Abstract
Familial combined hyperlipidemia (FCHL) is a complex trait leading to cardiovascular disease (CVD) risk. Elevated levels and size of apolipoprotein B (apoB) and low-density lipoprotein (LDL) are associated with FCHL, which is genetically heterogeneous and is likely caused by rare variants. We carried out a linkage-based genome scan of four large FCHL pedigrees for apoB level that is independent of LDL: apoB level that is adjusted for LDL level and size. Follow-up included SNP genotyping in the region with the strongest evidence of linkage. Several regions with the evidence of linkage in individual pedigrees support the rare variant model. Evidence of linkage was strongest on chromosome 4q, with multipoint analysis in one pedigree giving LOD = 3.1 with a parametric model, and a log Bayes Factor = 1.5 from a Bayesian oligogenic approach. Of the 293 SNPs spanning the implicated region on 4q, rs6829588 completely explained the evidence of linkage. This SNP accounted for 39% of the apoB phenotypic variance, with heterozygotes for this SNP having a trait value that was ~30% higher than that of the high-frequency homozygote, thus identifying and considerably refining a strong candidate region. These results illustrate the advantage of using large pedigrees in the search for rare variants: reduced genetic heterogeneity within single pedigrees coupled with the large number of individuals segregating otherwise-rare single variants leads to high power to implicate such variants.
Introduction
Familial combined hyperlipidemia (FCHL) is one of the most common genetic causes of hyperlipidemia (Breslow 2000; Eurlings et al. 2001; Zambon et al. 2006). It was first described in the Seattle Myocardial Infarction Study in 1973 (Goldstein et al. 1973), where at least 11% of individuals who were myocardial infarction (MI) survivors and less than 60 years of age had FCHL. The disease was initially characterized by variable lipoprotein phenotype (increased triglycerides, TG, and/or increased cholesterol, TC) both in the proband and in the relatives of the proband, such that these lipid levels could also vary within an individual over time. Lipid variables, including apoB level, LDL level, and LDL size have been identified as part of the phenotype (Ayyobi et al. 2003; Breslow 2000; Jarvik et al. 1994; Pauciullo et al. 2009). FCHL has been estimated to contribute to ≥20% of coronary artery disease (CAD) in males under the age of 60 years, and presumably the same percentage in women 10 years older (Babirak et al. 1992; Brown et al. 1990; Zambon et al. 2006). In a more recent study, 38% of subjects with MI age <40 years were found to have FCHL: a 24-fold risk (Wiesbauer et al. 2009).
FCHL is genetically heterogeneous (Babirak et al. 1989, 1992; Bosse et al. 2004b; Goldstein et al. 1975; Shoulders et al. 2004; Wierzbicki et al. 2008). Such heterogeneity complicates the identification of underlying genes, regardless of the study design. However, two strategies can mitigate the effects. The first involves the use of quantitative traits since it is well known that the use of quantitative traits underlying a trait is a more powerful strategy than the use of dichotomized phenotypes based on the same data (Wijsman and Amos 1997) in the search of genetic contributions to a trait, with such higher power necessary to overcome the effects of the heterogeneity. The second strategy focuses on individual rather than joint component phenotypes, under the assumption that this allows the definition of individual aspects of the pathway leading to the phenotype, thereby simplifying the mode of inheritance, and thus reducing the problem of heterogeneity without the complexity of a multivariate analysis. Toward this goal of gene identification in FCHL, genetic defects in lipoprotein lipase (LpL) (Babirak et al. 1989, 1992), hepatic lipase (Pajukanta et al. 1997), the APOA1/C3/A5 gene cluster (Wojciechowski et al. 1991), and upstream transcription factor 1 (USF1) (Pajukanta et al. 2004) have been implicated a minority of FCHL. In these studies, little evidence was found for apoB level, especially after adjustment for the lipid levels. In addition, there is stronger evidence for Mendelian segregation of related continuous lipid phenotypes, including apoB level, HDL level, and LDL density, than for FCHL as a dichotomous phenotype (Austin et al. 1990; Badzioch et al. 2004; Bredie et al. 1996; Vakkilainen et al. 2002). Finally, each of these component traits is a well-established risk factor for atherosclerosis in subjects with and without FCHL.
The search for genetic contributions to complex traits can be achieved with a number of different study designs. Pedigree-based designs are well established, and range from the use of small nuclear to large extended families. Such designs are the basis behind identification of large numbers of genomic regions that have been implicated in lipid variation (Bosse et al. 2004a; Dastani et al. 2006; Eurlings et al. 2001; Wang and Paigen 2005). Whole genome-wide association studies (GWAS), based on the use of unrelated individuals, have also recently become common and have identified a number of potential low-penetrance polymorphisms that may be associated with serum lipids (Kathiresan et al. 2008, 2009; Kooner et al. 2008; Willer et al. 2008). However, GWAS studies are not designed to identify rare variants that cause larger lipid effects, as illustrated by the large number of rare variants in LDLR that cause familial hypercholesterolemia (Day et al. 1997; Fouchier et al. 2001) or in the lipoprotein lipase gene (Brunzell and Deeb 2000; Nickerson et al. 1998). It is also likely that rare polymorphisms with large effects are more likely than common variants to cause heart attacks and strokes, leading causes of death in the developed world. In the search for rare variants for a complex trait with multiple contributing loci, the use of pedigree-based designs based on large pedigrees is particularly advantageous since such pedigrees have sufficient power, individually, for gene localization, unlike study designs based on either small pedigrees or unrelated individuals, which require combining information across many unrelated individuals. In addition, the use of single large pedigrees also tends to reduce the problem of genetic heterogeneity within individual pedigrees, thus further increasing power to localize genes.
ApoB level is a risk factor for FCHL that has to date yielded few clues regarding the underlying genetic basis. ApoB levels are disproportionately elevated in FCHL compared to age, sex, and weight-matched individuals for the degree of insulin resistance (Brunzell et al. 1995; Fujimoto et al. 1994; Purnell and Brunzell 1997; Zambon and Brunzell 1993) and obesity (Purnell et al. 1997). Reductions of apoB (Brown et al. 1990) or increase in LDL density (Zambon et al. 1999) were the best predictors of regression of CAD in a treatment trial. FCHL is characterized by increased numbers of small very-low-density lipoprotein (VLDL) (Brunzell et al. 1983) and small, dense LDL (Hokanson et al. 1993), and VLDL apoB secretion and LDL apoB turn-over are altered in FCHL (Chait et al. 1980; Venkatesan et al. 1993). ApoB levels are thus correlated with both LDL size and density. However, since apoB level is an independent predictor of FCHL over both LDL size (Jarvik et al. 1994) and level (Pauciullo et al. 2009), a portion of variation in apoB level may be influenced by variation in genes that do not directly affect these other two traits. Thus, genetic loci controlling apoB level independently of LDL size and level may also be etiologic in FCHL. Consistent with this, apoB level shows strong evidence for a genetic basis (Coresh et al. 1993; Hasstedt et al. 1987; Jarvik et al. 1993; Pairitz et al. 1988). Additionally, elevated apoB levels may be responsible for a larger portion of cardiovascular disease (CVD) than that in patients recognized as having FCHL (Brown et al. 1990; Zambon et al. 2006). Notwithstanding a few examples of Mendelian inheritance (Neuman et al. 2002; Schonfeld et al. 2005; Yuan et al. 2000), however, the relative paucity of studies that report loci contributing to the inheritance of apoB level as well as the diverse, non-overlapping locations of the few such loci reported to date (Allayee et al. 2002; Cantor et al. 2004; Feitosa et al. 2005; Sherva et al. 2007; Yuan et al. 2000) suggests that apoB level may be affected largely by rare variants in multiple genes.
Here, we address the genetic basis of apoB level in families with FCHL. Our general strategy is the identification of rare variants through the use of modern linkage methods that accommodate multilocus trait models, combined with use of large pedigrees that maximize power to detect such variants within individual pedigrees. The families in use show evidence of inheritance of high-penetrance hyperlipidemia (Jarvik et al. 1994). A subset have, individually and jointly, previously provided evidence for relatively simple inheritance of HDL (Gagnon et al. 2003, 2005) and LDL size (Badzioch et al. 2004), supporting the hypothesis that rare variants with relatively large effect are segregating in these families. We focus here on apoB level that is independent of LDL size and level, and we present strong evidence for the existence of rare variants segregating in these families, with strong evidence for a locus on chromosome 4 that is additionally supported by association with SNP genotypes.
Methods
Subjects and phenotypes
Pedigrees
The available pedigrees consisted of four large pedigrees, as described elsewhere (Badzioch et al. 2004; Gagnon et al. 2003, 2005; Goldstein et al. 1973; Jarvik et al. 1994; Wijsman et al. 1998) and summarized in Table 1. In brief, this data set comprises four families ascertained through a proband with FCHL. The ascertainment event for families F1 and F2 was early myocardial infarction (Goldstein et al. 1973), and for families F3 and F4 was presence of severe hypertriglyceridemia in the proband. In family F2, an extension of the pedigree ascertainment in a second individual was also through the most extreme TG values (TG < 1,000 mg/ml) (Brunzell et al. 1983; Chait et al. 1980) with combined hyperlipidemic patterns in the families of both parents. For purposes of the analyses reported here, the size range of these pedigrees was 48–87 individuals in 4–6 generations, for a total of 253 individuals.
Table 1.
