Abstract
The transcription factor Mef2 has well established roles in muscle development in Drosophila and in the differentiation of many cell types in mammals, including neurons. Here, we describe a role for Mef2 in the Drosophila pacemaker neurons that regulate circadian behavioral rhythms. We found that Mef2 is normally produced in all adult clock neurons and that Mef2 overexpression in clock neurons leads to long period and complex rhythms of adult locomotor behavior. Knocking down Mef2 expression via RNAi or expressing a repressor form of Mef2 caused flies to lose circadian behavioral rhythms. These behavioral changes are correlated with altered molecular clocks in pacemaker neurons: Mef2 overexpression causes the oscillations in individual pacemaker neurons to become desynchronized, while Mef2 knockdown strongly dampens molecular rhythms. Thus, a normal level of Mef2 activity is required in clock neurons to maintain robust and accurate circadian behavioral rhythms.
Introduction
Circadian rhythms of animal locomotor activity are controlled by molecular clocks located in central brain pacemaker neurons. These endogenous clocks allow animals to anticipate daily environmental changes and, as a result, animals retain behavioral rhythms even without any environmental cues. Genetic studies in Drosophila identified a number of core clock genes that form intracellular molecular clocks. In these clocks, two transcription factors, Clock (CLK) and Cycle (CYC) activate expression of period (per) and timeless (tim). PER and TIM proteins heterodimerize and then enter the nucleus after a substantial delay, where PER inhibits CLK/CYC activity, thus repressing per and tim expression. In a second interlocked loop, CLK and CYC activate expression of vrille (vri) and Par domain protein 1 (Pdp1), whose protein products feed back to regulate expression of Clk. Together with abundant posttranslational regulation, these transcriptional feedback loops lead to rhythmic clock gene RNA and protein accumulation (for review, see Hardin, 2005).
Recent studies in Drosophila indicate that clock neurons form a neural network that underlies robust circadian behavior (Grima et al., 2004; Stoleru et al., 2004, 2005). Two lines of evidence point to the small ventral lateral neurons (s-LNvs), which produce the neuropeptide pigment dispersing factor (PDF), as the most important pacemaker neurons. First, robust behavioral rhythms are lost when LNvs are either ablated or hyperpolarized (Renn et al., 1999; Nitabach et al., 2002). Second, accelerating the clock only in s-LNvs speeds up the clocks in the dorsal lateral neurons (LNds) and some dorsal neuron (DN) subgroups (Stoleru et al., 2005), indicating that s-LNvs can set the pace of other clock neurons. However, other clock neurons signal back to LNvs and can drive rhythmic outputs via LNvs, even if the LNvs lack a functional clock, at least in light/dark (LD) cycles (Stoleru et al., 2004). Indeed, coupling of clock neurons seems essential for robust circadian rhythms in mammals and can even override clock gene mutations that give phenotypes in dissociated cells (Liu et al., 2007). However, the molecular pathways by which fly and mammalian clock neurons communicate with each other to generate robust and self-sustaining behavioral rhythms are unclear.
To identify clock regulatory factors that could either help send or receive signals for intercellular communication, we used a Gal4/UAS mis-expression screen in clock neurons and assayed the behavior of flies in constant darkness (DD). The Pdf-Gal4 and tim(UAS)-Gal4 drivers were used to express genes either only in LNvs or in all clock neurons respectively.
Here we describe EP insertion line 1751 which caused long rhythms when expressed in LNvs and long periods or complex rhythms (more than one period) of locomotor activity when expressed in all clock neurons. Line 1751 is inserted in the Myocyte enhancer factor 2 (Mef2) locus, which encodes a transcription factor involved in muscle development in Drosophila (Bour et al., 1995; Lilly et al., 1995; Ranganayakulu et al., 1995). In mammals, four Mef2 genes are expressed in diverse cell types and Mef2 transcriptional activity is regulated by signal transduction pathways to control aspects of cell differentiation (for review, see Potthoff and Olson, 2007). Of particular relevance, Mef2 regulates synapse development in an activity-dependent manner in mammals (Flavell et al., 2006; Shalizi et al., 2006). Genes involved in synaptic development are among the Mef2 target genes in neurons and include a number of genes mis-regulated in epilepsy and autism spectrum disorder (Flavell et al., 2008). Although Mef2 is also produced in some embryonic and adult Drosophila brain neurons (Taylor et al., 1995; Schulz et al., 1996), its function in fly neurons has not been addressed. Here we show that Mef2 is normally produced in all fly clock neurons and that interfering with Mef2 activity in clock neurons causes flies to become behaviorally arrhythmic. At the molecular level, Mef2 overexpression slows down the molecular clock and causes individual s-LNvs to desynchronize from each other, while knocking down Mef2 expression via RNAi strongly dampens molecular clock oscillations. These phenotypes are consistent with a role for Mef2 in sustaining circadian rhythms by regulating core clock gene expression and/or by influencing communication between clock neurons.
Materials and Methods
Screen for altered circadian behavior.
An EP element (Rørth, 1996) inserted on the X-chromosome (EP55; Bloomington Stock Center, Indiana University, Bloomington, IN) was mobilized to the autosomes using standard procedures to generate novel EP insertions, and the resulting w+ male progeny were individually crossed to virgin female flies carrying an X-linked Pdf-Gal4 transgene (Park et al., 2000). The locomotor behavior of six male adult progeny from each cross was assayed in constant darkness at 25°C as described below. Lines were established for EP insertions that altered behavioral rhythms. DNA flanking the insertion was isolated and sequenced using standard procedures to determine their location.
Fly culture and strains.
Flies were reared on a standard cornmeal/agar/yeast medium and maintained at 25°C unless otherwise indicated. To make UAS-Mef2-EnR flies, DNA encoding the first 128aa of Mef2, which includes the MADS box and Mef2 domain, was fused by PCR N-terminal to DNA encoding amino acids 2-298 of Engrailed, which includes its repressor domain. This fragment was sequenced, inserted into pUAST and transgenic flies made according to standard procedures. Other fly lines have been described previously: Pdf2.4-Gal4 (Renn et al., 1999), Pdf0.5-Gal4 (Park et al., 2000), tim(UAS)-Gal4 (Blau and Young, 1999), Mef2-Gal4 (Ranganayakulu et al., 1998) and tubulin-Gal80ts (McGuire et al., 2003). Four UAS-Mef2 constructs were previously described as UAS-Mef2 I-IV (Gunthorpe et al., 1999), which correspond to Mef2 isoforms A–D respectively on FlyBase. Here, we refer to UAS-Mef2 III as UAS-Mef2 III(low) to distinguish it from the UAS-Mef2 construct of Bour et al. (1995), which we call UAS-Mef2 III(high) since it expresses higher levels of Mef2 isoform C (Gunthorpe et al., 1999). UAS-Mef2RNAi (transformant ID 15549) and UAS-Dicer-2 were described by Dietzl et al. (2007) and obtained from the Vienna Drosophila RNAi Center. Clock mutant flies were provided by Mike Young (Rockefeller University, New York, NY), Michael Rosbash (Brandeis University, Waltham, MA), and Ravi Allada (Northwestern University, Evanston, IL). These mutants were crossed to include a Pdf-RFP transgene that contains the Pdf promoter fused directly to RFP to help visualize LNvs in cyc0 and Clkar mutants when endogenous PDF levels are low. This transgene will be described in detail later (M. Ruben, M.D. Drapeau, and J. Blau, unpublished observations). UAS-nuclear LacZ flies were obtained from the Bloomington Stock Center. Control flies were generated by crossing UAS and EP lines to y w flies.
Measuring the period of circadian locomotor activity.
The locomotor activity of individual flies was recorded at 25°C (except where noted) for between 6 and 12 d in constant darkness (DD) using the TriKinetics infrared beam-crossing system and 5 min bins. Raw activity histograms were analyzed for circadian rhythms using Matlab and Clocklab software (Actimetrics). Lomb–Scargle periodograms were constructed according to Van Dongen et al. (1999) and rhythmicity was defined as the presence of a peak above the 0.05 significance line as described by Nitabach et al. (2006). A simple and robust rhythm was recorded if Lomb–Scargle analysis revealed one peak above the significance line, while a complex rhythm was defined by a fly having more than one peak above the significance line. Arrhythmicity was defined by the absence of any peak above the significance line. We used χ2 analysis to measure period length and power for the temperature shift experiment (see Fig. 5B). Period length and power were calculated for the initial days in DD, for the last 4 d at 31.5°C and the subsequent 4 d at 25°C. We used χ2 periodograms here since they seemed to more accurately reflect period length over the short timeframes at each temperature.
Figure 5.
