-HMPV P polyclonal antiserum and comparing band densities with those of HMPV N and P from co-immunoprecipitations with c-Myc–HMPV N and c-Myc–HMPV P. Less than 1 % of radiolabelled hMVP N or P co-immunoprecipitated with their partner viral protein in the experiments shown in Fig. 2.
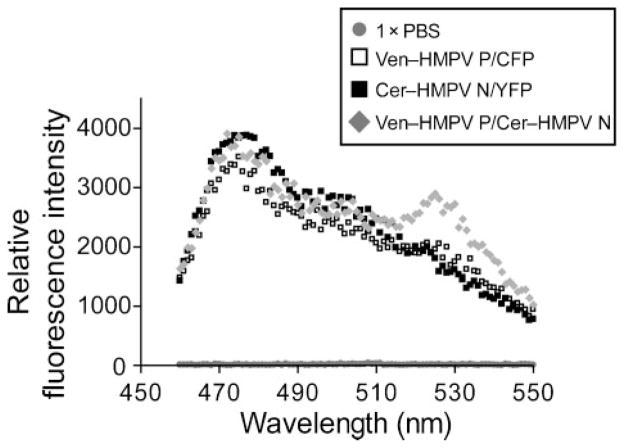
Fig. 2.
Analysis of HMPV P–N interactions by FRET fluorometry. Plasmid-based proteins were expressed in 293T cells by transient transfection and analysed by scanning cuvette fluorometry. The resulting fluorescence intensity curves are shown between the wavelength values of 450 and 550 nm. The emission peak for Cerulean (Cer)/CFP is 475 nm and for Venus (Ven)/YFP is 527 nm.
To study the interactions between HMPV N and P in mammalian cells, we employed a FRET fluorometry assay. Given that interactions between HMPV N and P occurred when additional amino acid sequences were appended to the N termini of these proteins in Y2H experiments, we hypothesized that HMPV N and P expressed as fusion proteins with N-terminal fluorescent protein molecules would interact in mammalian cells. In these experiments, we used Cerulean and Venus proteins, which are variants of the cyan fluorescent protein (CFP)/yellow fluorescent protein (YFP) FRET pair (Nagai et al., 2002; Rizzo et al., 2004). Efficient energy transfer occurs when Venus emission (527 nm) is observed following Cerulean excitation (433 nm). Expression of Venus–HMPV P and Cerulean–HMPV N resulted in efficient energy transfer, revealing that HMPV N and P interact in mammalian cells (Fig. 2). Whole cells were analysed in a scanning cuvette fluorometer 48 h after plasmid transfection, and spectrofluorometric analysis showed a Venus emission peak at 527 nm in cells expressing both proteins. This Venus emission peak was not seen in cells expressing Venus–HMPV P with CFP alone or Cerulean–HMPV N with YFP alone (Fig. 2). Venus and YFP expression levels were similar in all samples, verifying that emission was indeed due to FRET and not to variable levels of Venus or YFP expression (data not shown). These data confirmed that HMPV N and P interact in mammalian cells, and established the usefulness of FRET fluorometry for analysis of protein–protein interactions in this viral system.
HMPV N and P localize to and are the minimal requirements for cytoplasmic inclusion-like complex formation
A common feature of paramyxovirus infection is the formation of punctate areas of concentrated viral proteins termed viral inclusion bodies (Compans et al., 1966; Howe et al., 1967; Nakai et al., 1969; Norrby et al., 1970). To assess the components required for the formation of these viral inclusions, LLC-MK2 cells were infected with HMPV, fixed and stained with HMPV N- and P-specific antisera and analysed by fluorescence microscopy. HMPV N and P were detected as early as 4 h post-infection and localized to discrete punctate cytoplasmic complexes (Fig. 3a). To analyse further the fate of HMPV N and P in HMPV-infected and uninfected cells, fluorophore-tagged HMPV N and P were generated. Confocal microscopy analysis revealed that when GFP–HMPV N and RFP–HMPV P fusion proteins were expressed individually in uninfected LLC-MK2 cells, the proteins were distributed diffusely throughout the cytoplasm (Fig. 3b, c). However, when these proteins were expressed in HMPV-infected cells, the fluorescence patterns of GFP–HMPV N and RFP–HMPV P were altered dramatically. In each case, GFP–HMPV N or RFP–HMPV P was recruited to punctate, cytoplasmic viral inclusion bodies or inclusion-like complexes, as shown by anti-HMPV N or anti-HMPV P staining (Fig. 3d, e). These results demonstrated that plasmid-expressed, fluorophore-tagged HMPV N and HMPV P could be visualized and distinguished from native HMPV proteins in infected cells by using fluorescence microscopy, and that these altered proteins could be recruited to HMPV inclusion bodies during viral infection.