Sample and data characteristics of pedigrees used for analysis
| Pedigree |
|||||
|---|---|---|---|---|---|
| All | F1 | F2 | F3 | F4 | |
| Markers | |||||
| N | 253 | 63 | 48 | 87 | 55 |
| Obs | 198 | 49 | 39 | 68 | 42 |
| MGS | 213 | 55 | 41 | 73 | 44 |
| DCD | 149 | 37 | 35 | 60 | 17 |
| SNP | 148 | 38 | 34 | 54 | 22 |
| apoB-adj [mean (SD)] | –0.16 (17.9) | –5.63 (12.57) | 8.00 (18.96) | 0.30 (18.25) | –2.08 (18.62) |
| apoB-raw [mean (SD)] | 116.1 (35.5) | 115.8 (31.1) | 132.7 (39.0) | 112.5 (34.2) | 107.0 (34.1) |
| LDL [mean (SD)] | 120.7 (39.7) | 127.4 (35.3) | 132.3 (44.5) | 114.5 (39.9) | 112.1 (35.2) |
| LDL PPD [mean (SD)] | 260.8 (9.9) | 260.2 (9.2) | 260.3 (6.3) | 260.3 (11.7) | 262.9 (9.8) |
| BMI [mean (SD)] | 24.5 (5.1) | 24.0 (4.6) | 25.1 (5.5) | 24.3 (4.8) | 24.6 (5.7) |
N total number in pedigree; Obs total number with observed, adjusted apoB phenotype; MGS number of genotyped individuals in STR genome-scan; DCD number of genotyped individuals for STR follow-up markers; SNP number of genotyped individuals for SNP markers; apoB-adj apoB adjusted for covariates; apoB-raw original unadjusted apoB measurements
Phenotypes
We used apoB level, adjusted for covariates, as the primary variable of interest. Covariates used for adjustment included body mass index (BMI), age, gender, and two quantitative traits that are correlated with traits that define FCHL: LDL size (peak particle diameter, PPD) and LDL-C level. Covariate adjustments were determined by linear regression, using the complete sample but without specifically accounting for relationships among individuals. Inclusion of relatives in such an analysis affects the variance but not the expectation of parameter estimates (Durbin and Watson 1950; Tregouet et al. 1997), and so should have little effect on the resulting parameter values used for adjusting apoB levels. Residuals were used for further genetic analysis, after extreme values in two subjects were Winsorized (Dixon and Tukey 1968; Rivest 1994) to 4 SD from the mean. Such Winsorization has previously been shown to improve the robustness of results of genetic analysis (Igo et al. 2006) since it retains the existence of extreme values without such values being unduly influential.
Standard methods were used to determine plasma apoB (Warnick 1986) and LDL cholesterol (Warnick et al. 1990) levels, measured in mg/dl. LDL size, measured as peak particle diameter (PPD) in nanometers, was determined by gradient gel electrophoresis of whole plasma (Jarvik et al. 1994; Krauss and Burke 1982) with PPD measurements normalized to the same internal standards. BMI was computed using self-reported height and weight.
Genotyping
A total of 213 individuals were genotyped by the NHLBI Mammalian Genotyping service (MGS), using genome screening set 9, which comprised a total of 387 highly polymorphic STR markers spaced at ~10 cM intervals. This set of markers will henceforth be referred to as the MGS scan. Dense STR follow-up genotypes were obtained on 247 individuals for key regions from deCode (DCD markers), including samples for which whole-genome amplification was used to augment scarce samples. Finally, the Illumina HumanCVD Bead chip, consisting of a cardiovascular gene-centric SNP panel comprised of 48,742 SNPs, was used to provide dense SNP genotyping on 147 individuals. For key regions of interest as determined by the STR linkage analyses, the SNPs were used for further analyses to narrow the regions of interest, as described below. SNP genotypes were not used for genome-scan analyses for three reasons. First, there was already evidence from the STR analyses that no further information for linkage detection was expected by adding more markers in key regions. Second, computational demands on these large pedigrees would have been extreme (Wijsman et al. 2006; Wilcox et al. 2005). Third, the MCMC-based methods used (see below) are the only available approaches that allow the analysis of large pedigrees with large numbers of markers, but do not account for linkage disequilibrium (LD) among markers. It is necessary to account for LD in order to avoid the undesirable effect of inflated evidence of linkage that occurs when LD is not taken into account (Huang et al. 2004; Sieh et al. 2007).
Data cleaning and quality control procedures are described elsewhere (Gagnon et al. 2005). The average percentage of MGS markers for which genotypes were obtained on an individual was 98.4%. For the DCD markers, genotypes for 26 individuals were removed from analysis because joint consideration of low genotype completion rates (<90%) and/or high homozygosity rates (>35%) suggested incomplete amplification of their DNA and thus unreliable genotypes. These 26 subjects represented older DNA samples that had been subjected to whole-genome amplification. For the SNP markers, any cases of Mendelian inconsistencies in the pedigrees were resolved by deleting genotypes from the parents and all descendents of the parents in such pedigrees.
Statistical analysis
General strategy
Because of the concerns of probable underlying genetic heterogeneity, the pedigrees were analyzed in two groupings. First, all pedigrees were analyzed jointly to identify trait models for model-based analyses. Pedigrees were also analyzed, jointly, in a genome scan to identify regions with evidence of linkage in the full data set. Second, genome-scan linkage analyses were carried out on individual pedigrees since each pedigree was sufficiently large to provide evidence of linkage as an individual sample. This separate set of analyses on individual pedigrees allowed a deeper exploration of possible genetic heterogeneity among pedigrees than would be possible with only a joint analysis, including the possibility of locus heterogeneity, with different loci segregating in different families, and allelic heterogeneity, leading to different inheritance patterns at the same locus in separate pedigrees. Finally, additional markers and analyses were added to the genome-scan results, and used to refine results in the regions of greatest interest.
Trait model
Both complex segregation analysis (CSA) and Bayesian oligogenic segregation analysis (OSA) were used for trait model estimation. PAP, v. 5 (http://hasstedt.genetics.utah.edu) (Hasstedt and Cartwright 1981) was used to maximize the likelihood under different possible models for CSA, using combinations of models containing possible major gene, polygenic, and environmental effects, as well as estimation of the major-gene heterozygous transmission probability. Likelihood ratio (LR) tests were used for model comparison under the assumption of asymptotic equivalence to a chi-square distribution with degrees of freedom as the difference in number of free parameters between nested models. In the context of multiple segregating loci with sizeable effects, the single-locus model that can be modeled via CSA may not identify all the underlying major-gene models (Igo et al. 2006). In this context, comparison of the model obtained by CSA with the posterior distribution of models obtained from OSA with the Bayesian approach implemented in Loki (Heath 1997) can provide useful insights into the model complexity and parameters for additional models (Igo et al. 2006). Therefore, we also compared the results obtained with CSA to those obtained from posterior model distributions with OSA, and also with the posterior model distributions for the models with location parameters on chromosomes with evidence of linkage in the Bayesian oligogenic joint segregation and linkage analyses, as described further below.
Oligogenic segregation and linkage analysis
Bayesian oligogenic segregation analysis with and without joint linkage analysis was carried out with the program Loki, ver. 2.4.7 (Heath 1997), with analysis details as described elsewhere (Wijsman and Yu 2004). In brief, this approach uses prior distributions coupled with reversible-jump (Green 1995) MCMC-based sampling to obtain posterior distributions of parameters of interest, including parameters for models with variable dimensionality, including the number of diallelic quantitative trait loci (QTLs) affecting the trait. For analyses, we used 1,000 sweeps for burn-in, 100,000 sweeps for analysis (unless otherwise noted), and a thinning interval of 2, where a sweep is one round of updating all relevant parameters. For prior distributions, we assumed a Poisson distribution with mean = 2 for the number of QTLs and with a maximum of 17 QTLs, and a prior variance of 1,280 on the genotypic effects. This provided a relatively uniform, and therefore uninformative, prior distribution on the genotypic effects. The posterior probability (PP) of a trait model was computed as the proportion of all QTL models in a run with a particular bivariate distribution of genotype effects, defined by a ~99% confidence interval around the mean for each genotype effect. Evidence of linkage was evaluated with the Bayes’ factor (BF) for linkage (Wijsman and Yu 2004), which is the ratio of the posterior to the prior odds of linkage, with results presented on a base 10 logarithmic scale. Calibration of log10 BF as supporting evidence of linkage is approximate, but log10 BF > 1.5 corresponds to strong evidence and log10 BF > 2 to very strong evidence in favor of the alternative hypothesis of linkage (Kass and Raftery 1995). In addition, in our experience, most cases with log BF > 1.5 based on a prior mean = 2 for the number of QTLs appear to be equivalent to p < 0.001, either by reference to a different calibrated linkage statistic, or by carrying out computationally expensive simulations to provide calibration (Igo and Wijsman 2008). Genome-scan analyses were carried out on the set of four pedigrees, jointly, as well as on each individual pedigree, using all MGS markers on each chromosome in a multipoint analysis of that chromosome. No ascertainment correction was used, both because the complex ascertainment of the pedigrees violated the assumptions needed (Cannings and Thompson 1977) to apply an ascertainment correction, and because QTL localization is not affected by the absence of such a correction (Ma et al. 2007).
Follow-up analyses
We carried out additional analyses in key regions. These initially included long runs with Loki (1,000,000 sweeps) and parametric lod score analyses based on QTL models extracted from CSA of all pedigrees, as well as additional multipoint analyses with Loki that included the additional DCD markers. Parametric lod scores provide an interpretation of the strength of evidence of linkage that is widely used, provide an upper bound on the significance level (Morton 1955), which generally provides a conservative linkage test (Rao et al. 1978). These follow-up analyses were primarily directed toward chromosome 4 because this chromosome provided the strongest and most consistent evidence of linkage across families, and because evidence of linkage increased across families in the joint analysis over the single-family analyses, which is consistent with support from more than one family.