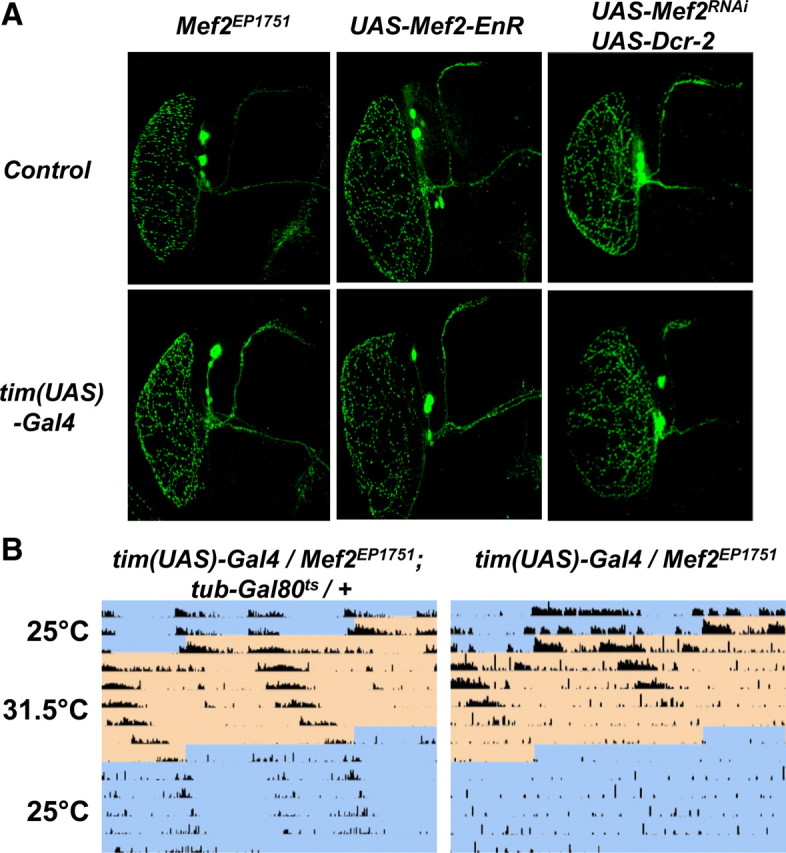
A, Mef2 is not required for the normal development of LNvs. Comparison of LNvs labeled with an antibody against PDF for Mef2EP1751 flies (left column), UAS-Mef2-EnR flies (center) and UAS-Mef2RNAi and UAS-Dcr-2 flies (right column) crossed to either y w (top row) or tim(UAS)-Gal4 (bottom row). B, Circadian phenotypes can be induced when Mef2 overexpression is restricted to adult clock neurons. Representative actograms of tim(UAS)-Gal4/Mef2EP1751 flies either with a tubulin-Gal80ts transgene (left) or without this transgene (right). Flies were maintained at 25°C until after eclosion and for the first 2.5 d in DD (shown in blue on the actogram), before shifting to the restrictive temperature for Gal80ts (31.5°C, shown in orange) for 6 d before returning to 25°C for 5 d. These results are representative of 15 flies assayed with or without the tubulin-Gal80ts transgene respectively. Average periods for tim(UAS)-Gal4/Mef2EP1751; tubulin-Gal80ts/+ flies were 26.0 ± 0.8 h and 24.6 ± 0.6 h for the last 4 d at 31.5°C and the next 4 d at 25°C respectively. Average periods for tim(UAS)-Gal4/Mef2EP1751 flies were 26.1 ± 0.7 h for the last 4 d at 31.5°C. On returning to 25°C, the average power of rhythms was higher for tim(UAS)-Gal4/Mef2EP1751; tubulin-Gal80ts/+ flies than for tim(UAS)-Gal4/Mef2EP1751 flies (p < 0.004), making period estimates for the latter very unreliable.
Immunocytochemistry.
Standard immunocytochemistry procedures were used to localize Mef2 protein in the fly brain and to study rhythms of clock proteins in s-LNvs as described by Cyran et al. (2003). For time course experiments, flies were first entrained to a 12:12 light-dark (LD) cycle for 3 d, then shifted to DD and dissected on either the second day (DD2) or the eighth day (DD8). Primary antibodies and their dilutions used were as follows: PER at 1/10,000 (from J. Hall, Brandeis University, Waltham, MA); VRI at 1/10,000 (from P. Hardin, Texas A&M University, College Station, TX); TIM at 1/2500 (from M. Young, Rockefeller University, New York, NY); PDP1 at 1/1000; Mef2 at either 1/200 or 1/1000 (from B. Patterson, National Cancer Institute, Bethesda, MD; and H. Nguyen, University of Erlangen-N̈urnberg, Erlangen, Germany); mouse anti-β-gal at 1/1000 (Promega); and mouse anti-PDF at 1/20. The PDF monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the Department of Biological Sciences, University of Iowa.
Microscopy and image analysis.
Images were acquired on a Leica confocal microscope with either a 40× or 20× oil-immersion lens by collecting z-sections at ∼1 and 2 μm intervals respectively and projecting the images onto a single plane for expression pattern studies, or by collecting single z-sections for time course analysis and localization of clock proteins. All samples in one experiment were processed in parallel except where indicated: i.e., they were collected at time points on the same day and then fixed and immunostained using master mixes for all solutions. Mef2 levels (see Fig. 3) were quantified using FIJI software (http://pacific.mpi-cbg.de). Desynchronization was assayed in the following manner: (1) The s-LNvs in a single cluster were scored for either the presence or absence of clock proteins and an s-LNv cluster was termed desynchronized if at least 1 s-LNv scored differently from the other s-LNvs. (2) PER levels were quantified for the s-LNvs in a given cluster and plotted to show the standard deviation (SD) between s-LNvs in the same hemisphere (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). Statistical analyses were done using Student's t tests.
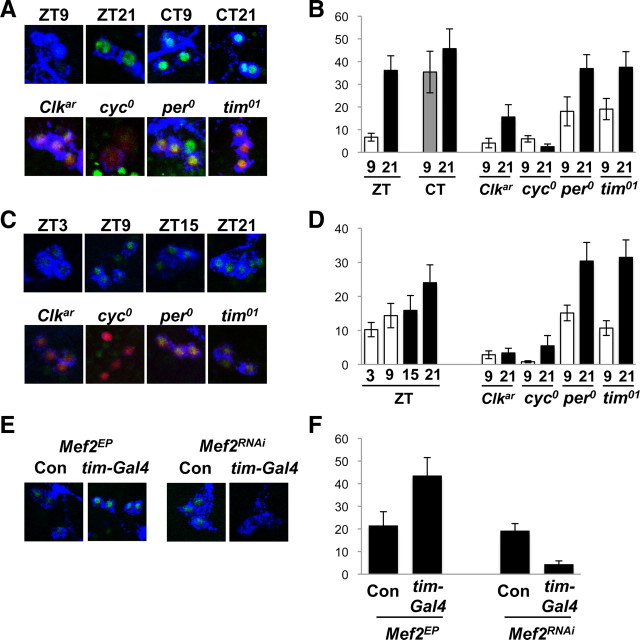
Figure 3.
Mef2 expression in LNvs is regulated by light and by clock genes. A, B, Representative images (A) and quantification (B) of Mef2 levels in adult s-LNvs. y w control brains were stained with antibodies to Mef2 (green) and PDF (blue) at ZT9 or ZT21 in LD and CT9 or CT21 on the second day of DD. Clkar, cyc0, per0 and tim01 brains were also stained with antibodies to Mef2 at ZT9 and ZT21. The clock mutant images are from ZT21. A Pdf-RFP transgene was included to help mark LNvs in clock mutants and the red channel was used to record RFP fluorescence since endogenous PDF levels are low in Clkar and cyc0 mutants. The strongly Mef2-staining cells visible in cyc0 mutants are not LNvs. Mef2 levels in adult s-LNvs were higher at ZT21 than at ZT9 (p < 0.0001) but not different between CT9 and CT21 (p = 0.42). Mef2 levels at ZT21 are higher in per0 and tim01 mutants than in Clkar and cyc0 mutants (p < 0.01). C, D, Representative images (C) and quantification (D) of Mef2 levels in larval LNs. y w (control) and clock mutant brains were stained as in A. The clock mutant images are from ZT21. The strongly Mef2+ cells visible in cyc0 mutants are not LNvs. Mef2 levels at ZT21 are higher than ZT3 (p < 0.01). Mef2 levels are higher in per0 and tim01 mutants than in Clkar and cyc0 mutants (p < 0.002) and show time-dependency in per0 and tim01 mutants (p < 0.05). E, F, Representative images (E) and quantification (F) of Mef2 levels in larval LNs at ZT21. tim(UAS)-Gal4/Mef2EP1751 had higher Mef2 levels than Mef2EP1751/+ controls (Con, p < 0.05). tim(UAS)-Gal4/+; UAS-Mef2RNAi/ UAS-Dcr-2 larvae had lower Mef2 levels than UAS-Mef2RNAi/+ control larvae (p < 0.0005).
