Abstract
BACKGROUND:
DNA mutations resulting in the McCoy and Swain-Langley polymorphisms have been identified on complement receptor 1 (CR1)—a ligand for rosetting of Plasmodium falciparum-infected RBCs. The molecular identification of the Kna/Knb polymorphism was sought to develop a genotyping method for use in the study of the Knops blood group and malaria.
STUDY DESIGN AND METHODS:
CR1 deletion constructs were used in inhibition studies of anti-Kna. PCR amplification of Exon 29 was followed by DNA sequencing. A PCR-RFLP was developed with NdeI, BsmI, and MfeI for the detection of Kna/Knb, McCa/McCb, and Sl1/Sl2, respectively. Knops phenotypes were determined with standard serologic techniques.
RESULTS:
A total of 310 Malian persons were phenotyped for Kna with 200 (64%) Kn(a+) and 110 (36%) Kn(a−). Many of the Kn(a−) exhibited the Knops-null phenotype, that is, Helgeson. The Kna/b DNA polymorphism was identified as a V1561M mutation with allele frequencies of Kna (V1561) 0.9 and Knb (M1561) 0.1.
CONCLUSION:
The high frequency (18%) of Knb in West African persons suggests that it is not solely a Caucasian trait. Furthermore, because of the high incidence of heterozygosity as well as amorphs, accurate Knops typing of donors of African descent is best accomplished by a combination of molecular and serologic techniques.
The Knops-Helgeson antigen was first reported by Helgeson and coworkers1 in an article describing a weak, antiglobulin-reactive antibody that was found in a 37-year-old Caucasian woman. A family study of the only compatible donor (MH) showed that the antigen was independent from the loci for ABO, MNSs, Rh, Lu, Jkm and Se; hence it was given the name Knops-Helgeson and abbreviated as Kna. A serum sample that reacted with 4.5 percent of Caucasian donors was later reported as being anti-Knb; however, this antigen was not found among African American blood donors.2
During the initial period when the Knops blood group system was being investigated, a number of antibodies with similar characteristics were described and placed in a group known as “high-titer, low avidity” (HTLA) antibodies. The group included Chido, Rodgers, York, Csa, JMH, Knops, McCoy, and Swain-Langley which have now been assigned to separate blood group systems. One of the characteristics of these antibodies was their weak and variable reactivity. Accurate phenotyping of RBCs was difficult and reference laboratories were often unable to duplicate each other's results. The identification of the Knops antigens on complement receptor 1 (CR1) began to explain some of these findings but also revealed the complexity of this system.3,4 Moulds and colleagues5 reported that low expression of RBC-CR1 often resulted in weak or negative Knops typings. Consequently, better methods were needed to correctly identify antigen-positive versus antigen-negative individuals.
A concerted effort to identify the molecular basis of the Knops alleles began following the report that CR1 was a ligand in the rosetting of Plasmodium-infected RBCs with noninfected cells.6 Early work by Molthan7 and by Moulds8 indicated that significant differences in several Knops antigen frequencies existed between Caucasian and African American persons. It was postulated that some Knops blood group phenotypes, for example, Sl(a−), might be protective against rosetting and severe malaria.6 Thus, the CR1 gene was systematically investigated to find mutations that might correlate with the blood group antigen specificities.
The most common form of the CR1 gene, that is, CR1*1, is composed of 38 exons spanning 133 kb producing a protein of 220 kDa. Large insertions and deletions have given rise to four structural variant genes but these are unrelated to the blood group antigens.9 Approximately 60 amino acids comprise a short consensus repeat (SCR), and seven SCRs are further arranged into four long homologous repeats. Because of this high degree of homology, locating the blood group antigens was not an easy task, but the single-nucleotide changes resulting in the McCa, McCb, Sla, and Vil antigens have now been identified in SCR25.9 The Sla antigen has recently been subdivided10 and, hence, Sla has been renamed Sl1 and Vil has been redesignated as Sl2.11 We now report the molecular identification of the third allelic pair, Kna/Knb, and outline a simple method for genotyping these polymorphisms.