Fig. 3.
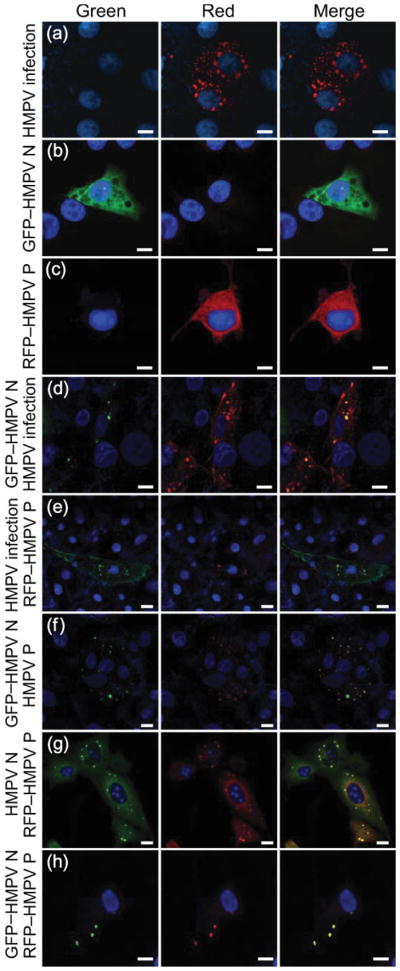
Analysis of HMPV N and P localization by fluorescence microscopy. LLC-MK2 cells were transfected with HMPV proteins or infected with HMPV and images were obtained by using confocal microscopy. Nuclear staining was obtained by treatment with TO-PRO-3 iodide. (a) HMPV-infected cells stained with anti-HMPV antisera followed by Alexa Fluor 546-labelled (red) goat anti-rabbit secondary antibody. (b) Cells transfected with GFP–HMPV N. (c) Cells transfected with RFP–HMPV P. (d) HMPV-infected cells stained with an anti-HMPV P polyclonal antibody followed by Alexa Fluor 546-labelled (red) goat anti-rabbit secondary antibody and transfected with GFP–HMPV N. (e) HMPV-infected cells stained with an anti-HMPV N polyclonal antibody followed Alexa Fluor 488-labelled (green) goat anti-rabbit secondary antibody and transfected with RFP–HMPV P. (f) Cells co-transfected with GFP–HMPV N and HMPV P and stained with an anti-HMPV P polyclonal antibody followed by Alexa Fluor 546-labelled (red) goat anti-rabbit secondary antibody. (g) Cells co-transfected with HMPV N and RFP–HMPV P and stained with anti-HMPV N polyclonal antibody followed by Alexa Fluor 488-labelled (green) goat anti-rabbit secondary antibody. (h) Cells co-transfected with GFP–HMPV N and RFP–HMPV P. Bars, 10 μm.
Once it was determined that plasmid-expressed, fluorescent-tagged HMPV proteins could be recruited to viral inclusion bodies in infected cells, we sought to test whether HMPV N and P expressed together were sufficient to form inclusion-like complexes in the absence of viral infection. HMPV N lacking a fluorescent tag was expressed in LLC-MK2 cells and immunofluorescence staining was performed using an HMPV N-specific antiserum. Expression of this protein alone displayed a diffuse pattern similar to that of GFP–HMPV N in Fig. 3(b) (data not shown). When HMPV P was expressed in cells and stained using an HMPV P-specific antiserum, a diffuse pattern similar to Fig. 3(c) was observed. In contrast, we found that co-expression of HMPV N (native or GFP-tagged forms) with HMPV P (native or RFP-tagged forms) formed punctate, cytoplasmic inclusion-like complexes (Fig. 3f–h). In order to verify that HMPV N and P were the minimal requirements for the formation of viral inclusions, we performed a series of additional experiments expressing N and P individually with HMPV M, M2-1 or M2-2 protein. Indeed, cytoplasmic inclusions were only observed when a combination of HMPV N and P were co-expressed either by transfection or by infection. Additionally, the co-expression of fluorescent protein alone (GFP or RFP) with HMPV N or P (native or fluorescently tagged) did not induce the formation of inclusion-like complexes (data not shown). These data showed that HMPV N and P are the minimal requirements for the formation of HMPV inclusion-like complexes in uninfected cells.
