Abstract
Background
Methylation profiling of tumor suppressor gene (TSGs) promoters is quickly becoming a powerful diagnostic tool for the early detection, prognosis, and even prediction of clinical response to treatment. Few studies address this in salivary gland tumors (SGTs); hence the promoter methylation profile of various TSGs was quantitatively assessed in primary SGT tissue to determine if tumor-specific alterations could be detected.
Methodology
DNA isolated from 78 tumor and 17 normal parotid gland specimens was assayed for promoter methylation status of 19 TSGs by fluorescence-based, quantitative methylation-specific PCR (qMSP). The data were utilized in a binary fashion as well as quantitatively (using a methylation quotient) allowing for better profiling and interpretation of results.
Principal Findings
The average number of methylation events across the studied genes was highest in salivary duct carcinoma (SDC), with a methylation value of 9.6, compared to the normal 4.5 (p<0.0003). There was a variable frequency and individual methylation quotient detected, depending on the TSG and the tumor type. When comparing normal, benign, and malignant SGTs, there was a statistically significant trend for increasing methylation in APC, Mint 1, PGP9.5, RAR-β, and Timp3.
Conclusions/Significance
Screening promoter methylation profiles in SGTs showed considerable heterogeneity. The methylation status of certain markers was surprisingly high in even normal salivary tissue, confirming the need for such controls. Several TSGs were found to be associated with malignant SGTs, especially SDC. Further study is needed to evaluate the potential use of these associations in the detection, prognosis, and therapeutic outcome of these rare tumors.
Introduction
Salivary gland tumors (SGTs) represent a diverse group of tumor types with a wide range of biological behaviors and histopathologic characteristics, which complicates their diagnosis and management [1], [2]. Approximately 40% of these tumors are malignant, and SGTs comprise about 5% of all head and neck malignancies, which equals only 0.3% of all malignant neoplasms [2], [3], [4].
The variable nature of SGTs creates difficulty in determining prognosis. Outcomes for patients with SGTs depend on the site of tumor, histology, extent of disease, completeness of surgery, and/or adjuvant radiation therapy, though there are many exceptions [5]. The classic example is adenoid cystic carcinoma where, despite thorough resection, up to 60% of patients experience locoregional or distant metastases. The median survival in the presence of distant metastases is around three years, though surprisingly, up to 10% of these patients may survive 10 years or longer with their metstases [6]. Thus, the behavior of this malignancy is quite variable and leads to much uncertainty for the patients afflicted with this disease. Even the most common ‘benign’ salivary tumor, pleomorphic adenoma, has a propensity for malignant transformation and possible recurrence.
The pathogenesis of human cancer is a heterogeneous process involving several pathways, and it has been proposed that the genotype may affect the clinical behavior and prognosis of the tumor [7], [8]. Recent studies have shown that development and progression of human malignancy is associated with accumulation of alterations in tumor suppressor genes (TSGs) and proto-oncogenes, and this appears to be true for some SGTs [9], [10], [11].
It is well documented that epigenetic alterations, such as DNA methylation and histone acetylation are important factors in human carcinogenesis. Methylation of cytosines in cytosine-guanine (CpG) islands contained within gene promoters can lead to transcriptional inactivation by blocking the RNA polymerase complex from binding to the promoter region. Inactivation of TSGs by hypermethylation of these CpG islands is a common feature of human carcinogenesis across many tumor types as it is associated with a partial or complete transcriptional block [12]. Methylation profiling of TSGs is quickly becoming a powerful diagnostic tool for the early detection, prognosis, and even prediction of clinical response to treatment of various cancers [13].
The etiology of most SGTs has not been determined, and little is known about the epigenetic alterations occurring within this class of neoplasms. Previous studies looking at the clinical significance of TSG promoter hypermethylation in SGTs used non-quantitative MSP and focused on a limited number of genes. There is only one other quantitative study done by Lee et al. that looked at promoter hypermethylation in salivary gland carcinomas using pyrosequencing, but surveyed the methylation status of only 3 genes [14]. In addition, the results from these studies have not been reproduced in a second independent study [15], [16], [17], [18]. Thus, there is a real need for more clarity regarding the association of TSG hypermethylation and SGTs, as this could lead to functional studies that could further elucidate the biology of this group of neoplasms [7]. An improved understanding of the molecular alterations associated with salivary gland tumors has the potential to improve the diagnosis, management, and outcomes seen in this patient population.
In this study we used quantitative MSP to determine the methylation frequencies and quantitative methylation values of nineteen known tumor suppressor genes in 78 SGT patients and 17 normal parotid tissue samples. Quantitative MSP has been successfully used in other tumor models and has the benefit of providing accurate and precise data regarding the level of methylation in the various tumors. Six of the genes we evaluated, including cyclin-dependent kinase inhibitor 2A (p16), stratifin 14-3-3σ, RAS-associated domain family protein 1A (RASSF1A), retinoic acid receptor β (RAR-β), death-associated protein kinase (DAPK), and O6-methylguanine-DNA-methyltransferase (MGMT), were previously shown to be aberrantly methylated in salivary gland cancer [5], [7], [8], [11], [18], [19], [20]. The remaining thirteen genes we studied have been implicated in other cancer types; these genes include absent in melanoma-1 (AIM1), adenomatous polyposis coli (APC), β-catenin, deleted in colorectal carcinoma (DCC), fragile histidine triad (FHIT), glutathione S-transferase P1 (GSTP1), hypermethylated in cancer-1 (HIC1), methylated in tumor-1 (Mint1), mismatch repair protein (MLH1), protein gene product 9.5 (PGP9.5), thrombospondin-1 (THBS1), tissue inhibitor of metalloproteinase-3 (TIMP3), and target of methylation induced silencing-1 (TMS1) [21], [22], [23], [24]. The aim of this study was to compare the promoter methylation profiles of benign SGTs (PA), and malignant SGTs (MEC, ACC, and SDC) along with normal parotid gland tissue in order to better understand the role of epigenetic silencing in salivary gland tumorigenesis.