Follow-up analysis also included evaluation of evidence for association of the trait with genotypes of all available 293 SNPs in the region with the strongest evidence of linkage on chromosome 4. Association is indicative of probable refined trait QTL localization. For these analyses, each SNP was included, individually, as a major-gene covariate in the Bayesian oligogenic segregation and linkage analysis, similar to the measured genotype approach (Boerwinkle et al. 1986). As a major-gene covariate, genotype effects are estimated separately for each of the three possible genotypes. In addition, the SNP has a location on the map so that together with the observed SNP genotypes and pedigree structure, information on other markers can be used to impute SNP genotypes for unsampled individuals, thereby increasing the sample size for use in determining evidence for association. The goal was to determine whether the SNP could explain most or all of the segregating trait variance, with a SNP that explains all of the segregating trait variance representing one that is perfectly associated with the causal site (Almasy and Blangero 2004). Evidence for the role of such a SNP was obtained both by estimation of its contribution to the genetic variance, and also by the average difference in the log BF achieved when the SNP was included as a covariate compared to an analysis without inclusion of the SNP. This was computed over the region spanning 153–189 cM, which was the region in the genome scan with log BF > 1 for all positions. Linkage disequilibrium between pairs of SNPs was estimated from the HapMap CEU samples, using HaploView (http://www.broad.mit.edu/mpg/haploview) (Barrett et al. 2005), and was used for interpretation of results obtained across analyses of individual SNPs. To reduce the computational burden for the large number of SNP-covariate analyses, markers included as the framework panel for this purpose were restricted to the STRs from the original MGS scan that spanned the region of interest.
Parametric linkage analysis
We carried out parametric LOD score analyses, including additional follow-up markers, for selected regions, based on the genome-scan results. These analyses required a prespecified model of inheritance, but trait model misspecification typically only reduces evidence of linkage over use of a more accurate trait model, and is therefore unlikely to give false positive evidence of linkage (Amos and de Andrade 2001; Clerget-Darpoux et al. 1986; Greenberg et al. 1998; Ott 1999). The trait model used was based on the most parsimonious dominant mixed model obtained from CSA for all families, using the major-gene parameter values, but defining the within-genotype residual variance as the total of the polygenic and environmental variance. In all cases, we carried out such analyses for the total data set, and for individual pedigrees, which, in this sample, are large enough to be analyzed individually. For these analyses, we used VITESSE (http://watson.hgen.pitt.edu/register/soft_doc.html) (O'Connell and Weeks 1995) for single-marker analyses, and lm_markers (Wijsman et al. 2006) from the MORGAN package (http://www.stat.washington.edu/thompson/Genepi/pangaea.shtml) for multipoint analyses, in all cases using the entire intact pedigree structures for analysis. For multipoint analyses, the use of the MCMC-based program lm_markers allowed simultaneous analysis with all available markers on a chromosome, including additional dense follow-up markers. To our knowledge, the program lm_markers is the only option available for computation of parametric lod scores with a quantitative trait model on large pedigrees and many markers (Wijsman et al. 2006).
Maps and marker allele frequencies
We used sex-average map positions based on the Marsh-field map (Broman et al. 1998), with map positions based on the Haldane map function. In the initial genome scan, maximum likelihood estimates of marker allele frequencies were estimated from the complete observed pedigree and marker data. For subsequent analyses, marker allele frequencies were forced to be those initially estimated from the data to eliminate marker allele frequency variation across analyses based on subsets of the data. For analysis of key SNPs, allele frequencies were based on either direct allele counting or on maximizing the likelihood given the pedigree data as indicated, and map positions were based on interpolation from the sequence map position to a meiotic map position.
Results
Sample characteristics
Summaries of the marker completion and lipid measurements are in Table 1. The fraction of all individuals in the pedigrees, including deceased and unsampled individuals, with observed phenotypic data was 78% overall, with little variability among pedigrees. Genotype completion with the MGS panel was 84% overall, but because of scarce sample, was only 59% for follow-up genotyping with DCD and SNP markers. The four families have similar, but not identical, lipid distributions. Mean BMI and PPD were very similar across the four pedigrees although the variability of PPD was lower for pedigree F2 and higher for pedigree F3 than for the other two pedigrees. The mean apoB level was somewhat higher for pedigree F2 than for the other three pedigrees, and the mean LDL level was lower for pedigree F3 and F4 than for the other two pedigrees. After adjustment for covariates, pedigree F1 had the lowest mean and least variable adjusted apoB, while F2 had the highest mean adjusted apoB. All further analyses were carried out only on the adjusted apoB trait, and all references here to apoB level henceforth refer to this covariate-adjusted apoB.
QTL models
Segregation analysis identified genetic factors as important for explaining the pattern of inheritance of apoB (Supplementary Table 1). Models identified as part of CSA that had a genetic component, including polygenic and Mendelian models, had statistically significantly (p < 0.05) higher likelihoods than did a simple single-mode environmental model. A dominant, mixed model (i.e., including polygenic variance) was most parsimonious although a major-gene general model with or without polygenic background factors had likelihoods that were similar to that of the dominant, mixed model (Supplementary Table 1). The parameters from this model used for parametric linkage analysis were a major allele frequency of 0.71 with heterozygotes and homozygotes for this allele having adjusted apoB values of −5.68, the homozygote for the minor allele having a mean value of 28.73, and a within-genotype standard deviation of 12.56.
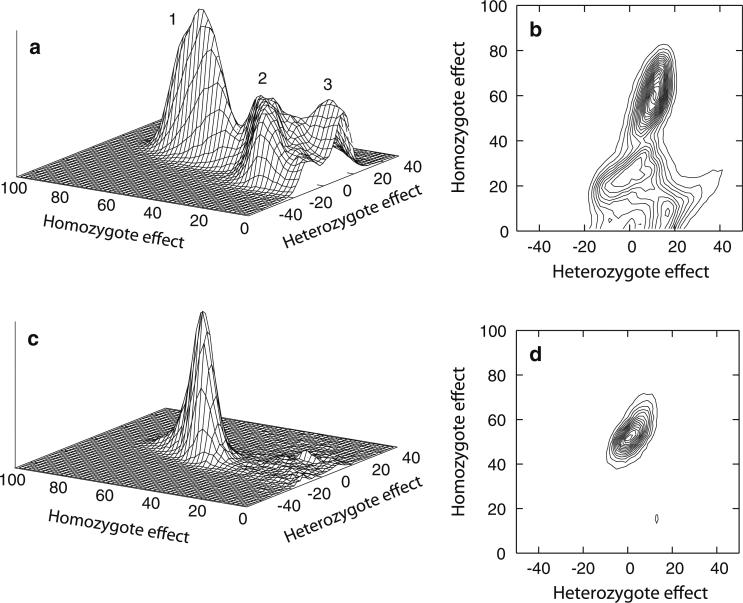
The posterior distribution of QTL model parameters identified a relatively simple model space. The genotypic means for a diallelic QTL can be summarized as two differences between mean values of genotypes, denoting the homozygote effect as ε22 = μ22 - μ11, and the heterozygote effect as ε12 = μ12 - μ11 (Fig. 1), where μ11 is the mean trait value of genotypes of individuals homozygous for allele 1, with similar terms for the other mean trait values. Analysis of all four families identified three modes in the oligogenic QTL model space (Fig. 1a, b). Relative to the baseline homozygote genotype mean, μ11, the model with the highest posterior probability, PP (PP = 0.38) is model 1 (Fig. 1a), and represents a small value for ε12 and a large value for ε22 (Fig. 1b). Model 1, which is dominant for allele 1, is most similar to the general Mendelian model identified for the full data set by CSA (Supplementary Table 1), with a high allele frequency, p1, and a large increase the mean genotype of the rare homozygote relative to the common homozygote, ε22. Model 1 was also the model that explained the pattern of inheritance and the evidence of linkage for family F3 (Fig. 1c, d), as described further below. Model 2 had lower overall support (PP = 0.28), a heterozygote effect that is centered on zero, and a more modest ε22 effect. This model is similar to both the dominant mixed and general mixed models identified by maximizing the likelihood under CSA, and explains only slightly less (31%) of the total variance than does model 1 (39%). Finally, model 3 is slightly overdominant, explained only about half as much of the total variance as did Model 2, and had still lower support (PP = 0.19). Model 3 describes a model that is essentially recessive for allele 1. Models 2 and 3, jointly, were the models that also were identified in analysis of the three families F1, F2, and F4, when family F3 was excluded (results not shown).
Fig. 1.