HMPV N and P interact in inclusion-like complexes
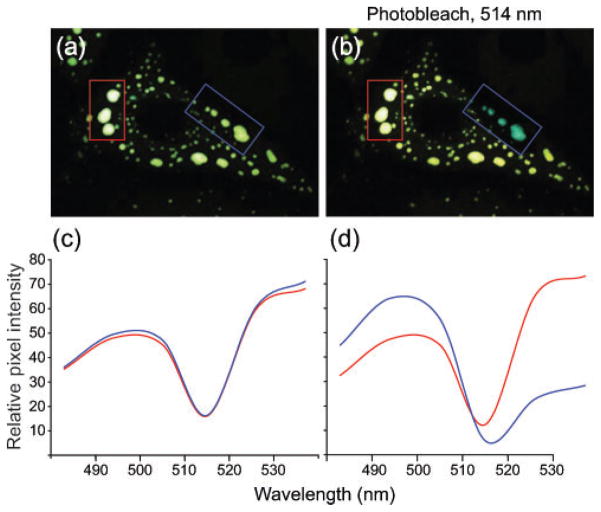
The experiments outlined above demonstrated that HMPV N and P interact and that they are the minimal requirements for inclusion-like complex formation. We next wanted to determine whether interactions between HMPV N and P were occurring within viral inclusion complexes. To do this, we used FRET microscopy, which allows the determination of protein–protein interactions at specific sites within cells expressing specific proteins of interest. Cerulean–HMPV N and Venus–HMPV P were co-expressed in LLC-MK2 cells by plasmid transfection, fixed after 36 h and the image was taken by using confocal microscopy. Results from this co-transfection revealed a specific overlap of HMPV N and P in inclusion-like complexes (Fig. 4a). Two specific regions of interest containing viral inclusion-like complexes were chosen (Fig. 4a, b, red and blue boxes). As protein interactions detected by FRET are represented by efficient energy transfer between the donor fluorophore (Cerulean) and the acceptor fluorophore (Venus), specific photobleaching of Venus results in a visual decrease in the Venus signal with a corresponding increase in the Cerulean signal (Fig. 4b, blue box). We then examined the spectral data collected in the region of interest pre- and post-bleaching by using the multichannel META detector of a Zeiss LSM 510 confocal microscope. Similar levels of CFP and YFP emission were observed for each region of interest before photobleaching (Fig. 4c, blue and red lines corresponding to the blue and red boxes). Following specific photobleaching of the region of interest marked (blue box), the emission intensity of Cerulean greatly increased together with a decrease in the emission intensity of Venus (Fig. 4d, blue line). Additionally, the emission spectrum of the non-photo-bleached region of interest remained consistent with the pre-bleach data (Fig. 4d, red line). These studies provided further evidence for HMPV N–P interaction and allowed detection of specific viral protein interactions within viral inclusion-like complexes.
Fig. 4.
Analysis of HMPV N–P interactions by FRET microscopy. (a) LLC-MK2 cells were co-transfected with Venus–HMPV P and Cerulean–HMPV N and images were obtained by confocal microscopy. (b) Co-transfected cells following photobleaching at 514 nm at a specific region of interest (blue box), and a non-bleached region (red box). (c, d) Emission scans were obtained from each region of interest before (c) or after (d) photobleaching at 514 nm. The colour of each line in the graph corresponds to the specific region of interest in the image.
The extreme N- and C-terminal regions of HMPV N are not required for interaction with HMPV P in Y2H experiments
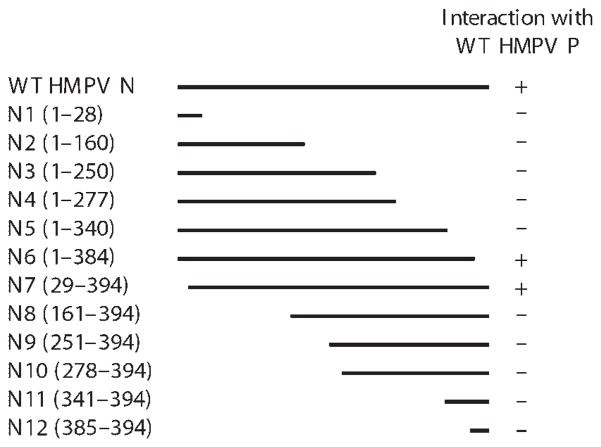
As HMPV N and P are both necessary and sufficient to form inclusion-like complexes, we sought to determine the specific regions of the viral proteins necessary for this interaction, and in turn the formation of inclusions. To this end, we performed mutational analysis of HMPV N to identify domains required for binding to HMPV P. Y2H experiments revealed that large regions of HMPV N are required for this interaction (Fig. 5). We constructed a series of N- or C-terminal deletion mutants of the N gene from HMPV wild-type strain TN/96-12 by PCR amplification using internal primers (see Supplementary Table S1). Design of this series of mutants was informed by published HMPV N sequence analysis (van den Hoogen et al., 2002). In addition to nutritional screening of yeast clones, positive results from nutritional selection were confirmed by β-galactosidase reporter expression. Interestingly, almost the entire HMPV N sequence was required for genetic interaction with HMPV P, with only the extreme N and C termini dispensable for binding (Fig. 5).
