Abstract
Precise manipulation of single molecules has already led to remarkable insights in physics, chemistry, biology, and medicine. However, widespread adoption of single-molecule techniques has been impeded by equipment cost and the laborious nature of making measurements one molecule at a time. We have solved these issues by developing an approach that enables massively parallel single-molecule force measurements using centrifugal force. This approach is realized in an instrument that we call the centrifuge force microscope in which objects in an orbiting sample are subjected to a calibration-free, macroscopically uniform force-field while their micro-to-nanoscopic motions are observed. We demonstrate high-throughput single-molecule force spectroscopy with this technique by performing thousands of rupture experiments in parallel, characterizing force-dependent unbinding kinetics of an antibody-antigen pair in minutes rather than days. Additionally, we verify the force accuracy of the instrument by measuring the well-established DNA overstretching transition at 66 ± 3 pN. With significant benefits in efficiency, cost, simplicity, and versatility, single-molecule centrifugation has the potential to expand single-molecule experimentation to a wider range of researchers and experimental systems.
Main Text
Single-molecule research has advanced greatly in the last decade, fueled in part by the development of technologies such as the atomic force microscope (AFM) and optical and magnetic tweezers, which enable precise physical manipulation of single molecular constructs (1). Remarkable studies with these instruments have already yielded new insight into such diverse areas as protein folding and unfolding dynamics, motor proteins, dynamic strength of receptor ligand interactions, enzymatic activity, and DNA mechanics (1–5). Widespread use of these powerful techniques, however, has been impeded by the laborious nature of making measurements one molecule at a time, the typically costly equipment, and the requisite technical expertise to perform these measurements. Recently these issues have received some attention with innovations such as multiplexed magnetic tweezer systems (6,7) to increase efficiency and more cost-effective designs for optical tweezers systems (8).
We have developed an approach to solve these problems: massively parallel single-molecule force measurements using centrifugal force. The basic concept is that by rapidly rotating a high-resolution detection system, a centrifugal force field can be applied to an ensemble of objects while simultaneously observing their micro-to-nanoscopic motions. This is implemented in a new instrument that we call the centrifuge force microscope (CFM) (Fig. 1), where an entire miniaturized video light microscope is mounted to a rotary stage. High-throughput single-molecule force spectroscopy is achieved by linking beads to a coverslip with single-molecule tethers and orienting the coverslip normally to the applied centrifugal force. By pulling the tethered particles directly away from the substrate, lever arm effects are minimized and control over surface-surface interactions is increased, enabling precise single-molecule force measurements. This differs from previous centrifuge microscope instruments in which the centrifugal force is applied parallel to the coverslip/substrate (9,10).
Figure 1.
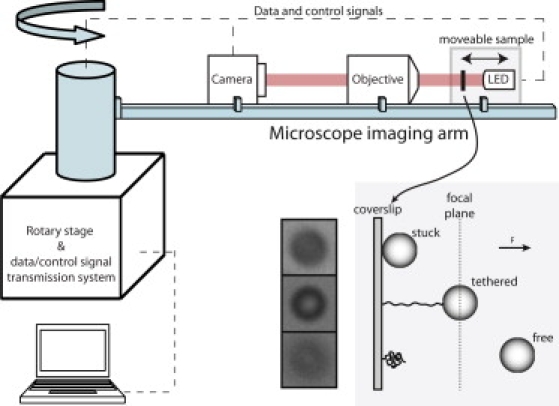
The centrifuge force microscope. A rotary stage spins a miniaturized microscope, imparting a centrifugal force on beads interacting with a coverslip (inset, right). Transmitted light microscope images are sufficient to clearly distinguish between stuck, tethered, and untethered beads (inset, left) using a 20× lens, 24kB DNA, and 2.8 micron beads. Dynamic readout and control of the CCD, LED, and piezo translator during rotation is enabled by an integrated fiber optic rotary joint with electrical slipring (not shown).