Materials and Methods
Tissue samples
Tumor samples from 17 normal salivary gland specimens, and 78 paraffin embedded tumor specimens were obtained from patients surgically treated at the Department of Otolaryngology-Head and Neck Surgery (Johns Hopkins Medical Institution, Baltimore, MD, USA) using appropriate written informed consent obtained after approval by the Johns Hopkins Institutional Review Board. There were a total of 26 benign tumor samples and 52 malignant tumor samples. All 26 benign samples were pleomorphic adenomas (PA). The malignant samples included 17 adenoid cystic carcinomas (ACC), 18 salivary ductal carcinomas (SDC), and 17 mucoepidermoid carcinomas (MEC).
DNA extraction
Paraffin embedded tissues sections were made and microdissected to ensure tumor purity. After de-paraffinization by xylene treatment, total genomic DNA was extracted by digestion with 50 µg/ml proteinase K (Boehringer, Mannheim, Germany) in the presence of 1% SDS at 48°C overnight, followed by phenol/chloroform extraction and ethanol precipitation. Genomic DNA was eluted in low-salt Tris-EDTA (LoTE) buffer and stored at −20°C.
Bisulfite Treatment
DNA from primary tumors and normal controls were subjected to bisulfite treatment, which modifies CpG islands including those of TSG promoters, using the Epitect Bisulfite Kit from Qiagen (Valencia, California). The bisulfite-modified genomic DNA was resuspended in 120–150 uL of H2O and stored at −80°C.
Quantitative Methylation-specific PCR (qMSP)
The bisulfite-modified DNA was used as a template for fluorescence-based real-time polymerase chain reaction (PCR), as previously described [25]. In brief, we evaluated the promoter methylation profiles of TSGs by fluorescence-based quantitative methylation-specific PCR (qMSP). Methylation specific primers and probes were designed to specifically amplify the bisulfite-modified promoters of the gene of interest (Table 1).
Table 1. Primer and probe sequences for candidate tumor suppressor genes.
| Gene | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) | Probe sequence (5′Fam-3′Tamra) |
| Aim1 | CGC GGG TAT TGG ATG TTA GT | CCG ACC CAC CTA TAC GAA AA | GGG AGC GTT GCG GAT TAT TCG TAG |
| APC | GAA CCA AAA CGC TCC CCA T | TTA TAT GTC GGT TAC GTG CGT TTA TAT | CCC GTC GAA AAC CCG CCG ATT A |
| β-catenin | GGA AAG GCG CGT CGA GT | TCC CCT ATC CCA AAC CCG | CGC GCG TTT CCC GAA CCG |
| DAP-K | GGA TAG TCG GAT CGA GTT AAC GTC | CCC TCC CAA ACG CCG A | TTC GGT AAT TCG TAG CGG TAG GGT TTG G |
| DCC | TTG TTC GCG ATT TTT GGT TTC | ACC GAT TAC TTA AAA ATA CGC G | GCG CTA AAC AAA AAA ACT CCG AAA A |
| FHIT | GGG CGC GGG TTT GGG TTT TTA C | GAA ACA AAA ACC CAC CGC CCC G | AAC GAC GCC GAC CCC ACT AAA CTC C |
| GSTP1 | AGT TGC GCG GCG ATT TC | GCC CCA ATA CTA AAT CAC GAC G | CGG TCG ACG TTC GGG GTG TAG CG |
| HIC1 | GTT AGG CGG TTA GGG CGT C | CCG AAC GCC TCC ATC GTA T | CAA CAT CGT CTA CCC AAC ACA CTC TCC TAC G |
| MGMT | CGA ATA TAC TAA AAC AAC CCG CG | GTA TTT TTT CGG GAG CGA GGC | AAT CCT CGC GAT ACG CAC CGT TTA CG |
| Mint1 | ATT TTC GAA GCG TTT GTT TGG C | ACA AAA AAC CTC AAC CCC GC | GCG AAA CTC CCC TAC TCT CCA AC |
| MLH1 | CGT TAT ATA TCG TTC GTA GTA TTC GTG TTT | CTA TCG CCG CCT CAT CGT | CGC GAC GTC AAA CGC CAC TAC G |
| p16 | TTA TTA GAG GGT GGG GCG GAT CGC | GAC CCC GAA CCG CGA CCG TAA | AGT AGT ATG GAG TCG GCG GCG GG |
| PGP 9.5 | CGG CGA GTG AGA TTG TAA GGT T | GAA CGA TCG CGA CCA AAT AAA TAC | TTC GGT CGT ATT ATT TCG CGT TGC GTA C |
| RAR-β | GGG ATT AGA ATT TTT TAT GCG AGT TGT | TAC CCC GAC GAT ACC CAA AC | TGT CGA GAA CGC GAG CGA TTC G |
| RASSF1A | GCG TTG AAG TCG GGG TTC | CCC GTA CTT CGC TAA CTT TAA ACG | ACA AAC GCG AAC CGA ACG AAA CCA |
| Stratifin 14-3-3σ | GAA GGT TAA GTT GGT AGA GTA GGT CGA AC | AAC TAC TAA AAA CAA ATT TCG CTC TTC G | CTC GCC CTT CTC CAC GAC GCC |
| THBS1 | CGA CGC ACC AAC CTA CCG | GTT TTG AGT TGG TTT TAC GTT CGT T | ACG CCG CGC TCA CCT CCC T |
| Timp3 | GCG TCG GAG GTT AAG GTT GTT | CTC TCC AAA ATT ACC GTA CGC G | AAC TCG CTC GCC CGC CGA A |
| TMS1 | TTG GAG GGT AAC GGA TCG GGG C | CCC GCT ACA ACC GCC GAC CAA A | GAC TCC GAA ACG AAA CCT AAA CTC CCC |
| β-actin | TGG TGA TGG AGG AGG TTT AGT AAG T | AAC CAA TAA AAC CTA CTC CTC CCT TAA | ACC ACC ACC CAA CAC ACA ATA ACA AAC ACA |
Fluorogenic PCRs were carried out in a reaction volume of 20 µL consisting of 600 nM of each primer; 200µM of probe; 0.75 U of platinum Taq polymerase (Invitrogen, Carlsbad, CA); 200 µM of each dATP, dCTP, dGTP, and dTTP; 200nM of ROX dye for reference (Invitrogen, Carlsbad, CA); 16.6 mmol/L of ammonium sulfate; 67 mmol/L of Trizma (Sigma, St Louis, MO); 6.7 mmol/L of magnesium chloride; 10 mmol/L of mercaptoethanol; and 0.1% dimethylsulfoxide. Three microliters of treated DNA solution were used in each real-time MSP reaction. Amplifications were carried out in 384-well plates in a 7900 Sequence Detector System (Perkin-Elmer Applied Biosystems, Norwalk, CT). Thermal cycling was initiated with a first denaturation step at 95°C for 2 minutes, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. Leukocytes from a healthy individual were methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc, Beverly, MA) to generate completely methylated DNA, and serial dilutions of this DNA were used for constructing the calibration curves on each plate. Each reaction was performed in triplicate to ensure consistent results.