MATERIALS AND METHODS
Samples
These studies were approved by the institutional review boards from the University of Texas-Houston Medical School, Drexel University, the University of Maryland, and the University of Bamako. The sample population consisted of 145 Caucasian persons from New Jersey, 76 healthy African American persons from Texas, and 339 individuals from Bandiagara, Mali (West Africa). The Malian population is composed of ~30 different ethnic groups but the predominant group in Bandiagara are the Dogon (80%) and Peuhl (10%-15%).
Following informed consent, 3 to 10 mL of EDTA blood was drawn by venipuncture from individuals ranging in age from 1 to 65 years old without regard to sex. RBC studies were performed within 48 hours from the time of venipuncture because it has been found that CR1 levels deteriorate upon storage. Following centrifugation, the buffy coat was removed to prepare genomic DNA with a DNA extraction kit (Gentra, Minneapolis, MN). A DAT was performed on all samples and any samples sensitized with IgG were omitted from the final serologic data interpretation.
RBC studies
Serologic testing
Antisera were obtained from blood donors previously identified by AABB reference laboratories as having anti-Kna. We were unable to obtain sufficient anti-Knb to use for population studies. The plasma sample from one donor (KN038), who was available for extensive testing, was chosen for population testing followed by confirmation of apparent negatives with a second anti-Kna (KN039). The donor is a 54-year-old woman of Caucasian, Mexican, and Native American extraction. She received several units of blood when she was 19 years old and had two pregnancies. The serum sample contained an anti-K and an anti-Kna that reacted 2 to 3+ when using a 1-hour, 37∞C incubation in saline followed by an anti-human globulin test and indicator cells having normal to high levels of RBC-CR1. The donor's RBCs had normal levels of CR1 and typed as K− and Kn(a−) with the original Knops serum. In addition, she had the Knops phenotype Kn(b+), McC(a−b+) and Sl1,−2 which were confirmed by both DNA sequencing and RFLP analyses.
DNA studies
CR1 constructs and inhibition tests
CR1 deletion constructs for LHR-A (1-7), LHR-B (8-14), LHR-C (15-21), and LHR-D+ (SCRs 22-30) were prepared as previously described by Krych-Goldberg and colleagues.12 A construct introducing the amino acid methionine in place of valine at position 1561 was made by incorporating into the LHR-D+ cDNA the single-nucleotide mutation G>A at bp 4708. For the inhibition studies, 1 μg of soluble CR1 was used to inhibit 75 μL of antisera at 24°C for 15 minutes.13 Test indicator cells were chosen to be ABO-compatible and antigen-positive and having median to high CR1 levels.
DNA sequencing
Based on the observation that the LHR-D+ construct uniquely inhibited the anti-Kna sera, and that the McC and Sl blood group antigens were found to occur in CR1 exon 29, encoding SCR 24-25 within LHR-D+, we focused our analysis on Exon 29. Genomic DNA (GenBank Accession No. L17390-L17430), from three donors who had produced anti-Kna, was amplified with primers for the introns immediately 5′ (24Li, CATTGGAT TATTTGCATTTGG) and 3′ (25Ri, TCTATACGTACCCTCAC ACCC) of Exon 29 resulting in a 540-bp amplicon. The amplicons were directly sequenced in a genetic analyzer (ABI Prism 3100, Applied Biosystems, Foster City, CA), with the 24Li primer. The sequences were analyzed with a DNA sequence analysis program (Omiga, Oxford Molecular Ltd., Oxford, England).