Results
Isolation of Mef2 in a circadian mis-expression screen
To identify novel genes involved in Drosophila circadian behavior, ∼2000 novel lines containing random EP insertions on either the second or third chromosome were generated and screened for altered behavioral rhythms in the presence of the Pdf-Gal4 driver. In adult flies, this construct drives expression in: (1) the 8 pacemaker s-LNvs that are crucial for circadian rhythms; (2) the 8 large LNvs (l-LNvs) that are probably more important for regulating arousal and sleep than circadian timing (Chung et al., 2009; Donlea et al., 2009; Parisky et al., 2008; Shang et al., 2008; Sheeba et al., 2008); and (3) in 4–6 cells of unknown function at the tip of the abdominal ganglion (Park et al., 2000). In our behavioral screen, we identified five lines with long-period rhythms, and the characterization of one of these is described here. Flies with both Pdf-Gal4 and EP insertion 1751 had a 24.7 h period, while flies containing EP1751 alone had a 23.9 h period (Fig. 1A, Table 1). Sequencing the region of insertion of the EP revealed that EP1751 is inserted on the second chromosome, 107bp upstream of the start site of transcription of Mef2 isoform F according to FlyBase. We refer to this allele hereafter as Mef2EP1751.
Figure 1.
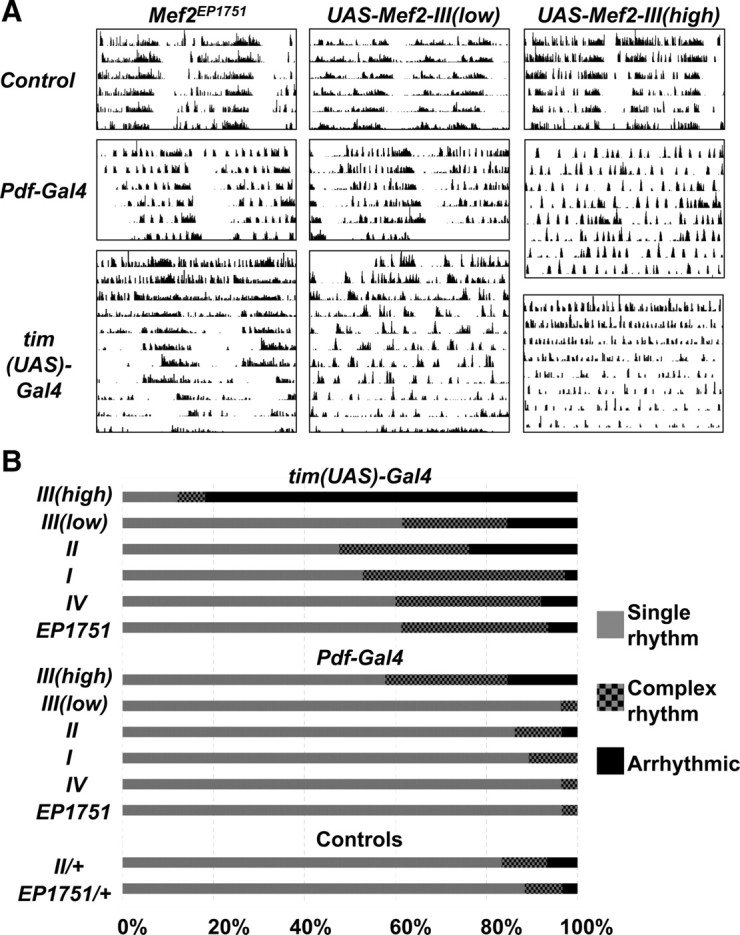
Overexpressing Mef2 in clock neurons disrupts normal circadian locomotor activity. A, Representative double-plotted actograms in DD from Mef2EP1751 flies (left column), UAS-Mef2 III(low) flies (center column), and UAS-Mef2 III(high) flies (right column). Actograms are shown for these flies crossed to y w flies (top row), Pdf-Gal4 (center row) and tim(UAS)-Gal4 flies (bottom row). B, Graph shows percentage of flies exhibiting either a single rhythm (gray), complex rhythms (checked) or arrhythmicity (black). A complex rhythm is defined as a rhythm exhibiting more than one significant period above the significance level at 95% confidence when analyzed with Lomb–Scargle analysis. Single rhythm and arrhythmicity are defined by the presence or the absence of a peak above the significance level respectively. I, II, and IV refer to UAS-Mef2 constructs for isoforms A, B, and D, respectively. III(low) and III(high) refer to two different UAS-Mef2 transgenes for Mef2 isoform C that express low and high levels of Mef2 protein respectively.
Table 1.
Adult locomotor activity rhythms with Mef2 overexpression
| Genotype | Period (h) | Power | n | Single rhythm (%) | Complex rhythm (%) | AR (%) |
|---|---|---|---|---|---|---|
| Pdf-Gal4 /UAS-Mef2-III(high) | 24.3 ± 0.6 | 24.8 ± 3.7 | 26 | 57.8 | 26.9 | 7.7 |
| tim(UAS)-Gal4/+;UAS-Mef2-III(high)/+ | 22.4 ± 1.4 | 11.5 ± 0.7 | 33 | 12.1 | 6.1 | 81.8 |
| Pdf-Gal4/UAS-Mef2-III(low) | 25.1 ± 0.1 | 57.2 ± 5.9 | 27 | 96.3 | 3.7 | 0.0 |
| tim(UAS)-Gal4/+;UAS-Mef2-III(low/+ | 24.6 ± 0.3 | 28.5 ± 4.7 | 26 | 61.5 | 23.1 | 15.4 |
| Pdf-Gal4/UAS-Mef2-II | 25.1 ± 0.1 | 58.1 ± 8.7 | 29 | 86.2 | 10.3 | 3.5 |
| tim(UAS)-Gal4/+;UAS-Mef2-II/+ | 25.6 ± 0.3 | 21.5 ± 2.1 | 21 | 47.6 | 28.6 | 23.8 |
| Pdf-Gal4/UAS-Mef2-I | 25.5 ± 0.1 | 58.3 ± 6.6 | 28 | 89.3 | 10.7 | 0.0 |
| tim(UAS)-Gal4/+;UAS-Mef2-I/+ | 25.4 ± 0.5 | 37.4 ± 6.7 | 18 | 52.8 | 44.4 | 2.8 |
| Pdf-Gal4/UAS-Mef2-IV | 25.5 ± 0.1 | 69.4 ± 9.7 | 28 | 96.4 | 3.6 | 0.0 |
| tim(UAS)-Gal4/+;UAS-Mef2-IV/+ | 25.1 ± 0.1 | 69.0 ± 10.1 | 25 | 60.0 | 32.0 | 8.0 |
| Pdf-Gal4/Mef2EP1751;Pdf-Gal4/+ | 24.7 ± 0.1 | 80.8 ± 4.9 | 29 | 96.5 | 3.5 | 0.0 |
| tim(UAS)-Gal4/Mef2EP1751 | 25.2 ± 0.2 | 47.0 ± 5.2 | 31 | 61.3 | 32.3 | 6.5 |
| UAS-Mef2-II/+ | 23.3 ± 0.2 | 53.5 ± 7.3 | 30 | 83.3 | 10.0 | 6.7 |
| Mef2EP1751/+ | 23.9 ± 0.1 | 51.8 ± 6.2 | 24 | 88.4 | 8.3 | 3.3 |
Lomb–Scargle analysis of adult locomotor activity showing period (in hours with SEM), power (to indicate the strength of the rhythm), number of flies analyzed (n), and the percentages exhibiting either a single rhythm, complex rhythm, or arrhythmicity (AR). The period is the average of single rhythmic flies and the highest peak detected (i.e., the dominant period) for flies with complex rhythms.