The TSG promoter methylation level in each sample was calculated and normalized with respect to an internal reference gene, β-actin. This measure, which we will further refer to as methylation quotient (MQ), represents the relative level of methylation in a particular sample [(gene of interest/reference gene) ×1000] and was used for direct comparison of samples.
In order for a tumor sample to be considered methylated at a specific TSG, it had to meet two specific criteria. Amplification must have been present in at least 2 of the 3 reaction wells in the triplicate run, and the mean methylation quotient must have fallen within the range of the serial standard curve dilutions. The lack of DNA contamination was verified by the absence of amplification of a distilled water negative control for each qMSP run.
Statistical Analysis
The primary objective in this study was to describe the methylation patterns of 19 tumor suppressor genes in normal tissue and four salivary gland tumor types: ACC, SDC, MEC and PA. To compare overall methylation levels by tumor type, the number of methylated genes for each patient was calculated as the sum of genes with any degree of methylation and the average value for normal samples was compared to the average for each tumor type using an analysis of variance.
Continuous distributions of qMSP ratios are usually skewed, often with a clump of zeros in the lower tail of the distribution. To determine if increasing methylation of any of the selected genes was seen across three categories of samples: normal, benign and malignant, two types of analyses were considered. To evaluate presence or absence of methylation for each gene, an exact version of the Cochran-Armitage trend test [26] was used to determine if the probability of methylation increased across the three categories. To evaluate the continuous methylation distributions of each gene across normal, benign and malignant categories, the non-parametric Cuzick test [27] for trend was used. All statistical computations were performed using the SAS system [28], StatXact [29] or R [30]. All p values reported are two sided.
Results
Clinical and Pathological Data
A total of 19 gene promoter regions were analyzed for methylation of CpG islands in 78 patients with salivary gland tumors and 17 normal salivary gland samples. The clinical and pathological characteristics of these patients, including age and smoking status, are depicted in Table 2. Of the 78 patients with salivary gland tumors, 35 were male and 43 were female and ages ranged from 12 to 85 years at the time of diagnosis. Of the 17 patients with ACC, 2 (11.8%) eventually developed local recurrence and 10 (58.8%) developed distant metastases. Of the 17 patients with MEC, 1 (5.9%) developed local recurrence and another 2 (11.8%) distant metastasis. As expected, none of the 26 patients with PA developed distant metastases, but 1 (3.8%) developed local recurrence. Of the 18 patients with SDC, 8 (44.4%) developed local recurrence and the same number developed distant metastases. The mean follow-up time for patients with malignancy (ACC, MEC, or SDC) was 3.2 years (range 0.01–18 years).
Table 2. Clinical and pathologic characteristics of patient populations.
| Category | Subcategory | Normal | PA | ACC | MEC | SDC |
| Patients, n | 17 | 26 | 17 | 17 | 18 | |
| Age, yr, median (range) | 58.7 (42–77) | 48.5 (12–74) | 58 (25–83) | 43 (15–77) | 61 (31–85) | |
| Sex, n (%) | Male | 10 (58.8%) | 9 (34.6%) | 9 (53%) | 6 (35.3%) | 11 (61.1%) |
| Female | 7 (41.2%) | 17 (65.4%) | 8 (47%) | 11 (64.7%) | 7 (38.9%) | |
| Smoking Status | Never | 3 (17.6%) | 13 (50%) | 10 (58.8%) | 11 (64.7%) | 4 (44.4%) |
| Former | 4 (23.6%) | 8 (30.8%) | 4 (23.5%) | 2 (11.8%) | 1 (5.6%) | |
| Current | 7 (41.2%) | 5 (19.2%) | 3 (17.6%) | 2 (11.8%) | 6 (33.3%) | |
| Unknown | 3 (17.6%) | - | - | 2 (11.8%) | 3 (16.7%) | |
| Salivary Gland Involvement, n (%) | Parotid | 22 (84.6%) | 5 (29.4%) | 10 (58.8%) | 16 (88.9%) | |
| Submandibular | 1 (3.8%) | 2 (11.8%) | 2 (11.8%) | 1 (5.6%) | ||
| Minor | 3 (11.5%) | 10 (58.8%) | 5 (29.4%) | 1 (5.6%) | ||
| Stage, n (%) | I | n/a | 2 (11.8%) | 9 (53%) | 0 (0%) | |
| II | n/a | 4 (23.5%) | 4 (23.5%) | 2 (11.1%) | ||
| III | n/a | 1 (5.9%) | 3 (17.6%) | 0 (0%) | ||
| IV | n/a | 7 (41.2%) | 0 (0%) | 16 (88.9%) | ||
| Unknown | n/a | 3 (17.6%) | 1 (5.9%) | 0 (0%) | ||
| Local Recurrence, n (%) | Yes | 1 (3.8%) | 2 (11.8%) | 1 (5.9%) | 8 (44.4%) | |
| No | 25 (96.2%) | 15 (88.2%) | 16 (94.1%) | 10 (55.6%) | ||
| Metastasis, n (%) | Yes | 0 (0%) | 10 (58.8%) | 2 (11.8%) | 8 (44.4%) |
PA - Pleomorphic Adenoma, ACC - Adenoid Cystic Carcinoma, MEC - Mucoepidermoid Carcinoma, SDC - Salivary Ductal Carcinoma.