Fig. 5.
Yeast two-hybrid mapping of HMPV N–P interactions. Full-length wild-type (WT) HMPV P was used as prey in pairwise crosses with each mutant HMPV N protein as bait. Interactions were scored as positive only if the transformed yeast grew on medium with the highest stringency nutritional selection (His−/Leu−/Ade−/Trp−) and negative if there was no growth on this medium.
The N terminus of HMPV N is required for localization to cytoplasmic inclusions
Following Y2H analysis and specific mapping of HMPV N and P binding, we sought to verify the requirement for HMPV N–P interactions in the formation of cytoplasmic inclusion-like complexes. HMPV mutants N1–N12 (Fig. 5) were expressed as GFP fusion constructs with HMPV P in LLC-MK2 cells. Cytoplasmic inclusions were absent in all cases where the HMPV N truncation mutants were unable to bind HMPV P by Y2H (data not shown). For example, in accordance with the Y2H data, HMPV N6 displayed a diffuse pattern when expressed alone (Fig. 6a), but was relocalized to cytoplasmic inclusions in combination with transfection of RFP–HMPV P (Fig. 6b), transfection of HMPV P with specific anti-P immunofluorescence staining (Fig. 6c) or HMPV infection with specific anti-P immunofluorescence staining (Fig. 6d). Surprisingly, GFP–HMPV N7 was unable to form or localize to cytoplasmic inclusion-like complexes under any experimental conditions, even though it was found to bind to HMPV P by Y2H analysis (Fig. 6e–h). The finding that the N terminus of HMPV N is required for the formation of cytoplasmic inclusion-like complexes revealed that the mechanism of formation of viral inclusion bodies is more complex than the simple protein–protein interactions between HMPV N and P, suggesting the role of an unknown cellular factor to facilitate this process.
Fig. 6.
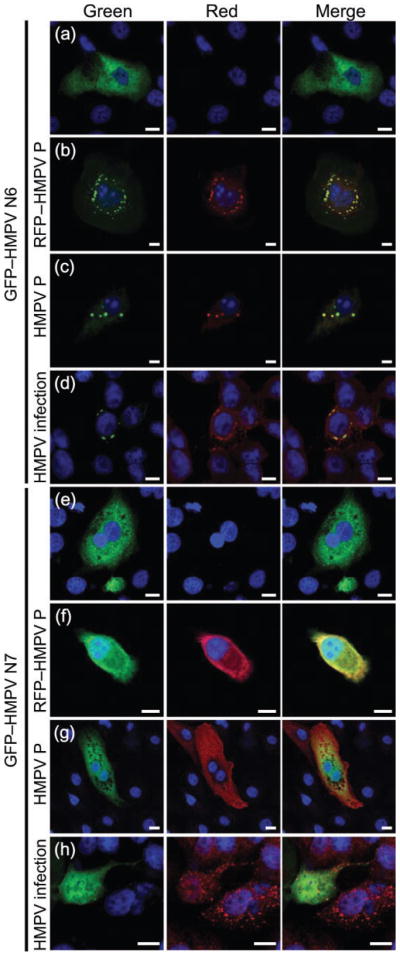
Formation of inclusion-like complexes by HMPV N truncation mutants. LLC-MK2 cells were transfected with HMPV proteins or infected with HMPV and images were obtained using confocal microscopy. Cells were transfected with GFP–HMPV N6 (a–d) or GFP–HMPV N7 (e–h) and co-transfected with RFP–HMPV P (b, f), co-transfected with HMPV P and stained with an anti-HMPV P polyclonal antibody followed by Alexa Fluor 546-labelled (red) goat anti-rabbit secondary antibody (c, g) or infected with HMPV and stained with an anti-HMPV P polyclonal antibody followed by Alexa Fluor 546-labelled (red) goat anti-rabbit secondary antibody (d, h). Bars, 10 μm.