Posterior distributions of QTL genotype effects for apoB in analysis of all four families in oligogenic segregation analysis only (a, b) and for family F3 alone for QTLs that visited chromosome 4 (c, d). Model numbers identified by OSA (Supplementary Table 1) are noted in a
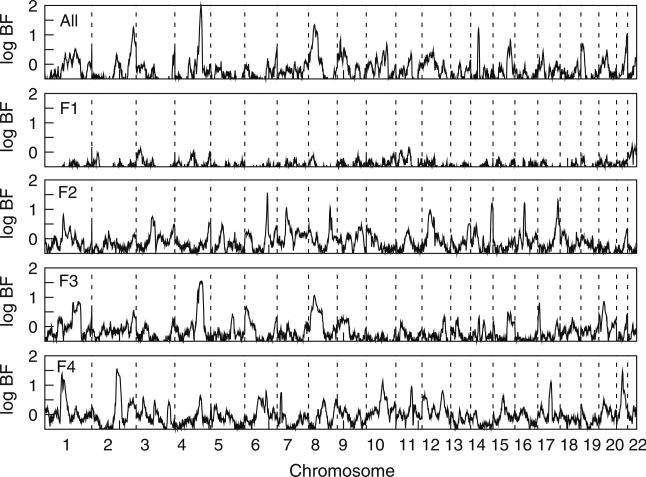
Genome scan
The genome scan identified one region with strong evidence of linkage to apoB in analysis of all families, as well as strong evidence of linkage in one individual family (Fig. 2; Table 2). A very strong log BF = 2 (posterior odds ratio of 98.5:1 supporting linkage) was obtained for the complete sample at 175 cM on chromosome (chr) 4q (Fig. 2), with no other regions with support for linkage as strong as this. There was particularly strong support for this region (log BF = 1.5) in family F3 alone, with additional modest support (log BF = 0.7) in family F4, and a relatively neutral signal (log BF = −0.15) in family F1. The overall strength of the evidence of linkage in the full sample likely reflects this support from more than one family since evidence of linkage in Bayesian methods increases non-linearly with increasing positive support from multiple families as the methods borrow strength across the evidence. The trait model that mapped to the region on chr 4q between 159 and 195 cM in analysis of the full sample, as well as analysis of family F3 alone, was Model 1 from the OSA (Fig. 1c, d). This model explained 78% of the total genetic variance segregating in family F3 and 60% of the genetic variance in analysis of all families. Additional follow-up analyses of this region is described below, and provided further support for linkage to this region.
Fig. 2.
Genome scan for adjusted apoB for all four families jointly (All), and for each individual family (F1–F4). Vertical dotted lines delimit the chromosome boundaries
Table 2.
Bayes’ factors for linkage for positions where the maximum log BF > 1 in at least one pedigree (F1-F4) or in the joint analysis (All)
| Chromosome | Position (cM) | All | F1 | F2 | F3 | F4 |
|---|---|---|---|---|---|---|
| 1 | 115 | –0.097 | –0.523 | –0.097 | 0.146 | 1.386 |
| 2 | 167 | 0.279 | –0.523 | 0.041 | –0.222 | 1.501 |
| 2 | 279 | 1.255 | –0.523 | 0.079 | 0.431 | 0.477 |
| 4 | 171 | 1.768 | –0.523 | –0.097 | 1.534 | 0.653 |
| 4 | 175 | 1.993 | –0.523 | –0.155 | 1.470 | 0.519 |
| 6 | 151 | 0.041 | –0.523 | 1.567 | –0.398 | 0.380 |
| 7 | 63 | –0.222 | –0.398 | 1.064 | –0.398 | –0.097 |
| 8 | 41 | 1.344 | –0.301 | –0.155 | 1.061 | –0.523 |
| 14 | 143 | –0.523 | –0.523 | 1.212 | –0.398 | –0.301 |
| 16 | 65 | –0.046 | –0.301 | 1.207 | –0.523 | 0.079 |
| 17 | 135 | –0.155 | –0.523 | 1.307 | 0.000 | –0.155 |
| 21 | 41 | 0.041 | –0.301 | –0.398 | –0.523 | 1.481 |
Positions shown are only those for which a maximum BF occurred at that position in at least one pedigree or in the full data set. Bold: log BF > 1
Several additional genomic regions also provided support for linkage to apoB either in the full sample or in individual families (Table 2). In addition to the region on chr 4, analysis of all families identified two other regions with strong evidence of linkage: a strong log BF = 1.3 was obtained on chr 2q at 279 cM and on chr 8p at 41 cM. The region on chr 2q appears to be a region in which the total support across pedigrees provides evidence of linkage, since there is modest support in three pedigrees, but no individual pedigree provides strong support. The region on chr 8p has reasonably strong support from pedigree F3 (log BF = 1.1), along with modest additional support from other pedigrees, to give an overall stronger signal in the analysis of all the families than in analysis of any individual family. Several additional regions with notable evidence of linkage only appeared in individual families. These included: in family F2, regions on chr 6 (log BF = 1.6 at 151 cM), chr 17 (log BF = 1.3 at 135 cM), chr 16 (log BF = 1.2 at 65 cM), and chr 14 (log BF = 1.2 at 143 cM); in family F3, a second region on chr 1 (log BF = 0.8); and in family F4, regions on chr 2 (log BF = 1.5 at 171 cM) and on chr 21 (log BF = 1.5 at 40 cM). In total, each of families F2–F4 yielded at least one region with very strong evidence of linkage (log BF > 1.5), and at least one additional region with more modest evidence of linkage (log BF > 1). Of these three families, the mode of inheritance in family F3 appeared simplest with only one strong and one modest signal, while the mode of inheritance in the other two families appeared potentially more complex with several additional notable signals. Family F1 yielded no regions with evidence of linkage anywhere in the genome.
Chromosome 4 follow-up analyses
STR markers
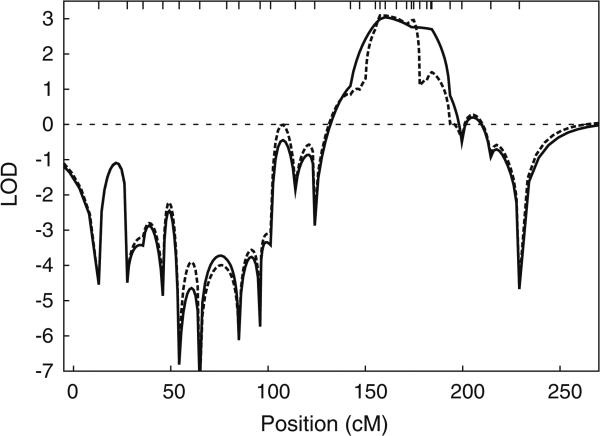
Linkage analysis with a parametric lod score approach provided statistically significant evidence of linkage of apoB to chr 4q, with family F3 achieving a maximum lod score, Zmax > 3 (Fig. 3). In this family, the multipoint Zmax = 3.05, based on the MGS markers, occurred at 161.8 cM, and the interval over which the lod score was within 1 of the maximum spanned 148.45–191.34 cM. Inclusion of DCD markers in the multipoint analysis slightly increased Zmax to 3.09 in a long MCMC-based run of 1 × 106 sweeps, while also narrowing the interval over which the lod score was within 1 of the maximum to 152.88–176.85 cM. In addition, all genome-scan markers within this same interval provided positive evidence of linkage in single-marker analyses, with several attaining Zmax near 3.0, typically at a recombination fraction of 0 (Supplementary Table 2).
Fig. 3.
LOD score curves on chromosome 4 for adjusted apoB and family F3. Solid line analysis based on 21 genome-scan STRs (D4S2366, D4S403, D4S2639, D4S2397, D4S2632, D4S1627, D4S3248, D4S2367, D4S3243, D4S2361, D4S1647, D4S2623, D4S2394, D4S1644, D4S1625, D4S1629, D4S2368, D4S2431, D4S2417, D4S408, and D4S1652). Dashed line analysis that also included nine additional STRs (D4S1575, GATA150B10, D4S1520, D4S2962, D4S2976, D4S3354, D4S1603, D4S3339, and D4S620). Tic marks on top horizontal axis denote positions of all 30 MGS and DCD markers
SNP markers
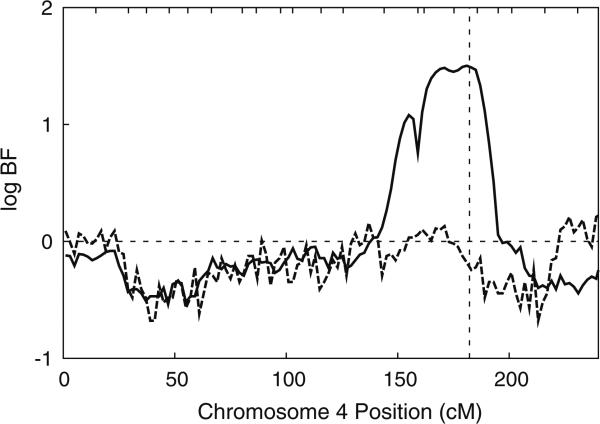
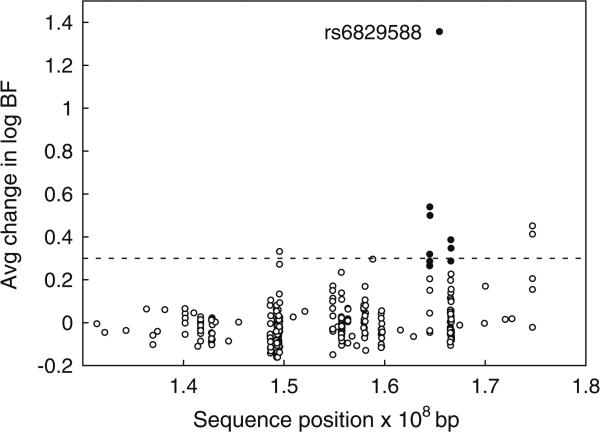
Further evaluation of 293 SNPs across this region identified a smaller region with evidence of association with apoB in family F3. Fifty-two of the 293 SNPs each explained >5% of the total genetic variance, and another 10 SNPs each explained >10% of the total genetic variance (Supplementary Figure 1). SNP rs6829588, in particular, explained ~39% of the total genetic variance and 50% of the genetic variance attributable to QTLs with a location in this region for family F3. Use of this SNP as a major-gene covariate virtually eliminated the evidence of linkage (Fig. 4), reducing the evidence of linkage to a trivial max log BF = 0.17, as would be expected if this SNP were in strong linkage disequilibrium with a causal variant (Almasy and Blangero 2004). Adjustment for flanking SNPs also reduced the evidence of linkage but to a lesser extent (Fig. 5). SNP rs6829588 is not in linkage disequilibrium with other SNPs used for analysis in this region (r2 < 0.02 with all other SNPs), and genotyping quality appeared to be excellent, in that there were no failed genotypes and no Mendelian inconsistencies. SNP rs6829588 was a single SNP on the CVD SNP panel, in an intergenic region, but SNPs in the flanking two gene clusters also had an effect on reducing the log BF when similarly investigated (Fig. 5).