Quantitative Methylation-Specific PCR in Salivary Gland Tissues
The average number of methylated genes per tissue type is shown in Table 3. As demonstrated, the number of methylated TSGs were significantly higher in SDC compared to normal salivary tissue (p<0.0003), but there was no apparent difference between normal and PA, MEC, or ACC.
Table 3. Average number of methylated genes per tumor type.
| Tissue Type | Sample Size | Average # of Methylated Genes† (Std Dev) |
| Normal | 17 | 4.53 (2.0) |
| PA | 26 | 5 (2.2) |
| ACC | 17 | 5.41 (2.4) |
| MEC | 17 | 4.47 (2.6) |
| SDC | 18 | 9.61* (3.0) |
†Calculated as any level of methylation detected within the 19 genes tested.
*significantly different than normal, p<0.0003.
The individual methylation frequency (percentage of all samples showing some degree of methylation) and mean methylation quotient (MQ as defined previously) values for the 19 TSG loci are listed in Table 4. At least one TSG locus was methylated in 77/78 tumor samples (98.7%), and a total of 73/78 tumor samples (93.6%) showed methylation at 3 or more of the loci. The methylation frequencies varied from 0 to over 100% for different TSGs in all four tumor types, and interestingly, this was also true of the normal salivary tissue.
Table 4. Frequency of positive cases [n (%)] and distribution of Methylation Quotient levels [Mean (range)] in normal parotid tissue, PA, ACC, MEC, and SDC.
| Gene | Normal (n = 17) | ACC (n = 17) | MEC (n = 17) | PA (n = 26) | SDC (n = 18) | |||||
| n (%) | Mean (range) | n (%) | Mean (range) | n (%) | Mean (range) | n (%) | Mean (range) | n (%) | Mean (range) | |
| Aim1 | 2 (11.8) | 101.5 (0–1570.9) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 3 (11.5) | 0.9 (0–14.7) | 2 (11.1) | 10.3 (0–177.7) |
| APC | 0 (0) | 0 (0-0) | 6 (35.3) | .6 (0–3.6) | 2 (11.8) | 0.4 (0–4.9) | 9 (34.6) | 39.2 (0–915.9) | 15 (83.3) | 1606.5 (0–9289.6) |
| β-catenin | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 | 0 (0-0) | 4 (22.2) | 135.9 (0–969.9) |
| DAP-K | 7 (41.2) | 4.8 (0–20.4) | 6 (35.3) | 3.2 (0–27.3) | 4 (23.5) | 1.8 (0–10.3) | 3 (11.5) | 0.5 (0–8.8) | 12 (66.7) | 148.1 (0–727.1) |
| DCC | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) |
| FHIT | 15 (88.2) | 168.0 (0–309.9) | 4 (23.5) | 19.4 (0–103.3) | 3 (17.6) | 15.7 (0–116.7) | 7 (26.9) | 15.4 (0–86.7) | 6 (33.3) | 133.6 (0–1168.5) |
| GSTP1 | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 8 (44.4) | 110.9 (0–633.7) |
| HIC1 | 17 (100) | 530.3 (182.3–1164.3) | 16 (94.1) | 461.6 (0–1150.7) | 17 (100) | 422.4 (72.8–1036.7) | 26 (100) | 907.8 (492.7–1843.0) | 18 (100) | 563.0 (162.0–1735.2) |
| MGMT | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 2 (11.1) | 117.2 (0–1248.6) |
| Mint1 | 3 (17.6) | 6.0 (0–37.5) | 9 (52.9) | 19.9 (0–111.9) | 5 (29.4) | 147.2 (0–2199.0) | 10 (38.5) | 6.4 (0–24.3) | 11 (61.1) | 2898.5 (0–44012.6) |
| MLH1 | 0 (0) | 0 (0-0) | 2 (11.8) | 1.6 (0–20.8) | 1 (5.9) | 0.3 (0–4.9) | 3 (11.5) | 7.6 (0–128.1) | 2 (11.1) | 0.8 (0–8.9) |
| p16 | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 2 (11.8) | 28.3 (0–463.3) | 1 (3.8) | 19.8 (0–514.2) | 6 (33.3) | 633.1 (0–5761.5) |
| PGP 9.5 | 3 (17.6) | 0.5 (0–4.1) | 5 (29.4) | 13.7 (0–156.8) | 2 (11.8) | 1.0 (0–13.2) | 10 (38.5) | 28.7 (0–274.0) | 16 (88.9) | 583.2 (0–2133.5) |
| RAR-β | 0 (0) | 0 (0-0) | 3 (17.6) | 8.0 (0–77.7) | 4 (23.5) | 342.0 (0–2934.0) | 2 (7.7) | 12.6 (0–313.2) | 14 (77.8) | 1587.2 (0–6284.7) |
| RASSF1A | 0 (0) | 0 (0-0) | 6 (35.3) | 87.6 (0–788.7) | 1 (5.9) | 31.8 (0–539.8) | 14 (53.8) | 103.6 (0–606.6) | 12 (66.7) | 969.7 (0–3952.4) |
| Stratifin 14-3-3σ | 17 (100) | 1407.8 (674.4–2439.8) | 17 (100) | 930.8 (0–2622.9) | 17 (100) | 1248.7 (0–2478.7) | 26 (100) | 1732.3 (607.7–6199.2) | 18 (100) | 1262.9 (440.1–2235.9) |
| THBS1 | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 0 (0) | 0 (0-0) | 2 (11.1) | 139.7 (0–1365.2) |
| Timp3 | 6 (35.3) | 6.4 (0–22.7) | 10 (58.8) | 14.7 (0–220.2) | 12 (70.6) | 10.3 (0–89.4) | 7 (26.9) | 1.7 (0–18.3) | 12 (66.7) | 220.0 (0–1274.0) |
| TMS1 | 7 (41.2) | 25.2 (0–88.9) | 9 (52.9) | 15.0 (0–189.8) | 7 (41.2) | 3.1 (0–9.4) | 9 (34.6) | 1.6 (0–9.1) | 13 (72.2) | 153.9 (0–838.6) |
*Note: Frequency of positive cases is expressed as number and (%). Distribution of MQ levels is the ratio of the methylation of the gene to β-actin ×1,000.