Knops genotyping
A PCR-RFLP was developed that allowed for the detection of all three single-nucleotide polymorphisms (SNPs). The forward primer sequence (24KnNde, No. 4664-4707) was 5′-ACCAGTGCCACACTGGACCAGATGGAGAACAGCTGTT TGAGCAT-3′ and the reverse primer sequence (25Rb, No. 4948-4967) was 5′-GGAGGAGTGTGGCAGCTTG-3′. The forward primer contains a deliberate mismatch (bolded) that can be used for the detection of the Kna polymorphism. The PCR was set up in a 100 ml total volume with ~500 ng of genomic DNA, 10 μL of 10× Taq polymerase buffer containing MgCl2, 10 μL of each primer, and 1 unit of Taq polymerase. The samples were hot started and amplified with the following program: denaturation at 94°C for 1 minute, annealing at 58°C for 1 minute, extension at 72°C for 1 minute for 44 cycles, and a final extension at 72°C for 10 minutes. The PCR product resulting from amplification with these primers was 305 bp.
For Kna/Knb detection, 1.6 mL of NdeI (20 U) and 2.4 μL of NdeI 10× buffer was added to 20 μL of PCR product and incubated at 37°C overnight. For McCa/McCb, 2 μL of BsmI (20 U) was added to 28 μL of PCR product and incubated at 65°C for 2 hours. For Sl1/Sl2, 2 μL of MfeI (20 U) was added to 28 μL of PCR product and incubated at 37°C for 2 hours. The digestion products were analyzed by 2 percent agarose gel containing ethidium bromide and band size compared to a Phi X marker. Samples heterozygous for each SNP were run on the gels as a positive restriction enzyme control.
RESULTS
Serologic studies
A total of 76 A-A and 310 samples from Bandiagara were phenotyped for Kna, McCa, McCb, and Sl1. Twenty-nine samples could not be tested by serologic methods because they had a positive DAT. Unexpectedly, it was found that 123 of the samples from Bandiagara typed as Kn(a−). More importantly, 47 of these Kn(a−) samples could be considered false-negatives because they expressed the Helgeson phenotype, that is, they typed negative for all four antigens. A similar high frequency of the Kn(a−) and Helgeson phenotypes has been noted in Kenyans (J.M. Moulds and D. Roberts, unpublished data) but was not observed among African American persons. The Helgeson phenotype did not appear to be associated with any one particular Knops genotype.
The frequencies for McCa, McCb, and Sl1 in the samples from persons from both African American and Bandiagara were comparable to those previously reported.14 False-negative serologic typings occurred in 20 to 25 percent of the Bandiagara, again owing to the Helgeson phenotype. Nevertheless, 66 percent of the McC(a−) and 81 percent of the Sl1 were heterozygous by genotyping which has previously been reported as a cause of discrepancies between genotype and phenotype.9
DNA studies
Identification of Kna/Knb
With our previously described sCR1 inhibition test,9,13 a total of seven sera containing anti-Kna were studied including the original Knops (DK) serum. All were completely inhibited with the full-length sCR1 and LHR-D+ but not the deletion constructs containing only LHR-A, B, or C. Two sera samples containing anti-Knb were also investigated with the various constructs with a Kn(a+b+) indicator cell. As expected, they were not inhibited with LHR-D+.
Based on the results of the inhibition studies, DNA from three Kn(a−) donors and a Kn(a+b+) donor were amplified for exon 29 and sequenced. The three Kn(a−) samples exhibited a homozygous mutation of G>A at bp 4708 encoding a methionine at amino acid at 1561. The sample from the heterozygous donor had both a G (valine) and an A (methionine) at this position.
These data suggested that the Kna/Knb polymorphism was based on the specific amino acids at residue 1561 with valine (V) being present in Kn(a+) and methionine (M) in Kn(b+). To further confirm this, constructs were prepared containing M in place of V and repeated the inhibition studies. The mutant construct was no longer able to inhibit the anti-Kna sera but now inhibited the anti-Knb, thus confirming that these amino acids were part of the Kna/Knb specificity.
Genotyping
The expected RFLP digestion fragments are shown in Table 1 and illustrated in Figs. 1 to 3. With these PCR-RFLP methods, a total of 339 individual DNA samples from Bandiagara were successfully amplified and the results compared to our previous population studies of African-derived donors. The results are shown in Table 2.