The centrifugal force applied to each molecular tether can be easily determined using F = mω2R, where m is the mass of the bead (minus the mass of the medium displaced to account for buoyancy), ω is the magnitude of its angular velocity, and R is its distance from the axis of rotation. Since R is a macroscopic length much larger than the motion of the particles and the region of observation, the force field is conveniently uniform over the sample and as stable as the constancy of ω. For monodisperse beads of known size and density (available commercially or by processing (11)) the centrifugal force on each particle is identical and can be calculated directly without calibration. Detection of molecular transitions, such as bond rupture or tether extension, is also straightforward. Since the force is normal to the coverslip and the whole system rotates, the beads appear relatively stationary in the field of view, but are pulled out of focus as a molecular tether stretches or detaches. Although a variety of bead detection schemes are possible, image focus provides the simplest way to determine if a bead is connected to the surface or not. For example, when measuring bond dissociation kinetics under constant force, one simply needs to measure the times at which singly tethered beads abruptly detach from the coverslip and disappear from view.
We demonstrate this method by performing thousands of single-molecule measurements in parallel to characterize the force-dependent unbinding kinetics of digoxigenin and its antibody (Fig. 2). Antigen molecules were tethered to monodisperse 2.8 μm beads by DNA tethers and brought into contact with the anti-digoxigenin coated coverslip. The sample was then accelerated within a few seconds to a constant velocity to apply a uniform force field to all of the beads. Singly-tethered beads responded by moving away from the glass substrate by a distance consistent with the compliance of double-stranded DNA, whereas beads that were not tethered to the surface extended far out of focus, and nonspecifically bound beads remained at or near the glass surface. Thus, the DNA tether provided a molecular signature for the discrimination of single molecular tethers, which could easily be observed by bead focus. Bond rupture events, indicated by detachment of tethered beads, were visually dramatic, and could be distinguished automatically by standard image processing algorithms. Instrument specifications, including spatial and temporal resolution are discussed in the Supporting Material.
Figure 2.
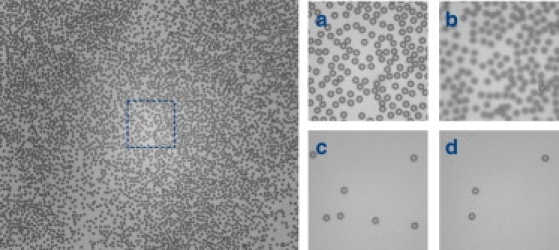
Time lapse images show the progression of a bond rupture experiment. Thousands of receptor-functionalized beads (diameter 2.8 μm) are against the ligand-functionalized coverslip (left). Zoom-in of a smaller region at four different times (right). (a) Beginning of the experiment with beads resting against the coverslip (b) The objective is focused one-tether-length away from the coverslip into the sample, so all beads in the wrong focal plane are blurry. (c) A centrifugal force field is generated that pulls the beads away from the coverslip. Beads tethered by a single DNA molecule move into focus, whereas unattached beads leave the field of view. (d) Beads detach from the coverslip as single receptor-ligand bonds rupture, resulting in fewer visible beads over time.
By applying various force clamps, we determined the force-dependent off-rate koff(f) = k0exp(f/fβ) (12,13) for the interaction of digoxigenin and its antibody. We found a stress-free off-rate of k0 = 0.015 ± 0.002 s−1 and a force scale of fβ = 4.6 ± 1.3 pN (Fig. 3). Using the same construct stretched between two beads, we applied force clamps using our micropipette-based optical trap force probe (instrument and methodology described previously in Zhang et al. (14)) and recorded rupture times, finding near perfect agreement with CFM measurements. Additionally, these results agree within error with previous AFM experiments (15). As an additional verification of the instrument, we used 25 micron beads to overstretch DNA, and found that overstretching occurred at 66 ± 3 pN, in agreement with previous measurements (5).
Figure 3.
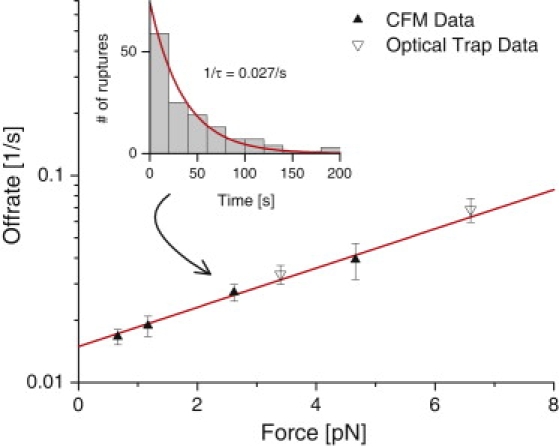
Force-dependent unbinding of digoxigenin and its antibody. Force clamps ranging from hundreds of femtoNewtons to several picoNewtons were applied using the CFM (solid triangles), as well as with the optical trap (open triangles). Each CFM data point was obtained from a single experiment lasting a few minutes, whereas optical trap data was collected serially over a period of many hours. Histograms of the rupture times with a 20 s bin width (10 s for the highest optical trap force) were fit with a decaying exponential to obtain the off-rate at each force (Inset). Plotted error bars in the off-rate result from the uncertainty in the least squares fit.