To test whether expressing Mef2 in all clock cells would affect circadian rhythms more strongly than expression only in LNvs, we assayed the locomotor activity of flies in which the tim(UAS)-Gal4 driver was crossed to Mef2EP1751. Visual inspection of the actograms of tim(UAS)-Gal4/Mef2EP1751 flies showed weak rhythms for the first 3 d in DD followed by superimposed short and long rhythms for days 4–12 (Fig. 1A). To quantify this phenomenon, we used Lomb–Scargle analysis of periodograms (see Materials and Methods). This analysis identified that 32% of tim(UAS)-Gal4/Mef2EP1751 flies had more than one rhythmic period detectable above the 95% significance line and we classify these flies as having complex rhythms. Additionally, 6% of tim(UAS)-Gal4/Mef2EP1751 flies were arrhythmic. In our opinion, Lomb–Scargle analysis is a conservative estimate of complex rhythms since very few tim(UAS)-Gal4/Mef2EP1751 flies had the normal single period circadian rhythm characteristic of wild-type flies when visually inspecting the actograms. In contrast, very few control flies exhibited complex rhythms (8%) and only 3% were arrhythmic. The average period of the most significant peaks for the rhythmic tim(UAS)-Gal4/Mef2EP1751 flies was 25.2 h, which is slightly longer than when Mef2EP1751 was activated only in LNvs by Pdf0.5-Gal4 (Fig. 1B, Table 1). These phenotypes were dependent on a Gal4 driver since Mef2EP1751 flies show normal rhythms without a Gal4 driver (Fig. 1A, Table 1), as do tim(UAS)-Gal4 flies without an EP element (Blau and Young, 1999).
Multiple Mef2 isoforms recapitulate the Mef2EP1751 phenotypes
Mef2 encodes a transcription factor required in Drosophila embryos for the development of muscle precursor cells. The single Drosophila Mef2 gene spans 45 kb. Five different transcripts have been described, each encoding distinct protein isoforms, which share the same DNA binding and dimerization domains (MADS and Mef2 box domains), but differing slightly in their C-terminal activation domains (FlyBase) (Taylor et al., 1995; Schulz et al., 1996; Gunthorpe et al., 1999). Four of these isoforms have been shown to rescue the Mef2 mutant phenotype in embryonic muscle precursor cells with the level of expression of these isoforms being the most important factor in determining the extent of rescue (Gunthorpe et al., 1999). We used UAS constructs for each of these to test whether expression of different Mef2 isoforms in clock neurons would recapitulate the phenotypes seen with expressing Mef2 via the Mef2EP1751 insertion, or whether functional differences exist between Mef2 isoforms in adult flies.
Flies containing UAS transgenes for Mef2 isoforms A–D (called UAS-Mef2 I–IV respectively) were first crossed to flies with a Pdf-Gal4 driver. We found that expression of UAS-Mef2 transgenes I–IV only in LNvs caused long rhythms with periods ranging between 25.1 and 25.5 h (Fig. 1, Table 1), which is slightly longer than the periods observed with Mef2EP1751. High-level expression of Mef2 isoform C via the UAS-Mef2 III(high) transgene caused ∼27% of the flies to exhibit complex rhythms, much higher than observed for the other isoforms or for control flies without a Gal4 driver (Fig. 1B). Gunthorpe et al. (1999) estimated that the UAS-Mef2 III(high) transgene produces ∼10-fold more Mef2 protein than UAS-Mef2 III(low) when expressed via an embryonic Gal4 driver, and this higher level of Mef2 expression correlates with the increased number of flies having complex rhythms in our assay.
When expressed in all clock cells using the tim(UAS)-Gal4 driver, UAS-Mef2 I, II, III(low) and IV produced similar phenotypes to tim(UAS)-Gal4/Mef2EP1751 flies with between 23 and 44% of flies showing complex rhythms, depending on the transgene. Flies expressing UAS-Mef2 II and UAS-Mef2 III(low) also increased the frequency of arrhythmicity (Fig. 1B, Table 1). Strikingly, 82% of flies expressing UAS-Mef2 III(high) in all clock neurons were arrhythmic, which supports the idea that the more strongly Mef2 is expressed, the greater the disruption to circadian rhythms. The most significant period of the rhythmic flies with the tim(UAS)-Gal4 driver was longer than in wild-type flies (25.1–25.6 h, Table 1), except for UAS-Mef2 III(high) where the high incidence of arrhythmicity and weak power rhythms prevented a reliable assessment of period length. In summary, expression of individual Mef2 isoforms in clock neurons causes phenotypes similar to those caused by Mef2EP1751 and there were no clear differences between the four isoforms used, with the differences in phenotypes likely attributable to different levels of expression of Mef2 in clock neurons.
Mef2 is normally expressed in clock neurons
Although Mef2 is expressed in vertebrate and fly brains (Leifer et al., 1994; Ikeshima et al., 1995; Lyons et al., 1995; Lin et al., 1996; Schulz et al., 1996), its expression has not previously been reported in clock neurons. To test whether the phenotypes described above arise from altering normal Mef2 levels in clock neurons, or from ectopic expression, we used a previously described Mef2 antibody (Lilly et al., 1995) to test whether clock neurons normally produce Mef2.
Clock neurons were visualized in wild-type adult fly brains using antibodies to PDF and TIM. These experiments were performed at ZT17 (ZT = Zeitgeber time, time in a 12 h:12 h LD cycle) when TIM is mainly cytoplasmic in clock neurons. Mef2 was clearly detected in the nucleus of all of the adult clock neuron groups: the LNv principal pacemaker neurons, the LNds and the DN1–3 subgroups (Fig. 2A). There were no obvious differences in Mef2 levels between the different clock neuron groups, although Mef2 levels were lower in clock neurons than in mushroom bodies. We also detected Mef2 staining in photoreceptor cells and many non-clock-containing neurons in agreement with previous reports (Schulz et al., 1996).
Figure 2.
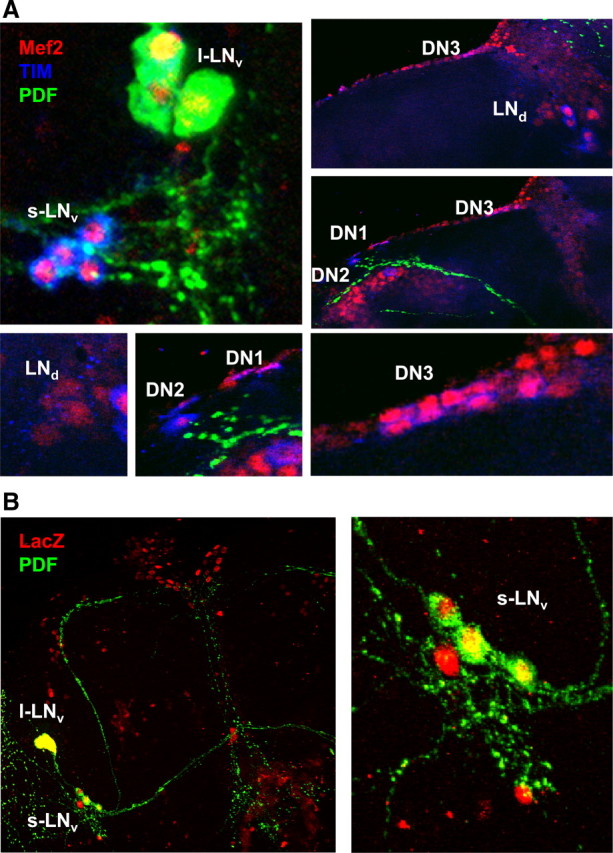
Mef2 is normally expressed in clock neurons. A, Whole-mount brains from y w flies at ZT17 stained with antibodies to Mef2 (red), TIM (blue, mainly cytoplasmic at ZT17) and PDF (green). Mef2 is detectable in the nucleus of all clock neurons groups at ZT17. B, Whole-mount brains from flies with Mef2-Gal4 and UAS-nLacZ transgenes stained with antibodies to LacZ (red) and PDF (green). The right-hand panel shows a close-up of the s-LNvs in the left panel.
To test which regulatory regions direct Mef2 expression to clock neurons, we used a Mef2-Gal4 line (Ranganayakulu et al., 1998) to express a UAS-nuclear LacZ transgene. LacZ was detected in the PDF-expressing LNvs and other brain cells, but overall LacZ was present in a much narrower range of cells than Mef2 protein (Fig. 2B). LacZ was not detected in the mushroom bodies since a more distal Mef2 enhancer is responsible for mushroom body expression (Schulz et al., 1996). Therefore Mef2-Gal4 includes regulatory information for expression in clock neurons in addition to the previously characterized enhancers that give expression in muscle and cardiac precursor cells (Gajewski et al., 1997; Cripps et al., 1998).
Next we tested whether Mef2 levels are constant or show time dependence in clock neurons. Focusing on the master s-LNv pacemaker neurons, we found significantly higher Mef2 levels toward the end of the night (ZT21) than the end of the day (ZT9, p < 0.0001; Fig. 3A,B). To test for any regulation of Mef2 by the core clock, we measured Mef2 levels in clock gene mutants. We found that Mef2 levels were lower in the s-LNvs of Clkar and cyc0 mutants than in per0 and tim01 mutants (Fig. 3A,B). We also found regulation of Mef2 levels by light and by clock genes in larval LNvs (Fig. 3C,D).