TABLE 1.
Expected band sizes (bp) for Knops genotyping with PCR-RFLP*
| Polymorphism | Base pair change | Amino acid change | Enzyme | Homozygous wild type | Heterozygous | Homozygous mutant |
|---|---|---|---|---|---|---|
| Kna/Knb | 4708G>A | V1561M | NdeI | 305 | 305, 261, 44 | 261, 44 |
| McCa/McCb | 4795A>G | K1590E | BsmI | 305 | 305, 166, 139 | 166, 139 |
| Sl1/Sl2 | 4828A>G | R1601G | MfeI | 305 | 305, 161, 144 | 161, 144 |
Mutant alleles (Knb, McCb, Sl2) were cut by the restriction enzyme indicated.
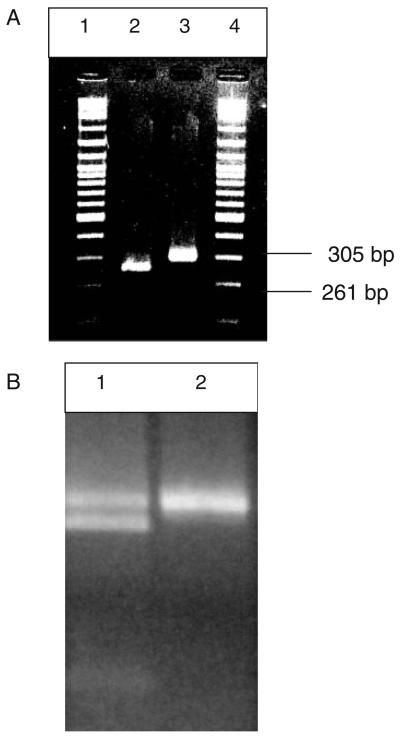
Fig. 1.
PCR-RFLP genotyping for Kna/Knb with NdeI. (A) A Kn(a− b+) donor (Lane 2) and a Kn(a+b−) donor (Lane 3). Note that the 44-bp band is not visible on this gel. (B) A Kn(a+b+) sample is observed in Lane 1 and a Kn(a+b−) is seen in Lane 2.
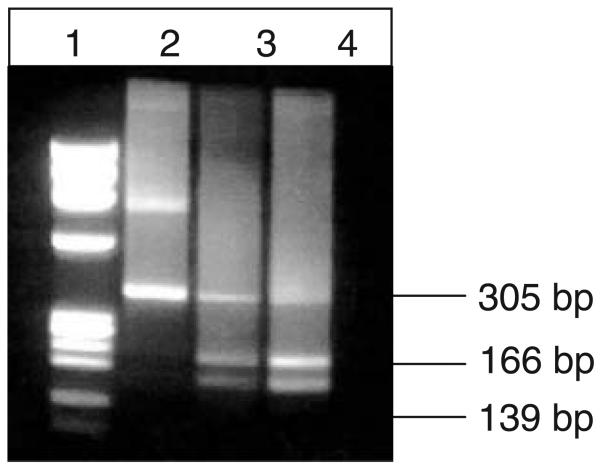
Fig. 3.
PCR-RFLP genotyping for Sl1 and Sl2 with MfeI. A Sl1,1 donor is observed in Lane 2, a Sl1,2 donor in Lane 3, and a Sl2,2 donor in Lane 4.
TABLE 2.
Knops allele frequencies based on PCR-RFLP genotyping
| Ethnicity | Number | Kna | Knb | McCa | McCb | Sl1 | Sl2 |
|---|---|---|---|---|---|---|---|
| Caucasian persons | 145 | 0.99 | 0.01 | 0.99 | <0.01 | 0.99 | <0.01 |
| African American persons | 76 | 0.98 | 0.02 | 0.78 | 0.22 | 0.48 | 0.52 |
| Total from Mali | 491 | 0.69 | 0.31 | 0.28 | 0.72 | ||
| Bandiagara persons | 342 | 0.90 | 0.10 | 0.69 | 0.31 | 0.31 | 0.69 |
| Other Mali sites* | 149 | NT | NT | 0.69 | 0.31 | 0.23 | 0.77 |
Mostly Bambaran and Melinke ethnicities.