Fig. 4.
log BF for linkage in joint oligogenic segregation and linkage analysis for family F3 for the original panel of 21 genome-scan STRs without (solid line) and with (dashed line) inclusion of SNP rs6829588 as a major-gene covariate. Vertical dotted line depicts position of rs6829588 at 182.17 cM
Fig. 5.
QTL size versus position on chromosome 4 for each of 293 SNPs used as a major-gene covariate. Solid circles represent the same SNPs as those indicated with solid circles in Supplementary Figure 1. Points above the dashed line represent more than a twofold change in the BF
The estimated parameter values for rs6829588 are consistent with the presence of association with a tightly linked QTL in family F3 (Table 3). In the full sample, the estimate of the rare homozygote effect was ~1.9 SD greater than zero, with a posterior probability of 0.97 exceeding zero. In family F3, the effects were much stronger, with both the heterozygote and the rare homozygote effects exceeding zero: the rare homozygote effect was more than 6 SD above zero with a posterior probability >0.99 that the effect exceeds zero; the heterozygote effect was 1.6 SD above zero with a posterior probability of 0.94 exceeding zero. For family F3, the estimated major allele frequency, pC = 0.83, and genotypic effects, εAA = 56.8 and εAC = 5.6, are remarkably similar to the genotypic effects for OSA model 1 (p1 = 0.83, ε22 = 61.05, e12 = 12.44), estimated over the entire sample in the absence of marker data, and to the equivalent model on chromosome 4 obtained for family F3 for the STR-only analysis (Fig. 1c, d; Table 3). It is worth noting that the allele frequency estimated for the minor allele of this SNP among founders in this FCHL sample is somewhat higher (0.15 in the whole sample) than in the reference Hapmap CEU sample (pA = 0.085, N = 118), and that in the two families with the greatest positive contribution to the evidence of linkage (F3, F4), the observed proportion of the A allele among all individuals was even higher (pA = 0.24 in family F3 and pC = 0.18 in family F4), suggesting chance excess of informative transmissions of the high risk but rarer A allele.
Table 3.
Estimated allele frequencies and genotype effects (SD) for QTLs on chromosome 4
| Parametera | Estimate | QTL at 153–189 cM |
rs6829588 |
||
|---|---|---|---|---|---|
| All | F3 | All | F3 | ||
| p1 | Posterior mean (SD) | 0.82 (0.09) | 0.75 (0.11) | 0.86 (0.02) | 0.85 (0.05) |
| ε 12 | Posterior mean (SD) | 6.97 (5.15) | 2.40 (6.65) | 1.58 (3.4) | 6.48 (4.1) |
| ε 22 | Posterior mean (SD) | 53.65 (11.04) | 52.39 (11.8) | 18.99 (9.98) | 57.16 (9.1) |
For rs6829588, subscripts 1 and 2 correspond to alleles C and A, respectively
Discussion
Here, we have provided strong evidence for a locus on chromosome 4q that contributes to variation in apoB level in a subset of FCHL families, with evidence for additional contributing loci on other chromosomes, including chromosomes 2q and 8p. The specific covariate adjustments used to define the trait were designed to isolate variation contributing to apoB level that is independent of that contributing to LDL cholesterol levels and size, with the goal of improving power to detect QTLs through reduction in the complexity of the trait. To our knowledge, this is the first report of the genetic basis of apoB level that is not confounded by the strong correlation between APOB and LDL cholesterol levels and size. The evidence for a locus on chromosome 4 derives from multiple sources. This includes genome-scan linkage analysis with more than one approach, inclusion of additional markers in the region with evidence of linkage, and demonstration that for the region with strongest evidence of linkage, inclusion of SNPs as measured genotypes explains much of the segregating variance.
The evidence for QTLs affecting apoB levels at particular genomic locations derives primarily from individual families. This pattern of evidence of linkage in single pedigrees has been observed in genome scans of other lipid traits in these large pedigrees (Badzioch et al. 2004; Gagnon et al. 2005), with little overlap in the regions identified for different traits/family combinations, consistent with a multilocus model for FCHL. It is also consistent with the marked heterogeneity in results of genome scans for many other complex traits, and is the expected pattern in the context of genetic heterogeneity, where it is harder to replicate evidence for any particular genomic region than it is to obtain evidence for at least one genomic region (Suarez et al. 1994). Different ascertainment criteria are also likely to complicate such replication studies: in the current analysis, the three pedigrees with initial ascertainment or extension through high TG levels all produced multiple genomic regions with evidence of linkage to apoB level, with some contribution to the same regions from the two pedigrees that had identical ascertainment criteria. Family F1, which had a very different ascertainment, did not produce even suggestive evidence of linkage to any genomic regions. This absence of evidence of linkage is unlikely to be explained through low power: this pedigree was the second largest and well typed, and yielded strong evidence of linkage to other lipid traits in previous analyses (Badzioch et al. 2004).
There is some overlap between our results and those of two previous genome scans for apoB level, despite the overall relative paucity of reports of genomic scans for apoB QTLs (Bosse et al. 2004a). There is modest statistical support from previous studies for a QTL in the same region of chromosome 4q as we identify here, also in FCHL families, but without the covariate adjustments used here (Cantor et al. 2004). There is also overlap between the region on chr 6q identified in our analysis and a region that provided suggestive evidence of linkage in a previous genome scan of apoB in pedigrees with familial hypobetalipoproteinemia (Sherva et al. 2007). Because almost all previous genome scans of apoB level have been analyzed with methods that do not easily allow evaluation of the contributions of individual pedigrees, it is not possible from the published information to determine whether there are any other large pedigrees with evidence of linkage to the regions we report here. However, it would be useful to individually re-analyze the existing large pedigrees to determine if there is evidence of linkage that has been missed in joint analyses.
The strong association between apoB level and genotypes at rs6829588 on chromosome 4 has two implications. First, it narrows, considerably, the size of the region on chromosome 4 that is likely to contain the variant(s) of interest. This is supported by the reduced contribution to apoB variance of SNPs in flanking genes, and suggests that the region of interest is bounded by genes that flank rs6829588. Such a ~2 cM region is vastly reduced in size over the 24 cM region defined by the linkage analysis, and is also much smaller than that could reasonably be expected to be obtainable from linkage analysis, alone, in a single, albeit large, pedigree (Boehnke 1994). Second, the location of this SNP in an intergenic region, the modest minor allele frequency in the HapMap CEU sample (0.085), high variation in allele frequency among HapMap reference samples (0.085–0.880), and the evidence for a strong effect only in one pedigree suggest that it is unlikely to be the causal locus. More likely, the causal variant occurred on a haplotype bearing the minor allele at this SNP, and other independent copies of the minor allele do not also carry the causal variant. This is supported by the observation that although this SNP explains a substantial fraction of the genetic variance in one pedigree, it neither accounts for all of the genetic variance in pedigree F3 nor does it appear to explain the genetic variance in the rest of the pedigrees, as suggested by the attenuation of the genotype effects when estimated on the whole sample. Explanations that could explain the current observations include (1) that there are multiple contributing loci in the region, and that this SNP is only one of them or only tags only one of them, (2) that this SNP is in strong but imperfect LD with the causal variant in the family with strong evidence of linkage, or (3) that the mode of inheritance of apoB level in this region is multiallelic, and this SNP only captures some of the segregating variance.
There are several possibilities regarding the causal loci contributing to the QTL on chromosome 4 identified here. The associated SNP, rs6829588, is in an intergenic region with the closest annotated genes 0.32 Mb (ANP32C) and 0.45 Mb (TRIM61) away. Neither of these genes has to date been implicated in CVD. The closest genes with published evidence for CVD implications, and specifically dyslipidemias, are NPY5R (Arnett et al. 2009; Blumenthal et al. 2002; Coletta et al. 2007) and carboxypeptidase E (CPE) (Jeffrey et al. 2008; Jia et al. 2008, 2009), which are 1.2 and 0.87 Mb from rs6829588, respectively. LD between SNPs in these genes and rs6829588 is low in the HapMap sample (r2 < 0.02) as would be expected from the large physical distances. No SNP in or near an annotated coding region with r2 > 0.1 with rs6829588 was identifiable in the HapMap sample, and all SNPs with r2 > 0.1 are in intergenic regions. The possibility remains that variation in either non-coding DNA or one of the other genes in the region on chromosome 4 may affect apoB levels, including TRIM61, MARCH1 or TKTL2. Further experimental work will be needed to identify the causal SNP, since there is no in silico information on this SNP in published eQTL databases (Dixon et al. 2007; Myers et al. 2007; Schadt et al. 2008).
