Abstract
Background information
JAM-C (junctional adhesion molecule C) has been implicated in the regulation of leukocyte migration, cell polarity, spermatogenesis, angiogenesis and nerve conduction. JAM-C has been also reported to concentrate at TJs (tight junctions) and desmosomes, although detailed localization studies remain incomplete.
Results
Monoclonal (LUCA14, MAB1189, Gi11, and PACA4) and polyclonal (40–9000) antibodies were employed to evaluate JAM-C expression/localization in various epithelial cell lines. However, RT–PCR (reverse transcription– PCR) assays revealed no JAM-C mRNA in SK-CO15, HeLa and HPAF-II cells, whereas abundant mRNA was detected in platelets, Caco-2 and ARPE cells. Interestingly, immunofluorescence localization in all cells revealed strong intercellular junctional staining with all of the above antibodies, except PACA4. Given the positive staining results in cells lacking JAM-C mRNA, immunoblot analyses were performed. Western blots revealed a prominent protein band at 52 kDa in all cells tested with all antibodies except PACA4. However, the correct size of JAM-C (37 kDa) was only detected in cells containing JAM-C mRNA. Immunofluorescence staining of JAM-C mRNA-expressing Caco-2 cells using mAb PACA4 revealed co-localization with occludin and ZO-1 (zonula occludens 1) at TJs. Analyses by MS identified the cross-reactive 52 kDa protein band as K8 (keratin 8). Furthermore, siRNA (small interfering RNA)-mediated downregulation of K8 in JAM-C mRNA-negative cells resulted in diminished junctional staining along with a reduction in the intensity of the 52 kDa protein band. Using an antibody specific for K8 phosphorylated at Ser73, the 52 kDa protein was identified as this phosphorylated form of K8.
Conclusions
The results from the present study demonstrate that a majority of available anti-human JAM-C antibodies cross-react with phosphorylated K8 and suggest that cellular localization studies using these reagents should be interpreted with caution. Of the JAM-C antibodies tested, only mAb PACA4 is monospecific for human JAM-C. Analyses using PACA4 reveal that JAM-C expression is variable in different epithelial cell lines with co-localization at TJs.
Keywords: antibody specificity, epithelial cell, junctional adhesion molecule C (JAM-C), keratin, tight junction
Introduction
JAMs (junctional adhesion molecules) are a novel subset of immunoglobulin superfamily proteins that belong to the larger CTX (cortical thymocyte Xenopus) family of proteins (Aurrand-Lions et al., 2001a). JAMs are expressed at intercellular junctions of endothelial and epithelial cells, as well as on the surface of leukocytes, platelets and erythrocytes (Naik et al., 1995; Malergue et al., 1998; Martin-Padura et al., 1998; Williams et al., 1999; Ebnet et al., 2004; Mandell and Parkos, 2005). JAM proteins have been implicated in regulation of cell-cell adhesion, barrier function, leukocyte migration, platelet activation, angiogenesis and retrovirus binding (Ebnet et al., 2004; Mandell and Parkos, 2005). So far, three members of the JAM family have been described: JAM-A (Martin-Padura et al., 1998), JAM-B (Cunningham et al., 2000; Palmeri et al., 2000) and JAM-C (Aurrand-Lions et al., 2000; Arrate et al., 2001). However, JAMs are closely related to other divergent proteins such as JAM4 (Hirabayashi et al., 2003), JAML (JAM-like) (Moog-Lutz et al., 2003), CLMP [CAR (coxsackie and adenovirus receptor)-like membrane protein] (Raschperger et al., 2004), CAR (Bergelson et al., 1997) and ESAM (endothelial cell adhesion molecule) (Hirata et al., 2001).
Human JAM-C cDNA encodes a 310 residue precursor protein with a molecular mass of 35 kDa, which includes an extracellular domain consisting of an N-terminal signal peptide, two Ig-like domains, two potential N-linked glycosylation sites, a single membrane-spanning region and a short cytoplasmic tail enclosing a PDZ-binding motif and a PKC (protein kinase C) phosphorylation consensus site (Arrate et al., 2001).
JAM-C has been shown to interact with PAR (protease-activated receptor)-3 and ZO-1 (zonula occludens 1) in endothelial cells and with PAR-6, Cdc42, PKCλ, PATJ (Pals1-associated TJ protein), and RA175 in spermatids, where JAM-C acts as scaffold to recruit polarity complexes to TJs (tight junctions) (Ebnet et al., 2003; Gliki et al., 2004; Mirza et al., 2006; Fujita et al., 2007). Furthermore, targeted disruption of JAM-C revealed a crucial role for this protein in spermatogenesis and maintaining the integrity and function of myelinated peripheral nerves, since JAM-C deficient mice are infertile due to an inability to produce mature spermatozoa (Gliki et al., 2004). Additionally, JAM-C knockout mice exhibit loss of myelin sheath integrity and display nerve conduction deficits (Scheiermann et al., 2007).
It has been reported that JAM-C is widely expressed in endothelial cells, platelets, T-cells and NK (natural killer) cells, and in tissues including intestine, skin, heart, lymph nodes, testis, thymus, lung, kidney, liver, placenta, retina, brain and peripheral nerves (Arrate et al., 2001; Aurrand-Lions et al., 2001a, 2005; Liang et al., 2002; Santoso et al., 2002; Ebnet et al., 2003; Gliki et al., 2004; Ludwig et al., 2005; Daniele et al., 2007; Scheiermann et al., 2007). In epithelial cells, JAM-C has been reported to be expressed at TJs of RPE (retinal pigment epithelium) and at the specialized AJs (adherent junctions) between Müller and photoreceptor cells (Daniele et al., 2007; Mandell et al., 2007). In addition, JAM-C has been reported in several carcinoma cell lines; NCI-H522, SUIT-2 and AZ-P7a. In addition, it is concentrated at primordial AJs in undifferentiated F9 cells and co-localizes with occludin at TJs of differentiated F9 cells (Santoso et al., 2005; Satohisa et al., 2005; Fuse et al., 2007). In CHO, MDCK and KLN205 cells transfected with full-length JAM-C, the protein concentrates at the plasma membrane, which results in better organization of cellular monolayers and improves barrier function (Aurrand-Lions et al., 2001a; Lamagna et al., 2005; Mandicourt et al., 2007). In contrast, in T84 cells and in human colonic mucosa, JAM-C was reported as having abundant basolateral expression with localization to desmosomes (Zen et al., 2004).
Given the differences in JAM-C expression reported for epithelia, JAM-C antibodies were used to assess JAM-C expression/localization in epithelial cell lines of varying origins (colon, pancreas, cervix and retina) and the expression was confirmed by PCR from mRNA. Of the epithelial cells lines tested, only Caco-2 and ARPE cells expressed JAM-C protein. Interestingly, the majority of JAM-C antibodies tested cross-reacted with K8 (keratin 8) and demonstrated an intercellular junctional staining pattern. Only one mAb, PACA4, was found to specifically recognize human JAM-C. The impact of these observations with respect to interpretation of localization studies is discussed.