The TSGs promoters were variably methylated in normal salivary tissue. Ten genes (APC, GSTP, DCC, MLH1, β -Catenin, MGMT, p16, RAR-β, RASSF1A and THBS1) displayed no methylation in the 17 normal tissue samples, while 2 genes (HIC1 and Stratifin) showed methylation in all 17 normal tissue samples.
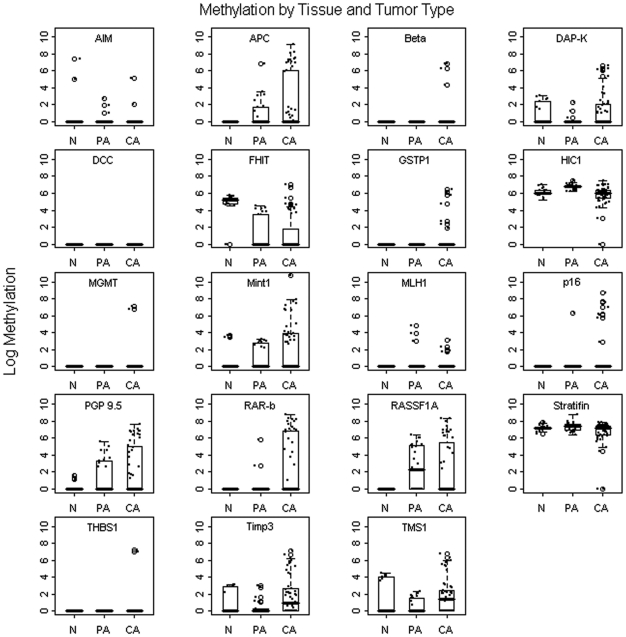
The most significantly methylated tumor type was SDC, with at least 5 loci methylated in all of the 18 samples and a total of 15 (83.3%) samples methylated at 7 or more loci. The other two types of malignant tumor tested, ACC and MEC, demonstrated less methylation than SDC. For ACC, at least three loci were methylated in 15 (88.2%) of the 17 samples but only 4 (23.5%) samples were methylated at 7 or more loci. Because of the relatively small sample size and high number of variables, we could not demonstrate statistical significance in the tumor cohorts when taken individually. Figure 1 depicts a graphic illustration of the methylation quotient distribution plots by gene and tumor type.
Figure 1. Methylation by tissue and tumor type.
Overview of the log methylation quotients of normal (N), benign pleomorphic adenoma (PA), and cancerous (CA) salivary gland tumors from the 19 tumor suppressor genes tested.
Methylation Levels Across Tumor Type
When dividing the groups into normal, benign (PA), and malignant (ACC, MEC, SDC), one would expect to see an increasing incidence of methylation or MQ across these three categories. The Cochran-Armitage test was used to test for increasing methylation frequency (in a binary fashion), and 7 genes met statistical significance (APC, GSTP1, Mint1, P16, PGP 9.5, RAR-β, and Timp3). The Cuzick test accounted for the MQ level as a continuous variable, and 8 genes met significance (APC, HIC1, Mint1, PGP 9.5, RAR-β, and Timp3). P16 and RASSF1A are interesting as they could be specific for tumors as all of the normal samples showed no methylation. However, there was methylation in benign and malignant SGTs, though they failed to meet statistical significance on both of the statistical tests (p16 was significant on the Cochran-Armitage test). Five genes showed overlapping significance in both tests (APC, Mint1, PGP9.5, RAR-β, and Timp3). Interestingly, FHIT showed a reverse correlation, with higher levels and frequency of methylation seen in the normal samples than the benign or malignant tumors. The summary data is shown in Table 5. There were no apparent correlations between any of these genes' methylation status or level and clinical or pathologic findings, including environmental factors such as age and smoking, perhaps due to the sample size limitations.
Table 5. Cochran-Armitage and Cuzick tests of trend across sample groups.
| Gene | Normal | Benign | Carcinoma | C-A | Cuzick |
| PA | ACC/MEC/SDC | p-Value | p-Value | ||
| n (%) | n (%) | n (%) | |||
| Aim1 | 2 (11.8) | 3 (11.5) | 2 (3.9) | 0.19 | 0.54 |
| APC * | 0 | 9 (34.6) | 23 (44.2) | 0.002 | 0.006 |
| β-Catenin | 0 | 0 | 4 (7.7) | 0.09 | 0.56 |
| DAP-K | 7 (41.2) | 3 (11.5) | 22 (42.3) | 0.36 | 0.42 |
| DCC | 0 | 0 | 0 | na | na |
| FHIT * | 15 (88.2) | 7 (26.9) | 13 (25) | <.001 | <.001 |
| GSTP1 | 0 | 0 | 8 (15.4) | 0.02 | 0.25 |
| HIC1 | 17 (100) | 26 (100) | 51 (98.1) | 0.41 | 0.03 |
| MGMT | 0 | 0 | 2 (3.9) | 0.24 | 0.77 |
| Mint1 * | 3 (17.7) | 10 (38.5) | 25 (48.1) | 0.03 | 0.01 |
| MLH1 | 0 | 3 (11.5) | 5 (9.6) | 0.32 | 0.66 |
| P16 | 0 | 1 (3.9) | 8 (15.4) | 0.03 | 0.28 |
| PGP 9.5 * | 3 (17.7) | 10 (38.5) | 23 (45.1) | 0.05 | 0.03 |
| RAR-β * | 0 | 2 (7.7) | 21 (40.4) | <.001 | 0.003 |
| RASSF1A | 0 | 14 (53.9) | 19 (36.5) | 0.06 | 0.06 |
| Stratifin 14-3-3σ | 17 (100) | 26 (100) | 50 (96.2) | 0.24 | 0.06 |
| THBS1 | 0 | 0 | 2 (3.9) | 0.24 | 0.77 |
| Timp3 * | 6 (35.3) | 7 (26.9) | 34 (65.4) | 0.004 | 0.02 |
| TMS1 | 7 (41.1) | 9 (34.6) | 29 (55.8) | 0.15 | 0.28 |
| Total (n) | 17 | 26 | 52 |
C-A: Cochran-Armitage tests for trend were used for binary methylation values.
Cuzick test for trend: a non-parametric test for continuous data values.