Rhythms in LD cycles and regulation by the clock suggested that Mef2 levels would also be rhythmic in DD like PER, TIM and other clock proteins. Surprisingly, we found that Mef2 levels were similar at CT9 and CT21, suggesting that Mef2 expression is inhibited by light and/or that Mef2 protein is unstable in the presence of light (Fig. 3A,B). Therefore, although Mef2 levels are altered by core gene mutations, this is not circadian regulation. We also noticed variation in Mef2 levels between s-LNvs in different brains, which could reflect a difference in exposure to light and/or other stimuli that regulate Mef2 levels. Since Mef2 levels are reduced in response to light independent of the circadian clock, Mef2 could provide a novel mechanism by which environmental information feeds into the molecular clock.
The experiments described here indicate that Mef2 is normally expressed in clock neurons. We quantified Mef2 levels in tim(UAS)-Gal4/Mef2EP1751 larval s-LNvs and found that Mef2 levels at ZT21 are approximately twice the levels in wild-type LNvs (Fig. 3E,F). Thus the altered behavioral rhythms seen in Figure 1 presumably result from overexpression of Mef2 in clock neurons rather than from ectopic expression. Since this overexpression of Mef2 alters locomotor rhythms, we propose that 24 h behavioral rhythms in wild-type flies require normal levels of Mef2 activity.
Mef2 regulates Drosophila circadian locomotor activity
Since Mef2 mutants affect many tissues and since many also cause embryonic lethality, we used the Gal4/UAS system to interfere with Mef2 function only in clock neurons in two mechanistically distinct ways. First, we used a Mef2-EnR construct, which has the MADS box and Mef2 DNA-binding and dimerization domains fused to the Engrailed transcriptional repression domain (EnR). Replacing the activation domain of transcription factors has been widely used to dominantly interfere with endogenous transcription factor function both in Drosophila (Han et al., 2002) and in mammals, including investigations of mammalian Mef2 function (Karamboulas et al., 2006; Arnold et al., 2007).
We found that 39% of flies in which UAS-Mef2-EnR was expressed via Pdf-Gal4 were arrhythmic and the remainder exhibited weaker rhythms than control flies (Table 2, Fig. 4). When UAS-Mef2-EnR was expressed using tim(UAS)-Gal4, flies had a ∼25.4 h period, although half of these flies died within the first 5 d of the assay. The high incidence of lethality is unusual in our experience and probably reflects a small amount of non-clock cell expression of the tim(UAS)-Gal4 driver. Overall, the results with UAS-Mef2-EnR indicate that downregulation of Mef2 target genes in clock neurons prevents robust circadian behavior.
Table 2.
Adult locomotor activity rhythms with UAS-Mef2-EnR and UAS-Mef2RNAi
| Genotype | Period (h) | Power | n | AR | Loss of rhythms |
|---|---|---|---|---|---|
| UAS-Mef2-EnR/+ | 23.7 ± 0.1 | 52.4 ± 5.7 | 33 | 1 | |
| Pdf-Gal4/+;Pdf-Gal4/UAS-Mef2-EnR | 23.6 ± 0.4 | 32.9 ± 6.9 | 18 | 7 | |
| tim(UAS)-Gal4/+;UAS-Mef2-EnR/+ | 25.4 ± 0.7 | 29.2 ± 7.4 | 14 | 5 | |
| UAS-Mef2RNAi/+ | 23.8 ± 0.2 | 80.7 ± 16.9 | 23 | 0 | |
| Pdf-Gal4/+;UAS-dcr-2/UAS-Mef2RNAi | 25.0 ± 0.4 | 32.6 ± 5.2 | 25 | 3 | |
| tim(UAS)-Gal4/+;UAS-dcr-2/UAS-Mef2RNAi | 23.7 ± 0.5 | 50 ± 4.7 | 36 | 25 |
Lomb–Scargle analysis of adult locomotor activity showing period (in hours) with SEM, power (to indicate the strength of the rhythm), the number of flies analyzed (n), and the numbers of either arrhythmic flies (AR) or flies that were arrhythmic from days 7 to 12 in the assay despite starting off as rhythmic.
Figure 4.
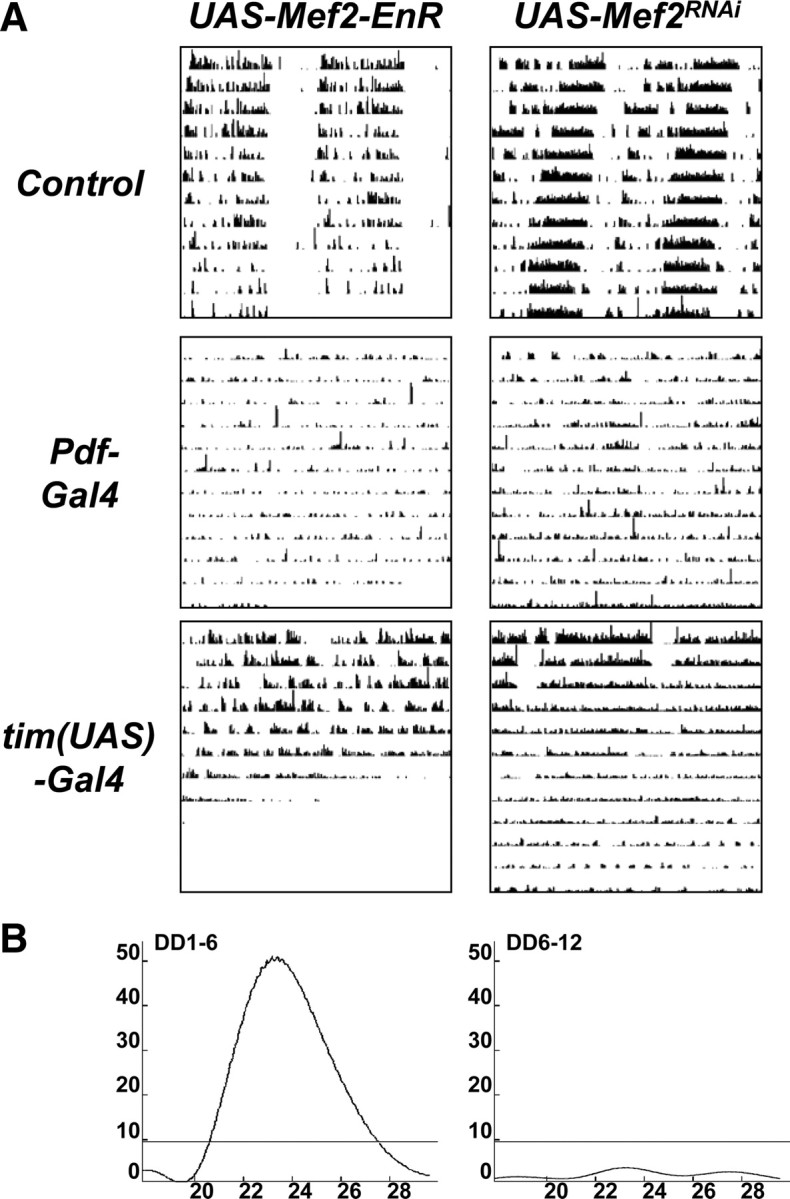
Expression of a Mef2 repressor or knocking down Mef2 expression in clock neurons disrupts circadian behavioral rhythms. A, Representative double-plotted actograms from UAS-Mef2-EnR (left panels) and UAS-Mef2RNAi (right panels) flies in DD crossed to either y w control flies (top row), Pdf-Gal4 (center panel) and tim(UAS)-Gal4 flies (bottom panels). The Pdf-Gal4 line crossed to UAS-Mef2-EnR had two copies of the Pdf0.5-Gal4 driver. The Pdf-Gal4 and tim(UAS)-Gal4 flies crossed to UAS-Mef2RNAi flies also had a UAS-Dcr-2 transgene. B, Representative Lomb–Scargle periodograms for tim(UAS)-Gal4/+; UAS-Dcr-2/UAS-Mef2RNAi flies from day 1 to 6 (left) and from day 6 to 12 (right) show the decrease in power of the rhythm in the second half of the assay.
For a second and independent way to interfere with Mef2 function, we used transgenic RNA interference (RNAi) to reduce Mef2 RNA levels and thus Mef2 activity. We used a UAS-Mef2RNAi construct in combination with UAS-Dicer-2 (UAS-Dcr-2) to knock down Mef2 expression and we confirmed that this reduced Mef2 protein levels in LNvs (Fig. 3E,F).