Results
JAM-C mRNA is expressed in restricted subsets of epithelial cells
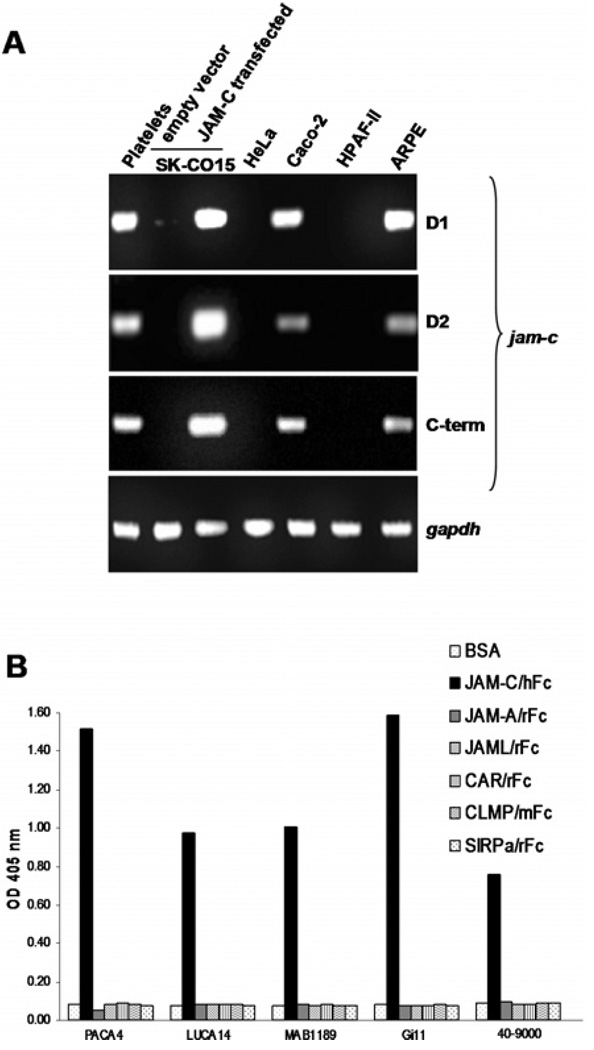
To further characterize JAM-C expression in epithelia, we evaluated several types of human epithelial cell lines derived from colon (SK-CO15 and Caco-2), cervix (HeLa), pancreas (HPAF-II) and retina (ARPE). As a positive control, human platelets, which express high levels of JAM-C, were used (Santoso et al., 2002). In platelets, JAM-C has been shown to be a counter-receptor for the leukocyte integrin Mac-1 and mediates leukocyte–platelet interactions. By RT (reverse transcription)–PCR assays, we used three independent sets of exon-spanning primers specific for the Ig-like domain 1 (D1), the Ig-like domain 2 (D2) or the cytoplasmic tail (C-term) of human JAM-C. As shown in Figure 1(A), with all JAM-C primers, JAM-C mRNA was expressed exclusively in Caco-2 and ARPE cells. No JAM-C mRNA was detected in SK-CO15, HeLa or HPAF-II cells. SK-CO15 cells transfected with full-length human JAM-C (SK-CO15/JAM-C) confirmed the expected sizes of JAM-C transcripts.
Figure 1. JAM-C expression is restricted to specific epithelial cell lines.
(A) Using RT–PCR, JAM-C mRNA is variably expressed in epithelial cell lines. As positive controls, platelets and SK-CO15/JAM-C were used. GAPDH was used as loading control. (B) Using ELISA assay, JAM-C antibodies were tested to specifically recognize the recombinant protein JAM-C/hFc or other human recombinant proteins of the JAM family or structurally similar proteins. BSA-coated wells in the absence of recombinant protein were used as control.
Cell labelling experiments with anti-human JAM-C antibodies demonstrate desmosomal localization in cells lacking JAM-C mRNA
Given the restricted expression of mRNA for JAM-C in the epithelial cell lines tested, we surveyed protein expression/localization using a panel of several monoclonal (PACA4, LUCA14, MAB1189 and Gi11) and polyclonal (40–9000) antibodies. Binding and specificity of the anti-human JAM-C antibodies was evaluated in an ELISA employing human recombinant proteins fused to human Fc. By ELISA, antibody binding to JAM-C/Fc and several recombinant proteins of the JAM family (JAM-A) or structurally similar molecules [JAML, CAR, CLMP and SIRPα (signal-regulatory protein α) was tested (Figure 1B). As expected, all monoclonal and polyclonal JAM-C antibodies bound to JAM-C/Fc, but not to other recombinant proteins.
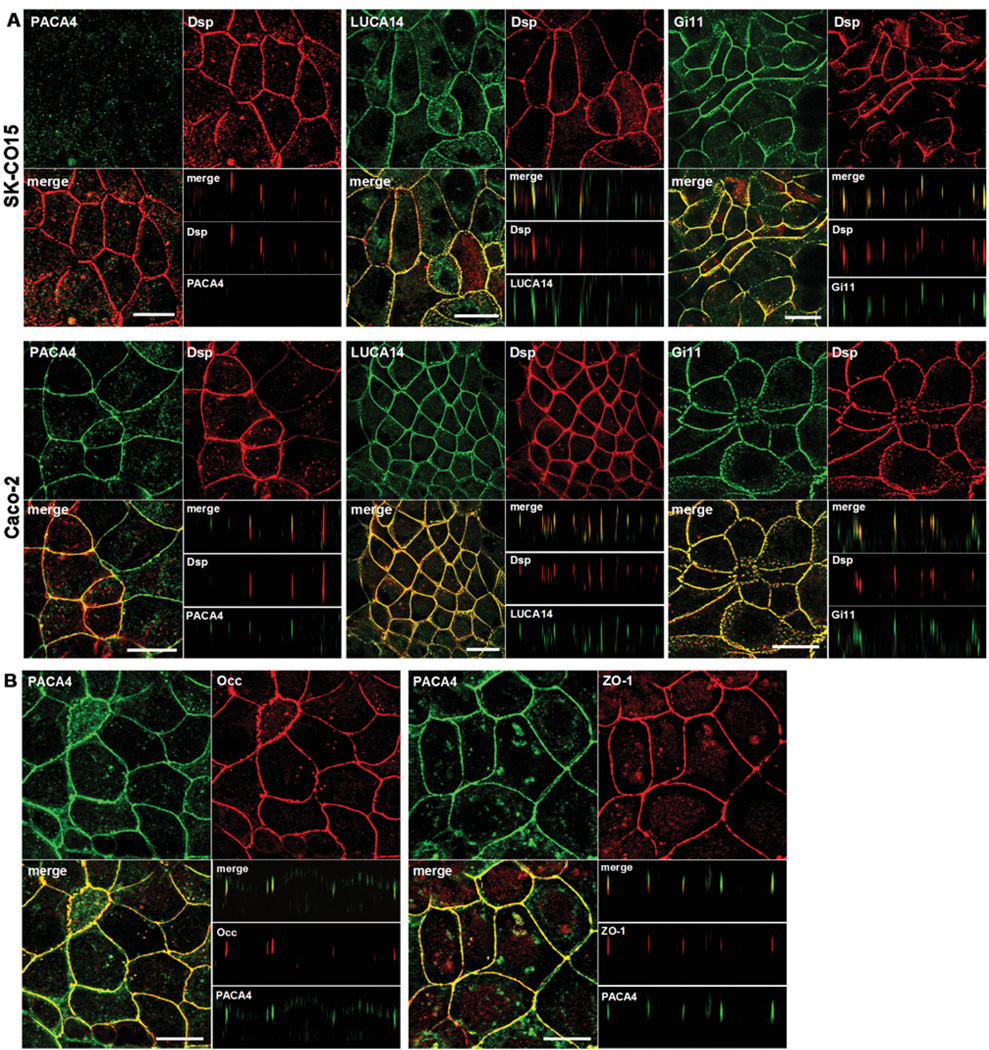
Next, IF (immunofluorescence) experiments were conducted to study JAM-C protein localization. Cell monolayers were fixed and permeabilized under routine conditions using ethanol or paraformaldehyde/Triton X-100 (results not shown). In SK-CO15 cells, where JAM-C mRNA was not detected, LUCA14 and Gi11 antibodies labelled intercellular junctions and perfectly co-localized with the specific desmosomal marker desmoplakin (Figure 2A, upper panel). Conversely, using a recently described monoclonal antibody termed PACA4 (Mandell et al., 2007; Sircar et al., 2007), no staining was detected. We observed similar results in HPAF-II and HeLa cells, where JAM-C is also not expressed (results not shown). Interestingly, in Caco-2 cells that express JAM-C, PACA4 labelled the most apical lateral membrane and did not co-localize with desmoplakin (Figure 2A, lower panel, see z-sections). Accordingly, PACA4 staining co-localized with the TJ markers, occludin and ZO-1 (Figure 2B). Analogous results in ARPE and SK-CO15/JAM-C cells were observed (results not shown). Thus, we can conclude that JAM-C co-localizes with TJ markers and not desmosomes in the epithelial cell lines tested.
Figure 2. JAM-C localizes with TJs of Caco-2 cells.
(A) Epithelial cell lines were double labelled with PACA4, LUCA14 or Gi11 (green) and with anti-desmoplakin (Dsp, red) antibodies.
