Abstract
Microfluidic technologies have been applied extensively in rapid sample analysis. Some current challenges for standard microfluidic systems are relatively high detection limits, and reduced resolving power and peak capacity compared to conventional approaches. The integration of multiple functions and components onto a single platform can overcome these separation and detection limitations of microfluidics. Multiplexed systems can greatly increase peak capacity in multidimensional separations and can increase sample throughput by analyzing many samples simultaneously. On-chip sample preparation, including labeling, preconcentration, cleanup and amplification, can all serve to speed up and automate processes in integrated microfluidic systems. This paper summarizes advances in integrated multi-process microfluidic systems for automated analysis, their benefits and areas for needed improvement.
INTRODUCTION
Microfluidic analysis systems have advanced rapidly since the early 1990s,1, 2 providing new capabilities for chemistry, biology, and medicine. For instance, microfluidic devices offer low sample and reagent consumption3 (which is critical for expensive pharmaceutical characterization or trace samples), small dead volume,4 fast mixing,5–7 rapid analysis speed,8 high throughput,9 and valveless flow control.10 Consequently, these advantages of microfabricated devices have been exploited widely in bioanalysis, and reviews cover areas such as protein separation,2, 11 cell analysis,12–14 genomics,15, 16 and biomarker assays.17, 18 Because the field of microfluidics has become so broad, our focus here is on integrated microfluidic methods in separation-based analysis with strong automation potential.
To date, many microfluidic designs have made, but they are generally tested with low complexity samples. For actual biological specimens, which are mixtures with wide analyte concentration ranges, it remains a challenge to directly separate even tens of components on microdevices. The small microchip platform size usually results in a short separation length, limiting the resolving power and peak capacity, which are critical for separating complex mixtures.19 For instance, the peak capacity of a polydimethylsiloxane (PDMS) microchip for micellar electrokinetic chromatography (MEKC) was ~12 for protein separation.20 Importantly, to completely isolate a 20-component mixture with 95% probability, the peak capacity must be ~800.21 Clearly, resolving power and peak capacity in microfluidic systems could be improved. In addition, tiny sample volumes (usually in the microliter range)22 are placed on microdevices, and often nanoliter or smaller volumes are injected. Furthermore, microchips generally have a short optical detection path,23 such that the detection limit is another aspect of microfluidic devices that could be improved.
Fortunately, these separation and detection limitations can be overcome by integrating multiple functions and components at the chip scale. Methods for microfluidic device fabrication are generally based on photolithographic processes, which make complex designs possible.24 Moreover, fabrication techniques have been developed to transfer these complex designs into low-cost materials like plastics.25, 26 By integrating sample preparation processes into a single microdevice, trace samples can be preconcentrated before analysis. Multi-dimensional separations on-chip can significantly improve the sample capacity. Importantly, because the samples in many integrated microdevices are manipulated by voltages, these microfluidic systems can be readily automated. Compared with traditional methods, automated sample analysis can be more economical, requiring less human intervention, and enabling increased sample throughput.27 Consequently, these advantages make integrated microdevices especially attractive for automating the characterization of complex mixtures.
Since the applications and principles of integrated microdevices have been reviewed elsewhere,2, 24 we focus this review on integrated microfluidic methods with high potential for automating analysis. Multiplexed separation and on-chip sample preparation will be emphasized in this work. We note that on-chip sample preparation is a broad topic, encompassing cell analysis,14 sample purification28 and other technologies. Hence, to provide an in-depth discussion, we limit the scope of this review to the sample preparation areas of labeling, preconcentration, and PCR amplification.
MULTIPLEXED SEPARATION
(a) Multidimensional Systems
Because the overall peak capacity of multidimensional separations is the product of the peak capacities of the individual, orthogonal one-dimensional methods,29, 30 these systems are of great interest for complex mixture analysis. For example, two-dimensional gel electrophoresis (2DE) is an established approach for high-resolution profiling of proteins,31 separating analytes according to isoelectric point in the first dimension (isoelectric focusing, IEF), and then by mass-to-charge ratio in the second dimension (polyacrylamide gel electrophoresis, PAGE). Despite its enormously successful application in biochemistry and clinical studies,32 the downsides of 2DE are also significant: extensive hands-on labor (gel preparation, staining, etc.) and slow separation (approximately one day).33 To increase throughput and facilitate automation, 2DE has been transferred into a microfluidic platform. For instance, MEKC coupled with capillary electrophoresis (CE) was demonstrated for peptide separation in 2000.34 However, this approach used different buffers for the two dimensions, which in turn increased the complexity of device operation. More recently, Herr et al.30 developed a microchip IEF-CE system which used the same buffer for both dimensions. Microchip IEF-PAGE systems have now been automated, providing a separation time of <2 hours.35 Another approach for 2DE involves using a gel for the first dimension, and a solution electrophoresis method for the second dimension, as implemented by Osiri et al.36 with a capillary gel electrophoresis-MEKC system that was used to profile fetal calf serum proteins. Chen et al.37 recently reviewed two-dimensional microchip separations, and the reader can refer to this reference for additional information.
Future multidimensional electrophoresis microdevices will need to further increase peak capacity through more effective coupling of separation dimensions. Improvements to device fabrication and operation should enhance the interface between the first and second separation techniques, potentially providing additional information.
(b) Parallel separations
Although multidimensional separations can increase peak capacity, usually only one sample is analyzed per run. Sample throughput or capacity can be increased by performing separations in parallel columns, in a manner similar to what is done in slab gel electrophoresis. In a microchip format, forming parallel capillaries is readily achieved via photolithographic patterning. The first capillary array system for microchip electrophoresis was demonstrated in 1997 with 12 separation channels in parallel (Fig. 1A).38 In this design, channels terminate at one cathode reservoir while the other end of each lane has a cross injection design with 3 reservoirs; this device was successfully utilized in human HLA-H genotyping. Mathies’ group further explored a 48-channel design with 96 sample reservoirs.39 The unique rectilinear layout facilitated sample loading via a multichannel micropipettor. However, detection constraints made scaling to more channels difficult; moreover, in this design two samples shared one separation channel, which increased the possibility of cross contamination.
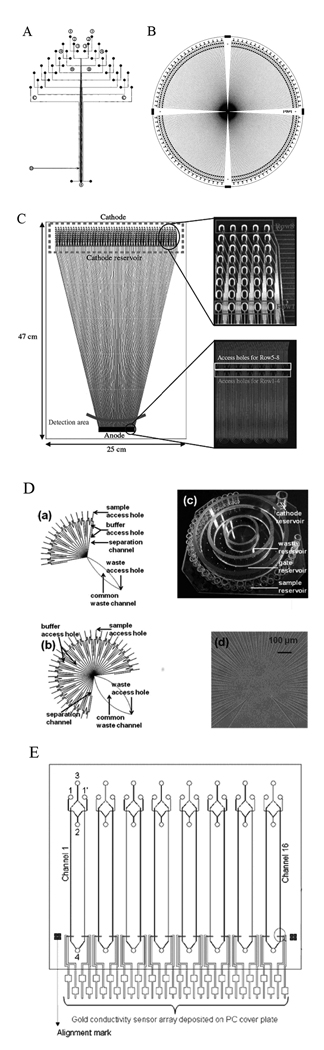
Figure 1.
Parallel separations in microdevices. (A) 12-channel microdevice with linear fluorescence scan detection; chip size was 50 mm × 75 mm. (B) 384-lane radial microdevice for rotary fluorescence scanning detection on a 200-mm-diameter wafer. Lanes are ~60 µm wide and 30 µm deep, and the effective separation length is 8.0 cm. (C) 384-lane fan-shaped microdevice. The plate dimensions are 47 cm × 25 cm × 1.4 mm, and the dimensions of each channel are 40 µm (depth) × 90 µm (top width). The upper inset shows oval sample holes connected directly to the separation channels. The lower inset shows access holes for an anode reservoir. (D) 16 or 36 parallel separation channels used with CCD detection. (a) Design of a microfluidic network containing 16 parallel separation channels on a 76 × 76 mm2 Borofloat glass substrate. (b) Design of a 36-channel network. (c) Photograph of a 36-channel chip. (d) Image of the detection area on a 36-channel chip. (E) 16-channel device with an integrated contact conductivity sensor array detector. The lengths of the injection channel and separation channel were 9 mm and 54 mm, respectively. Channel sizes were 60 µm (width) × 40 µm (depth). Topographical layout of the lithographically printed Au conductivity sensor array (gray); the outlet end consisted of 16 Au electrodes (7.62 mm long × 500 µm wide) serving as the conductivity sensors. Adapted and reproduced from Refs. 9, 38, 44, 45, and 47, with permission from ACS and Wiley.