DISCUSSION
Previous serologic studies found that Kna was a high-incidence antigen in Caucasian and African American persons. Our earlier studies of native African persons indicated that Kna was also a high-incidence antigen in this group.14 Thus, it was surprising to find that 36 percent of the samples from Bandiagara, Mali, typed as Kn(a−). Furthermore, 14 percent of the Bandiagara samples exhibited the Knops-null or Helgeson phenotype. These observations had not been previously made in African American persons or in other test sites in Mali where the incidence of the Helgeson phenotype was approximately 1 percent.2,7,14 Because we were unable to serologically type or Knb, it is unknown whether these samples were heterozygotes. It has previously been reported that heterozygosity at the McC and Sl loci can result in false-negative phenotypes.9 Alternatively, the phenotype differences may relate to the ethnicity of the current study group. The other Mali sites have ethnic groups composed predominately of Bambaran and Melinke persons whereas the major ethnic groups in Bandiagara are Dogon and Peuhl persons.
To identify potential regions of the CR1 gene that might encode the Kna/Knb polymorphism, inhibition studies were performed with various CR1 deletion constructs. Tamasauskas and colleagues15 reported that three examples of anti-Kna were mostly inhibited with LHR-C whereas three others were inhibited by LHR-D. It was found that only the LHR-D+-containing constructs were able to completely neutralize all of our anti-Kna including the original (DK) serum sample. DNA sequencing of this region identified a SNP at bp 4708 that changed the wild-type amino acid valine (No. 1561) to methionine. Interestingly, the corresponding amino acid in LHRs A, B, and C also carry valine. Because antibodies can recognize a series of six amino acids for binding, it is possible that adjoining amino acids are also important in the antigenic site. When analyzing the six amino acids on either side of 1561, we find that heterogeneity exists between the LHRs (Fig. 4). This may explain some of the differences in the inhibition studies between the previous reported work and the current study.
Fig. 4.
Alignment of amino acid sequences in homologous regions of human CR1 where the *Kna/Knb polymorphism is located. Bold letters indicate differences between the LHRs.
Because the Kna/Knb mutation did not introduce a RFLP, primers were designed that would introduce a restriction enzyme site into the mutant DNA. The McC and Sl SNPs are also in the same exon; thus, the PCR allowed for the genotyping at all three loci. With the PCRRFLP method, the allele frequencies were 0.90 for Kna and 0.10 for Knb in the Bandiagara population. By direct allele count the projected phenotype frequency of Knb in Bandiagara would be 18.3 percent, which is considerably higher than previously reported for any group. Other serologic studies have reported that the frequency of Knb ranged from 1.2 percent among African American persons living in Philadelphia2 to 4.7 percent among 63 random Caucasian donors.16 Its role in CR1 function as well as rosetting and malaria are presently under investigation.
The PCR-RFLP was also used to genotype for the McC and Sl loci in several areas of Mali as well as among African American persons. As shown in Table 2, the gene frequencies for Sl1 and Sl2 in African American persons are almost equal (0.48 vs. 0.52) whereas Sl2 is greatly increased in Africa, reaching its highest frequency among the Bambaran people (0.77). The Sl2 gene is almost completely absent in Caucasian and Asian persons.17 Adding our data to that previously published, over 1800 samples from Western Africa (Gambia, Senegal, Guinea, Sierra Leone, Ivory Coast, Ghana, and Mali) have been genotyped.9,17 The combined gene frequencies for McCa range from 0.63 to 0.69 and for McCb 0.31 to 0.37 whereas the frequencies for Sl1 are 0.19 to 0.23 and 0.77 to 0.81 for Sl2.9,14 This is in contrast to studies of more than 1200 Kenyan persons who exhibited a lower frequency of McCb (0.16) and Sl2 (0.67) (P. Zimmerman, personal communication). The data suggest that the McCb and Sl2 genes may be under selective pressure in Africa.