(B) Caco-2 cells were double labelled with PACA4 (green) and occludin or ZO-1 (red). z-sections show localization of proteins at the TJ (most apical lateral staining) or at the lateral membrane. Bar=20 µm.
JAM-C antibodies cross-react with a 52 kDa protein identified as K8
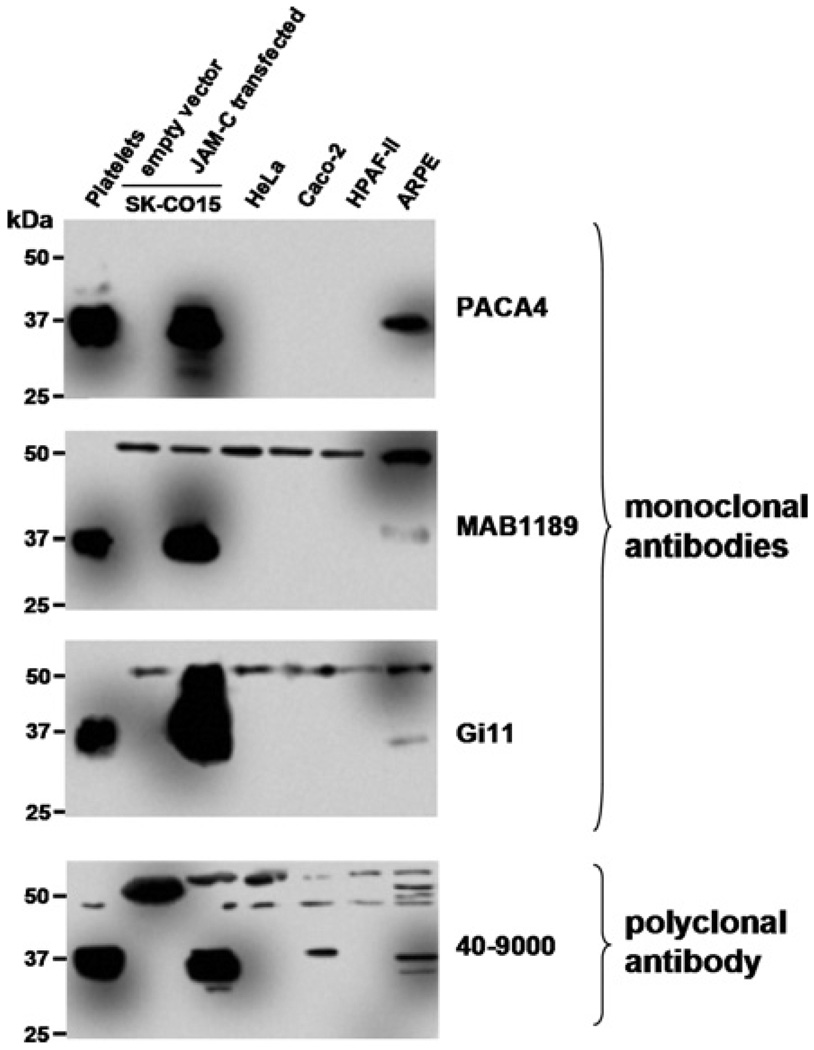
In parallel experiments, the various epithelial cell lines were subjected to SDS/PAGE followed by Western blotting. Surprisingly, in Western blots of epithelial cell lysates, MAB1189, Gi11 and 40–9000 antibodies detected a larger protein (52 kDa) in all cells tested (Figure 3), in addition to an expected 37 kDa band for JAM-C. The staining pattern for the expected 37 kDa JAM-C band was weak in ARPE cells, compared with a robust signal in controls (platelets and SK-CO15/JAM-C). In Caco-2 cells, only the 40–9000 antibody was able to faintly recognize the 37 kDa band. We observed similar results with other JAM-C polyclonal antibodies (40–8900, Zymed and AF1189, R&D; results not shown). However, PACA4 antibody only revealed a single 37 kDa band corresponding to the correct size of JAM-C in platelets, ARPE and SK-CO15/JAM-C cells but not in Caco-2, SK-CO15, HPAF-II and HeLa cells. Although Caco-2 cells express JAM-C mRNA and demonstrated intercellular junction staining with PACA4, Western blots of Caco-2 cells with the same antibody failed to detect the 37 kDa JAM-C protein band. It is likely that the failure of PACA4 to visualize JAM-C in Western blots of these cells is due to limited antibody sensitivity under conditions with low concentrations of denatured epitopes.
Figure 3. JAM-C protein is expressed in Caco-2 and ARPE cells.
Western blots of epithelial cell lysates using monoclonal and polyclonal JAM-C antibodies. Platelets and SK-CO15/JAM-C lysates were used as a positive control.
To elucidate the identity of the 52 kDa epithelial protein recognized by the panel of JAM-C antibodies, immunoprecipation studies with LUCA14 were performed. JAM-C mRNA-deficient SK-CO15 cells were solubilized in a buffer containing 1% Triton X-100, followed by immunoprecipitation with LUCA14 and separated by SDS/PAGE. A prominent 52 kDa band was apparent in the Coomasie Blue-stained polyacrylamide gel that was subjected to proteomic analysis. MS analysis identified the 52 kDa protein as K8 (mascot score=104). Alignment of human JAM-C and K8 protein sequences revealed several regions of homology between K8 peptides identified with JAM-C (Figure 4). However, the complete sequences of JAM-C and K8 are only 12.2% identical and 21.7% similar. These observations suggest that JAM-C antibodies may recognize a complex epitope not apparent in the linear sequence.
Figure 4. Alignment of protein sequences of JAM-C and K8.
Human JAM-C and K8 protein sequences were aligned. The 52 kDa protein band recognized by mAb LUCA14 was sequenced by MALDI–TOF and identified as K8. Resulting sequence peptides from MS analysis are highlighted.
Down-regulation of K8 expression in epithelial cells diminishes expression of the JAM-C cross-reactive 52 kDa protein
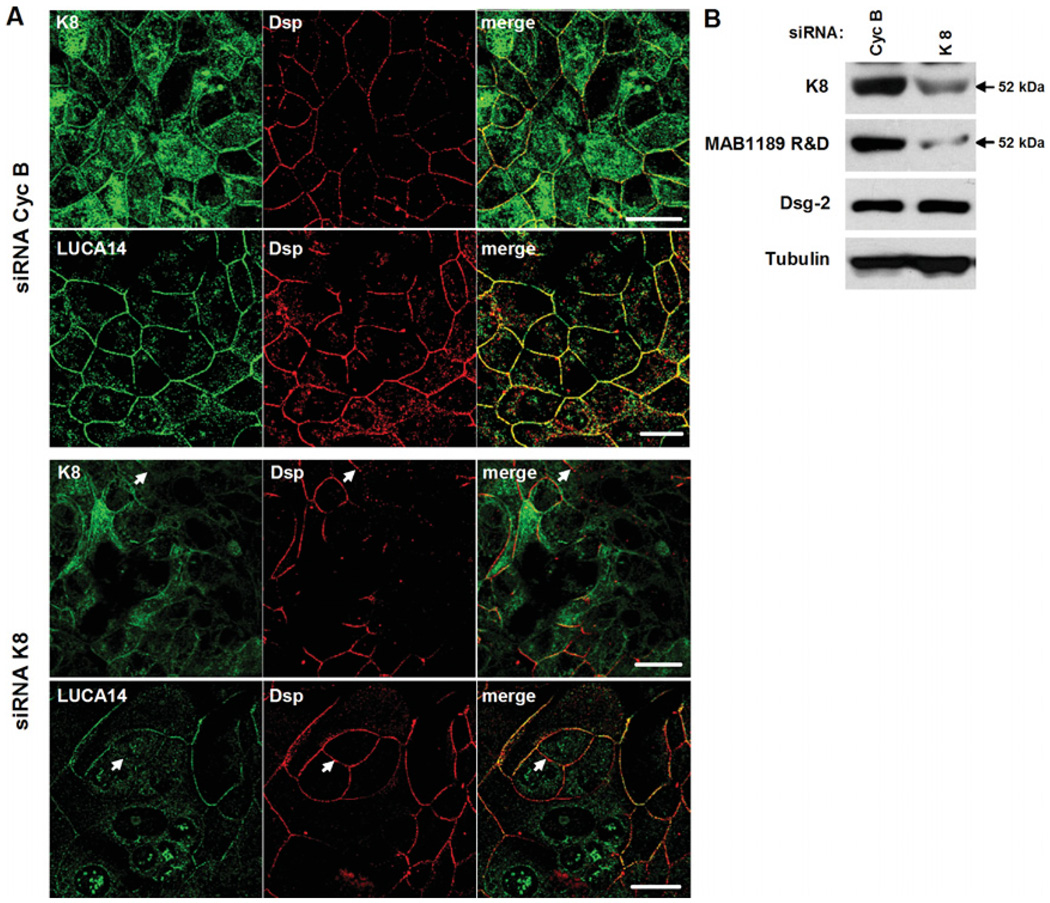
To confirm identify of the 52 kDa protein recognized by JAM-C antibodies as K8, siRNA (small interfering RNA)-mediated downregulation experiments were performed. SK-CO15 cells, which do not express JAM-C, were transfected with siRNAs specific for K8 or the unrelated protein Cyc B (cyclophilin B) as a control. 72 h post-transfection, localization and immunoblotting experiments were performed using LUCA14 and MAB1189 antibodies respectively. In the cells transfected with Cyc B siRNA, the staining pattern observed with K8 and LUCA14 antibodies were similar and highlighted intercellular junctions (Figure 5A). However, K8 also localized diffusely in the cytosol. As expected, in cells transfected with K8 siRNA, K8 expression was decreased (Figure 5A, upper panel). As can be seen, staining with LUCA14 antibody was also reduced after downregulation of K8, especially at the plasma membrane (Figure 5A, lower panel). Desmoplakin staining was also diminished after K8 downregulation, which is not surprising since K8 is a component of intermediate filaments and may play a role in regulating localization of desmoplakin. However, decreased staining of K8 and LUCA14 observed at cellular borders did not co-localize with desmoplakin, suggesting independent mechanisms of regulation (Figure 5A, arrows). By immunoblot (Figure 5B), the 52 kDa K8 band was diminished after treatment with siRNA. A similar reduction was observed in the 52 kDa protein detected with the MAB1189 antibody. However, the desmosomal transmembrane protein desmoglein-2 and tubulin were not affected. A Western blot of desmoplakin was not obtained due to a lack of suitable blotting antibodies.