Because of limitations of the rectilinear format, a radial microplate design with 96 channels was developed.40 The radial design used a rotary confocal fluorescence scanning system to probe each channel. Importantly, with an increase in wafer size, more lanes or longer separation channels could be obtained. For instance, 384 lane microchip DNA analyzers were constructed in 8 inch (200 mm) diameter substrates (Fig. 1B).9 Separation length and resolution are somewhat limited in the radial design; a 6 inch (150 mm) diameter wafer only offers a 5.5 cm separation length,41 which is not ideal for DNA sequencing. For a fixed device size, the separation length can be increased by folding the channel in a serpentine fashion. However, because the inside track of a turn will be shorter than the outside track, a “U” shaped turn will cause band dispersion, reducing resolution. Paegel et al.42 found that this dispersion can be significantly reduced by narrowing the width of a channel in a turn, which extended the separation length to ~16 cm on a 6 inch diameter wafer, yielding an average sequencing read of 430 bases.43 Recently, Kumagai et al.44 developed a microfabricated DNA sequencing device with 384 lanes in a fan shape (Fig. 1C). The microplate formed a part of a much larger automated apparatus which yielded a throughput of 5×106 bases per day per instrument.
CCD-based fluorescence detection is also being pursued as a simpler setup than the scanning confocal arrangement. Pei et al.45 developed a multichannel system for enzyme assays using a CCD detector (Fig. 1D). In this radial system, parallel separation channels were directed to a common waste reservoir via a channel, and the CCD captured light at the center to obtain fluorescence images of all the channels. Recently, Dishinger et al.46 used a similar system to monitor insulin secretion using 15 parallel channels.
Because fluorescence detection requires complex equipment, making miniaturization a challenge, alternative approaches to parallel lane detection are being pursued. Shadpour et al.47 developed a 16-channel device with an integrated contact conductivity sensor array (Fig. 1E). In this system, a gold conductivity sensor array was first patterned on a polycarbonate film and then aligned with a substrate having hot-embossed microchannels, thus interfacing each separation channel with a pair of Au electrodes. Separations were carried out for amino acids, peptides, proteins, and oligonucleotides. However, the limit of detection (LOD) was poorer than for conductivity detection in a single channel, and the LOD was considerably worse than for fluorescence detection. Moreira et al.48 developed a multichannel system with a single electrode for amperometric detection, which worked when the channels were operated serially, rather than in parallel.
A major emphasis for future work in parallel separations should be enhancing and simplifying detection, since fabrication capabilities have become quite advanced. Simpler alternatives to scanning confocal fluorescence detection, such as CCD fluorescence detection and electrochemical methods, are attractive targets.
ON-CHIP SAMPLE PREPARATION
(a) Labeling
Due to small channel dimensions, a good detector is an essential part of microfluidic systems. Demonstrated detection methods in microdevices include UV absorbance,49 laser induced fluorescence (LIF),50 mass spectrometry,51 electrochemistry,52 and chemiluminescence.53 Of these methods, LIF is the most popular because of its low LOD.54 Although many proteins can be directly detected via native fluorescence from tryptophan, an uncommon deep-UV light source is needed for optimal excitation.55 Therefore, derivatization of proteins with a fluorophore is typically needed for LIF detection. Fluorescein isothiocyanate (FITC) is widely employed in off-chip labeling due to its high quantum yield (~0.7) and water solubility.56 However, because of the slow rate of reaction for FITC with amine groups (12–24 hours at room temperature),56 off-chip FITC labeling limits throughput and prolongs the analysis time. Thus, integrating the labeling process into a microfluidic system can automate and speed up analysis. Typically, the derivatization of sample on-chip can be performed either before (precolumn) or after (postcolumn) separation.
In precolumn labeling, the dye and sample are generally mixed for a controlled time in a diffusion-based reaction chamber before separation. The first precolumn labeling in glass microchips was demonstrated by Jacobson et al.,57 and similar results have also been shown in polymeric microdevices.58, 59 Yu et al.59 demonstrated a precolumn labeling system using fluorogenic “chameleon” dyes. These labels offer fast reaction times, and the net charge on molecules is unchanged after reaction. Recently, Mair et al.60 developed a periodic monolith microfluidic system which yielded a modest improvement in the mixing efficiency, leading to a 22% greater fluorescence level than in an open channel design. Digital (droplet) microfluidics show promise for precolumn labeling; for instance, two droplets can be manipulated and mixed readily using this approach.7
Fluorescence tagging often results in multiply labeled analytes having different separation properties that can result in multiple peaks for a single analyte.61 In addition, fluorescent labeling can change the analyte charge or size, which can also negatively impact separation with precolumn tagging. Because of these limitations, postcolumn labeling is a desirable strategy. In this format, the fluorescent tag is added at the end of column, such that the numbers and positions of attached dyes have little time to influence separation. For postcolumn labeling, the reaction kinetics must be fast, and the mixing efficiency of sample and dye streams must be thorough to reduce band broadening. In the first microfluidic system with postcolumn labeling,62 significant band broadening occurred, and separation efficiency was rather low. To improve efficiency, Fluri et al.63 found that channel widths should be narrow (~45 µm) to facilitate rapid diffusion, and pH differences in the mixing solutions should be minimized. Liu et al.64 used a noncovalent label, NanoOrange, to bind hydrophobic regions of proteins and provide fluorescence emission. Their results indicated that the labeling reaction rate was close to the diffusion limit, which resulted in little band broadening. Sieben et al.65 developed devices that have two injection systems, one for the sample and the other for the label. By using pinched injection,22 a controlled plug of label could be loaded into the separation channel. An additional purge reservoir was integrated into the device to clean out the separation channel after labeling. A similar protocol has also been utilized for chemiluminescence detection.66 In this approach, the detection system was simplified compared with LIF, since no light source was needed.
A key area for future development in on-chip labeling will be to improve detection limits to near what is achieved with off-chip labeling. Another important future direction will be to implement on-chip labeling in parallel analysis systems.
(b) Traditional online preconcentration techniques
Sample concentration techniques in CE such as sweeping and stacking have been proven effective for pharmaceutical species,67 herbicides,68 steroids,69 and peptides.70 These methods have also been applied in microfluidic formats. For instance, Jung et al.71 developed a porous polymer plug in a microchannel to create a high conductivity buffer zone, and enriched fluorescent analytes 1000-fold using field-amplified sample stacking. The same group developed CE microchips coupled with isotachophoresis, which could enrich Alexa Fluor 488 nearly two million fold,72 and under optimized conditions the detection limit of Alexa Fluor 488 was ~100 aM.73 However, it is important but difficult to find suitable leading and terminating electrolytes for isotachophoresis. A review on stacking and sweeping in microchip systems was recently published.74
Desirable future emphases in microchip usage of traditional capillary preconcentration methods are apparent. First, direct comparisons of the performance of microchip vs. capillary methods should be done. Additionally, microchip experiments should be carried out on real samples in complex matrixes.
(c) Solid phase extraction
Solid phase extraction (SPE) is a widely used method for sample preparation. It can be fully automated with commercial systems like SPE-DEX (Horizon Technology), OSP2 (Merck), and MicroLab SPE (Hamilton).75 In SPE, sample is retained on a solid medium, allowing the matrix to be rinsed away and the retained material to be eluted for analysis.76 The promise of sample enrichment and cleanup by SPE has led researchers to apply this approach in microdevices. A SPE column has been fabricated by coating microchip walls with silanes, and 80-fold preconcentration of coumarin C460 was observed;77 however, due to the limited surface area, the loading capacity of this approach was relatively low.