DISCUSSION
A complete understanding of early events in paramyxovirus nucleocapsid and polymerase complex assembly and HMPV replication may provide new insights into paramyxovirus replication. We undertook experiments to determine which HMPV proteins interact, starting with a Y2H interaction analysis. We elected to use this technique in initial studies of HMPV because, in spite of limitations, Y2H analysis has been used successfully to explore interactions between replication complex proteins in HRSV (Hengst & Kiefer, 2000). These published reports provided a basis of comparison for our HMPV studies and an opportunity to compare interaction patterns among replication complex proteins for two closely related members of the paramyxovirus subfamily Pneumovirinae. We found that HMPV N and P proteins interacted with each other, but did not interact with HMPV M, M2-1 or M2-2 protein in Y2H experiments. The interaction of HMPV N and P in Y2H experiments has not been demonstrated previously. The interaction of N and P proteins in infected cells has been observed for other paramyxoviruses including Sendai virus (Curran et al., 1995; Ryan & Kingsbury, 1988; Ryan & Portner, 1990), MeV (Huber et al., 1991) and HRSV (Castagne et al., 2004; García et al., 1993; García-Barreno et al., 1996; Lu et al., 2002; Murphy et al., 2003; Murray et al., 2001; Stokes et al., 2003; Tran et al., 2007). The limitations of Y2H analysis in the study of protein interactions required for paramyxovirus replication are significant. Our findings that HMPV N and P do not interact with M, M2-1 or M2-2 when expressed as fusion proteins in yeast nuclei cannot be taken as definitive evidence that these proteins do not interact during HMPV replication in the mammalian cell cytoplasm. Additionally, our studies did not extend to analysis of the HMPV major polymerase protein (L). It is likely that HMPV replication depends on complex interactions between N, P, M, M2-1, M2-2 and L that do not occur when these proteins are expressed as fusion proteins in the absence of viral RNA and mammalian cytoplasmic proteins.
Unexpectedly, we found that HMPV N and P did not homodimerize when expressed as fusion proteins in Y2H experiments, which contrasts with Y2H analysis of HRSV N and P (Hengst & Kiefer, 2000) in experiments also using the commercial Clontech Matchmaker 2 Y2H system. An important goal for our research programme is an assessment of the extent to which our understanding of HRSV replication will guide our understanding of the newly discovered and closely related human pathogen HMPV. We surmised that these discrepant outcomes in Y2H experiments may reflect structural differences between HMPV and HRSV proteins. Given that N and P are expressed with yeast proteins fused at the N terminus in Y2H analysis, our unexpected findings guided subsequent experiments to determine the importance of N-terminal sequences in dimerization of HMPV N and P. Y2H experiments that fail to show HMPV N and P homo-dimerization are not necessarily evidence that these proteins do not interact during viral replication, as would be expected in the light of findings in HRSV (Castagne et al., 2004; García-Barreno et al., 1996; Murphy et al., 2003; Murray et al., 2001; Stokes et al., 2003).
We found that interactions between HMPV N and P, and HMPV P homodimerization, could be detected consistently in co-immunoprecipitation experiments using in vitro-transcribed and -translated proteins, albeit with low sensitivity. Again, the limitations of in vitro co-immunoprecipitation analysis are significant. We present these findings in the context of published work on HRSV, given interest in comparing these closely related pathogens. Other investigators have used in vitro-transcribed and -translated proteins to study HRSV N, P and M2-1 interactions and found no evidence for complex formation, contradictory to their findings in mammalian cells (García et al., 1993). Our findings are interesting to consider as we compare the behaviour of HMPV N and P with published studies of HRSV, but underscore the limitations of these experimental approaches. It is likely that optimal interaction between these HMPV proteins requires the presence of additional factors such as cellular proteins or RNA.
We found that HMPV N and P are the minimum viral protein requirements for the formation of inclusion-like complexes in mammalian cells (Fig. 3). A clear determination of the relevance and role of cytoplasmic inclusion bodies for the replication of HMPV will provide insights into early paramyxovirus replication. It has been shown in studies of other paramyxoviruses that viral inclusion bodies in infected cells probably contain aggregates of viral nucleocapsids (García et al., 1993; García-Barreno et al., 1996). The formation of inclusion-like complexes in uninfected cells expressing HRSV proteins requires both the N and P proteins (García et al., 1993; García-Barreno et al., 1996). In contrast, the individual expression of the N protein is sufficient to induce inclusion-like complex formation in MeV (Spehner et al., 1991). We hypothesize that a requirement for both the N and P proteins for the formation of inclusion bodies may be a distinguishing characteristic of viruses of the subfamily Pneumovirinae in the family Paramyxoviridae and an important event in early viral replication for these pathogens. Interestingly, investigators have found significant differences in nucleocapsid morphology between viruses of the subfamilies Pneumovirinae and Paramyxovirinae (Bhella et al., 2002).