Figure 5. Knock-down of K8 decreases the expression of the 52 kDa protein recognized by JAM-C antibodies.
SK-CO15 cells were transfected with siRNAs for K8 or Cyc B (control). (A) Cells were double stained with K8 or LUCA14 (green) and anti-desmoplakin (Dsp, red) antibodies. Arrows indicate a plasma membrane localization. Bar=20 µm. (B) Western blot analysis of cell lysates after K8 or Cyc B (as a control) siRNA transfection.
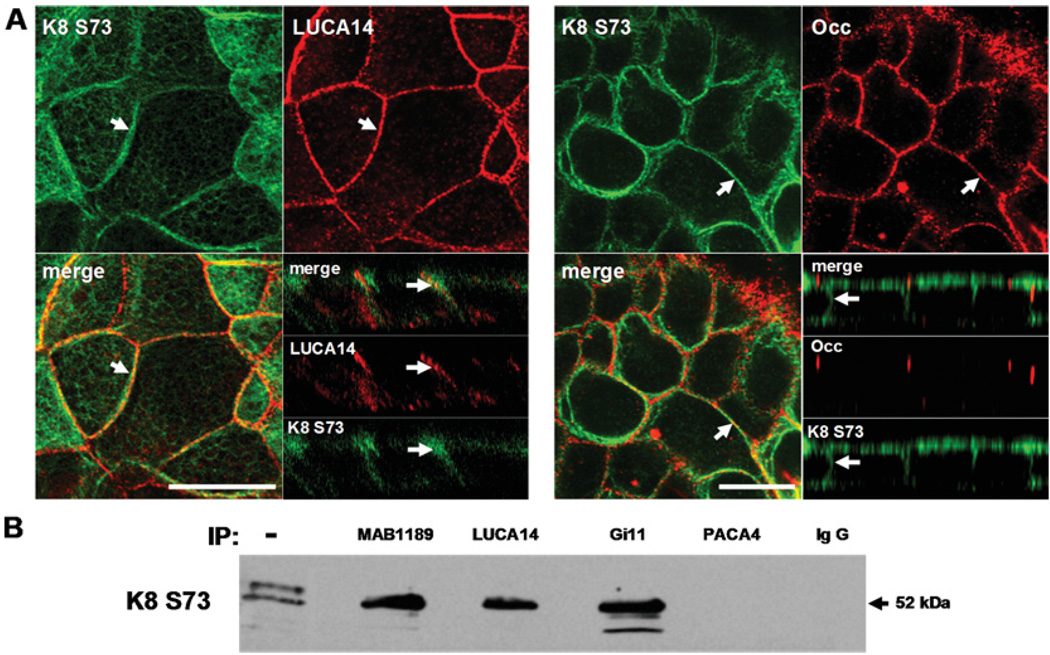
Since the staining pattern for K8 and LUCA14 antibodies was similar but not identical, we hypothesized that JAM-C antibodies may cross-react with a post-transcriptionally modified form of K8. It is well known that K8 is phosphorylated (Omary et al., 1998), thus experiments were performed to examine whether JAM-C antibodies recognize a phosphorylated form of K8. K8 possesses three major phospho-serine sites, two in the N-terminal (Ser23 and Ser73) and one in the C-terminal region of the molecule (Ser431) (Omary et al., 1998). However, only the amino region of K8 aligns with JAM-C (see Figure 4). Of both amino serine phosphorylated residues (Ser23 and Ser73), we tested anti-K8 (phospho-Ser73) due to its commercial availability. Interestingly, IF assays using this anti-K8 (phospho-Ser73) antibody revealed a similar staining pattern as was observed with LUCA14 along the lateral membrane (Figure 6A). Such staining was distinct from that observed with anti-occludin. Additionally, in Western blots of immunoprecipitates using the cross-reacting antibodies (MAB1189, LUCA14 and Gi11), the anti-K8 (phospho Ser73) antibody recognized the same 52 kDa band (Figure 6B). However, this was not the case when the JAM-C specific antibody (PACA4) or mouse IgG were used for immunoprecipitation (see Figure 3). These results suggest that the 52 kDa intercellular protein recognized by JAM-C antibodies is K8 that is phosphorylated at Ser73.
Figure 6. JAM-C antibodies cross-react with phosphorylated K8.
(A) SK-CO15 cells were double labelled with K8 phospho-Ser73 (K8 S73) (green) and LUCA14 or anti-occludin (red) antibodies. Arrows indicate a plasma membrane localization. Bar=20 µm. (B) Immunoprecipitates with JAM-C antibodies from SK-CO15 were blotted with K8 S73 antibody. Negative controls consisted of immunoprecipitation using mAb PACA or mouse IgG. (−) indicates the original cell lysate.