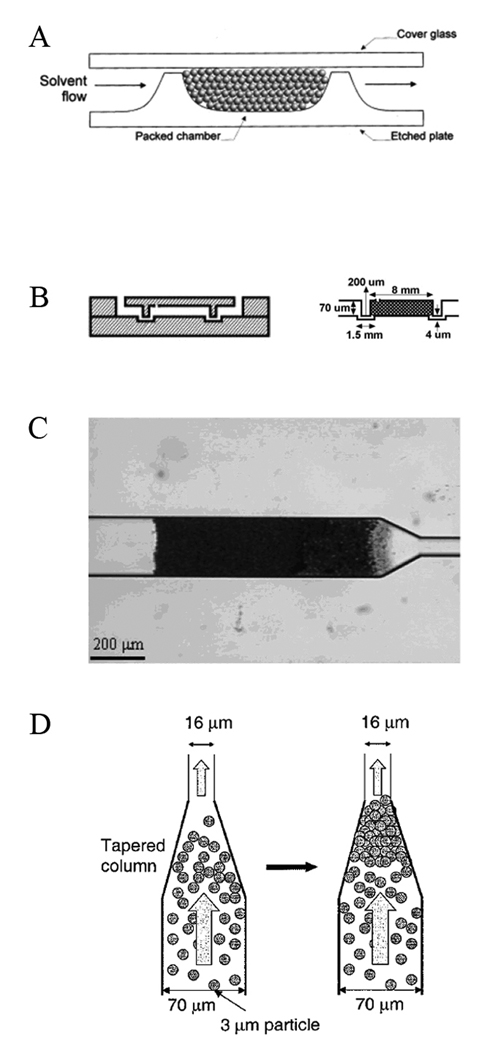
Silica bead78 and polymer monolith79, 80 SPE columns with high surface area and greater loading capacity have been integrated into microchips. Because silica beads are commercially available and their properties are well characterized, microchip SPE columns made by packed beads are attractive. However, it is necessary to localize these particles in targeted regions of microchips using physical barriers. For example, a sol-gel structure was fabricated to retain silica beads, and this system was tested in on-chip DNA purification.81 In an alternate format, a two-weir design which created a cavity to trap beads was explored (Fig. 2A).82 Two photomasks were used in device fabrication, one to pattern the tops of the weirs for etching, and the other to pattern the channels for etching to a different depth. In this manner, a 1-µm gap was formed to prevent beads from passing out from the SPE bed.78 Zhong et al.83 developed a two-side etching and alignment protocol to construct weirs in a different manner (Fig. 2B). A top plate containing weirs and a bottom plate having the connection channels were aligned, and a 4-µm gap was created by sealing the plates together. Instead of a microfabricated weir structure, a physical barrier can be prepared on-chip with a photopolymerized frit (Fig. 2C).84 In a different approach, beads can be packed through a tapered geometry by the keystone effect (Fig. 2D).85 The channel which contained the beads tapered from a 70-µm to a 16-µm width. When beads flowed through the channel, the density of the particles increased in the taper, such that they aggregated without a physical barrier.
Figure 2.
Retaining packed beads in microchannels. (A) Two-weir design, showing weir heights in relation to channel depth and particle size. Electroosmotic flow is driven by walls and by free silanol groups on particles. Solvent flow direction is indicated for a preconcentration step. (B) Two-side etching/alignment protocol. Two prefabricated plates were aligned and thermally bonded. On the right is a drawing of the cross section of a packed chip with its dimensions. (C) Photopolymerized frits retaining 3-µm-diameter beads. (D) The keystone effect. A suspension of particles is flowed toward the taper by vacuum. At the taper, the density of the particles increases, and these first particles act as “keystones”, blocking the others and allowing the packed segment to grow. Adapted and reproduced from Refs. 82–85, with permission from ACS and Wiley.
Packed-bead columns have disadvantages in terms of packing procedures and frit fabrication, which complicate microdevice preparation. On the other hand, monoliths are an attractive alternative to packed particles because of low backpressure and high surface area.86 Thermally polymerized monolith materials have been successfully applied as SPE columns.87 In 2001 Yu et al.79 photopolymerized a monolith column in a microfluidic system and performed SPE. Enrichment of peptides and proteins up to 1000 fold was achieved on this column. More importantly, due to low backpressure, the linear flow rate in these monoliths could reach 10 µL/min, which far exceeded flow in packed microchip columns.
Monolith columns have also been applied for DNA enrichment in complex mixtures like blood. However, nonspecific binding hindered elution of nucleic acids and decreased sample loading capacity due to competitive adsorption; the presence of proteins lowered the monolith extraction efficiency from ~80% to <40%.88 Therefore, Wen et al.89 developed a two-stage microchip SPE system. Before monolith column extraction, a C18 reversed-phase column was used to remove proteins in the sample. Although the procedure was more complex, whole blood DNA extraction capabilities were significantly improved. For a 10-µL whole blood sample, ~70% of the protein was removed by the C18 column, affording more interaction between DNA and the monolithic material. This two-stage system enriched DNA ~20 fold in the reversed-phase portion, and the overall DNA extraction efficiency was ~70%.
In the previous applications described, microchip monolith SPE enriched analytes based on general interactions like hydrophobic absorption. To improve the selectivity, affinity elements can be immobilized on a monolith. Glycoproteins were retained on a monolith with immobilized agglutinin, and then eluted in several fractions due to different affinities.90 Yang et al.91 prepared anti-FITC modified sample pretreatment monoliths in microfluidic devices. FITC-labeled amino acids were enriched 20-fold and purified from a mixture containing a contaminant protein. However, the extraction and separation were performed on separate devices, which hindered automation. To circumvent this, Sun et al.92 coupled anti-FITC affinity monoliths with electrophoretic analysis on a single device. Sample loading, rinsing, elution, and separation were all performed in an automated manner by controlling the potentials applied to various reservoirs. FITC-tagged species were selectively retained by the immunoaffinity column and separated from other contaminants. The retained proteins were then eluted from the monolith with 200 mM acetic acid.
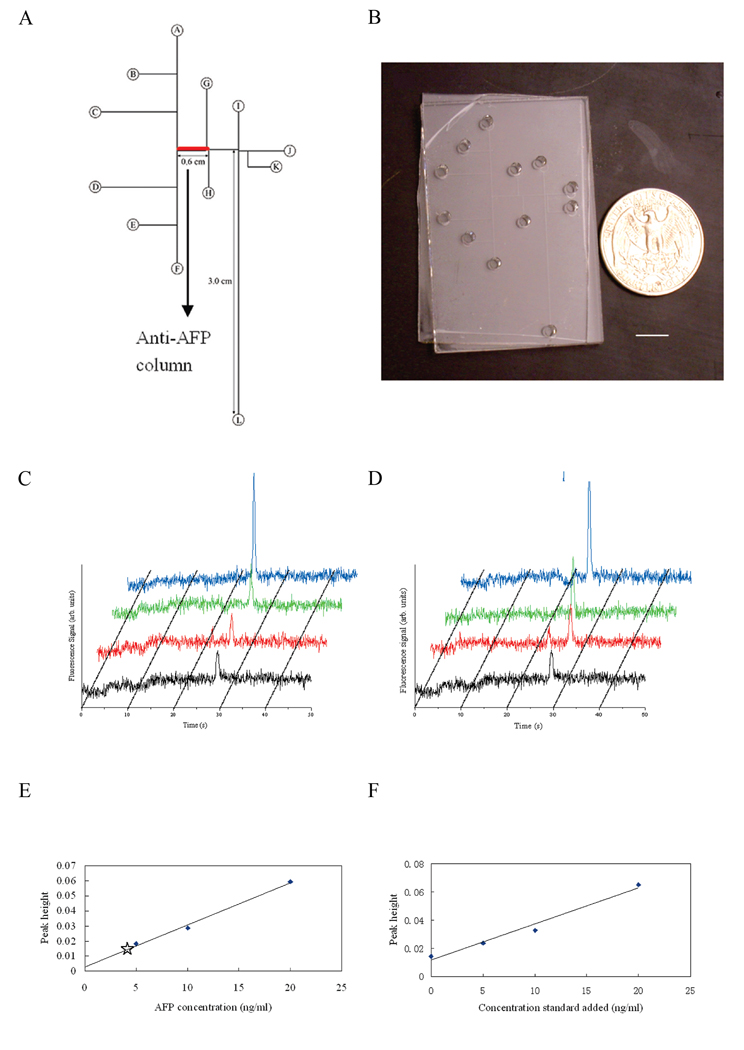
In addition to monoliths, other affinity columns have also been integrated into a microfluidic format. For instance, microchannel surfaces were coated with silane and then protein A via physisorption.93 Rabbit immunoglobulin G was concentrated and detected at 50 nM levels. However, this approach, as well as the other monolith work discussed previously, was only tested for capturing target analytes in buffer solutions instead of complex matrixes like tissue or blood. Recently, Yang et al.94 developed an integrated microfluidic system to perform quantitative determination of alpha-fetoprotein (AFP) in human serum, using both the method of standard addition and a calibration curve (Figure 3). The microdevices were made of poly(methyl methacrylate) (PMMA), and a photo-defined polymer was formed on the microchannel surface, which allowed antibody immobilization. All assay steps, including affinity extraction, elution, separation, and quantification, were performed on-chip in an automated manner via changing the applied potentials in the reservoirs. AFP concentrations in human serum measured in these microdevices using both calibration curve and standard addition methods compared favorably with those determined using a commercial assay kit. Phillips et al.95, 96 have utilized immunoaffinity CE to measure biomarkers and neuropeptides in human biopsies. The analytes were captured by a replaceable immunoaffinity disk having attached antibodies. After removing non-target materials, the captured analyte was labeled in situ, released, and then separated by microchip CE. The system was semi-automated, and the separation step was completed within 5 min.
Figure 3.