*statistically significant for both tests for increasing frequency in malignant tumors.
Discussion
Promoter methylation has emerged as one of the key mechanisms of TSG silencing in many cancers, and we sought to further elucidate its role in SGTs. The aims of this study were to identify and characterize the methylation status of a broad panel of TSG promoters in several different SGTs. In order to accomplish this, we used qMSP to evaluate a panel of 19 TSGs among true SGT malignancies (ACC, MEC, SDC), one benign SGT type (PA), as well as in normal salivary tissue.
This is the first study applying qMSP to evaluate SGT promoter hypermethylation. This is also the first study to include an extensive panel of TSGs (19 in our study), whereas most of the previous studies focused only on a single gene or a small panel of genes.
Previous studies did indicate a role for hypermethylation of TSG promoters in SGT carcinogenesis. Williams et al. evaluated 102 tumor samples and 29 normal salivary glands using non-quantitative MSP for four TSGs (DAPK, MGMT, RAR-β, and RASSF1A). This study detected hypermethylation of RAR-β (29%) and RASSF1A (48%) for SDC and hypermethylation of RASSF1A (43%) for ACC [18]. In a study by Guo et al., hypermethylation induced inactivation of the p16 gene was reported in 34.2% (13 of 38) of the MEC studied [19]. Uchida et al. demonstrated that downregulation of 14-3-3 σ via hypermethylation may be critical in the development of ACC. A similar study by Li et al., showed promoter methylation of p16 (47%), RASSF1A (41%) and DAPK (21%) in a cohort of 60 patients with ACC and indicated that RASSF1A may be linked to metastasis potential in ACC [7], [8].
Our results showed considerable heterogeneity in frequency and quantity of methylation at individual tumor suppressor genes in different SGT types. Smoking and age were not found to globally affect any methylation profiles in this cohort. We found SDC to be the most methylated tumor type, which showed significantly higher methylation frequency and level across our 19-gene panel. Interestingly, when looking at the average levels across the 19 genes tested, the normal tissue values were comparable to ACC, MEC, and PA. This finding highlights the importance of including normal controls, particularly when there are likely tissue-specific differences in methylation.
Five genes were found to have significantly increasing methylation status (frequency and level) when comparing normal, benign, and malignant salivary tissue: APC, Mint1, PGP 9.5, RAR-β, and Timp3. Of these, only RAR-β has been previously described to exhibit promoter methylation in SGTs [18]. Loss of expression of APC, or adenomatous polyposis coli, via promoter hypermethylation has been previously described in head and neck squamous cell carcinoma, bladder cancer, prostate cancer, lung cancer, and several other cancer types [31], [32], [33], [34]. This TSG encodes a large protein with multiple cellular functions including signal transduction in the Wnt-signaling pathway. In one study, there were sequence mutations of APC in 2/20 ACC cases, but promoter methylation was not studied [35].
Mint1 is a protein trafficking molecule, and its methylation has been implicated in many other tumor types [36], [37], [38], [39]. The mechanism of how it suppresses growth is not known. PGP9.5, also known as UCHL1 or ubiquitin carboxyl-terminal hydrolase L1, is a neuro-specific peptide that functions to remove ubiquitin from ubiquinated proteins and prevents them from targeted degradation by proteosomes [40]. This gene has also been implicated in the carcinogenesis of many tumor types, including head and neck squamous cell carcinoma, as well as pancreatic, lung, colorectal, and ovarian carcinomas [40], [41], [42], [43]. Timp3, or tissue inhibitor of metalloproteinases-3, has been found to inhibit angiogenesis through a VEGF mediated pathway [44], and has been found to be silenced through promoter hypermethylation in a variety of tumor types [45], [46], [47], [48]. To our knowledge, this is the first time that these three genes (Mint1, PGP 9.5, and Timp3) have been implicated in salivary gland tumorigenesis.
RASSF1A and p16 both seem to be specific for SGTs and not normal parotid tissue although they did not meet robust statistical significance. They could be further tested in a larger cohort for potential biomarker development for SGTs as none of the normal tissue samples showed promoter methylation. These two gene promoters may not have met statistical significance for differential methylation due to the limited samples examined and the relatively low frequency of methylation in these tissues. Previous reports have shown RASSF1A to ne hypermethylated and correlating with metastatic potential as well as tumor grade and 3-year survival [8], [14], [18].
While the significance of promoter methylation in cancer is a relatively recent discovery, it already has a wide range of possible clinical applications. It can serve as an excellent means of molecular detection of cancer in serum, saliva, and urine samples [49], [50]. In prostate cancer, promoter hypermethylation is an independent prognostic factor for relapse in cancer patients following radical prostatectomy [51]. In some instances, hypermethylation of certain TSG promoters predicts the response of tumors to therapy, as is the case with MGMT hypermethylation and response of primary gliomas to 1,2-bis(2-chloroethyl)-1-nitrosourea (BCNU) and temozolomide [52], [53]. It is our hope that similar findings in methylation of TSG promoters in primary SGTs might convey comparable prognostic and therapeutic implications.
In conclusion, our study is the first QMSP analysis of multiple TSGs in salivary gland tumors. The relatively high frequency and degree of methylation of some TSGs in normal salivary tissue highlights the tissue specificity of these genes, as well as the need for controlled experiments. Our results indicate that APC, Mint1, PGP 9.5, RAR-β, and Timp3 are particularly important in SGTs, and may contribute to salivary gland carcinogenesis. Further study is needed to elucidate the mechanisms by which they contribute to tumorigenesis. Larger sample cohorts are required to determine their possible roles as markers of detection or prognosis.