We used FRET fluorometry and microscopy techniques in our studies of HMPV N–P interactions (Figs 2 and 5). To our knowledge, the use of this experimental approach has not been reported for the study of paramyxovirus proteins important for the nucleocapsid or polymerase in the nucleocapsid complex. Using these techniques, we showed interactions between viral proteins within mammalian cells permissive for viral infection and within discrete sub-cellular regions. We anticipate that this approach will also be useful in the identification of cellular factors required for inclusion body formation and early HMPV replication and will, in turn, shed light on possible inhibitors of HMPV infection. We expect that co-immunoprecipitation experiments performed in mammalian cells expressing these proteins will confirm these results as antibody reagents are developed in the future for the study of this newly discovered paramyxovirus.
Mutational analysis has been employed extensively in studies of paramyxovirus N and P protein interactions, and HRSV N regions required for its interaction with HRSV P have been identified (Castagne et al., 2004; García-Barreno et al., 1996; Hengst & Kiefer, 2000; Murphy et al., 2003; Murray et al., 2001; Slack & Easton, 1998; Stokes et al., 2003). Our finding that large segments of HMPV N are required for binding to HMPV P, and that the extreme N-and C-terminal sequences of HMPV N are not required for N–P interaction, are consistent with findings for the other members of the subfamily Pneumovirinae (Barr & Easton, 1995; García-Barreno et al., 1996; Hengst & Kiefer, 2000; Krishnamurthy & Samal, 1998). However, our finding that the N terminus of HMPV N is required for the formation of viral inclusion bodies is unique among paramyxoviruses studied to date (Fig. 6). Further support for these findings in the form of co-immunoprecipitation experiments using untagged viral proteins expressed in mammalian cells awaits development and characterization of monoclonal antibodies specific for HMPV N and P sequences that are retained in deletion mutants.
Given that HMPV N–P interactions are retained by a mutant of HMPV N deficient in extreme N-terminal amino acids, it is interesting to speculate that this region is required for interaction with a cellular protein or nucleic acid to allow inclusion body formation. It is also interesting to note that a high-probability sumoylation motif is located within the 28 aa that we have shown to be required for the formation of inclusion-like complexes. Further studies may reveal cellular factors required for the formation of paramyxovirus inclusion bodies.
In summary, we found that HMPV N and P interact and provide the minimal viral protein requirements for inclusion-like complex formation in mammalian cells. In addition, we have identified a specific region within HMPV N that is required for the formation of viral inclusions. HMPV shares with HRSV the requirement for both N and P for viral inclusion complex formation. However, our experiments suggest that these related human pathogens may have significant differences in viral protein structure and interaction characteristics. Further study of the role of HMPV N–P inclusion bodies in viral replication and the identification of cellular factors required for their formation may provide important insights into paramyxovirus replication and assembly.
Supplementary Material
Acknowledgments
This work was directly supported by grants NIH K08 AI58006 (T. R. P), NIH T32-HL07526 (A. D.) and K08 AI56170 (J. V. W.). This work was also supported by the Howard Hughes Medical Institute Postdoctoral Fellowships for Physicians Program and the Howard Hughes Medical Institute Community of Scholars Program. J. E. C. holds a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund. Confocal and FRET microscopy experiments were performed in the VUMC Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, HD15052, DK59637 and EY08126). We thank Sharon Tollefson for expert technical assistance, the Piston and Goldenring laboratories for reagents and technical support, and Melissa Maginnis for a critical review of the manuscript.