JAM-C expression in natural human tissues
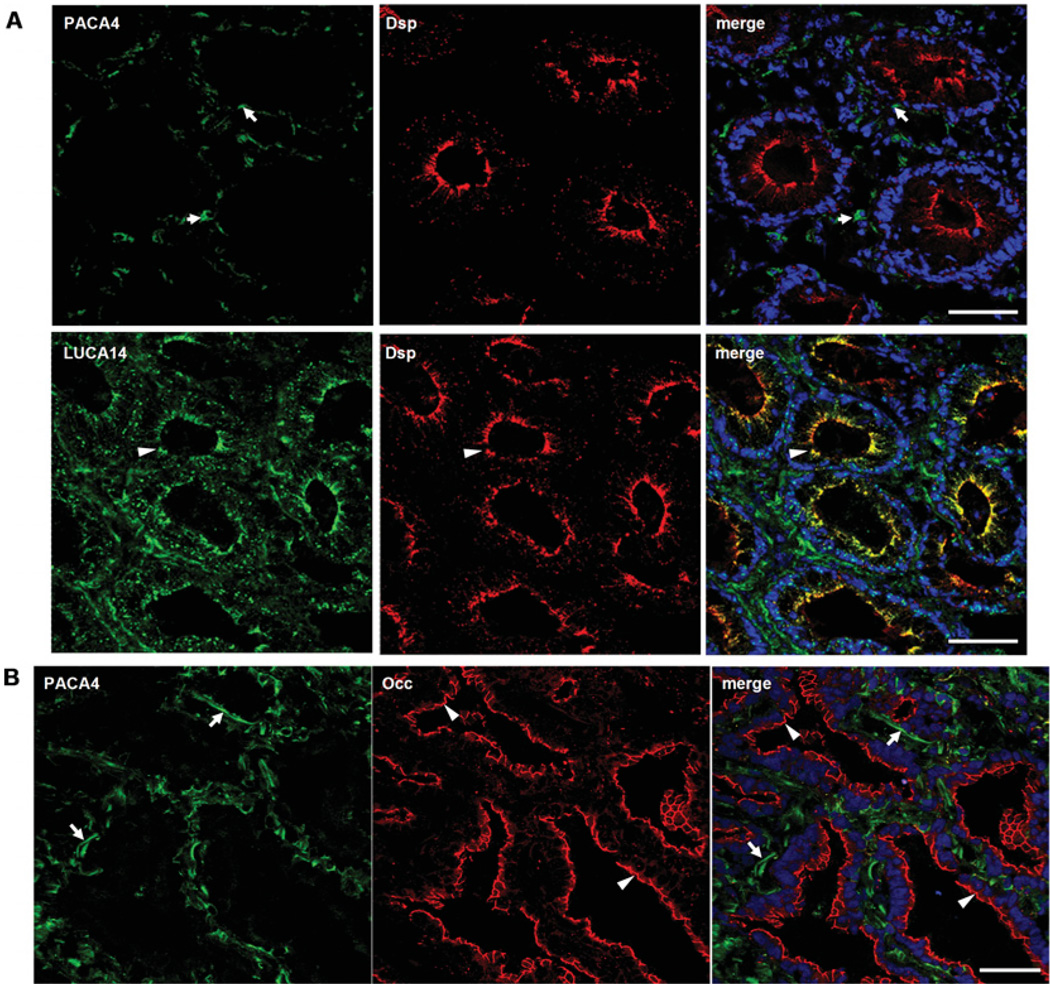
Given the above results, the expression of JAM-C was evaluated using different JAM-C antibodies in several normal human tissues including colon, lung, liver and kidney. Interestingly, in colon, PACA4 did not significantly label epithelial cells but appeared to stain vascular structures (arrows, Figure 7A, upper panel). Conversely, as previously described, LUCA14 staining colocalized with desmoplakin in epithelial crypts (arrowheads, Figure 7A, lower panel). In lung, there was no colocalization of PACA4 with the epithelial TJ marker occludin (arrows, Figure 7B). Instead, PACA4 labelling was prominent on subepithelial structures consistent with blood vessels (arrowheads, Figure 7B). Similar results were observed in normal human tissues derived from kidney and liver (results not shown). These results suggest that in natural human colonocytes, K8 recognized by JAM-C cross-reacting antibodies is localized at desmosomes. These data also suggest that JAM-C protein is not abundantly expressed in epithelia from colon, liver, lung and kidney.
Figure 7. Expression of JAM-C in human colonic and lung mucosa.
(A) Frozen sections of normal human colonic mucosa were double labelled with PACA4 or LUCA14 (green) and anti-desmoplakin (Dsp; red) antibodies. (B) Frozen sections of normal human lung epithelium were double labelled with PACA4 (green) and anti-occludin (Occ; red) antibodies. Arrows indicate vascular structures and arrowheads show epithelial localization. Nuclei were labelled with TOPRO (blue). Bar=50 µm.
Discussion
JAM-C has been reported to be expressed at intercellular junctions of endothelial cells, where it co-localizes with the TJ component, ZO-1 (Aurrand-Lions et al., 2001a, 2001b; Ebnet et al., 2003; Chavakis et al., 2004). JAM-C contains a PDZ-binding motif at the C-terminus (Aurrand-Lions et al., 2001b), which might mediate its junctional localization through interactions with TJ scaffolding proteins, as has been demonstrated for other JAM proteins (Liu et al., 2000; Amieva et al., 2003). In addition, JAM-C has been shown to interact with polarity molecules such as PAR-3, PAR-6, Cdc42, PKCλ and PATJ, where it has been proposed as an anchor to recruit polarity complexes to TJ (Ebnet et al., 2003; Gliki et al., 2004; Mirza et al., 2006; Fujita et al., 2007). These findings suggest that JAM-C plays a role in the formation and maintenance of endothelial contacts.
In contrast with endothelia, studies of JAM-C expression in epithelia are limited to reports of both TJ and desmosomal localization in different cell types. Furthermore, JAM-C expression has been reported in several tissues (Arrate et al., 2001; Aurrand-Lions et al., 2001a; Gliki et al., 2004; Ludwig et al., 2005; Daniele et al., 2007; Scheiermann et al., 2007). However, in these studies, total tissue lysates were assayed and not specific cell-type expression patterns. We performed IF assays in different human tissues (colon, liver, lung and kidney) and also examined several human epithelial cell lines of various origins in order to characterize the expression of JAM-C in a variety of different epithelia. Interestingly, we observed variable JAM-C expression that was limited to Caco-2 and ARPE cells. In contrast, some colonic (SK-CO15), cervical (HeLa) and pancreatic (HPAF-II) epithelial cell lines, in addition to normal colonic crypt, lung, liver and kidney epithelia, appeared to lack expression of JAM-C. In the latter tissues that were tested, JAM-C staining was prominent in vascular structures. These observations suggest that JAM-C may play different functional roles in some epithelia. However, it is also possible that variable JAM-C expression in epithelial cells could be compensated for differential expression of other JAMs or related proteins. For example, JAM-C, but not JAM-A, was detected at AJs of retina, however in Jam-C−/− mice, JAM-A is expressed instead (Daniele et al., 2007). Interestingly, we have observed that JAM-A is abundantly expressed in cell lines that are now shown to lack JAM-C protein expression (Mandell et al., 2005). Perhaps up-regulation of JAM-A expression may compensate for the lack of JAM-C. Given these observations, we hypothesize that the absence of JAM-C in some epithelial cells is compensated for by the expression of other proteins with similar roles. Nevertheless, more experiments are required to verify this possibility.
In Caco-2 and ARPE cells, we have shown that JAM-C is expressed at intercellular junctions, where it co-localizes with TJ markers, occludin and ZO-1, but not with the desmosomal marker desmoplakin. These findings are consistent with previous studies reporting that JAM-C co-localizes with ZO-1 and occludin at TJs of RPE and in F9 epithelial cells (Satohisa et al., 2005; Daniele et al., 2007). Interestingly, in another cell line (T84), we reported JAM-C expression in desmosomes, but not in TJs (Zen et al., 2004), which prompted the present study. We thus performed experiments examining expression and localization of JAM-C in epithelia with a panel of several monoclonal and polyclonal antibodies against JAM-C that have been used by others to analyse its biochemistry and expression. Although all antibodies specifically recognized recombinant JAM-C, it was surprising that MAB1189, Gi11 and 40–900 antibodies recognized a band of 52 kDa. Only in Caco-2 and ARPE cells, was a protein band corresponding to the correct size for JAM-C (37 kDa) detected. As shown in this report, mAbs LUCA14 and Gi11 labelled a protein at desmosomes in epithelial cells lacking JAM-C, and this staining was independent of fixation and permeabilization conditions, since similar results were obtained using ethanol or paraformaldehyde and Triton X-100 solubilization (results not shown). The only antibody that exclusively recognized the 37 kDa JAM-C protein and stained the TJ region was PACA4. These findings suggest that the JAM-C antibodies MAB1189, LUCA14, Gi11 and 40–900 cross-react with a 52 kDa desmosomal-associated protein while also recognizing JAM-C. Conversely, our findings suggest that PACA4 is the only antibody that specifically detects JAM-C. Thus, localization studies employing JAM-C antibodies used in this study may require re-interpretation. Our localization results obtained with PACA4 suggest that JAM-C is a TJ-associated protein in some epithelia. Given the limited expression pattern we observed for JAM-C in several epithelial cell lines of diverse origin, further detailed analyses of expression using PCR and mAbs such as PACA4 are warranted.