The results obtained here are consistent with the hypothesis that rare variants contribute to the traits of interest segregate in these unusual pedigrees. The evidence is also consistent with the growing evidence that rare variants may be more important than common variants in explaining the genetic variance of some complex traits (Cohen et al. 2004; Hamsten and Eriksson 2008; Manolio et al. 2009). In this context, our results illustrate an important advantage of large pedigrees: such pedigrees may be particularly useful for identifying some trait loci of interest since individual large pedigrees can be analyzed as individual units of study, and may represent near-Mendelian inheritance for any single trait. Identification of the causal site(s) is likely to require evaluation of sequence data in a future analysis. Finally, our results here coupled with other results reported elsewhere on the same families (Badzioch et al. 2004; Gagnon et al. 2005) suggest that one possible explanation for FCHL is that of an oligogenic mode of inheritance, even within these large families. The diagnosis of FCHL may reflect the existence of more than one QTL within a family, with the different extreme lipid levels explained by segregating variants for the different component phenotypes at different loci, as suggested by the results obtained in the extended families used here.
Supplementary Material
Supplementary Table 1. Trait mode-of-inheritance models for apoB: selected models evaluated in the case of CSA, and sub-models with highest posterior probabilities from OSA.
Supplementary Table 2. Lod scores obtained for analysis with individual markers for family F3, using a parametric lod score approach with model parameters from complex segregation analysis indicated in Supplementary Table 1.
Acknowledgments
Supported by National Institutes of Health grants HL30086 and GM46255, and by the University of Washington General Clinical Research Center, MO1-RR-00037. The authors thank Hiep Nguyen for computer support, and the family members for their participation. The study complies with the current laws of the United States of American, in which it was performed, and was approved by the University of Washington Institutional Review Board.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-010-0819-2) contains supplementary material, which is available to authorized users.
Conflict of interest statement The authors declare that they have no conflict of interest.
Contributor Information
Ellen M. Wijsman, Division of Medical Genetics, Department of Medicine, University of Washington, 4333 Brooklyn Ave NE, Box 359460, Seattle, WA 98195-9460, USA Department of Biostatistics, University of Washington, Seattle, WA, USA.
Joseph H. Rothstein, Division of Medical Genetics, Department of Medicine, University of Washington, 4333 Brooklyn Ave NE, Box 359460, Seattle, WA 98195-9460, USA
Robert P. Igo, Jr., Department of Epidemiology and Biostatistics, Case Western Reserve University, Cleveland, OH, USA
John D. Brunzell, Division of Metabolism, Endocrinology, and Nutrition, Department of Medicine, University of Washington, Seattle, WA, USA
Arno G. Motulsky, Department of Genome Sciences, University of Washington, Seattle, WA, USA
Gail P. Jarvik, Department of Genome Sciences, University of Washington, Seattle, WA, USA
References
- Allayee H, Krass KL, Pajukanta P, Cantor RM, van der Kallen CJH, Mar R, Rotter JI, de Bruin TWA, Peltonen L, Lusis AJ. Locus for elevated apolipoprotein B levels on chromosome 1p31 in families with familial combined hyperlipidemia. Circ Res. 2002;90:926–931. doi: 10.1161/01.res.0000015885.27134.f0. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Exploring Positional candidate genes: linkage conditional on measured genotype. Behav Genet. 2004;34:173–177. doi: 10.1023/B:BEGE.0000013731.03827.69. [DOI] [PubMed] [Google Scholar]
- Amos CI, de Andrade M. Genetic linkage methods for quantitative traits. Stat Methods Med Res. 2001;10:3–25. doi: 10.1177/096228020101000102. [DOI] [PubMed] [Google Scholar]
- Arnett DK, Devereux RB, Rao DC, Li N, Tang WH, Kraemer R, Claas SA, Leon JM, Broeckel U. Novel genetic variants contributing to left ventricular hypertrophy: the HyperGEN study. J Hypertens. 2009;27:1585–1593. doi: 10.1097/HJH.0b013e32832be612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MA, Brunzell JD, Fitch WL, Krauss RM. Inheritance of low density lipoprotein subclass patterns in familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 1990;10:520–530. doi: 10.1161/01.atv.10.4.520. [DOI] [PubMed] [Google Scholar]
- Ayyobi AF, McGladdery SH, McNeely MJ, Austin MA, Motulsky AG, Brunzell JD. Small, dense LDL and elevated apolipoprotein B are the common characteristics for the three major lipid phenotypes of familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 2003;23:1289–1294. doi: 10.1161/01.ATV.0000077220.44620.9B. [DOI] [PubMed] [Google Scholar]
- Babirak SP, Iverius PH, Fujimoto WY, Brunzell JD. Detection and characterization of the heterozygote state for lipoprotein lipase deficiency. Arterioscler Thromb Vasc Biol. 1989;9:326–334. doi: 10.1161/01.atv.9.3.326. [DOI] [PubMed] [Google Scholar]
- Babirak SP, Brown BG, Brunzell JD. Familial combined hyperlipidemia and abnormal lipoprotein lipase. Arterioscler Thromb Vasc Biol. 1992;12:1176–1183. doi: 10.1161/01.atv.12.10.1176. [DOI] [PubMed] [Google Scholar]
- Badzioch MD, Igo RP, Jr, Gagnon F, Brunzell JD, Krauss RM, Motulsky AG, Wijsman EM, Jarvik GP. LDL particle size loci in familial combined hyperlipidemia: evidence for multiple loci from a genome scan. Arterioscler Thromb Vasc Biol. 2004;24:1942–1950. doi: 10.1161/01.ATV.0000143499.09575.93. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Blumenthal JB, Andersen RE, Mitchell BD, Seibert MJ, Yang H, Herzog H, Beamer BA, Franckowiak SC, Walston JD. Novel neuropeptide Y1 and Y5 receptor gene variants: associations with serum triglyceride and high-density lipoprotein cholesterol levels. Clin Genet. 2002;62:196–202. doi: 10.1034/j.1399-0004.2002.620302.x. [DOI] [PubMed] [Google Scholar]
- Boehnke M. Limits of resolution of genetic linkage studies: implications for positional cloning of human disease genes. Am J Hum Genet. 1994;55:379–390. [PMC free article] [PubMed] [Google Scholar]
- Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. Ann Hum Genet. 1986;50:181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- Bosse Y, Chagnon YC, Despres JP, Rice T, Rao DC, Bouchard C, Perusse L, Vohl MC. Compendium of genome-wide scans of lipid-related phenotypes: adding a new genome-wide search of apolipoprotein levels. J Lipid Res. 2004a;45:2174–2184. doi: 10.1194/jlr.R400008-JLR200. [DOI] [PubMed] [Google Scholar]
- Bosse Y, Perusse L, Vohl MC. Genetics of LDL particle heterogeneity: from genetic epidemiology to DNA-based variations. J Lipid Res. 2004b;45:1008–1026. doi: 10.1194/jlr.R400002-JLR200. [DOI] [PubMed] [Google Scholar]
- Bredie SJH, Kiemeney LA, deHaan AFJ, Demacker PNM, Stalenhoef AFH. Inherited susceptibility determines the distribution of dense low-density lipoprotein subfraction profiles in familial combined hyperlipidemia. Am J Hum Genet. 1996;58:812–822. [PMC free article] [PubMed] [Google Scholar]
- Breslow JL. Genetics of lipoprotein abnormalities associated with coronary heart disease susceptibility. Annu Rev Genet. 2000;34:233–254. doi: 10.1146/annurev.genet.34.1.233. [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G, Albers JJ, Fisher LD, Schaefer SM, Lin JT, Kaplan C, Zhao XQ, Bisson BD, Fitzpatrick VF, Dodge HT. Regression of coronary-artery disease as a result of intensive lipid-lowering therapy in men with high-levels of apolipoprotein B. N Engl J Med. 1990;323:1289–1298. doi: 10.1056/NEJM199011083231901. [DOI] [PubMed] [Google Scholar]
- Brunzell JD, Deeb SS. Familial lipoprotein lipase deficiency, apoCII deficiency and hepatic lipase deficiency. In: Scriver CR, Beaudet AI, Sly WS, Vale D, editors. The metabolic and molecular bases of inherited disease. 8th edn. McGraw-Hill; New York: 2000. pp. 2789–2816. [Google Scholar]
- Brunzell JD, Albers JJ, Chait A, Grundy SM, Groszek E, McDonald GB. Plasma lipoproteins in familial combined hyperlipidemia and monogenic familial hypertriglyceridemia. J Lipid Res. 1983;24:147–155. [PubMed] [Google Scholar]
- Brunzell JD, Austin MA, Deeb SS, Hokanson JE, Jarvik GP, Nevin DN, Wijsman E, Zambon A, Motulsky A. Familial combined hyperlipidemia and genetic risk for atherosclerosis. In: Woodford FP, Davignon J, Sniderman A, editors. Atherosclerosis X. Elsevier; New York: 1995. pp. 624–627. [Google Scholar]
- Cannings C, Thompson EA. Ascertainment in the sequential sampling of pedigrees. Clin Genet. 1977;12:208–212. doi: 10.1111/j.1399-0004.1977.tb00928.x. [DOI] [PubMed] [Google Scholar]
- Cantor RM, de Bruin T, Kono N, Napier S, van Nas A, Allayee H, Lusis AJ. Quantitative trait loci for apolipoprotein B, cholesterol, and triglycerides in familial combined hyperlipidemia pedigrees. Arterioscler Thromb Vasc Biol. 2004;24:1935–1941. doi: 10.1161/01.ATV.0000142358.46276.a7. [DOI] [PubMed] [Google Scholar]