Pdf-Gal4/+; UAS-Dcr-2/UAS-Mef2RNAi flies have ∼25 h rhythms, with a much lower power than control flies (32.6 vs 80.7; Fig. 4A, Table 2). Expression of UAS-Mef2RNAi and UAS-Dcr-2 in all clock neurons via tim(UAS)-Gal4 driver made 70% of flies become progressively arrhythmic over a 12 d period (Fig. 4). Similar, but weaker, phenotypes were seen without UAS-Dcr-2 (data not shown). In summary, the results with expression of Mef2-EnR and Mef2RNAi in clock neurons indicate that normal levels of Mef2 activity are required to sustain circadian behavioral rhythms in DD. Although Mef2RNAi might have been expected to give the opposite of Mef2 overexpression (i.e., short period behavioral rhythms), the long period seen here may be the result of weak rhythms, which is the true phenotype. Together with the results of Mef2 overexpression described in Figure 1, and the detection of Mef2 protein in adult clock neurons in Figures 2 and 3, we conclude that normal Mef2 activity is essential for the maintenance of robust 24 h rhythms in DD.
Mef2 has a functional role in adult clock neurons
Alterations to s-LNv morphology have previously been associated with a change in the period of behavioral rhythms (Berni et al., 2008). Given the widespread roles of Mef2 in development and differentiation, we tested whether LNv morphology is altered in flies with altered Mef2 activity levels. We used antibodies to PDF to label the LNv cell bodies and projections. The results in Figure 5A indicate that adult LNvs are present and project normally when tim(UAS)-Gal4 was used to express either Mef2EP1751, Mef2-EnR or Mef2RNAi. Therefore normal levels of Mef2 are not required for correct LNv morphology at least at the level measured here. However, we did notice increased PDF levels in when tim(UAS)-Gal4 was used to express Mef2EP1751 (discussed later).
Next we tested whether overexpressing Mef2 in clock neurons in adulthood is sufficient to alter circadian behavioral rhythms. For this experiment, we used flies with a tubulin-Gal80ts transgene (tub-Gal80ts, McGuire et al., 2003) in addition to tim-(UAS)-Gal4 and Mef2EP1751 transgenes. tub-Gal80ts gives ubiquitous expression of a temperature-sensitive Gal80 protein, which represses Gal4 activity at permissive temperatures (McGuire et al., 2003). These flies were raised at the permissive temperature of 25°C and locomotor activity of adult flies was assayed in DD first at 25°C and then at the restrictive temperature of 31.5°C (Fig. 5B). Since the first time that flies were shifted to the restrictive temperature was during the locomotor assay, Gal4 activity and thus Mef2 overexpression in clock neurons would only be initiated in adulthood after clock neurons are functional.
The representative actogram in Figure 5B shows that at 25°C, tim(UAS)-Gal4/Mef2EP1751 flies with tub-Gal80ts had normal rhythms, consistent with no Mef2 overexpression. After shifting to 31.5°C, their periods lengthened, reaching an average of 26.0 ± 0.8 h. Importantly after returning to 25°C, all of the flies remained rhythmic (n = 15) and their periods shortened again, consistent with Gal80 reactivation preventing Mef2 overexpression. However, there was a residual long-period (average 24.6 ± 0.6 h), presumably due to persistence of Mef2 RNA and/or protein.
As a control, we also assayed tim(UAS)-Gal4/Mef2EP1751 flies without tub-Gal80ts with the same temperature shifts. On shifting to 31.5°C, these flies also had long periods (26.1 ± 0.7 h), but these degenerated into very weak rhythms, complex rhythms or arrhythmicity. After returning to 25°C, these flies had significantly weaker power rhythms than flies with the tub-Gal80ts transgene (p < 0.004). The periods of tim(UAS)-Gal4/Mef2EP1751 flies are longer at 31.5°C than at 25°C (Fig. 1A), whether or not they have the tub-Gal80ts transgene and this is probably due to increased Gal4 activity at higher temperatures (Duffy, 2002). Control flies with tim(UAS)-Gal4, tub-Gal80ts and UAS-nuclear LacZ had normal period lengths at both 25°C and 31.5°C (data not shown). In summary, the ability of Mef2 to lengthen period when overexpressed only in adulthood indicates that normal Mef2 levels are required in adult clock neurons for 24 h rhythms.
Altered s-LNv molecular clock oscillations in flies overexpressing Mef2
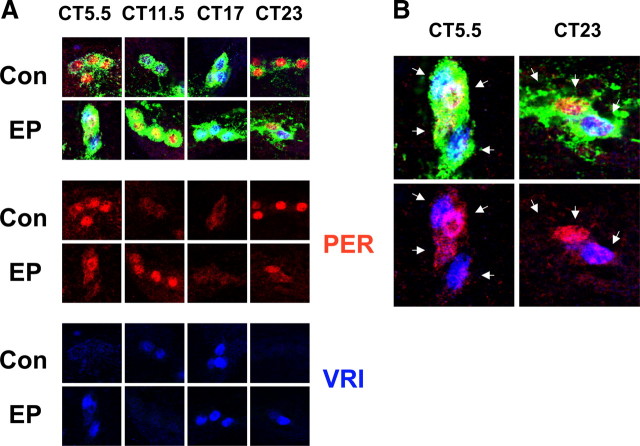
Since Mef2 affects circadian behavior, we expected changes in the molecular clocks of their master pacemaker neurons, the s-LNvs. We examined the s-LNv molecular clocks at different times of day on either the second day (DD2) or the eighth day (DD8) in DD in flies that were previously entrained to LD cycles.
In control flies, the s-LNv molecular clocks show robust oscillations at DD2 with PER at high levels at CT23 and CT7 (Fig. 6A), VRI at high levels at CT11 and CT18 (Fig. 6A), and PDP1 and TIM at high levels at CT18 and CT23 (Fig. 6B). On DD2, PER, TIM, PDP1 and VRI all show clear oscillations in the s-LNvs of tim(UAS)-Gal4/Mef2EP1751 flies, since there were always one or two time points at which a particular clock protein was undetectable (Fig. 6A,B). However, VRI was detected in all four s-LNvs in tim(UAS)-Gal4/Mef2EP1751 flies at CT23 as opposed to being undetectable in control flies at that time. Similarly PER was still detectable at CT11 in tim(UAS)-Gal4/Mef2EP1751 flies, but not in control flies. Therefore, PER and VRI disappearance is slightly delayed in tim(UAS)-Gal4/Mef2EP1751 s-LNvs relative to controls. The timing of TIM and PDP1 accumulation was similar in both tim(UAS)-Gal4/Mef2EP1751 and control flies on day 2 in DD (Fig. 6B), although levels may have been slightly lower in the latter. Overall, these data indicate that the s-LNv molecular clocks in tim(UAS)-Gal4/Mef2EP1751 flies are functional but are slightly delayed in phase compared with wild-type flies.
Figure 6.
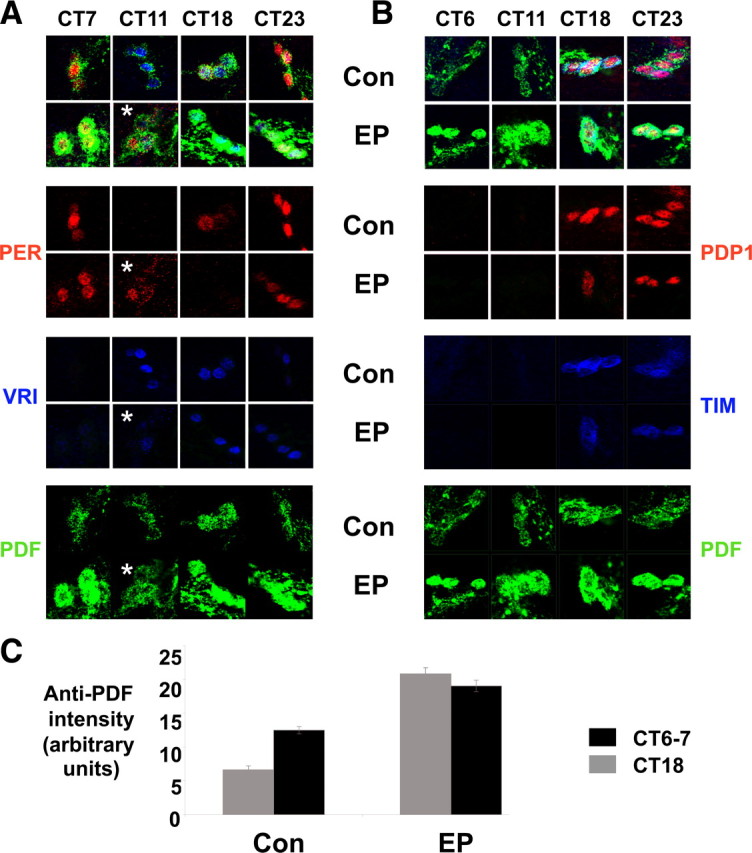
Molecular clock oscillations in s-LNvs of flies overexpressing Mef2 on day 2 in DD. A, B, Time series immunocytochemistry on whole mount brains from either Mef2EP1751/+ control flies (labeled Con) or tim(UAS)-Gal4/Mef2EP1751 flies (EP) showing s-LNvs stained with antibodies to PER (red), VRI (blue) and PDF (green) in A or PDP1 (red), TIM (blue), and PDF (green) in B. The asterisk indicates one image taken from an independent experiment from a different day. These images are representative of at least 10 brains stained in each experiment. Each experiment was performed 3 times with very similar results. C, Quantification of anti-PDF staining in s-LNv cell bodies performed at CT6 or CT7 and CT18.