Immunoprecipitation of a 52 kDa protein identified by Western blots of epithelial cells by a majority of the JAM-C antibodies was confirmed by MS to be K8. This keratin has been described as a 53.5 kDa polypeptide that belongs to the type B of keratins and exists in combination with K18 at intermediate filaments (Moll et al., 1982; Lu et al., 2005). K8 is primarily found in simple, glandular, pseudo-stratified and transitional epithelium and is also present in the majority of adenocarcinomas and ductal carcinomas (Oshima et al., 1996; Owens and Lane, 2003). We found that K8 localization at cell–cell junctions was very similar to the staining pattern observed using LUCA14 and Gi11 antibodies. Furthermore, knock-down of K8 diminished the intercellular pattern staining generated by LUCA14 and specifically reduced expression of the 52 kDa protein recognized by MAB1189 antibody. K8 is also subject to post-translational modifications such as glycosylation and phosphorylation in the N- and C-terminus (Omary et al., 1998; Coulombe and Omary, 2002). Since K8 localized to both the membrane and cytosol, we hypothesized that the 52 kDa protein may represent a phosphorylated version of K8 that is expressed predominantly at cell–cell junctions. Using a mAb that specifically recognizes K8 phosphorylated at Ser73, we observed a prominent junctional staining pattern. Indeed, expression of K8 at cellular borders has been previously described, where it co-localized with desmoplakin and desmoglein at desmosomes in hepatocytes (Loranger et al., 2006). Phosphorylated K8 has also been described at cell–cell contacts in human and mouse liver tissues (Tao et al., 2006). In our analyses, the anti-K8 (phospho-Ser73) antibody only recognized the 52 kDa band in imunoprecipitates using JAM-C cross-reacting antibodies. These findings suggest that the 52 kDa protein recognized by JAM-C antibodies that localizes at desmosomes is a phosphorylated form of K8.
In conclusion, this report highlights the importance of detailed molecular, biochemical and localization studies to verify specificity of antibodies. We have demonstrated specificity of JAM-C mAb PACA4, whereas other anti-human JAM-C antibodies should be carefully assessed when used in localization studies.
Materials and methods
Cells
SK-CO15 and Caco-2 (derived from colon), HeLa (derived from cervix) and ARPE (derived from retina) epithelial cell lines, were grown in high glucose DMEM (Dulbecco’s modified Eagle’s medium), supplemented with 10% FBS, 100 units/ml penicillin, 100 µg/ml streptomycin, 15 mM Hepes (pH 7.4), 2 mM L-glutamine, and 1% non-essential aminoacids. HPAF-II cells (derived from pancreas) were grown in RPMI supplemented with 10% FBS and 1 mM sodium pyruvate. Platelets were isolated from whole blood of normal human volunteers by centrifugation at 150 g for 15 min, washed with HBSS (Hanks balanced salt solution) devoid of calcium and stored at −80°C. Blood was drawn and handled according to protocols for the protection of human subjects, as approved by the Emory University Hospital Institutional Review Board, and all volunteer subjects gave informed consent in accordance with the Declaration of Helsinki (2000).
Antibodies
Mouse monoclonal antibodies that bind to the extracellular domain of human JAM-C were obtained from R&D Systems (MAB1189; Minneapolis, MN, U.S.A.), BD Biosciences (Gi11; San Jose, CA, U.S.A.) and as a gift from Raven Biotechnologies (LUCA14 and PACA4; San Francisco, CA, U.S.A.). A polyclonal antibody against the internal region of JAM-C was purchased from Zymed (40–9000; South San Francisco, CA, U.S.A.). Rabbit polyclonal anti-desmoplakin, and anti-occludin and anti-ZO-1 antibodies were obtained from AbD Serotec (Oxford, U.K.) and Zymed respectively. Mouse monoclonal anti-K8 and anti-tubulin antibodies were obtained from Sigma (Saint Louis, MO, U.S.A.). Rabbit monoclonal anti-K8 (phospho-Ser73) was purchased from Abcam (Cambridge, MA, U.S.A.). Mouse monoclonal anti-desmoglein-2 was generated in house as previously described in (Nava et al., 2007).
Cloning and transfection of full length JAM-C
cDNA encoding full length human JAM-C was amplified by PCR from a Marathon-ready colonic cDNA library (Qiagen). Primers used, including KpnI and XhoI restriction sites respectively, were: forward 5′-ATATGGTACCCCTCAGCTT-CCTCTGTCACC-3′ and reverse 5′-ATATCTCGAGTCAGA-TCACAAACGATGACTTGT-3′. JAM-C cDNA was cloned into pcDNA3 (Invitrogen) and transfected into SK-CO15 cells using Lipofectamine™ 2000 (Invitrogen), according to the manufacturer’s instructions.
RT-PCR
Total RNA isolation was performed using the RNeasy kit (Qiagen) according to the provided protocol. Subsequently, cDNA was synthesized by RT using oligo(dT) primer and Superscript II (Invitrogen). PCR was performed using Taq DNA polymerase (Roche) and three sets of exon-spanning primers located in different regions of human JAM-C: Ig-like domain 1 (D1), forward 5′-CTTCTTCCTGCTGCTGCTTT-3′ and reverse 5′-CAGCGATAAAGGGCTGAGTC-3′; Ig-like domain 2 (D2), forward 5′-GCCGAAGGCTGTACCAGTAG-3′ and reverse 5′-ATCAGGGCCAGTACAGCAAG-3′; and cytoplasmic tail (C-term), forward 5′-GTACTGGCCCTGATCA-CGTT-3′ and reverse 5′-TTTACCGGGTCCATCTTGAG-3′. As a control, primers of the constitutive gene gapdh were employed. The PCR protocol followed standard conditions: 95°C for 15 min, 40 cycles comprised of 95°C for 15 s, 60°C for 30 s and 72°C for 30 s and finally an incubation of 72°C for 10 min.
ELISA
Recombinant proteins containing extracellular domains of human JAM-C (R&D), JAM-A, JAML, CAR, CLMP and SIRPα [purified as previously described in (Barton et al., 2001; Liu et al., 2004)] fused to IgG1Fc fragment, were immobilized at a concentration of 5 µg/ml in 96-well plates at 4°C overnight, followed by blocking with 1% BSA for 1 h at room temperature (24°C). Monoclonal and polyclonal antibodies against JAM-C were added and incubated for 1 h at room temperature. Wells were washed with HBSS and incubated with HRP (horseradish peroxidase)-conjugated secondary antibodies. After addition of ABTS [2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid)] substrate, wells were analysed using a microtitre plate reader at 405 nm.
Knockdown of keratin 8
Two duplex siRNA oligonucleotides of K8 (si5 and si7, Qiagen) were transfected into SK-CO15 using HiPerFect (Qiagen), according to the manufacturer’s instructions. As control, the siRNA directed to the unrelated protein cyclophilin B (Dharmacon, Chicago, IL, U.S.A.) was used.
Western blot
Cells were lysed in RIPA buffer (20 mM Tris/HCl, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% sodium deoxycholate, 1% Triton X-100 and 0.1% SDS, pH 7.4) and boiled under reducing conditions. Samples were separated by SDS/PAGE and transferred onto PVDF membranes. Membranes were blocked for 1 h with 5% (w/v) skimmed non-fat dried milk in TTBS (Tween Tris-buffered saline; 50 mM Tris/HCl, pH 7.6, 150 mM NaCl and 0.05% Tween-20) and incubated overnight with primary antibodies. After washing with TTBS, membranes were incubated with HRP-conjugated secondary antibodies followed by ECL (enhanced chemiluminescence) detection.
IF and IH (immunohistochemistry)
For IF, cells were fixed with ethanol for 20 min at −20°C and blocked with 1% BSA. For IH, mucosal biopsies from normal patients were embedded in freezing medium (Tissue-Tek OCT compound) and snap-frozen in liquid nitrogen. Frozen tissues were sectioned (8 µm thick), ethanol-fixed and blocked with 1% BSA and 10% normal goat serum. Cells and tissue sections were incubated with primary antibodies for 1 h at room temperature. After washing with PBS, the samples were incubated with fluorescently labelled Alexa Fluor®-555 and Alexa Fluor®-488 antibodies for 1 h at room temperature. For tissues, nuclei were also stained with TOPRO-3 dye for 1 min at room temperature. Preparations were embedded in ProLong Gold antifade medium and analysed using a Zeiss LSM510 confocal laser microscope.