Footnotes
References
- Barr J, Easton AJ. Characterisation of the interaction between the nucleoprotein and phosphoprotein of pneumonia virus of mice. Virus Res. 1995;39:221–235. doi: 10.1016/0168-1702(95)00090-9. [DOI] [PubMed] [Google Scholar]
- Bhella D, Ralph A, Murphy LB, Yeo RP. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J Gen Virol. 2002;83:1831–1839. doi: 10.1099/0022-1317-83-8-1831. [DOI] [PubMed] [Google Scholar]
- Bitko V, Oldenburg A, Garmon NE, Barik S. Profilin is required for viral morphogenesis, syncytium formation, and cell-specific stress fiber induction by respiratory syncytial virus. BMC Microbiol. 2003;3:9. doi: 10.1186/1471-2180-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosis S, Esposito S, Niesters HG, Crovari P, Osterhaus AD, Principi N. Impact of human metapneumovirus in childhood: comparison with respiratory syncytial virus and influenza viruses. J Med Virol. 2005;75:101–104. doi: 10.1002/jmv.20243. [DOI] [PubMed] [Google Scholar]
- Burke E, Dupuy L, Wall C, Barik S. Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology. 1998;252:137–148. doi: 10.1006/viro.1998.9471. [DOI] [PubMed] [Google Scholar]
- Burke E, Mahoney NM, Almo SC, Barik S. Profilin is required for optimal actin-dependent transcription of respiratory syncytial virus genome RNA. J Virol. 2000;74:669–675. doi: 10.1128/jvi.74.2.669-675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagne N, Barbier A, Bernard J, Rezaei H, Huet JC, Henry C, Da Costa B, Eleouet JF. Biochemical characterization of the respiratory syncytial virus P–P and P–N protein complexes and localization of the P protein oligomerization domain. J Gen Virol. 2004;85:1643–1653. doi: 10.1099/vir.0.79830-0. [DOI] [PubMed] [Google Scholar]
- Chenik M, Chebli K, Gaudin Y, Blondel D. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J Gen Virol. 1994;75:2889–2896. doi: 10.1099/0022-1317-75-11-2889. [DOI] [PubMed] [Google Scholar]
- Compans RW, Holmes KV, Dales S, Choppin PW. An electron microscopic study of moderate and virulent virus–cell interactions of the parainfluenza virus SV5. Virology. 1966;30:411–426. doi: 10.1016/0042-6822(66)90119-x. [DOI] [PubMed] [Google Scholar]
- Crowe JE., Jr Human metapneumovirus as a major cause of human respiratory tract disease. Pediatr Infect Dis J. 2004;23:S215–S221. doi: 10.1097/01.inf.0000144668.81573.6d. [DOI] [PubMed] [Google Scholar]
- Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology. 1995;214:139–149. doi: 10.1006/viro.1995.9946. [DOI] [PubMed] [Google Scholar]
- Esper F, Martinello RA, Boucher D, Weibel C, Ferguson D, Landry ML, Kahn JS. A 1-year experience with human metapneumovirus in children aged <5 years. J Infect Dis. 2004;189:1388–1396. doi: 10.1086/382482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. J Infect Dis. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- García J, García-Barreno B, Vivo A, Melero JA. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology. 1993;195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- García-Barreno B, Delgado T, Melero JA. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J Virol. 1996;70:801–808. doi: 10.1128/jvi.70.2.801-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Kiefer P. Domains of human respiratory syncytial virus P protein essential for homodimerization and for binding to N and NS1 protein. Virus Genes. 2000;20:221–225. doi: 10.1023/a:1008188527858. [DOI] [PubMed] [Google Scholar]
- Herfst S, de Graaf M, Schickli JH, Tang RS, Kaur J, Yang CF, Spaete RR, Haller AA, van den Hoogen BG, et al. Recovery of human metapneumovirus genetic lineages A and B from cloned cDNA. J Virol. 2004;78:8264–8270. doi: 10.1128/JVI.78.15.8264-8270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe C, Morgan C, de Vaux St Cyr C, Hsu KC, Rose HM. Morphogenesis of type 2 parainfluenza virus examined by light and electron microscopy. J Virol. 1967;1:215–237. doi: 10.1128/jvi.1.1.215-237.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Cattaneo R, Spielhofer P, Orvell C, Norrby E, Messerli M, Perriard JC, Billeter MA. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology. 1991;185:299–308. doi: 10.1016/0042-6822(91)90777-9. [DOI] [PubMed] [Google Scholar]
- Kallewaard NL, Bowen AL, Crowe JE., Jr Cooperativity of actin and microtubule elements during replication of respiratory syncytial virus. Virology. 2005;331:73–81. doi: 10.1016/j.virol.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, Samal SK. Identification of regions of bovine respiratory syncytial virus N protein required for binding to P protein and self-assembly. J Gen Virol. 1998;79:1399–1403. doi: 10.1099/0022-1317-79-6-1399. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Kolakofsky D. Paramyxoviridae: the Viruses and Their Replication. Philadelphia: Lippincott Williams and Wilkins; 2001. [Google Scholar]