We also noted that levels of PDF are ∼2-fold higher in the cell bodies of tim(UAS)-Gal4/Mef2EP1751 s-LNvs compared with control flies at both CT6/7 and CT18 on DD2 (Fig. 6C). PDF levels are also higher in the dorsal projections of tim(UAS)-Gal4/Mef2EP1751 s-LNvs (data not shown). Previous studies have shown that overexpressing Pdf in the dorsal part of the brain causes complex rhythms in flies (Helfrich-Förster et al., 2000), therefore increased PDF levels in s-LNvs could contribute to the complex rhythms found in tim(UAS)-Gal4/Mef2EP1751 flies.
The timing of PER and VRI accumulation and turnover in tim(UAS)-Gal4/Mef2EP1751 s-LNvs on day 8 in DD was also delayed compared with control flies (Fig. 7A). PER and VRI levels were either very low or undetectable at one time point (CT17 and CT11.5 respectively) but present at other time points, indicating that the molecular clocks are still oscillating. However, on closer inspection we noticed different numbers of s-LNvs with high levels of VRI staining at CT5 and CT23 in tim(UAS)-Gal4/Mef2EP1751 flies. A higher-power magnification of the three s-LNvs shown at CT23 in Figure 7B revealed that (from left to right) one s-LNv had neither PER nor VRI, another s-LNv had high levels of PER (red) but no VRI, while the third had high levels of VRI (blue) and low levels of PER. Thus the molecular clocks in the s-LNvs in tim(UAS)-Gal4/Mef2EP1751 flies have different phases from each other and/or are running at different speeds. Desynchrony within one s-LNv cluster was detected in one third of the tim(UAS)-Gal4/Mef2EP1751 brains examined across all time points (18/54), but this was very rarely seen in control brains (2/43). To measure s-LNv synchrony more precisely, we quantified PER levels in individual in one cluster and plotted the average and the distribution of a single cluster (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). We found that PER levels in the s-LNvs of tim(UAS)-Gal4/Mef2EP1751 flies tend to have a much larger SD than in control flies. These data support the conclusion that the molecular clocks in tim(UAS)-Gal4/Mef2EP1751 s-LNvs have a higher incidence of desynchrony than in control flies and this could even underlie the complex rhythms that appear in DD. The overall delayed appearance of VRI and PER in tim(UAS)-Gal4/Mef2EP1751 s-LNvs is consistent with the ∼25 h period rhythm component seen in tim(UAS)-Gal4/Mef2EP1751 flies.
Figure 7.
Desynchronized molecular clock oscillations in s-LNvs of flies overexpressing Mef2 on day 8 in DD. Time series immunocytochemistry on whole mount brains from either Mef2EP1751/+ control flies (labeled Con) or tim(UAS)-Gal4/Mef2EP1751 flies (EP). Images show s-LNvs stained with antibodies to PER (red), VRI (blue) and PDF (green). A shows stainings across 4 time points from both genotypes. B shows higher-power magnification of s-LNvs from tim(UAS)-Gal4/Mef2EP1751 flies at CT5.5 (left panels) and CT23 (right panels). The top panels show staining for PER, VRI and PDF, while the green PDF channel has been removed from these same images for the bottom panels. Arrowheads point to the same individual s-LNvs on the top and bottom. Across all time points, desynchrony within an s-LNv group was detected in 18/54 tim(UAS)-Gal4/Mef2EP1751 brains, versus 2/43 control brains. These images are representative of at least 10 brains stained in each experiment. Each experiment was performed 3 times with very similar results.
Altered molecular clock oscillations in s-LNvs of flies expressing Mef2RNAi
We also examined the effect of knocking down Mef2 expression on the s-LNv molecular clock by assaying clock protein oscillations in tim(UAS)-Gal4/+; UAS-Mef2RNAi/UAS-Dcr-2 flies. Figure 8A shows that oscillations of PER and VRI were detected in both control and tim(UAS)-Gal4/+;UAS-Mef2RNAi/UAS-Dcr-2 s-LNvs on DD2. However, VRI was not detectable at CT12, indicating that VRI accumulation is delayed in these flies relative to control flies. By DD8, PER was barely detectable in tim(UAS)-Gal4/+;UAS-Mef2RNAi/UAS-Dcr-2 s-LNvs at any time point, and VRI was only detected at high levels at one time point (CT17; Fig. 8B). Mef2 knock down had no obvious effect on PDF levels in s-LNvs. Thus expression of Mef2RNAi in all clock cells dampens the molecular oscillator in the s-LNvs, and this parallels the behavior of these flies in which flies become arrhythmic during DD (Fig. 4).
Figure 8.
Knocking down Mef2 in clock neurons dampens molecular clock oscillations in s-LNvs. A, B, Time series immunocytochemistry on whole mount brains from either UAS-Mef2RNAi/+ control flies (labeled Con) or tim(UAS)-Gal4/+; UAS-Mef2RNAi/UAS-Dcr-2-2 flies (RNAi). Images show s-LNvs stained with antibodies to PER (red), VRI (blue) and PDF (green). Time series are shown from either the second day (DD2 in A) or the eighth day in DD (DD8, B). These images are representative of at least 10 brains stained in each experiment. Each experiment was performed 3 times with very similar results.
Discussion
Mef2 is required for robust circadian rhythms
This study establishes that normal Mef2 levels and transcriptional activity are required for the maintenance of robust and accurate Drosophila circadian locomotor rhythms. This conclusion is based on: (1) flies in which Mef2 was overexpressed in clock neurons via Mef2EP1751 or UAS-Mef2 transgenes; and (2) flies with a repressor form of Mef2 (Mef2-EnR) or RNAi to knockdown Mef2 expression. All of these manipulations altered normal rhythms, and some had strong effects when targeted only to the PDF-producing LNvs. Interestingly, some of the behavioral phenotypes developed over several days e.g., the appearance of complex rhythms in Mef2EP1751 flies and the loss of rhythms in Mef2RNAi flies.
Molecular correlates of the adult behavioral alterations also developed after multiple days in DD. Mef2 overexpression in all clock neurons dramatically increased the incidence of desynchronized s-LNv molecular clocks by day 8 in DD. Since there were always time points when a particular clock protein was absent, we conclude that the s-LNv clocks in Mef2EP1751 flies still oscillate but have lost their normal tight coupling to one another. Flies in which Mef2RNAi was expressed in all clock neurons had relatively normal molecular rhythms on day 2 in DD, but strongly dampened oscillations in their s-LNv molecular clocks by day 8. These results indicate that Mef2 is probably not a typical core clock gene in which the behavioral and molecular phenotypes are obvious immediately on transferring flies to DD (e.g., Price et al., 1998). Instead, Mef2 is required to maintain robust and synchronized molecular and behavioral circadian rhythms.
A functional role for Mef2 in clock neurons
Our results show that Mef2 is normally expressed in clock neurons and its levels are regulated by both light and clock genes. Importantly, the behavioral alterations seen with altered Mef2 activity only in clock neurons indicates that wild-type Mef2 activity is required for their normal function in circadian rhythms. Although we cannot rule out subtle alterations to s-LNv morphology with manipulation of Mef2 activity, the persistence of molecular oscillations in the s-LNvs on day 2 in DD in flies in which Mef2 expression was knocked down via Mef2RNAi indicates that these cells are largely functional. Furthermore, since overexpressing Mef2 in adult clock neurons after they have developed was sufficient to lengthen period, we conclude that Mef2 has an important regulatory role in adult pacemaker neurons. Below, we describe three possibilities for the normal role of Mef2 in clock neurons that are not mutually exclusive.