Immunoprecipitation
SK-CO15 cells were lysed in HO buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EGTA, 1.5 mM MgCl2, 10% glycerol and 1% Triton X-100) and subjected to immunoprecipitation employing mAb LUCA14, MAB1189 and Gi11 or mAb PACA4 and mouse IgG as control. Proteins were separated by SDS-PAGE and analysed by Western blot.
MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS
Immunoprecipitates with mAb LUCA14 were separated by SDS/PAGE and stained with Coomassie Blue. The 52 kDa protein was cut off and analysed by MS analysis through Pick’n POST analytical services (Alphalyse, Palo Alto, CA, U.S.A.). Database search was performed using Mascot version 2.1.03.
Acknowledgements
We thank Dr. Susan Voss for tissue culture expertise, Moshe Bachar for technical assistance, Dr. Jan-Michael Klapproth (Department of Gastroenterology, Emory University Hospital, Atlanta, GA, U.S.A.) and Dr. Asma Nusrat (Department of Pathology, Emory University Hospital, Atlanta, GA, U.S.A.) for providing tissue material. The PACA4 antibody was a gift from Raven Biotechnologies for research purposes only. One of the authors, Mr T. Liang, is employed by Raven Biotechnologies which may, as the commercial source of PACA4, stand to benefit from the conclusions reached by this study. At the time of this publication, there are no plans to commercialize this antibody and no monetary or financial incentive was given to the researchers in the current study regarding the use of this material. Investigators interested in obtaining PACA4 for research purposes only may contact Mr. T. Liang at Raven Biotechnologies.
Funding
This study was supported by National Institutes of Health [grant numbers R01-DK72564 and R01-DK61379].
Abbreviations used
- AJ
adherent junction
- CAR
coxsackie and adenovirus receptor
- CLMP
CAR-like membrane protein
- Cyc B
cyclophilin B
- IF
immunofluorescence
- IH
immunohistochemistry
- JAM
junctional adhesion molecule
- JAML
JAM-like
- HBSS
Hanks balanced salt solution
- HRP
horseradish peroxidase
- K8
keratin 8
- MALDI–TOF
matrix-assisted laser-desorption ionization–time-of-flight
- PAR
protease-activated receptor
- PATJ
Pals1-associated TJ protein
- PKC
protein kinase C
- RPE
retinal pigment epithelium
- RT
reverse transcription
- siRNA
small interfering RNA
- SIRPα
signal-regulatory protein α
- TJ
tight junction
- ZO-1
zonula occludens 1
References
- Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300:1430–1434. doi: 10.1126/science.1081919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrate MP, Rodriguez JM, Tran TM, Brock TA, Cunningham SA. Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J. Biol. Chem. 2001;276:45826–45832. doi: 10.1074/jbc.M105972200. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions MA, Duncan L, Du Pasquier L, Imhof BA. Cloning of JAM-2 and JAM-3: an emerging junctional adhesion molecular family? Curr. Top. Microbiol. Immunol. 2000;251:91–98. doi: 10.1007/978-3-642-57276-0_12. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions M, Johnson-Leger C, Wong C, Du Pasquier L, Imhof BA. Heterogeneity of endothelial junctions is reflected by differential expression and specific subcellular localization of the three JAM family members. Blood. 2001a;98:3699–3707. doi: 10.1182/blood.v98.13.3699. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions M, Duncan L, Ballestrem C, Imhof BA. JAM-2, a novel immunoglobulin superfamily molecule, expressed by endothelial and lymphatic cells. J. Biol. Chem. 2001b;276:2733–2741. doi: 10.1074/jbc.M005458200. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions M, Lamagna C, Dangerfield JP, Wang S, Herrera P, Nourshargh S, Imhof BA. Junctional adhesion molecule-C regulates the early influx of leukocytes into tissues during inflammation. J. Immunol. 2005;174:6406–6415. doi: 10.4049/jimmunol.174.10.6406. [DOI] [PubMed] [Google Scholar]
- Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. Junction adhesion molecule is a receptor for reovirus. Cell. 2001;104:441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- Chavakis T, Keiper T, Matz-Westphal R, Hersemeyer K, Sachs UJ, Nawroth PP, Preissner KT, Santoso S. The junctional adhesion molecule-C promotes neutrophil transendothelial migration invitro and invivo. J. Biol. Chem. 2004;279:55602–55608. doi: 10.1074/jbc.M404676200. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell. Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- Cunningham SA, Arrate MP, Rodriguez JM, Bjercke RJ, Vanderslice P, Morris AP, Brock TA. A novel protein with homology to the junctional adhesion molecule. Characterization of leukocyte interactions. J. Biol. Chem. 2000;275:34750–34756. doi: 10.1074/jbc.M002718200. [DOI] [PubMed] [Google Scholar]
- Daniele LL, Adams RH, Durante DE, Pugh EN, Jr, Philp NJ. Novel distribution of junctional adhesion molecule-C in the neural retina and retinal pigment epithelium. J. Comp. Neurol. 2007;505:166–176. doi: 10.1002/cne.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebnet K, Aurrand-Lions M, Kuhn A, Kiefer F, Butz S, Zander K, Meyer zu Brickwedde MK, Suzuki A, Imhof BA, Vestweber D. The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: a possible role for JAMs in endothelial cell polarity. J. Cell. Sci. 2003;116:3879–3891. doi: 10.1242/jcs.00704. [DOI] [PubMed] [Google Scholar]
- Ebnet K, Suzuki A, Ohno S, Vestweber D. Junctional adhesion molecules (JAMs): more molecules with dual functions? J. Cell Sci. 2004;117:19–29. doi: 10.1242/jcs.00930. [DOI] [PubMed] [Google Scholar]
- Fujita E, Tanabe Y, Hirose T, Aurrand-Lions M, Kasahara T, Imhof BA, Ohno S, Momoi T. Loss of partitioning-defective-3/isotype-specific interacting protein (par-3/ASIP) in the elongating spermatid of RA175 (IGSF4A/SynCAM)-deficient mice. Am. J. Pathol. 2007;171:1800–1810. doi: 10.2353/ajpath.2007.070261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse C, Ishida Y, Hikita T, Asai T, Oku N. Junctional adhesion molecule-C promotes metastatic potential of HT1080 human fibrosarcoma. J. Biol. Chem. 2007;282:8276–8283. doi: 10.1074/jbc.M608836200. [DOI] [PubMed] [Google Scholar]
- Gliki G, Ebnet K, Aurrand-Lions M, Imhof BA, Adams RH. Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature. 2004;431:320–324. doi: 10.1038/nature02877. [DOI] [PubMed] [Google Scholar]
- Hirabayashi S, Tajima M, Yao I, Nishimura W, Mori H, Hata Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol. Cell. Biol. 2003;23:4267–4282. doi: 10.1128/MCB.23.12.4267-4282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata K, Ishida T, Penta K, Rezaee M, Yang E, Wohlgemuth J, Quertermous T. Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. J. Biol. Chem. 2001;276:16223–16231. doi: 10.1074/jbc.M100630200. [DOI] [PubMed] [Google Scholar]
- Lamagna C, Meda P, Mandicourt G, Brown J, Gilbert RJ, Jones EY, Kiefer F, Ruga P, Imhof BA, Aurrand-Lions M. Dual interaction of JAM-C with JAM-B and αMβ2 integrin: function in junctional complexes and leukocyte adhesion. Mol. Biol. Cell. 2005;16:4992–5003. doi: 10.1091/mbc.E05-04-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang TW, Chiu HH, Gurney A, Sidle A, Tumas DB, Schow P, Foster J, Klassen T, Dennis K, DeMarco RA, et al. Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM 2 interacts with T, NK, and dendritic cells through JAM 3. J. Immunol. 2002;168:1618–1626. doi: 10.4049/jimmunol.168.4.1618. [DOI] [PubMed] [Google Scholar]