Integrated affinity column–microchip CE devices for biomarker quantitation. (A) Layout of an integrated AFP analysis microdevice. (B) Photograph of a microfluidic device with integrated anti-AFP affinity column. Scale bar is 1 cm. (C) Microchip CE of Alexa Fluor 488 labeled human serum and of AFP standard solutions after affinity column extraction. (D) Microchip CE of Alexa Fluor 488 labeled human serum after standard addition and affinity column extraction. (E) Calibration curve generated from peak heights in (C), with the unknown sample data point indicated with a star. (F) Standard addition plot of concentration of standard added vs. peak height generated from peak heights in (D). Adapted and reproduced from Ref. 94, with permission from ACS.
Non-electrically driven immunoassays can also be performed in microchip devices. For instance, Kong et al.97 formed elastomeric microvalves in 3-layer microchips to control flow, although the valves were actuated by a vacuum pump and a compressor. Using this system, clenbuterol was determined in pig urine samples in 30 min. Fan et al.98 designed a PDMS-on-glass microsystem to perform protein assays on blood samples. Plasma was separated from whole blood on chip, and selected proteins were detected by antigen-antibody interaction.
The variety of column materials used thus far (commercial silica particles, monoliths, etc.) indicates that consensus is still lacking as to the optimal column properties. Future efforts should focus on determining which type of column works best for a given analysis. In addition, further work is needed to streamline and simplify column fabrication.
(d) Membrane filtration
Another common method for sample preconcentration and cleanup is membrane filtration, which utilizes the size difference between analytes and buffer ions. Larger molecules cannot pass through a porous layer in a semipermeable hollow fiber,99 membrane,100 or joint,101 while smaller species are allowed to transit. In one design, a porous membrane was sandwiched between two PDMS pieces to create a three-dimensional microfluidic channel structure.102 This system achieved 300-fold concentration of fluorescein in around 40 min. The fluorescein was concentrated outside a 10-nm pore membrane (with openings larger than the molecular size of fluorescein), because the negatively charged diffuse layer on the interior of the membrane repelled anions. Song et al.103 used a laser to pattern a nanoporous membrane at the junction of a cross channel. This device could concentrate proteins over 100–fold in 2 min, and the degree of concentration was limited only by analyte solubility. Similarly, an anionic polyacrylamide gel preconcentrator was laser photopolymerized in one arm of a cross channel in a PMMA microdevice.104 The negatively charged sulfonate groups in the gel repelled negatively charged proteins, enabling concentration of proteins up to 100,000-fold. Foote et al.100 used a silicate membrane deposited between two adjacent microchannels, and a ~600-fold signal increase for proteins was achieved. Kim et al.105 developed a simple protocol to fabricate a nanoporous membrane in microdevices. They used razor blades to form a gap in microchannels in a PDMS substrate; Nafion 117 was then filled into the gap and a portion of the microchannels via capillary forces. In this protocol, preconcentration was achieved in large channels with dimensions up to 0.1 mm by 1 mm.
Semi-permeable membranes can also be integrated with other microchip functionalities. Herr et al.106 fabricated a size-exclusion membrane at the injection junction of a microdevice, allowing antibody enrichment at the membrane surface. Sample loaded on the membrane was captured via antigen-antibody interaction, and enriched species were eluted into a separation channel for electrophoretic immunoassay. This system measured a biomarker for periodontal disease in saliva in <10 min with comparable results to conventional methods. A similar design has been developed into a portable diagnostic format for rapid detection of biological toxins.107 A membrane can also be used for solid-phase extraction. Lion et al.108 integrated a poly(vinylidene difluoride) membrane to desalt and concentrate samples before analyzing with mass spectrometry. Kim et al.109 sandwiched an aluminum oxide membrane between PDMS pieces; when a blood sample passed through the membrane, DNA was selectively enriched and then eluted with buffer. The extraction time was <10 min while the recovery was ~40 ng of DNA per microliter of blood. A membrane has also been applied to enrich nonvolatile analytes by evaporation to reduce the amount of liquid phase.110 The membrane was located at the interface between a gas and liquid channel; sample was introduced into the liquid channel, and water evaporated into the gas channel through which nitrogen was flowing.
Presently, membranes are formed through sandwiching between channels or photopolymerization. Although these methods work acceptably for prototyping efforts, more straightforward fabrication techniques will be needed in the future. Moreover, the relatively slow rate of enrichment in membrane-based systems is an issue that could be addressed with highspeed membrane-based filtration devices.
(e) PCR amplification for DNA analysis
The polymerase chain reaction is an exponential amplification technique for DNA diagnostics. The method relies on thermally cycling samples in different temperature zones as follows: denaturation to single-stranded DNA, annealing primers to the single-stranded DNA template, and polymerase extension of the annealed duplex DNA. Miniaturization of PCR allows rapid thermal cycling, small sample quantities, and potential to integrate with other microfluidic methods. Since the first report of integrated PCR-CE microdevices,111 considerable progress has been made in the coupling of PCR with other technologies (CE, immunoassay, cell isolation, DNA arrays, etc.) on microdevices. Because Chen et al.112 reviewed integrated PCR microfluidic technologies in 2007, we focus here on more recent breakthroughs in the field.
Liu et al.113 developed a portable microsystem for forensic genetic analysis. All the electronics for temperature control, microfluidic manipulation and CE separation, as well as optics for four-color LIF detection, were integrated in a 12×10×4 in.3 box. Oral swabs and human bone extracts were successfully analyzed by this system in less than 1.5 h. To improve upon the efficiency of cross injectors, an inline-injection system114 with DNA-based affinity capture was developed. Immobilized oligonucleotides in a polyacrylamide gel provided more efficient injection of DNA fragments while cleaning up sequencing samples. Subsequently, a photo-defined, crosslinked polyacrylamide gel affinity matrix was integrated with PCR-CE microdevices.115 This affinity system increased the injection efficiency to nearly 100% and minimized band broadening. The devices were evaluated in detecting diluted Escherichia coli O157 in a high background of E. coli K12.
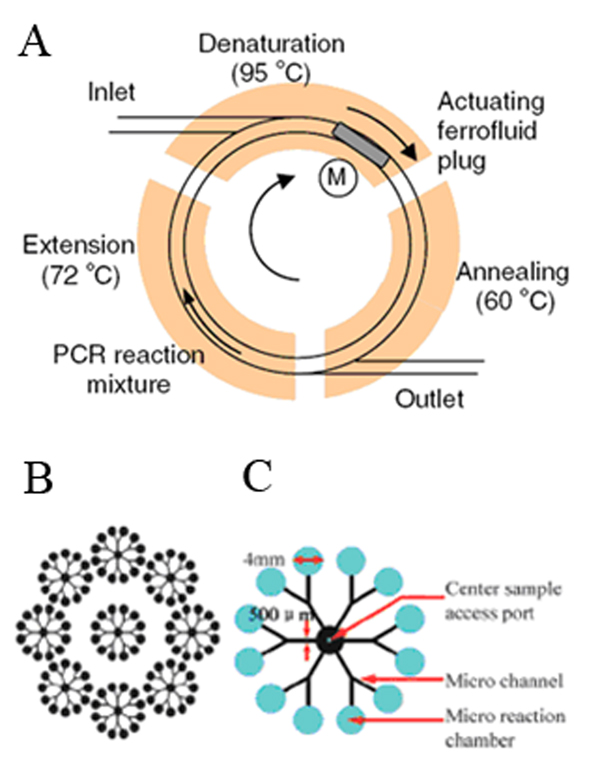
Continuous-flow PCR offers another format for integrating PCR into microdevices. In this approach, instead of heating and cooling a sample in a fixed location, three regions on a device are maintained at constant temperature, and the sample is passed repeatedly through these zones to thermally cycle. Recently, Sun et al. developed a system in which PCR solution was driven magnetically around a loop in either one channel116 or multiple channels.117 As shown in Figure 4A, the system had three temperature zones, a magnet, and a ferrofluid (a stable suspension of magnetic particles in oil). With the rotation of the magnet, the ferrofluid plug moved accordingly and cycled the PCR reaction mixture around the circular channel. This system was applied in amplifying genetically modified soy and maize samples in less than 13 min.
Figure 4.
Integrated PCR microdevices. (A) Continuous-flow PCR driven by the magnetic force. PCR reaction mixture, pushed by a ferrofluid plug, flows continuously through three temperature zones in the circular channel. (B) PCR array device layout for DNA methylation analysis. Nine functional units are arranged in a circular pattern. A total of 108 reactions can be done with nine sample injections, which significantly improves the throughput. (C) Functional unit layout. The sample inlet is 0.64-mm diameter and reaction chambers at the periphery are 4-mm diameter. Primers specific for tumor suppressor promoters are deposited onto the bottom surface of designated reaction chambers by pipetting. Adapted and reproduced from Refs. 116 and 118, with permission from Springer and RSC.