One explanation for the period alterations seen with Mef2 overexpression is that Mef2 direct target genes include one or more of the core clock genes whose altered expression can change period length. Mef2 has been reported to work in concert with basic Helix-Loop-Helix (bHLH) transcription factors in muscle development (Molkentin and Olson, 1996). This is intriguing because the core clock transcriptional regulators CLK and CYC are bHLH transcription factors. Indeed the low levels of VRI and PER seen when Mef2 expression was targeted by RNAi are consistent with reduced expression of CLK/CYC-activated genes.
It is also possible that Mef2 regulates PDF synthesis, maturation and/or release, which could in turn affect period length and/or the coherence of rhythms. Although simple overexpression of PDF in LNvs does not affect the overall strength of behavioral rhythms (Helfrich-Förster et al., 2000), complex rhythms have been observed in sine oculis mutants in which PDF levels are increased in LNvs (Wülbeck et al., 2008). Complex rhythms were also observed by Helfrich-Förster et al. (2000) when Pdf was ectopically expressed in the dorsal part of the brain, although this is likely from non-clock neurons that continuously release PDF and therefore disrupt the normal s-LNv-mediated rhythm in PDF signaling. The desynchrony of the s-LNv molecular clocks when Mef2 is overexpressed could also arise from altered PDF levels since PDF is required for s-LNvs to maintain synchrony (Lin et al., 2004).
A third possibility is that Mef2 regulates period length and rhythm coherence by altering the excitability of clock neurons and/or by affecting communication between clock neurons. This idea is based on the similarities in the behavioral phenotypes and increased PDF levels that we observe with Mef2 overexpression in all clock neurons and those observed by Nitabach et al. (2006) with LNv hyper-excitation via NaChBac, a bacterial voltage-gated Na+ channel. Indeed, a growing body of evidence indicates that events at the membrane of pacemaker neurons in both flies and mammals are intimately tied to their molecular clocks (Harmar et al., 2002; Nitabach et al., 2002; Lundkvist et al., 2005; Liu et al., 2007). A role for Mef2 in cellular communication in Drosophila is consistent with Mef2 regulating synapse formation in rat hippocampal and cerebellar neurons (Flavell et al., 2006; Shalizi et al., 2006). In hippocampal neurons, Ca2+ influxes in response to neuronal activity activate Calcineurin, which dephosphorylates and activates Mef2. In turn, increased Mef2 activity leads to reduced synapse numbers (Flavell et al., 2006). Although it is not known whether Drosophila Mef2 activity can be regulated in this manner, normal Ca2+ levels in LNvs are required for 24 h rhythms (Harrisingh et al., 2007). Since mammalian circadian pacemaker neurons show daily changes in Ca2+ levels (Colwell, 2000; Ikeda et al., 2003), it is conceivable that daily changes in Ca2+ levels in LNvs could influence Mef2 activity. Mef2 could even contribute to the recent description of circadian rhythms in the branching patterns of s-LNvs (Fernández et al., 2008). In the long-run, it will be important to identify the set of genes regulated by Mef2 in s-LNvs, uncover their function, and thus to understand how Mef2 contributes to robust circadian behavior. It will also be interesting to test a role for mammalian Mef2 in pacemaker neurons in the SCN.
Footnotes
This investigation was conducted in a facility constructed with support from Research Facilities Improvement Grant Number C06 RR-15518-01 from National Center for Research Resources, National Institutes of Health (NIH), and the work was supported by NIH Grant GM063911 to J.B. We are very grateful to Cyrille Alexandre, Ravi Allada, Jeff Hall, Zhe Han, Paul Hardin, Javier Morante, Hanh Nguyen, Eric Olson, Jae Park, Bruce Paterson, Michael Rosbash, Mike Young, the Bloomington Stock Center, the Vienna Drosophila RNAi Center, and the Developmental Studies Hybridoma Bank at the University of Iowa for supplying us with DNA, antibodies, and fly strains. We thank Meg Younger and Monica Dandapani for their help with the genetic screen. We also thank Rich Cripps, Claude Desplan, and Hanh Nguyen for helpful discussions and David Dahdal, Marc Ruben, and Javier Morante for comments on the manuscript.
References
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Berni J, Beckwith EJ, Fernández MP, Ceriani MF. The axon-guidance roundabout gene alters the pace of the Drosophila circadian clock. Eur J Neurosci. 2008;27:396–407. doi: 10.1111/j.1460-9568.2007.06010.x. [DOI] [PubMed] [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Bour BA, O'Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol. 2009;10:386–390. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS. Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur J Neurosci. 2000;12:571–576. doi: 10.1046/j.1460-9568.2000.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps RM, Black BL, Zhao B, Lien CL, Schulz RA, Olson EN. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science. 2009;324:105–108. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Fernández MP, Berni J, Ceriani MF. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 2008;6:e69. doi: 10.1371/journal.pbio.0060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski K, Kim Y, Lee YM, Olson EN, Schulz RA. D-mef2 is a target for Tinman activation during Drosophila heart development. EMBO J. 1997;16:515–522. doi: 10.1093/emboj/16.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Gunthorpe D, Beatty KE, Taylor MV. Different levels, but not different isoforms, of the Drosophila transcription factor DMEF2 affect distinct aspects of muscle differentiation. Dev Biol. 1999;215:130–145. doi: 10.1006/dbio.1999.9449. [DOI] [PubMed] [Google Scholar]
- Han Z, Fujioka M, Su M, Liu M, Jaynes JB, Bodmer R. Transcriptional integration of competence modulated by mutual repression generates cell-type specificity within the cardiogenic mesoderm. Dev Biol. 2002;252:225–240. doi: 10.1006/dbio.2002.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN. Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J Neurosci. 2007;27:12489–12499. doi: 10.1523/JNEUROSCI.3680-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C, Täuber M, Park JH, Mühlig-Versen M, Schneuwly S, Hofbauer A. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J Neurosci. 2000;20:3339–3353. doi: 10.1523/JNEUROSCI.20-09-03339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen CN. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 2003;38:253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Ikeshima H, Imai S, Shimoda K, Hata J, Takano T. Expression of a MADS box gene, MEF2D, in neurons of the mouse central nervous system: implication of its binary function in myogenic and neurogenic cell lineages. Neurosci Lett. 1995;200:117–120. doi: 10.1016/0304-3940(95)12092-i. [DOI] [PubMed] [Google Scholar]
- Karamboulas C, Swedani A, Ward C, Al-Madhoun AS, Wilton S, Boisvenue S, Ridgeway AG, Skerjanc IS. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J Cell Sci. 2006;119:4305–4314. doi: 10.1242/jcs.03185. [DOI] [PubMed] [Google Scholar]
- Leifer D, Golden J, Kowall NW. Myocyte-specific enhancer binding factor 2C expression in human brain development. Neuroscience. 1994;63:1067–1079. doi: 10.1016/0306-4522(94)90573-8. [DOI] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin X, Shah S, Bulleit RF. The expression of MEF2 genes is implicated in CNS neuronal differentiation. Brain Res Mol Brain Res. 1996;42:307–316. doi: 10.1016/s0169-328x(96)00135-0. [DOI] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundkvist GB, Kwak Y, Davis EK, Tei H, Block GD. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neurosci. 2005;25:7682–7686. doi: 10.1523/JNEUROSCI.2211-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci U S A. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, Griffith LC. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Elliott DA, Harvey RP, Olson EN. Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development. 1998;125:3037–3048. doi: 10.1242/dev.125.16.3037. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rørth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci U S A. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz RA, Chromey C, Lu MF, Zhao B, Olson EN. Expression of the D-MEF2 transcription in the Drosophila brain suggests a role in neuronal cell differentiation. Oncogene. 1996;12:1827–1831. [PubMed] [Google Scholar]
- Shalizi A, Gaudillière B, Yuan Z, Stegmüller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Taylor MV, Beatty KE, Hunter HK, Baylies MK. Drosophila MEF2 is regulated by twist and is expressed in both the primordia and differentiated cells of the embryonic somatic, visceral and heart musculature. Mech Dev. 1995;50:29–41. doi: 10.1016/0925-4773(94)00323-f. [DOI] [PubMed] [Google Scholar]
- Van Dongen HP, Olofsen E, VanHartevelt JH, Kruyt EW. A procedure of multiple period searching in unequally spaced time-series with the Lomb-Scargle method. Biol Rhythm Res. 1999;30:149–177. doi: 10.1076/brhm.30.2.149.1424. [DOI] [PubMed] [Google Scholar]
- Wülbeck C, Grieshaber E, Helfrich-Förster C. Pigment-dispersing factor (PDF) has different effects on Drosophila's circadian clocks in the accessory medulla and in the dorsal brain. J Biol Rhythms. 2008;23:409–424. doi: 10.1177/0748730408322699. [DOI] [PubMed] [Google Scholar]