Our data provide interesting insights into problems that often occur when trying to perform phenotyping of RBCs for the various Knops system antigens. It has previously been reported that false-negative results are obtained when the RBCs have low CR1 copy number either owing to an inherited or acquired deficiency or after prolonged storage of the blood sample.18 Accurate phenotypes can be obtained on most samples from Caucasian persons that have moderate to high CR1 levels and who are homozygous for Kna, McCa, and Sl1 alleles (assuming all alleles are expressed).
The situation in African persons, however, is much more complex. In Bandiagara, 66 percent of the apparent McC(a−) and 81 percent of Sl1 samples were heterozygous by genotyping and, thus, were false-negative phenotypes. Similar results have been found in an earlier study of Malian donors from Bamako and Tienequebougou.9 Further complicating the phenotype status is the unequal expression of alleles.9 For example, if an individual is positive for an allele but that gene is only weakly expressed, false-negative serologic typings may even though the total CR1 copy numbers are in the normal range. Some family studies have also indicated that an amorph may exist in the Knops blood group as has been described for most other systems (J.M. Moulds, unpublished data). The molecular mechanism for this is currently under investigation. Once identified, genotyping for expression as well as the Knops blood group-related SNPs will make it possible to accurately type samples for research or transfusion purposes.
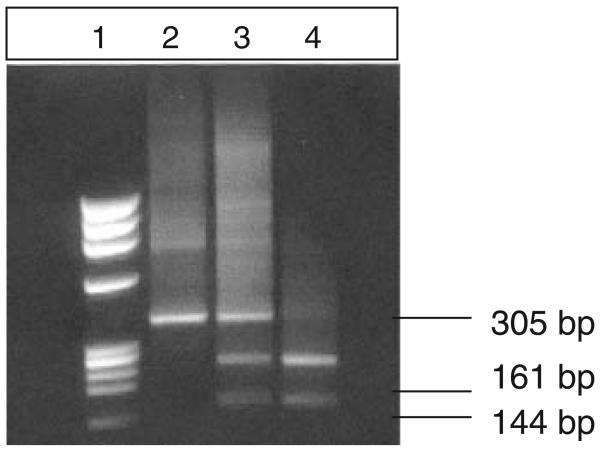
Fig. 2.
PCR-RFLP genotyping for McCa/McCb with BsmI. A McC(a+b−) donor is observed in Lane 2 and McC(a+b+) donors in Lanes 3 and 4.
ACKNOWLEDGMENTS
The authors thank Norbert Ahrens, Teresa Harris, Sue Johnson, John Moulds, Marilyn Moulds, Steve Pierce, and Sue Riester for supplying Knops antisera and/or RBCs. John Atkinson, Malgorzata Krych, and Richard Hauhart supplied the CR1 constructs. The authors are indebted to all the blood donors in this study but particularly M. Wallick, the anti-Kna donor used in the population and DNA sequencing studies, J. Henry the Kn(a+b+) donor, and M. Strickner a Kn(a−b+) donor.
Supported by a grant from the National Institutes of Health, R01 AI 042367 (J.M.M.) and contract No1-AI85346 (CUP).