Methylation-specific PCR is used to analyze DNA methylation, which is associated with tumorigenesis. Zhang et al.118 developed a droplet-in-oil microfluidic system for high-throughput DNA methylation detection. The design layout, which allowed 108 reactions to be performed in parallel, is shown in Figure 4B–C. The droplet-in-oil protocol reduced contamination and prevented evaporation during thermal cycling.
Integrated PCR methods have become rather advanced, such that many important areas for future work will be in the application of these high-performance systems to real-world problems. Large-scale implementation of integrated microchip PCR is envisioned in fields such as food safety testing and forensics.
SUMMARY
The advancement of microfluidic technologies has provided faster and smaller analysis systems. However, the detection limit and peak capacity of many microdevices are not ideal for trace analyses in complex mixtures. Integrating multiple sample preparation functions into microfluidic formats can improve throughput, simplify pretreatment, and most importantly, automate processes.
Multidimensional microdevices can significantly increase speed in complex sample analysis; however, the inability to analyze highly complex mixtures remains a limitation for these systems. Array platforms can either analyze one sample in parallel or many samples simultaneously; to date, 384 parallel lanes have been integrated into a microdevice. Online labeling can eliminate sample preparation steps and increase analysis speed while automating processes. Multiple dye attachment in precolumn labeling can result in band broadening, while choosing an appropriate dye is critical for post-column labeling. To preconcentrate sample or remove impurities before analysis, SPE and membrane filtration have been integrated into microdevices. Samples can be concentrated by 2 to 4 orders magnitude under optimized conditions. For on-chip SPE, packed beads and monolith columns are widely used as stationary phases. Packed particles in microchannels require a suitable retaining structure, while monoliths are attractive because of low backpressure and high surface area. Integrated affinity extraction can effectively capture target components from a complex matrix and therefore is desirable for biological sample analysis. Semipermeable membranes can enrich samples up to 100,000-fold, although further work is needed in developing straightforward fabrication methods. PCR can be integrated into portable microsystems for various genetic analyses, and progress continues in this area.
Multifunctional microdevices have allowed researchers to analyze biological samples in an automated manner. Importantly, integrated microchips have provided improvements to sample throughput, peak capacity, and limit of detection. With continued development, integrated microfluidic devices will be more robust and fully automated for high-throughput complex sample analysis in the near future.
ACKNOWLEDGEMENTS
We thank Elisabeth Pound and Chad Rogers for assistance with technical editing. This work was supported by a Presidential Early Career Award for Scientists and Engineers through the National Institutes of Health (R01 EB006124).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.West J, Becker M, Tombrink S, Manz A. Micro total analysis systems: latest achievements. Anal. Chem. 2008;80:4403–4419. doi: 10.1021/ac800680j. [DOI] [PubMed] [Google Scholar]
- 2.Dittrich PS, Tachikawa K, Manz A. Micro total analysis systems. Latest advancements and trends. Anal. Chem. 2006;78:3887–3908. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 3.Auroux PA, Iossifidis D, Reyes DR, Manz A. Micro total analysis systems. 2. Analytical standard operations and applications. Anal. Chem. 2002;74:2637–2652. doi: 10.1021/ac020239t. [DOI] [PubMed] [Google Scholar]
- 4.Fuentes HV, Woolley AT. Electrically actuated, pressure-driven liquid chromatography separations in microfabricated devices. Lab Chip. 2007;7:1524–1531. doi: 10.1039/b708865e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang X, Ng JM, Stroock AD, Dertinger SK, Whitesides GM. A miniaturized, parallel, serially diluted immunoassay for analyzing multiple antigens. J. Am. Chem. Soc. 2003;125:5294–5295. doi: 10.1021/ja034566+. [DOI] [PubMed] [Google Scholar]
- 6.Sudarsan AP, Ugaz VM. Fluid mixing in planar spiral microchannels. Lab Chip. 2006;6:74–82. doi: 10.1039/b511524h. [DOI] [PubMed] [Google Scholar]
- 7.Abdelgawad M, Watson MW, Wheeler AR. Hybrid microfluidics: a digital-to-channel interface for in-line sample processing and chemical separations. Lab Chip. 2009;9:1046–1051. doi: 10.1039/b820682a. [DOI] [PubMed] [Google Scholar]
- 8.Liu P, Mathies RA. Integrated microfluidic systems for high-performance genetic analysis. Trends Biotechnol. 2009;27:572–581. doi: 10.1016/j.tibtech.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Emrich CA, Tian H, Medintz IL, Mathies RA. Microfabricated 384-lane capillary array electrophoresis bioanalyzer for ultrahigh-throughput genetic analysis. Anal. Chem. 2002;74:5076–5083. doi: 10.1021/ac020236g. [DOI] [PubMed] [Google Scholar]
- 10.Huynh BH, Fogarty BA, Martin RS, Lunte SM. On-line coupling of microdialysis sampling with microchip-based capillary electrophoresis. Anal. Chem. 2004;76:6440–6447. doi: 10.1021/ac049365i. [DOI] [PubMed] [Google Scholar]
- 11.Peng Y, Pallandre A, Tran NT, Taverna M. Recent innovations in protein separation on microchips by electrophoretic methods. Electrophoresis. 2008;29:157–178. doi: 10.1002/elps.200700347. [DOI] [PubMed] [Google Scholar]
- 12.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 13.Ateya DA, Erickson JS, Howell PB, Jr, Hilliard LR, Golden JP, Ligler FS. The good, the bad, and the tiny: a review of microflow cytometry. Anal. Bioanal. Chem. 2008;391:1485–1498. doi: 10.1007/s00216-007-1827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spegel C, Heiskanen A, Skjolding LHD, Emnéus J. Chip based electroanalytical systems for cell analysis. Electroanalysis. 2008;20:680–702. [Google Scholar]
- 15.Kelly RT, Woolley AT. Microfluidic systems for integrated, high-throughput DNA analysis. Anal. Chem. 2005;77 96A-102A. [Google Scholar]
- 16.Ali I, Aboul-Enein HY, Gupta VK. Microchip-Based Nano Chromatography and Nano Capillary Electrophoresis in Genomics and Proteomics. Chromatographia. 2009;69:S13–S22. [Google Scholar]
- 17.Marko-Varga GA, Nilsson J, Laurell T. New directions of miniaturization within the biomarker research area. Electrophoresis. 2004;25:3479–3491. doi: 10.1002/elps.200406109. [DOI] [PubMed] [Google Scholar]
- 18.Hou C, Herr AE. Clinically relevant advances in on-chip affinity-based electrophoresis and electrochromatography. Electrophoresis. 2008;29:3306–3319. doi: 10.1002/elps.200800244. [DOI] [PubMed] [Google Scholar]
- 19.Bharadwaj R, Santiago JG, Mohammadi B. Design and optimization of on-chip capillary electrophoresis. Electrophoresis. 2002;23:2729–2744. doi: 10.1002/1522-2683(200208)23:16<2729::AID-ELPS2729>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 20.Roman GT, Carroll S, McDaniel K, Culbertson CT. Micellar electrokinetic chromatography of fluorescently labeled proteins on poly(dimethylsiloxane)-based microchips. Electrophoresis. 2006;27:2933–2939. doi: 10.1002/elps.200500795. [DOI] [PubMed] [Google Scholar]
- 21.Ramsey JD, Jacobson SC, Culbertson CT, Ramsey JM. High-efficiency, two-dimensional separations of protein digests on microfluidic devices. Anal. Chem. 2003;75:3758–3764. doi: 10.1021/ac0264574. [DOI] [PubMed] [Google Scholar]
- 22.Roddy ES, Xu H, Ewing AG. Sample introduction techniques for microfabricated separation devices. Electrophoresis. 2004;25:229–242. doi: 10.1002/elps.200305742. [DOI] [PubMed] [Google Scholar]