Footnotes
Competing Interests: Dr. Califano is the Director of Research of The Milton J. Dance Head and Neck Endowment. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: Dr. Ha is supported by the Johns Hopkins Clinician Scientist Award and the National Institute of Dental and Craniofacial Research (K08-DE018463). Dr. Sidransky is supported by the National Cancer Institute SPORE (5P50CA096784-05) and EDRN (U01CA084986). Dr. Califano is supported by the National Institute of Dental and Craniofacial Research, and the National Cancer Institute SPORE (5P50CA096784-05) and EDRN (U01CA084986). Dr. Mydlarz is supported, in part, by the National Institutes of Health (NIH) T32 training grant (T32DC000027). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Johns ME, Goldsmith MM. Incidence, diagnosis, and classification of salivary gland tumors. Part 1. Oncology (Williston Park) 1989;3:47–56; discussion 56, 58, 62. [PubMed] [Google Scholar]
- 2.Johns ME, Goldsmith MM. Current management of salivary gland tumors. Part 2. Oncology (Williston Park) 1989;3:85–91; discussion 94, 99. [PubMed] [Google Scholar]
- 3.Lalami Y, Vereecken P, Dequanter D, Lothaire P, Awada A. Salivary glands carcinomas, paranasal sinus cancers and melanoma of the head and neck: an update about rare but challenging tumors. Curr Opin Oncol. 2006;18:258–265. doi: 10.1097/01.cco.0000219255.30220.90. [DOI] [PubMed] [Google Scholar]
- 4.Zhang CY, Mao L, Li L, Tian Z, Zhou XJ, et al. Promoter methylation as a common mechanism for inactivating E-cadherin in human salivary gland adenoid cystic carcinoma. Cancer. 2007;110:87–95. doi: 10.1002/cncr.22758. [DOI] [PubMed] [Google Scholar]
- 5.Kishi M, Nakamura M, Nishimine M, Ikuta M, Kirita T, et al. Genetic and epigenetic alteration profiles for multiple genes in salivary gland carcinomas. Oral Oncol. 2005;41:161–169. doi: 10.1016/j.oraloncology.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Spiro RH, Huvos AG, Strong EW. Adenoid cystic carcinoma of salivary origin. A clinicopathologic study of 242 cases. Am J Surg. 1974;128:512–520. doi: 10.1016/0002-9610(74)90265-7. [DOI] [PubMed] [Google Scholar]
- 7.Uchida D, Begum NM, Almofti A, Kawamata H, Yoshida H, et al. Frequent downregulation of 14-3-3 sigma protein and hypermethylation of 14-3-3 sigma gene in salivary gland adenoid cystic carcinoma. Br J Cancer. 2004;91:1131–1138. doi: 10.1038/sj.bjc.6602004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, El-Naggar A, Mao L. Promoter methylation of p16INK4a, RASSF1A, and DAPK is frequent in salivary adenoid cystic carcinoma. Cancer. 2005;104:771–776. doi: 10.1002/cncr.21215. [DOI] [PubMed] [Google Scholar]
- 9.el-Naggar AK, Hurr K, Kagan J, Gillenwater A, Callender D, et al. Genotypic alterations in benign and malignant salivary gland tumors: histogenetic and clinical implications. Am J Surg Pathol. 1997;21:691–697. doi: 10.1097/00000478-199706000-00009. [DOI] [PubMed] [Google Scholar]
- 10.Cerilli LA, Swartzbaugh JR, Saadut R, Marshall CE, Rumpel CA, et al. Analysis of chromosome 9p21 deletion and p16 gene mutation in salivary gland carcinomas. Hum Pathol. 1999;30:1242–1246. doi: 10.1016/s0046-8177(99)90044-8. [DOI] [PubMed] [Google Scholar]
- 11.Nishimine M, Nakamura M, Kishi M, Okamoto M, Shimada K, et al. Alterations of p14ARF and p16INK4a genes in salivary gland carcinomas. Oncol Rep. 2003;10:555–560. [PubMed] [Google Scholar]
- 12.Esteller M. Dormant hypermethylated tumour suppressor genes: questions and answers. J Pathol. 2005;205:172–180. doi: 10.1002/path.1707. [DOI] [PubMed] [Google Scholar]
- 13.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 14.Lee ES, Issa JP, Roberts DB, Williams MD, Weber RS, et al. Quantitative promoter hypermethylation analysis of cancer-related genes in salivary gland carcinomas: comparison with methylation-specific PCR technique and clinical significance. Clin Cancer Res. 2008;14:2664–2672. doi: 10.1158/1078-0432.CCR-07-1232. [DOI] [PubMed] [Google Scholar]
- 15.Maruya S, Kim HW, Weber RS, Lee JJ, Kies M, et al. Gene expression screening of salivary gland neoplasms: molecular markers of potential histogenetic and clinical significance. J Mol Diagn. 2004;6:180–190. doi: 10.1016/s1525-1578(10)60508-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruya S, Kurotaki H, Shimoyama N, Kaimori M, Shinkawa H, et al. Expression of p16 protein and hypermethylation status of its promoter gene in adenoid cystic carcinoma of the head and neck. ORL J Otorhinolaryngol Relat Spec. 2003;65:26–32. doi: 10.1159/000068658. [DOI] [PubMed] [Google Scholar]
- 17.Maruya S, Kurotaki H, Wada R, Saku T, Shinkawa H, et al. Promoter methylation and protein expression of the E-cadherin gene in the clinicopathologic assessment of adenoid cystic carcinoma. Mod Pathol. 2004;17:637–645. doi: 10.1038/modpathol.3800104. [DOI] [PubMed] [Google Scholar]
- 18.Williams MD, Chakravarti N, Kies MS, Maruya S, Myers JN, et al. Implications of methylation patterns of cancer genes in salivary gland tumors. Clin Cancer Res. 2006;12:7353–7358. doi: 10.1158/1078-0432.CCR-06-1272. [DOI] [PubMed] [Google Scholar]
- 19.Guo XL, Sun SZ, Wang WX, Wei FC, Yu HB, et al. Alterations of p16INK4a tumour suppressor gene in mucoepidermoid carcinoma of the salivary glands. Int J Oral Maxillofac Surg. 2007;36:350–353. doi: 10.1016/j.ijom.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Augello C, Gregorio V, Bazan V, Cammareri P, Agnese V, et al. TP53 and p16INK4A, but not H-KI-Ras, are involved in tumorigenesis and progression of pleomorphic adenomas. J Cell Physiol. 2006;207:654–659. doi: 10.1002/jcp.20601. [DOI] [PubMed] [Google Scholar]