- Chait A, Albers JJ, Brunzell JD. Very low density lipoprotein overproduction in genetic forms of hypertriglyceridaemia. Eur J Clin Invest. 1980;10:17–22. doi: 10.1111/j.1365-2362.1980.tb00004.x. [DOI] [PubMed] [Google Scholar]
- Clerget-Darpoux F, Bonaiti-Pellie C, Hochez J. Effects of misspecifying genetic parameters in lod score analysis. Biometrics. 1986;42:393–399. [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Coletta DK, Schneider J, Stern MP, Blangero J, DeFronzo RA, Duggirala R, Jenkinson CP. Association of neuropeptide Y receptor Y5 polymorphisms with dyslipidemia in Mexican Americans. Obesity. 2007;15:809–815. doi: 10.1038/oby.2007.610. [DOI] [PubMed] [Google Scholar]
- Coresh J, Beaty TH, Kwiterovich PO., Jr Inheritance of plasma apolipoprotein B levels in families of patients undergoing coronary arteriography at an early age. Genet Epidemiol. 1993;10:159–176. doi: 10.1002/gepi.1370100303. [DOI] [PubMed] [Google Scholar]
- Dastani Z, Engert JC, Genest J, Marcil M. Genetics of high-density lipoproteins. Curr Opin Cardiol. 2006;21:329–335. doi: 10.1097/01.hco.0000231403.94856.cd. [DOI] [PubMed] [Google Scholar]
- Day INM, Whittall RA, Odell SD, Haddad L, Bolla MK, Gudnason V, Humphries SE. Spectrum of LDL receptor gene mutations in heterozygous familial hypercholesterolemia. Hum Mutat. 1997;10:116–127. doi: 10.1002/(SICI)1098-1004(1997)10:2<116::AID-HUMU4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Dixon WJ, Tukey JW. Approximate behavior of distribution of Winsorized T (Trimming/Winsorization 2). Technometrics. 1968;10:83–98. [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KCC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WOC. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Durbin J, Watson GS. Testing for serial correlation in least squares regression. I. Biometrika. 1950;37:409–428. [PubMed] [Google Scholar]
- Eurlings PMH, van der Kallen CJH, Geurts JMW, van Greevenbroek MMJ, de Bruin TWA. Genetic dissection of familial combined hyperlipidemia. Mol Genet Metab. 2001;74:98–104. doi: 10.1006/mgme.2001.3232. [DOI] [PubMed] [Google Scholar]
- Feitosa MF, Borecki IB, Rankinen T, Rice T, Despres JP, Chagnon YC, Gagnon J, Leon AS, Skinner JS, Bouchard C, Province MA, Rao DC. Evidence of QTLs on chromosomes 1q42 and 8q24 for LDL-cholesterol and apoB levels in the HERITAGE Family Study. J Lipid Res. 2005;46:281–286. doi: 10.1194/jlr.M400252-JLR200. [DOI] [PubMed] [Google Scholar]
- Fouchier SW, Defesche JC, Umans-Eckenhausen MAW, Kastelein JJP. The molecular basis of familial hypercholesterolemia in the Netherlands. Hum Genet. 2001;109:602–615. doi: 10.1007/s00439-001-0628-8. [DOI] [PubMed] [Google Scholar]
- Fujimoto WY, Kahn SE, Brunzell JD. Metabolic basis for coronary artery disease risk in central obesity and glucose intolerance multiple risk factors in cardiovascular disease.. The second symposium proceedings.; Churchill Livingstone, Tokyo. 1994. pp. 189–192. [Google Scholar]
- Gagnon F, Jarvik G, Motulsky A, Deeb S, Brunzell J, Wijsman E. Evidence of linkage of HDL level variation to APOC3 in two samples with different ascertainment. Hum Genet. 2003;113:522–533. doi: 10.1007/s00439-003-1006-5. [DOI] [PubMed] [Google Scholar]
- Gagnon F, Jarvik GP, Badzioch MD, Motulsky AG, Brunzell JD, Wijsman EM. Genome scan for quantitative trait loci influencing HDL levels: evidence for multilocus inheritance in familial combined hyperlipidemia. Hum Genet. 2005;117:494–505. doi: 10.1007/s00439-005-1338-4. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Schrott HG, Hazzard WR, Bierman EL, Motulsky AG. Hyperlipidemia in coronary heart disease. II. Genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J Clin Invest. 1973;52:1544–1568. doi: 10.1172/JCI107332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Dana SE, Brunschede GY, Brown MS. Genetic heterogeneity in familial hypercholesterolemia: evidence for two different mutations affecting functions of low-density lipoprotein receptor. Proc Natl Acad Sci USA. 1975;72:1092–1096. doi: 10.1073/pnas.72.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82:711–732. [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE. The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet. 1998;63:870–879. doi: 10.1086/301997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamsten A, Eriksson P. Identifying the susceptibility genes for coronary artery disease: from hyperbole through doubt to cautious optimism. J Intern Med. 2008;263:538–552. doi: 10.1111/j.1365-2796.2008.01958.x. [DOI] [PubMed] [Google Scholar]
- Hasstedt S, Cartwright P. PAP: pedigree analysis package. Department of Medical Biophysics and Computing, University of Utah; Salt Lake City: 1981. [Google Scholar]
- Hasstedt SJ, Wu L, Williams RR. Major locus inheritance of apolipoprotein B in Utah pedigrees. Genet Epidemiol. 1987;4:67–76. doi: 10.1002/gepi.1370040202. [DOI] [PubMed] [Google Scholar]
- Heath SC. Markov Chain Monte Carlo Segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson JE, Austin MA, Zambon A, Brunzell JD. Plasma triglyceride and LDL heterogeneity in familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 1993;13:427–434. doi: 10.1161/01.atv.13.3.427. [DOI] [PubMed] [Google Scholar]
- Huang QQ, Shete S, Amos CI. Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet. 2004;75:1106–1112. doi: 10.1086/426000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo RP, Wijsman EM. Empirical significance values for linkage analysis: Trait simulation using posterior model distributions from MCMC oligogenic segregation analysis. Genet Epidemiol. 2008;32:119–131. doi: 10.1002/gepi.20267. [DOI] [PubMed] [Google Scholar]
- Igo RP, Chapman NH, Wijsman EM. Segregation analysis of a complex quantitative trait: approaches for identifying influential data points. Hum Hered. 2006;61:80–86. doi: 10.1159/000093085. [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Beaty TH, Gallagher PR, Coates PM, Cortner JA. Genotype at a major locus with large effects on apolipoprotein B levels predicts familial combined hyperlipidemia. Genet Epidemiol. 1993;10:257–270. doi: 10.1002/gepi.1370100406. [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Brunzell JD, Austin MA, Krauss RM, Motulsky AG, Wijsman EM. Genetic predictors of FCHL in four large pedigrees: Influence of ApoB level major locus predicted genotype and LDL subclass phenotype. Arterioscler Thromb Vasc Biol. 1994;14:1687–1694. doi: 10.1161/01.atv.14.11.1687. [DOI] [PubMed] [Google Scholar]
- Jeffrey KD, Alejandro EU, Luciani DS, Kalynyak TB, Hu XK, Li H, Lin YL, Townsend RR, Polonsky KS, Johnson JD. Carboxypeptidase E mediates palmitate-induced beta-cell ER stress and apoptosis. Proc Natl Acad Sci USA. 2008;105:8452–8457. doi: 10.1073/pnas.0711232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia EZ, Wang J, Yang ZJ, Zhu TB, Wang LS, Chen B, Cao KJ, Huang J, Ma WZ. Molecular scanning of the human carboxypeptidase E gene for mutations in Chinese subjects with coronary atherosclerosis. Mol Cell Biochem. 2008;307:31–39. doi: 10.1007/s11010-007-9581-8. [DOI] [PubMed] [Google Scholar]
- Jia EZ, Wang J, Yang ZJ, Zhu TB, Wang LS, Wang H, Li CJ, Chen BO, Cao KJ, Huang J, Ma WZ. Association of the mutation for the human carboxypeptidase E gene exon 4 with the severity of coronary artery atherosclerosis. Mol Biol Rep. 2009;36:245–254. doi: 10.1007/s11033-007-9173-4. [DOI] [PubMed] [Google Scholar]
- Kass R, Raftery A. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PIW, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Perez FJG, Frazer KA, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- Krauss RM, Burke DJ. Identification of multiple subclasses of plasma low density lipoproteins in normal humans. J Lipid Res. 1982;23:97–104. [PubMed] [Google Scholar]
- Ma JZ, Amos CI, Daw EW. Ascertainment correction for Markov chain Monte Carlo segregation and linkage analysis of a quantitative trait. Genet Epidemiol. 2007;31:594–604. doi: 10.1002/gepi.20231. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton NE. Sequential tests for the detection of linkage. Am J Hum Genet. 1955;7:277–318. [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Awebster J, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Holmans P, Heward CB, Reiman EM, Stephan D, Hardy J. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Neuman RJ, Yuan B, Gerhard DS, Liu KY, Yue P, Duan S, Averna M, Schonfeld G. Replication of linkage of familial hypobetalipoproteinemia to chromosome 3p in six kindreds. J Lipid Res. 2002;43:407–415. [PubMed] [Google Scholar]
- Nickerson DA, Taylor SL, Weiss KM, Clark AG, Hutchinson RG, Stengard J, Salomaa V, Vartiainen E, Boerwinkle E, Sing CF. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat Genet. 1998;19:233–240. doi: 10.1038/907. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet. 1995;11:402–408. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- Ott J. Analysis of human genetic linkage. 3rd edn. Johns Hopkins University Press; Baltimore: 1999. [Google Scholar]