- Liu Y, Nusrat A, Schnell FJ, Reaves TA, Walsh S, Pochet M, Parkos CA. Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci. 2000;113:2363–2374. doi: 10.1242/jcs.113.13.2363. [DOI] [PubMed] [Google Scholar]
- Liu Y, O’Connor MB, Mandell KJ, Zen K, Ullrich A, Buhring HJ, Parkos CA. Peptide-mediated inhibition of neutrophil transmigration by blocking CD47 interactions with signal regulatory protein alpha. J. Immunol. 2004;172:2578–2585. doi: 10.4049/jimmunol.172.4.2578. [DOI] [PubMed] [Google Scholar]
- Loranger A, Gilbert S, Brouard JS, Magin TM, Marceau N. Keratin 8 modulation of desmoplakin deposition at desmosomes in hepatocytes. Exp. Cell Res. 2006;312:4108–4119. doi: 10.1016/j.yexcr.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Lu H, Hesse M, Peters B, Magin TM. Type II keratins precede typeI keratins during early embryonic development. Eur. J. Cell Biol. 2005;84:709–718. doi: 10.1016/j.ejcb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Ludwig RJ, Zollner TM, Santoso S, Hardt K, Gille J, Baatz H, Johann PS, Pfeffer J, Radeke HH, Schon MP, et al. Junctional adhesion molecules (JAM)-B and -C contribute to leukocyte extravasation to the skin and mediate cutaneous inflammation. J. Invest. Dermatol. 2005;125:969–976. doi: 10.1111/j.0022-202X.2005.23912.x. [DOI] [PubMed] [Google Scholar]
- Malergue F, Galland F, Martin F, Mansuelle P, Aurrand-Lions M, Naquet P. A novel immunoglobulin superfamily junctional molecule expressed by antigen presenting cells, endothelial cells and platelets. Mol. Immunol. 1998;35:1111–1119. doi: 10.1016/s0161-5890(98)00102-3. [DOI] [PubMed] [Google Scholar]
- Mandell KJ, Parkos CA. The JAM family of proteins. Adv. Drug Deliv. Rev. 2005;57:857–867. doi: 10.1016/j.addr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Mandell KJ, Babbin BA, Nusrat A, Parkos CA. Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on β1 integrins and Rap1 activity. J. Biol. Chem. 2005;280:11665–11674. doi: 10.1074/jbc.M412650200. [DOI] [PubMed] [Google Scholar]
- Mandell KJ, Berglin L, Severson EA, Edelhauser HF, Parkos CA. Expression of JAM-A in the human corneal endothelium and retinal pigment epithelium: localization and evidence for role in barrier function. Invest. Ophthalmol. Vis. Sci. 2007;48:3928–3936. doi: 10.1167/iovs.06-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandicourt G, Iden S, Ebnet K, Aurrand-Lions M, Imhof BA. JAM-C regulates tight junctions and integrin-mediated cell adhesion and migration. J. Biol. Chem. 2007;282:1830–1837. doi: 10.1074/jbc.M605666200. [DOI] [PubMed] [Google Scholar]
- Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A, Ruco L, Villa A, et al. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M, Hreinsson J, Strand ML, Hovatta O, Soder O, Philipson L, Pettersson RF, Sollerbrant K. Coxsackievirus and adenovirus receptor (CAR) is expressed in male germ cells and forms a complex with the differentiation factor JAM-C in mouse testis. Exp. Cell Res. 2006;312:817–830. doi: 10.1016/j.yexcr.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Moog-Lutz C, Cave-Riant F, Guibal FC, Breau MA, Di Gioia Y, Couraud PO, Cayre YE, Bourdoulous S, Lutz PG. JAML, a novel protein with characteristics of a junctional adhesion molecule, is induced during differentiation of myeloid leukemia cells. Blood. 2003;102:3371–3378. doi: 10.1182/blood-2002-11-3462. [DOI] [PubMed] [Google Scholar]
- Naik UP, Ehrlich YH, Kornecki E. Mechanisms of platelet activation by a stimulatory antibody: cross-linking of a novel platelet receptor for monoclonal antibody F11 with the FcγRII receptor. Biochem. J. 1995;310:155–162. doi: 10.1042/bj3100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava P, Laukoetter MG, Hopkins AM, Laur O, Gerner-Smidt K, Green KJ, Parkos CA, Nusrat A. Desmoglein-2: a novel regulator of apoptosis in the intestinal epithelium. Mol. Biol. Cell. 2007;18:4565–4578. doi: 10.1091/mbc.E07-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Liao J, Price D. Keratin modifications and solubility properties in epithelial cells and invitro. Subcell. Biochem. 1998;31:105–140. [PubMed] [Google Scholar]
- Oshima RG, Baribault H, Caulin C. Oncogenic regulation and function of keratins 8 and 18. Cancer Metastasis Rev. 1996;15:445–471. doi: 10.1007/BF00054012. [DOI] [PubMed] [Google Scholar]
- Owens DW, Lane EB. The quest for the function of simple epithelial keratins. Bioessays. 2003;25:748–758. doi: 10.1002/bies.10316. [DOI] [PubMed] [Google Scholar]
- Palmeri D, van Zante A, Huang CC, Hemmerich S, Rosen SD. Vascular endothelial junction-associated molecule, a novel member of the immunoglobulin superfamily, is localized to intercellular boundaries of endothelial cells. J. Biol. Chem. 2000;275:19139–19145. doi: 10.1074/jbc.M003189200. [DOI] [PubMed] [Google Scholar]
- Raschperger E, Engstrom U, Pettersson RF, Fuxe J. CLMP, a novel member of the CTX family and a new component of epithelial tight junctions. J. Biol. Chem. 2004;279:796–804. doi: 10.1074/jbc.M308249200. [DOI] [PubMed] [Google Scholar]
- Santoso S, Sachs UJ, Kroll H, Linder M, Ruf A, Preissner KT, Chavakis T. The junctional adhesion molecule 3 (JAM-3) on human platelets is a counterreceptor for the leukocyte integrin Mac-1. J. Exp. Med. 2002;196:679–691. doi: 10.1084/jem.20020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso S, Orlova VV, Song K, Sachs UJ, Andrei-Selmer CL, Chavakis T. The homophilic binding of junctional adhesion molecule-C mediates tumor cell-endothelial cell interactions. J. Biol. Chem. 2005;280:36326–36333. doi: 10.1074/jbc.M505059200. [DOI] [PubMed] [Google Scholar]
- Satohisa S, Chiba H, Osanai M, Ohno S, Kojima T, Saito T, Sawada N. Behavior of tight-junction, adherens-junction and cell polarity proteins during HNF-4α-induced epithelial polarization. Exp. Cell Res. 2005;310:66–78. doi: 10.1016/j.yexcr.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Scheiermann C, Meda P, Aurrand-Lions M, Madani R, Yiangou Y, Coffey P, Salt TE, Ducrest-Gay D, Caille D, Howell O, et al. Expression and function of junctional adhesion molecule-C in myelinated peripheral nerves. Science. 2007;318:1472–1475. doi: 10.1126/science.1149276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sircar M, Bradfield PF, Aurrand-Lions M, Fish RJ, Alcaide P, Yang L, Newton G, Lamont D, Sehrawat S, Mayadas T, et al. Neutrophil transmigration under shear flow conditions invitro is junctional adhesion molecule-C independent. J. Immunol. 2007;178:5879–5887. doi: 10.4049/jimmunol.178.9.5879. [DOI] [PubMed] [Google Scholar]
- Tao GZ, Nakamichi I, Ku NO, Wang J, Frolkis M, Gong X, Zhu W, Pytela R, Omary MB. Bispecific and human disease-related anti-keratin rabbit monoclonal antibodies. Exp. Cell Res. 2006;312:411–422. doi: 10.1016/j.yexcr.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Williams LA, Martin-Padura I, Dejana E, Hogg N, Simmons DL. Identification and characterisation of human junctional adhesion molecule (JAM) Mol. Immunol. 1999;36:1175–1188. doi: 10.1016/s0161-5890(99)00122-4. [DOI] [PubMed] [Google Scholar]
- Zen K, Babbin BA, Liu Y, Whelan JB, Nusrat A, Parkos CA. JAM-C is a component of desmosomes and a ligand for CD11b/CD18-mediated neutrophil transepithelial migration. Mol. Biol. Cell. 2004;15:3926–3937. doi: 10.1091/mbc.E04-04-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]