- 23.Yi C, Zhang Q, Li CW, Yang J, Zhao J, Yang M. Optical and electrochemical detection techniques for cell-based microfluidic systems. Anal. Bioanal. Chem. 2006;384:1259–1268. doi: 10.1007/s00216-005-0252-x. [DOI] [PubMed] [Google Scholar]
- 24.Reyes DR, Iossifidis D, Auroux PA, Manz A. Micro total analysis systems. 1. Introduction, theory, and technology. Anal. Chem. 2002;74:2623–2636. doi: 10.1021/ac0202435. [DOI] [PubMed] [Google Scholar]
- 25.Soper SA, Ford SM, Qi S, McCarley RL, Kelly K, Murphy MC. Polymeric microelectromechanical systems. Anal. Chem. 2000;72 doi: 10.1021/ac0029511. 642A-651A. [DOI] [PubMed] [Google Scholar]
- 26.Kelly RT, Woolley AT. Thermal bonding of polymeric capillary electrophoresis microdevices in water. Anal. Chem. 2003;75:1941–1945. doi: 10.1021/ac0262964. [DOI] [PubMed] [Google Scholar]
- 27.Hille JM, Freed AL, Wätzig H. Possibilities to improve automation, speed and precision of proteome analysis: a comparison of two-dimensional electrophoresis and alternatives. Electrophoresis. 2001;22:4035–4052. doi: 10.1002/1522-2683(200111)22:19<4035::AID-ELPS4035>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 28.Wen J, Legendre LA, Bienvenue JM, Landers JP. Purification of nucleic acids in microfluidic devices. Anal. Chem. 2008;80:6472–6479. doi: 10.1021/ac8014998. [DOI] [PubMed] [Google Scholar]
- 29.Giddings JC. Unified Separation Science. New York: John Wiley and Sons; 1991. [Google Scholar]
- 30.Herr AE, Molho JI, Drouvalakis KA, Mikkelsen JC, Utz PJ, Santiago JG, Kenny TW. On-chip coupling of isoelectric focusing and free solution electrophoresis for multidimensional separations. Anal. Chem. 2003;75:1180–1187. doi: 10.1021/ac026239a. [DOI] [PubMed] [Google Scholar]
- 31.Issaq HJ, Conrads TP, Janini GM, Veenstra TD. Methods for fractionation, separation and profiling of proteins and peptides. Electrophoresis. 2002;23:3048–3061. doi: 10.1002/1522-2683(200209)23:17<3048::AID-ELPS3048>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Mauri P, Scigelova M. Multidimensional protein identification technology for clinical proteomic analysis. Clin. Chem. Lab Med. 2009;47:636–646. doi: 10.1515/CCLM.2009.165. [DOI] [PubMed] [Google Scholar]
- 33.Penque D. Two-dimensional gel electrophoresis and mass spectrometry for biomarker discovery. Proteomics Clin. Appl. 2009;3:155–172. doi: 10.1002/prca.200800025. [DOI] [PubMed] [Google Scholar]
- 34.Rocklin RD, Ramsey RS, Ramsey JM. A microfabricated fluidic device for performing two-dimensional liquid-phase separations. Anal. Chem. 2000;72:5244–5249. doi: 10.1021/ac000578r. [DOI] [PubMed] [Google Scholar]
- 35.Hiratsuka A, Kinoshita H, Maruo Y, Takahashi K, Akutsu S, Hayashida C, Sakairi K, Usui K, Shiseki K, Inamochi H, Nakada Y, Yodoya K, Namatame I, Unuma Y, Nakamura M, Ueyama K, Ishii Y, Yano K, Yokoyama K. Fully automated two-dimensional electrophoresis system for high-throughput protein analysis. Anal. Chem. 2007;79:5730–5739. doi: 10.1021/ac070485a. [DOI] [PubMed] [Google Scholar]
- 36.Osiri JK, Shadpour H, Park S, Snowden BC, Chen ZY, Soper SA. Generating high peak capacity 2-D maps of complex proteomes using PMMA microchip electrophoresis. Electrophoresis. 2008;29:4984–4992. doi: 10.1002/elps.200800496. [DOI] [PubMed] [Google Scholar]
- 37.Chen H, Fan ZH. Two-dimensional protein separation in microfluidic devices. Electrophoresis. 2009;30:758–765. doi: 10.1002/elps.200800566. [DOI] [PubMed] [Google Scholar]
- 38.Woolley AT, Sensabaugh GF, Mathies RA. High-speed DNA genotyping using microfabricated capillary array electrophoresis chips. Anal. Chem. 1997;69:2181–2186. doi: 10.1021/ac961237+. [DOI] [PubMed] [Google Scholar]
- 39.Simpson PC, Roach D, Woolley AT, Thorsen T, Johnston R, Sensabaugh GF, Mathies RA. High-throughput genetic analysis using microfabricated 96-sample capillary array electrophoresis microplates. Proc. Natl. Acad. Sci. USA. 1998;95:2256–2261. doi: 10.1073/pnas.95.5.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Simpson PC, Scherer JR, Wexler D, Skibola C, Smith MT, Mathies RA. Radial capillary array electrophoresis microplate and scanner for high-performance nucleic acid analysis. Anal. Chem. 1999;71:5354–5361. doi: 10.1021/ac990518p. [DOI] [PubMed] [Google Scholar]
- 41.Medintz IL, Paegel BM, Blazej RG, Emrich CA, Berti L, Scherer JR, Mathies RA. High-performance genetic analysis using microfabricated capillary array electrophoresis microplates. Electrophoresis. 2001;22:3845–3856. doi: 10.1002/1522-2683(200110)22:18<3845::AID-ELPS3845>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 42.Paegel BM, Hutt LD, Simpson PC, Mathies RA. Turn geometry for minimizing band broadening in microfabricated capillary electrophoresis channels. Anal. Chem. 2000;72:3030–3037. doi: 10.1021/ac000054r. [DOI] [PubMed] [Google Scholar]
- 43.Paegel BM, Emrich CA, Wedemayer GJ, Scherer JR, Mathies RA. High throughput DNA sequencing with a microfabricated 96-lane capillary array electrophoresis bioprocessor. Proc. Natl. Acad. Sci. USA. 2002;99:574–579. doi: 10.1073/pnas.012608699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumagai H, Utsunomiya S, Nakamura S, Yamamoto R, Harada A, Kaji T, Hazama M, Ohashi T, Inami A, Ikegami T, Miyamoto K, Endo N, Yoshimi K, Toyoda A, Hattori M, Sakaki Y. Large-scale microfabricated channel plates for high-throughput, fully automated DNA sequencing. Electrophoresis. 2008;29:4723–4732. doi: 10.1002/elps.200800301. [DOI] [PubMed] [Google Scholar]
- 45.Pei J, Dishinger JF, Roman DL, Rungwanitcha C, Neubig RR, Kennedy RT. Microfabricated channel array electrophoresis for characterization and screening of enzymes using RGS-G protein interactions as a model system. Anal. Chem. 2008;80:5225–5231. doi: 10.1021/ac800553g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dishinger JF, Reid KR, Kennedy RT. Quantitative monitoring of insulin secretion from single islets of Langerhans in parallel on a microfluidic chip. Anal. Chem. 2009;81:3119–3127. doi: 10.1021/ac900109t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shadpour H, Hupert ML, Patterson D, Liu C, Galloway M, Stryjewski W, Goettert J, Soper SA. Multichannel microchip electrophoresis device fabricated in polycarbonate with an integrated contact conductivity sensor array. Anal. Chem. 2007;79:870–878. doi: 10.1021/ac0612168. [DOI] [PubMed] [Google Scholar]
- 48.Moreira NH, de Almeida AL, Piazzeta MH, de Jesus DP, Deblire A, Gobbi AL, da Silva JA. Fabrication of a multichannel PDMS/glass analytical microsystem with integrated electrodes for amperometric detection. Lab Chip. 2009;9:115–121. doi: 10.1039/b807409g. [DOI] [PubMed] [Google Scholar]
- 49.Ou J, Glawdel T, Ren CL, Pawliszyn J. Fabrication of a hybrid PDMS/SU-8/quartz microfluidic chip for enhancing UV absorption whole-channel imaging detection sensitivity and application for isoelectric focusing of proteins. Lab Chip. 2009;9:1926–1932. doi: 10.1039/b821438g. [DOI] [PubMed] [Google Scholar]
- 50.Lagally ET, Scherer JR, Blazej RG, Toriello NM, Diep BA, Ramchandani M, Sensabaugh GF, Riley LW, Mathies RA. Integrated portable genetic analysis microsystem for pathogen/infectious disease detection. Anal. Chem. 2004;76:3162–3170. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- 51.Meng Z, Qi S, Soper SA, Limbach PA. Interfacing a polymer-based micromachined device to a nanoelectrospray ionization Fourier transform ion cyclotron resonance mass spectrometer. Anal. Chem. 2001;73:1286–1291. doi: 10.1021/ac000984a. [DOI] [PubMed] [Google Scholar]