- Lu B, Brazas R, Ma CH, Kristoff T, Cheng X, Jin H. Identification of temperature-sensitive mutations in the phospho-protein of respiratory syncytial virus that are likely involved in its interaction with the nucleoprotein. J Virol. 2002;76:2871–2880. doi: 10.1128/JVI.76.6.2871-2880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JA, Erdman DD, Weinberg GA, Edwards K, Hall CB, Walker FJ, Iwane M, Anderson LJ. Human metapneumovirus infection among children hospitalized with acute respiratory illness. Emerg Infect Dis. 2004;10:700–705. doi: 10.3201/eid1004.030555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LB, Loney C, Murray J, Bhella D, Ashton P, Yeo RP. Investigations into the amino-terminal domain of the respiratory syncytial virus nucleocapsid protein reveal elements important for nucleocapsid formation and interaction with the phosphoprotein. Virology. 2003;307:143–153. doi: 10.1016/s0042-6822(02)00063-6. [DOI] [PubMed] [Google Scholar]
- Murray J, Loney C, Murphy LB, Graham S, Yeo RP. Characterization of monoclonal antibodies raised against recombinant respiratory syncytial virus nucleocapsid (N) protein: identification of a region in the carboxy terminus of N involved in the interaction with P protein. Virology. 2001;289:252–261. doi: 10.1006/viro.2001.1150. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nakai T, Shand FL, Howatson AF. Development of measles virus in vitro. Virology. 1969;38:50–67. doi: 10.1016/0042-6822(69)90127-5. [DOI] [PubMed] [Google Scholar]
- Norrby E, Marusyk H, Orvell C. Morphogenesis of respiratory syncytial virus in a green monkey kidney cell line (Vero) J Virol. 1970;6:237–242. doi: 10.1128/jvi.6.2.237-242.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Ryan KW, Kingsbury DW. Carboxyl-terminal region of Sendai virus P protein is required for binding to viral nucleocapsids. Virology. 1988;167:106–112. doi: 10.1016/0042-6822(88)90059-1. [DOI] [PubMed] [Google Scholar]
- Ryan KW, Portner A. Separate domains of Sendai virus P protein are required for binding to viral nucleocapsids. Virology. 1990;174:515–521. doi: 10.1016/0042-6822(90)90105-z. [DOI] [PubMed] [Google Scholar]
- Slack MS, Easton AJ. Characterization of the interaction of the human respiratory syncytial virus phosphoprotein and nucleocapsid protein using the two-hybrid system. Virus Res. 1998;55:167–176. doi: 10.1016/s0168-1702(98)00042-2. [DOI] [PubMed] [Google Scholar]
- Spehner D, Kirn A, Drillien R. Assembly of nucleocapsid like structures in animal cells infected with a vaccinia virus recombinant encoding the measles virus nucleoprotein. J Virol. 1991;65:6296–6300. doi: 10.1128/jvi.65.11.6296-6300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes HL, Easton AJ, Marriott AC. Chimeric pneumovirus nucleocapsid (N) proteins allow identification of amino acids essential for the function of the respiratory syncytial virus N protein. J Gen Virol. 2003;84:2679–2683. doi: 10.1099/vir.0.19370-0. [DOI] [PubMed] [Google Scholar]
- Tran TL, Castagne N, Bhella D, Varela PF, Bernard J, Chilmonczyk S, Berkenkamp S, Benhamo V, Grznarova K, et al. The nine C-terminal amino acids of the respiratory syncytial virus protein P are necessary and sufficient for binding to ribonucleoprotein complexes in which six ribonucleotides are contacted per N protein protomer. J Gen Virol. 2007;88:196–206. doi: 10.1099/vir.0.82282-0. [DOI] [PubMed] [Google Scholar]
- Ulloa L, Serra R, Asenjo A, Villanueva N. Interactions between cellular actin and human respiratory syncytial virus (HRSV) Virus Res. 1998;53:13–25. doi: 10.1016/s0168-1702(97)00121-4. [DOI] [PubMed] [Google Scholar]
- Ulloa-Gutierrez R, Skippen P, Synnes A, Seear M, Bastien N, Li Y, Forbes JC. Life-threatening human metapneumo-virus pneumonia requiring extracorporeal membrane oxygenation in a preterm infant. Pediatrics. 2004;114:e517–e519. doi: 10.1542/peds.2004-0345. [DOI] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, Bestebroer TM, Osterhaus AD, Fouchier RA. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002;295:119–132. doi: 10.1006/viro.2001.1355. [DOI] [PubMed] [Google Scholar]
- van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis. 2003;188:1571–1577. doi: 10.1086/379200. [DOI] [PubMed] [Google Scholar]
- Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE., Jr Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JV, Tollefson SJ, Johnson JE, Crowe JE., Jr The cotton rat (Sigmodon hispidus) is a permissive small animal model of human metapneumovirus infection, pathogenesis, and protective immunity. J Virol. 2005;79:10944–10951. doi: 10.1128/JVI.79.17.10944-10951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Davis PJ, Li J, Cavanagh D. Cloning and sequencing of the matrix protein (M) gene of turkey rhinotracheitis virus reveal a gene order different from that of respiratory syncytial virus. Virology. 1992;186:426–434. doi: 10.1016/0042-6822(92)90007-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.