- 21.Jeronimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, et al. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 22.Ha PK, Califano JA. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006;7:77–82. doi: 10.1016/S1470-2045(05)70540-4. [DOI] [PubMed] [Google Scholar]
- 23.Fukasawa M, Kimura M, Morita S, Matsubara K, Yamanaka S, et al. Microarray analysis of promoter methylation in lung cancers. J Hum Genet. 2006;51:368–374. doi: 10.1007/s10038-005-0355-4. [DOI] [PubMed] [Google Scholar]
- 24.Mandelker DL, Yamashita K, Tokumaru Y, Mimori K, Howard DL, et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963–4968. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- 25.Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9:1370–1375. [PubMed] [Google Scholar]
- 26.Armitage P. Test for linear trend in proportions and frequencies. Biometrics. 1955;11:375–396. [Google Scholar]
- 27.Cuzick J. A Wilcoxon-type test for trend. Stat Med. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
- 28.SAS User's Guide. Version 5 ed. Cary, NC: SAS Institute Inc; 1985. [Google Scholar]
- 29.StatXact 4 for windows. Version 4 ed. Cambridge, MA: Cytel Software Corporation; 1997. [Google Scholar]
- 30.R: A language and environment for statistical computing. Vienna, Austria: Foundation for Statistical Computing; 2004. [Google Scholar]
- 31.Ellinger J, El Kassem N, Heukamp LC, Matthews S, Cubukluoz F, et al. Hypermethylation of cell-free serum DNA indicates worse outcome in patients with bladder cancer. J Urol. 2008;179:346–352. doi: 10.1016/j.juro.2007.08.091. [DOI] [PubMed] [Google Scholar]
- 32.Chen K, Sawhney R, Khan M, Benninger MS, Hou Z, et al. Methylation of multiple genes as diagnostic and therapeutic markers in primary head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2007;133:1131–1138. doi: 10.1001/archotol.133.11.1131. [DOI] [PubMed] [Google Scholar]
- 33.Henrique R, Ribeiro FR, Fonseca D, Hoque MO, Carvalho AL, et al. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin Cancer Res. 2007;13:6122–6129. doi: 10.1158/1078-0432.CCR-07-1042. [DOI] [PubMed] [Google Scholar]
- 34.Safar AM, Spencer H, 3rd, Su X, Coffey M, Cooney CA, et al. Methylation profiling of archived non-small cell lung cancer: a promising prognostic system. Clin Cancer Res. 2005;11:4400–4405. doi: 10.1158/1078-0432.CCR-04-2378. [DOI] [PubMed] [Google Scholar]
- 35.Daa T, Kashima K, Kaku N, Suzuki M, Yokoyama S. Mutations in components of the Wnt signaling pathway in adenoid cystic carcinoma. Mod Pathol. 2004;17:1475–1482. doi: 10.1038/modpathol.3800209. [DOI] [PubMed] [Google Scholar]
- 36.Ogi K, Toyota M, Ohe-Toyota M, Tanaka N, Noguchi M, et al. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin Cancer Res. 2002;8:3164–3171. [PubMed] [Google Scholar]
- 37.Toyota M, Kopecky KJ, Toyota MO, Jair KW, Willman CL, et al. Methylation profiling in acute myeloid leukemia. Blood. 2001;97:2823–2829. doi: 10.1182/blood.v97.9.2823. [DOI] [PubMed] [Google Scholar]
- 38.Chan AO, Kim SG, Bedeir A, Issa JP, Hamilton SR, et al. CpG island methylation in carcinoid and pancreatic endocrine tumors. Oncogene. 2003;22:924–934. doi: 10.1038/sj.onc.1206123. [DOI] [PubMed] [Google Scholar]
- 39.An C, Choi IS, Yao JC, Worah S, Xie K, et al. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res. 2005;11:656–663. [PubMed] [Google Scholar]
- 40.Bittencourt Rosas SL, Caballero OL, Dong SM, da Costa Carvalho Mda G, Sidransky D, et al. Methylation status in the promoter region of the human PGP9.5 gene in cancer and normal tissues. Cancer Lett. 2001;170:73–79. doi: 10.1016/s0304-3835(01)00449-9. [DOI] [PubMed] [Google Scholar]
- 41.Tokumaru Y, Yamashita K, Osada M, Nomoto S, Sun DI, et al. Inverse correlation between cyclin A1 hypermethylation and p53 mutation in head and neck cancer identified by reversal of epigenetic silencing. Cancer Res. 2004;64:5982–5987. doi: 10.1158/0008-5472.CAN-04-0993. [DOI] [PubMed] [Google Scholar]
- 42.Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, et al. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–3742. [PubMed] [Google Scholar]
- 43.Okochi-Takada E, Nakazawa K, Wakabayashi M, Mori A, Ichimura S, et al. Silencing of the UCHL1 gene in human colorectal and ovarian cancers. Int J Cancer. 2006;119:1338–1344. doi: 10.1002/ijc.22025. [DOI] [PubMed] [Google Scholar]
- 44.Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med. 2003;9:407–415. doi: 10.1038/nm846. [DOI] [PubMed] [Google Scholar]
- 45.Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 46.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 47.Ninomiya I, Kawakami K, Fushida S, Fujimura T, Funaki H, et al. Quantitative detection of TIMP-3 promoter hypermethylation and its prognostic significance in esophageal squamous cell carcinoma. Oncol Rep. 2008;20:1489–1495. [PubMed] [Google Scholar]
- 48.Hoque MO, Begum S, Brait M, Jeronimo C, Zahurak M, et al. Tissue inhibitor of metalloproteinases-3 promoter methylation is an independent prognostic factor for bladder cancer. J Urol. 2008;179:743–747. doi: 10.1016/j.juro.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60:892–895. [PubMed] [Google Scholar]
- 50.Hoque MO, Begum S, Topaloglu O, Chatterjee A, Rosenbaum E, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 51.Rosenbaum E, Hoque MO, Cohen Y, Zahurak M, Eisenberger MA, et al. Promoter hypermethylation as an independent prognostic factor for relapse in patients with prostate cancer following radical prostatectomy. Clin Cancer Res. 2005;11:8321–8325. doi: 10.1158/1078-0432.CCR-05-1183. [DOI] [PubMed] [Google Scholar]
- 52.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 53.Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]