- 52.Dungchai W, Chailapakul O, Henry CS. Electrochemical detection for paper-based microfluidics. Anal. Chem. 2009;81:5821–5826. doi: 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- 53.Zhao S, Li X, Liu YM. Integrated microfluidic system with chemiluminescence detection for single cell analysis after intracellular labeling. Anal. Chem. 2009;81:3873–3878. doi: 10.1021/ac900391u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Götz S, Karst U. Recent developments in optical detection methods for microchip separations. Anal. Bioanal. Chem. 2007;387:183–192. doi: 10.1007/s00216-006-0820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulze P, Belder D. Label-free fluorescence detection in capillary and microchip electrophoresis. Anal. Bioanal. Chem. 2009;393:515–525. doi: 10.1007/s00216-008-2452-7. [DOI] [PubMed] [Google Scholar]
- 56.Mycek M, Pogue BW. Handbook of biomedical fluorescence. New York: Marcel Dekker Inc; 2003. [Google Scholar]
- 57.Jacobson SC, Hergenröder R, Moore AW, Ramsey JM. Precolumn Reactions with Electrophoretic Analysis Integrated on a Microchip. Anal. Chem. 1994;66:4127–4132. [Google Scholar]
- 58.Ro KW, Lim K, Kim H, Hahn JH. Poly(dimethylsiloxane) microchip for precolumn reaction and micellar electrokinetic chromatography of biogenic amines. Electrophoresis. 2002;23:1129–1137. doi: 10.1002/1522-2683(200204)23:7/8<1129::AID-ELPS1129>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 59.Yu M, Wang H-Y, Woolley AT. Polymer microchip capillary electrophoresis of proteins either off- or on-chip labeled with chameleon dye for simplified analysis. Electrophoresis. doi: 10.1002/elps.200900349. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mair DA, Schwei TR, Dinio TS, Svec F, Fréchet JM. Use of photopatterned porous polymer monoliths as passive micromixers to enhance mixing efficiency for on-chip labeling reactions. Lab Chip. 2009;9:877–883. doi: 10.1039/b816521a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krull IS, Strong R, Sosic Z, Cho BY, Beale SC, Wang CC, Cohen S. Labeling reactions applicable to chromatography and electrophoresis of minute amounts of proteins. J. Chromatogr. B. 1997;699:173–208. doi: 10.1016/s0378-4347(97)00157-6. [DOI] [PubMed] [Google Scholar]
- 62.Jacobson SC, Koutny LB, Hergenröder R, Moore AW, Ramsey JM. Microchip Capillary Electrophoresis with an Integrated Postcolumn Reactor. Anal. Chem. 1994;66:3472–3476. [Google Scholar]
- 63.Fluri K, Fitzpatrick G, Chiem N, Harrison DJ. Integrated capillary electrophoresis devices with an efficient postcolumn reactor in planar quartz and glass chips. Anal. Chem. 1996;68:4285–4290. doi: 10.1021/ac9604090. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y, Foote RS, Jacobson SC, Ramsey RS, Ramsey JM. Electrophoretic separation of proteins on a microchip with noncovalent, postcolumn labeling. Anal. Chem. 2000;72:4608–4613. doi: 10.1021/ac000625f. [DOI] [PubMed] [Google Scholar]
- 65.Sieben VJ, Backhouse CJ. Rapid on-chip postcolumn labeling and high-resolution separations of DNA. Electrophoresis. 2005;26:4729–4742. doi: 10.1002/elps.200500459. [DOI] [PubMed] [Google Scholar]
- 66.Hashimoto M, Tsukagoshi K, Nakajima R, Kondo K, Arai A. Microchip capillary electrophoresis using on-line chemiluminescence detection. J. Chromatogr. A. 2000;867:271–279. doi: 10.1016/s0021-9673(99)01169-3. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Y, McLaughlin K, Lunte CE. On-column sample preconcentration using sample matrix switching and field amplification for increased sensitivity of capillary electrophoretic analysis of physiological samples. Anal. Chem. 1998;70:4578–4585. doi: 10.1021/ac980427c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carabias-Martínez R, Rodríguez-Gonzalo E, Revilla-Ruiz P, Domínguez-Álvarez J. Solid-phase extraction and sample stacking-micellar electrokinetic capillary chromatography for the determination of multiresidues of herbicides and metabolites. J. Chromatogr. A. 2003;990:291–302. doi: 10.1016/s0021-9673(02)01969-6. [DOI] [PubMed] [Google Scholar]
- 69.Kim JB, Otsuka K, Terabe S. On-line sample concentration in micellar electrokinetic chromatography with cationic micelles in a coated capillary. J. Chromatogr. A. 2001;912:343–352. doi: 10.1016/s0021-9673(01)00599-4. [DOI] [PubMed] [Google Scholar]
- 70.Siri N, Riolet P, Bayle C, Couderc F. Automated large-volume sample stacking procedure to detect labeled peptides at picomolar concentration using capillary electrophoresis and laser-induced fluorescence detection. J. Chromatogr. B. 2003;793:151–157. doi: 10.1016/s1570-0232(03)00372-6. [DOI] [PubMed] [Google Scholar]
- 71.Jung B, Bharadwaj R, Santiago JG. Thousandfold signal increase using field-amplified sample stacking for on-chip electrophoresis. Electrophoresis. 2003;24:3476–3483. doi: 10.1002/elps.200305611. [DOI] [PubMed] [Google Scholar]
- 72.Jung B, Bharadwaj R, Santiago JG. On-chip millionfold sample stacking using transient isotachophoresis. Anal. Chem. 2006;78:2319–2327. doi: 10.1021/ac051659w. [DOI] [PubMed] [Google Scholar]
- 73.Jung B, Zhu Y, Santiago JG. Detection of 100 aM fluorophores using a high-sensitivity on-chip CE system and transient isotachophoresis. Anal. Chem. 2007;79:345–349. doi: 10.1021/ac060949p. [DOI] [PubMed] [Google Scholar]
- 74.Sueyoshi K, Kitagawa F, Otsuka K. Recent progress of online sample preconcentration techniques in microchip electrophoresis. J. Sep. Sci. 2008;31:2650–2666. doi: 10.1002/jssc.200800272. [DOI] [PubMed] [Google Scholar]
- 75.Hennion MC. Solid-phase extraction: method development, sorbents, and coupling with liquid chromatography. J. Chromatogr. A. 1999;856:3–54. doi: 10.1016/s0021-9673(99)00832-8. [DOI] [PubMed] [Google Scholar]
- 76.Berrueta LA, Gallo B, Vicente F. A Review of Solid-Phase Extraction - Basic Principles and New Developments. Chromatographia. 1995;40:474–483. [Google Scholar]
- 77.Kutter JP, Jacobson SC, Ramsey JM. Solid phase extraction on microfluidic devices. J. Microcolumn Sep. 2000;12:93–97. [Google Scholar]
- 78.Jemere AB, Oleschuk RD, Ouchen F, Fajuyigbe F, Harrison DJ. An integrated solid-phase extraction system for sub-picomolar detection. Electrophoresis. 2002;23:3537–3544. doi: 10.1002/1522-2683(200210)23:20<3537::AID-ELPS3537>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 79.Yu C, Davey MH, Svec F, Fréchet JMJ. Monolithic porous polymer for on-chip solid-phase extraction and preconcentration prepared by photoinitiated in situ polymerization within a microfluidic device. Anal. Chem. 2001;73:5088–5096. doi: 10.1021/ac0106288. [DOI] [PubMed] [Google Scholar]
- 80.Stachowiak TB, Rohr T, Hilder EF, Peterson DS, Yi M, Svec F, Fréchet JM. Fabrication of porous polymer monoliths covalently attached to the walls of channels in plastic microdevices. Electrophoresis. 2003;24:3689–3693. doi: 10.1002/elps.200305536. [DOI] [PubMed] [Google Scholar]
- 81.Wolfe KA, Breadmore MC, Ferrance JP, Power ME, Conroy JF, Norris PM, Landers JP. Toward a microchip-based solid-phase extraction method for isolation of nucleic acids. Electrophoresis. 2002;23:727–733. doi: 10.1002/1522-2683(200203)23:5<727::AID-ELPS727>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 82.Oleschuk RD, Shultz-Lockyear LL, Ning Y, Harrison DJ. Trapping of bead-based reagents within microfluidic systems: on-chip solid-phase extraction and electrochromatography. Anal. Chem. 2000;72:585–590. doi: 10.1021/ac990751n. [DOI] [PubMed] [Google Scholar]
- 83.Zhong R, Liu D, Yu L, Ye N, Dai Z, Qin J, Lin B. Fabrication of two-weir structure-based packed columns for on-chip solid-phase extraction of DNA. Electrophoresis. 2007;28:2920–2926. doi: 10.1002/elps.200600604. [DOI] [PubMed] [Google Scholar]
- 84.Ramsey JD, Collins GE. Integrated microfluidic device for solid-phase extraction coupled to micellar electrokinetic chromatography separation. Anal. Chem. 2005;77:6664–6670. doi: 10.1021/ac0507789. [DOI] [PubMed] [Google Scholar]