ABBREVIATIONS
- CR1
complement receptor 1
- SCR(s)
short consensus repeat(s)
- SNP(s)
single-nucleotide polymorphism(s)
REFERENCES
- 1.Helgeson M, Swanson J, Polesky HF. Knops-Helgeson (Kna), a high frequency erythrocyte antigen. Transfusion. 1970;10:137–8. doi: 10.1111/j.1537-2995.1970.tb00720.x. [DOI] [PubMed] [Google Scholar]
- 2.Molthan L. The serology of the York-Cost-McCoy-Knops red blood cell system. Am J Med Technol. 1983;49:49–56. [PubMed] [Google Scholar]
- 3.Moulds JM, Nickells MW, Moulds JJ, Brown MC, Atkinson JP. The C3b/C4b receptor is recognized by the Knops, McCoy, Swain-Langley, and York blood group antisera. J Exp Med. 1991;173:1159–63. doi: 10.1084/jem.173.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao N, Ferguson DJ, Lee SF, Telen MJ. Identification of human erythrocyte blood group antigens on the C3b/C4b receptor. J Immunol. 1991;146:3502–7. [PubMed] [Google Scholar]
- 5.Moulds JM, Moulds JJ, Brown M, Atkinson JP. Antiglobulin testing for CR1-related (Knops/McCoy/Swain-Langley/York) blood group antigens: negative and weak reactions are caused by variable expression of CR1. Vox Sang. 1992;62:230–5. doi: 10.1111/j.1423-0410.1992.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 6.Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997;388:292–5. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- 7.Molthan L. The status of the McCoy/Knops antigens. Med Lab Sci. 1983;40:59–63. [PubMed] [Google Scholar]
- 8.Moulds MK. Serological investigation and clinical significance of high-titer, low-avidity (HTLA) antibodies. Am J Med Technol. 1981;47:789–95. [PubMed] [Google Scholar]
- 9.Moulds JM, Zimmerman PA, Doumbo OK, et al. Molecular identification of Knops blood group polymorphisms found in long homologous region D of complement receptor 1. Blood. 2001;97(9):2879–85. doi: 10.1182/blood.v97.9.2879. [DOI] [PubMed] [Google Scholar]
- 10.Moulds JM, Zimmerman PA, Doumbo OK, et al. Expansion of the Knops blood group system and subdivision of Sla. Transfusion. 2002;42:251–6. doi: 10.1046/j.1537-2995.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 11.Daniels GL, Cartron JP, Fletcher A, et al. International Society of Blood Transfusion Committee on terminology for red cell surface antigens: Vancouver report. Vox Sang. 2003;84:244–7. doi: 10.1046/j.1423-0410.2003.00282.x. [DOI] [PubMed] [Google Scholar]
- 12.Krych-Goldberg M, Hauhart RE, Subramanian VB. Decay accelerating activity of complement receptor type one (CD35): two active sites are required for dissociating C5 convertases. J Biol Chem. 1999;274:31160–8. doi: 10.1074/jbc.274.44.31160. [DOI] [PubMed] [Google Scholar]
- 13.Moulds JM, Rowe KE. Neutralization of Knops system antibodies using soluble complement receptor 1. Transfusion. 1996;36:517–20. doi: 10.1046/j.1537-2995.1996.36696269510.x. [DOI] [PubMed] [Google Scholar]
- 14.Moulds JM, Kassambara L, Middleton JJ, et al. Identification of complement receptor one (CR1) polymorphisms in West Africa. Genes Immun. 2000;1:325–9. doi: 10.1038/sj.gene.6363676. [DOI] [PubMed] [Google Scholar]
- 15.Tamasauskas D, Powell V, Schawalder A, Yazdanbakhsh K. Localization of Knops system antigens in the long homologous repeats of complement receptor 1. Transfusion. 2001;41:1397–404. doi: 10.1046/j.1537-2995.2001.41111397.x. [DOI] [PubMed] [Google Scholar]
- 16.Mallan MT, Grimm W, Hindley L, et al. The Hall serum: detecting Knb, the antithetical allele to Kna (Abstract) Transfusion. 1980;20:630. [Google Scholar]
- 17.Zimmerman PA, Fitness J, Moulds JM, et al. CR1 Knops blood group alleles are not associated with severe malaria in the Gambia. Genes Immun. 2003;4:368–73. doi: 10.1038/sj.gene.6363980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moulds JM, Brown LL. Loss of complement receptor one (CR1) from stored blood and its effect on Knops blood group reactivity. Immunohematology. 1995;11:46–50. [Google Scholar]