- Pairitz G, Davignon J, Mailloux H, Sing CF. Sources of interindividual variation in the quantitative levels of apolipoprotein B in pedigrees ascertained through a lipid clinic. Am J Hum Genet. 1988;43:311–321. [PMC free article] [PubMed] [Google Scholar]
- Pajukanta P, Porkka KVK, Antikainen M, Taskinen MR, Perola M, Murtomäki-Repo S, Ehnholm S, Nuotio I, Suurinkeroinen L, Lahdenkari AT, Syvänen AC, Viikari JSA, Ehnholm C, Peltonen L. No evidence of linkage between familial combined hyperlipidemia and genes encoding lipolytic enzymes in Finnish families. Arterioscler Thromb Vasc Biol. 1997;17:841–850. doi: 10.1161/01.atv.17.5.841. [DOI] [PubMed] [Google Scholar]
- Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XQJ, Soro-Paavonen A, Naukkarinen J, Saarela J, Laakso M, Ehnholm C, Taskinen MR, Peltonen L. Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1). Nat Genet. 2004;36:371–376. doi: 10.1038/ng1320. [DOI] [PubMed] [Google Scholar]
- Pauciullo P, Gentile M, Marotta G, Baiano A, Ubaldi S, Jossa F, Iannuzzo G, Faccenda F, Panico S, Rubba P. Small dense low-density lipoprotein in familial combined hyperlipidemia: Independent of metabolic syndrome and related to history of cardiovascular events. Atherosclerosis. 2009;203:320–324. doi: 10.1016/j.atherosclerosis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Purnell JQ, Brunzell JD. The central role of dietary fat, not carbohydrate, in the insulin resistance syndrome. Curr Opin Lipidol. 1997;8:17–22. doi: 10.1097/00041433-199702000-00005. [DOI] [PubMed] [Google Scholar]
- Purnell JQ, Kahn SE, Schwartz RS, Brunzell JD. Evidence of genetic control of elevated lipid and apoB levels in addition to visceral obesity/insulin resistance, in familial combined hyperlipidemia. J Investig Med. 1997;45:105A. [Google Scholar]
- Rao DC, Keats BJB, Morton NE, Yee S, Lew R. Variability of human linkage data. Am J Hum Genet. 1978;30:516–529. [PMC free article] [PubMed] [Google Scholar]
- Rivest LP. Statistical properties of Winsorized means for skewed distributions. Biometrika. 1994;81:373–383. [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:1020–1032. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonfeld G, Lin X, Yue P. Familial hypobetalipoproteinemia: genetics and metabolism. Cell Mol Life Sci. 2005;62:1372–1378. doi: 10.1007/s00018-005-4473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Yue P, Schonfeld G, Neuman RJ. Evidence for a quantitative trait locus affecting low levels of apolipoprotein B and low density lipoprotein on chromosome 10 in Caucasian families. J Lipid Res. 2007;48:2632–2639. doi: 10.1194/jlr.M700078-JLR200. [DOI] [PubMed] [Google Scholar]
- Shoulders CC, Jones EL, Naoumova RP. Genetics of familial combined hyperlipidemia and risk of coronary heart disease. Hum Mol Genet. 2004;13:R149–R160. doi: 10.1093/hmg/ddh069. [DOI] [PubMed] [Google Scholar]
- Sieh W, Yu C-E, Bird TD, Schellenberg GD, Wijsman EM. Accounting for linkage disequilibrium among markers in linkage analysis: impact of haplotype frequency estimation and molecular haplotypes for a gene in a candidate region for Alzheimer's disease. Hum Hered. 2007;63:26–34. doi: 10.1159/000098459. [DOI] [PubMed] [Google Scholar]
- Suarez BK, Hampe CL, van Eerdewegh P. Problems of replicating linkage claims in psychiatry. In: Gerson ES, Cloninger CR, editors. Genetic approaches to mental disorders. American Psychiatric Press; Washington, DC: 1994. pp. 23–46. [Google Scholar]
- Tregouet DA, Ducimetiere P, Tiret l. Testing association between candidate-gene markers and phenotype in related individuals, by use of estimating equations. Am J Hum Genet. 1997;61(1):189–199. doi: 10.1086/513895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakkilainen J, Pajukanta P, Cantor RM, Nuotio IO, Lahdenpera S, Ylitalo K, Pihlajamaki J, Kovanen PT, Laakso M, Viikari JSA, Peltonen L, Taskinen MR. Genetic influences contributing to LDL particle size in familial combined hyperlipidaemia. Eur J Hum Genet. 2002;10:547–552. doi: 10.1038/sj.ejhg.5200844. [DOI] [PubMed] [Google Scholar]
- Venkatesan S, Cullen P, Pacy P, Halliday D, Scott J. Stable isotopes show a direct relation between VLDL apo B overproduction and serum triglyceride levels and indicate a metabolically and biochemically coherent basis for familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 1993;13:1110–1118. doi: 10.1161/01.atv.13.7.1110. [DOI] [PubMed] [Google Scholar]
- Wang XS, Paigen B. Genome-wide search for new genes controlling plasma lipid concentrations in mice and humans. Curr Opin Lipidol. 2005;16:127–137. doi: 10.1097/01.mol.0000162317.09054.9d. [DOI] [PubMed] [Google Scholar]
- Warnick GR. Enzymatic methods for quantification of lipoprotein lipids. Methods Enzymol. 1986;129:101–123. doi: 10.1016/0076-6879(86)29064-3. [DOI] [PubMed] [Google Scholar]
- Warnick GR, Knopp RH, Fitzpatrick VF, Branson L. Estimating low-density lipoprotein cholesterol by the Friedewald equation is adequate for classifying patients on the basis of nationally recommended cutpoints. Clin Chem. 1990;36:15–19. [PubMed] [Google Scholar]
- Wierzbicki AS, Graham CA, Young IS, Nicholls DP. Familial combined hyperlipidaemia: under-defined and under-diagnosed? Curr Vasc Pharmacol. 2008;6:13–22. doi: 10.2174/157016108783331268. [DOI] [PubMed] [Google Scholar]
- Wiesbauer F, Blessberger H, Azar D, Goliasch G, Wagner O, Gerhold L, Huber K, Widhalm K, Abdolvahab F, Sodeck G, Maurer G, Schillinger M. Familial-combined hyperlipidaemia in very young myocardial infarction survivors (< or =40 years of age). Eur Heart J. 2009;30:1073–1079. doi: 10.1093/eurheartj/ehp051. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Amos CI. Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: summary of GAW10 contributions. Genet Epidemiol. 1997;14:719–735. doi: 10.1002/(SICI)1098-2272(1997)14:6<719::AID-GEPI28>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Yu D. Joint oligogenic segregation and linkage analysis using Bayesian Markov chain Monte Carlo methods. Mol Biotechnol. 2004;28:205–226. doi: 10.1385/MB:28:3:205. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Brunzell J, Jarvik GP, Austin M, Motulsky A, Deeb S. Evidence against linkage of familial combined hyperlipidemia to the apolipoprotein AI-CIII-AIV gene complex. Arterioscler Thromb Vasc Biol. 1998;18:215–226. doi: 10.1161/01.atv.18.2.215. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Rothstein JH, Thompson EA. Multipoint linkage analysis with many multiallelic or dense diallelic markers: MCMC provides practical approaches for genome scans on general pedigrees. Am J Hum Genet. 2006;79:846–858. doi: 10.1086/508472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox MA, Pugh EW, Zhang H, Zhong X, Levinson DF, Kennedy GC, Wijsman EM. Comparison of single-nucleotide polymorphisms and microsatellite markers for linkage analysis in the COGA and simulated data sets for Genetic Analysis Workshop 14: Presentation groups 1, 2, and 3. Genet Epidemiol. 2005;29(Suppl 1):S7–S28. doi: 10.1002/gepi.20106. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen HQ, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski A, Farrall M, Cullen P, Wilson T, Bayliss D, Farren B, Griffin B, Caslake M, Packard C, Shepherd J, Thakker R, Scott J. Familial combined hyperlipidaemia linked to the apolipoprotein AI-CII-AIV gene cluster on chromosome 11q23-q24. Nature. 1991;349:161–164. doi: 10.1038/349161a0. [DOI] [PubMed] [Google Scholar]
- Yuan B, Neuman RJ, Duan SH, Weber JL, Kwok PY, Saccone NL, Wu JS, Liu KY, Schonfeld G. Linkage of a gene for familial hypobetalipoproteinemia to chromosome 3p21.1–22. Am J Hum Genet. 2000;66:1699–1704. doi: 10.1086/302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon A, Brunzell JD. The lipoprotein metabolism in the plurimetabolic syndrome. In: Crepaldi C, Tiengo A, Manzato E, editors. Diabetes, obesity and hyperlipidemia: the plurimetabolic syndrome. Elsevier; New York: 1993. pp. 197–200. [Google Scholar]
- Zambon A, Hokanson JE, Brown BG, Brunzell JD. Evidence for a new pathophysiological mechanism for coronary artery disease regression—hepatic lipase-mediated changes in LDL density. Circulation. 1999;99:1959–1964. doi: 10.1161/01.cir.99.15.1959. [DOI] [PubMed] [Google Scholar]
- Zambon A, Brown BG, Deeb SS, Brunzell JD. Genetics of apolipoprotein B and apolipoprotein AI and premature coronary artery disease. J Intern Med. 2006;259:473–480. doi: 10.1111/j.1365-2796.2006.01645.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Trait mode-of-inheritance models for apoB: selected models evaluated in the case of CSA, and sub-models with highest posterior probabilities from OSA.
Supplementary Table 2. Lod scores obtained for analysis with individual markers for family F3, using a parametric lod score approach with model parameters from complex segregation analysis indicated in Supplementary Table 1.