- 85.Ceriotti L, de Rooij NF, Verpoorte E. An integrated fritless column for on-chip capillary electrochromatography with conventional stationary phases. Anal. Chem. 2002;74:639–647. doi: 10.1021/ac0109467. [DOI] [PubMed] [Google Scholar]
- 86.Svec F, Huber CG. Monolithic materials: Promises, challenges, achievements. Anal. Chem. 2006;78:2101–2107. doi: 10.1021/ac069383v. [DOI] [PubMed] [Google Scholar]
- 87.Xie SF, Svec F, Fréchet JMJ. Porous polymer monoliths: Preparation of sorbent materials with high-surface areas and controlled surface chemistry for high-throughput, online, solid-phase extraction of polar organic compounds. Chem. Mater. 1998;10:4072–4078. [Google Scholar]
- 88.Wen J, Guillo C, Ferrance JP, Landers JP. DNA extraction using a tetramethyl orthosilicate-grafted photopolymerized monolithic solid phase. Anal. Chem. 2006;78:1673–1681. doi: 10.1021/ac051796t. [DOI] [PubMed] [Google Scholar]
- 89.Wen J, Guillo C, Ferrance JP, Landers JP. Microfluidic-based DNA purification in a two-stage, dual-phase microchip containing a reversed-phase and a photopolymerized monolith. Anal. Chem. 2007;79:6135–6142. doi: 10.1021/ac0703698. [DOI] [PubMed] [Google Scholar]
- 90.Mao X, Luo Y, Dai Z, Wang K, Du Y, Lin B. Integrated lectin affinity microfluidic chip for glycoform separation. Anal. Chem. 2004;76:6941–6947. doi: 10.1021/ac049270g. [DOI] [PubMed] [Google Scholar]
- 91.Yang W, Sun X, Pan T, Woolley AT. Affinity monolith preconcentrators for polymer microchip capillary electrophoresis. Electrophoresis. 2008;29:3429–3435. doi: 10.1002/elps.200700704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun X, Yang W, Pan T, Woolley AT. Affinity monolith-integrated poly(methyl methacrylate) microchips for on-line protein extraction and capillary electrophoresis. Anal. Chem. 2008;80:5126–5130. doi: 10.1021/ac800322f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dodge A, Fluri K, Verpoorte E, de Rooij NF. Electrokinetically driven microfluidic chips with surface-modified chambers for heterogeneous immunoassays. Anal. Chem. 2001;73:3400–3409. doi: 10.1021/ac0015366. [DOI] [PubMed] [Google Scholar]
- 94.Yang W, Sun X, Wang HY, Woolley AT. Integrated microfluidic device for serum biomarker quantitation using either standard addition or a calibration curve. Anal. Chem. 2009;81:8230–8235. doi: 10.1021/ac901566s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Phillips TM, Wellner EF. Analysis of inflammatory biomarkers from tissue biopsies by chip-based immunoaffinity CE. Electrophoresis. 2007;28:3041–3048. doi: 10.1002/elps.200700193. [DOI] [PubMed] [Google Scholar]
- 96.Phillips TM, Wellner EF. Chip-based immunoaffinity CE: application to the measurement of brain-derived neurotrophic factor in skin biopsies. Electrophoresis. 2009;30:2307–2312. doi: 10.1002/elps.200900095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kong J, Jiang L, Su X, Qin J, Du Y, Lin B. Integrated microfluidic immunoassay for the rapid determination of clenbuterol. Lab Chip. 2009;9:1541–1547. doi: 10.1039/b818430e. [DOI] [PubMed] [Google Scholar]
- 98.Fan R, Vermesh O, Srivastava A, Yen BK, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat. Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu XZ, Hosaka A, Hobo T. An On-Line Electrophoretic Concentration Method for Capillary Electrophoresis of Proteins. Anal. Chem. 1998;70:2081–2084. [Google Scholar]
- 100.Foote RS, Khandurina J, Jacobson SC, Ramsey JM. Preconcentration of proteins on microfluidic devices using porous silica membranes. Anal. Chem. 2005;77:57–63. doi: 10.1021/ac049136w. [DOI] [PubMed] [Google Scholar]
- 101.Wei W, Yeung ES. On-line concentration of proteins and peptides in capillary zone electrophoresis with an etched porous joint. Anal. Chem. 2002;74:3899–3905. doi: 10.1021/ac025612b. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Y, Timperman AT. Integration of nanocapillary arrays into microfluidic devices for use as analyte concentrators. Analyst. 2003;128:537–542. doi: 10.1039/b300102d. [DOI] [PubMed] [Google Scholar]
- 103.Song S, Singh AK, Kirby BJ. Electrophoretic concentration of proteins at laser-patterned nanoporous membranes in microchips. Anal. Chem. 2004;76:4589–4592. doi: 10.1021/ac0497151. [DOI] [PubMed] [Google Scholar]
- 104.Yamamoto S, Hirakawa S, Suzuki S. In situ fabrication of ionic polyacrylamide-based preconcentrator on a simple poly(methyl methacrylate) microfluidic chip for capillary electrophoresis of anionic compounds. Anal. Chem. 2008;80:8224–8230. doi: 10.1021/ac801245n. [DOI] [PubMed] [Google Scholar]
- 105.Kim SJ, Han J. Self-sealed vertical polymeric nanoporous-junctions for high-throughput nanofluidic applications. Anal. Chem. 2008;80:3507–3511. doi: 10.1021/ac800157q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc. Natl. Acad. Sci. USA. 2007;104:5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meagher RJ, Hatch AV, Renzi RF, Singh AK. An integrated microfluidic platform for sensitive and rapid detection of biological toxins. Lab Chip. 2008;8:2046–2053. doi: 10.1039/b815152k. [DOI] [PubMed] [Google Scholar]
- 108.Lion N, Gellon JO, Jensen H, Girault HH. On-chip protein sample desalting and preparation for direct coupling with electrospray ionization mass spectrometry. J. Chromatogr. A. 2003;1003:11–19. doi: 10.1016/s0021-9673(03)00771-4. [DOI] [PubMed] [Google Scholar]
- 109.Kim J, Gale BK. Quantitative and qualitative analysis of a microfluidic DNA extraction system using a nanoporous AlOx membrane. Lab Chip. 2008;8:1516–1523. doi: 10.1039/b804624g. [DOI] [PubMed] [Google Scholar]
- 110.Timmer BH, van Delft KM, Olthuis W, Bergveld P, van den Berg A. Micro-evaporation electrolyte concentrator. Sens. Actuators B. 2003;91:342–346. [Google Scholar]
- 111.Woolley AT, Hadley D, Landre P, deMello AJ, Mathies RA, Northrup MA. Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device. Anal. Chem. 1996;68:4081–4086. doi: 10.1021/ac960718q. [DOI] [PubMed] [Google Scholar]
- 112.Chen L, Manz A, Day PJ. Total nucleic acid analysis integrated on microfluidic devices. Lab Chip. 2007;7:1413–1423. doi: 10.1039/b708362a. [DOI] [PubMed] [Google Scholar]
- 113.Liu P, Seo TS, Beyor N, Shin KJ, Scherer JR, Mathies RA. Integrated portable polymerase chain reaction-capillary electrophoresis microsystem for rapid forensic short tandem repeat typing. Anal. Chem. 2007;79:1881–1889. doi: 10.1021/ac061961k. [DOI] [PubMed] [Google Scholar]
- 114.Blazej RG, Kumaresan P, Cronier SA, Mathies RA. Inline injection microdevice for attomole-scale sanger DNA sequencing. Anal. Chem. 2007;79:4499–4506. doi: 10.1021/ac070126f. [DOI] [PubMed] [Google Scholar]
- 115.Thaitrong N, Toriello NM, Del Bueno N, Mathies RA. Polymerase chain reaction-capillary electrophoresis genetic analysis microdevice with in-line affinity capture sample injection. Anal. Chem. 2009;81:1371–1377. doi: 10.1021/ac802057f. [DOI] [PubMed] [Google Scholar]
- 116.Sun Y, Kwok YC, Foo-Peng Lee P, Nguyen NT. Rapid amplification of genetically modified organisms using a circular ferrofluid-driven PCR microchip. Anal Bioanal. Chem. 2009;394:1505–1508. doi: 10.1007/s00216-009-2808-7. [DOI] [PubMed] [Google Scholar]
- 117.Sun Y, Nguyen NT, Kwok YC. High-throughput polymerase chain reaction in parallel circular loops using magnetic actuation. Anal. Chem. 2008;80:6127–6130. doi: 10.1021/ac800787g. [DOI] [PubMed] [Google Scholar]
- 118.Zhang Y, Bailey V, Puleo CM, Easwaran H, Griffiths E, Herman JG, Baylin SB, Wang TH. DNA methylation analysis on a droplet-in-oil PCR array. Lab Chip. 2009;9:1059–1064. doi: 10.1039/b821780g. [DOI] [PMC free article] [PubMed] [Google Scholar]