As demonstrated by these examples, the CFM offers a unique set of advantages, largely derived from the properties of the centrifugal force field, namely that it is macroscopically uniform, highly stable, calibration-free, and dynamically controllable in an essentially deterministic way. Thus, a desired force history can be applied to an ensemble of single molecules without the need for active feedback (passive force clamps have been similarly used in optical traps as described in Greenleaf et al. (16)). The force field conveniently couples to mass density, complimenting optical and magnetic tweezers that couple to polarizability and magnetic moment. Not only does this eliminate the possibility of radiative damage, but it also expands the range of systems that can be studied with force (e.g., beads or objects made of any material can be used, as long as they have a different mass density than their surroundings). Furthermore, the CFM can achieve an enormous force range by varying bead size, bead material, and rotation speed. For example, even our current prototype is capable of sub-femtoNewton (e.g., 1 micron polystyrene particles in water, at 60 revolutions per minute) to microNewton (e.g., 100 micron borosilicate glass particles in water, at 600 revolutions per minute) forces, covering a range that would typically require more than one instrument (1).
The most obvious benefit of this method is the ability to perform massively parallel measurements, enabling dramatic improvements in efficiency capable of reducing experimental time from days to minutes. More than simply speeding up progress, this efficiency also enables new experiments that would be all but impossible with other methods (e.g., near equilibrium measurements that observe interactions with hour-long lifetimes would by unfeasible with sequentially collected statistics). With larger statistical sets now easily attainable, more detailed characterizations and model testing is possible, as well as the observation of population heterogeneity. Additionally, parallel measurements can be used to test entire families of interactions simultaneously (e.g., multiple drugs candidates could be tested simultaneously against a target receptor).
The CFM is also versatile, able to incorporate many existing microscopy techniques to suit individual needs. For instance, reflection interference contrast microscopy imaging (17) could be incorporated to achieve sub-nm resolution bead tracking, and fluorescence imaging could be added to allow visualization of subtle molecular transitions during the experiment (2,3).
Importantly, this method is cost effective and simple to use. The single-molecule centrifugation experiments are straightforward, with a preprogrammed force protocol, minimal setup, and no need for user intervention. The material cost of our prototype was ∼$15,000, and could easily be reduced to ∼$5,000 by using a more cost effective rotary drive and camera. By contrast, it is not uncommon for research grade single-molecule instruments such as the optical trap and AFM to cost over $100,000 (including the optical trap system used in this study).
Centrifugal force has been overlooked in the single-molecule community for too long. We have shown that single-molecule centrifugation can be simple, inexpensive, and significantly more efficient than many methods, enabling new avenues of research and opening single-molecule experimentation to a wider scientific community.
Supporting Material
Supporting methods and materials are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00342-5.
Supporting Material
Acknowledgments
We thank Diane Schaak for help with sample preparation, discussions, and comments on the manuscript, C. Stokes for help integrating electronics, and D. Rogers for help with the fabrication of the instrument.
This work was supported by the Rowland Junior Fellows program.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References and Footnotes
- 1.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deniz A.A., Mukhopadhyay S., Lemke E.A. Single-molecule biophysics: at the interface of biology, physics and chemistry. J. R. Soc. Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritort F. Single-molecule experiments in biological physics: methods and applications. J. Phys. Condens. Matter. 2006;18:R531–R583. doi: 10.1088/0953-8984/18/32/R01. [DOI] [PubMed] [Google Scholar]
- 4.Lang M.J., Block S.M. Resource Letter: LBOT-1: Laser-based optical tweezers. Am. J. Phys. 2003;71:201–215. doi: 10.1119/1.1532323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustamante C., Bryant Z., Smith S.B. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 6.Danilowicz C., Greenfield D., Prentiss M. Dissociation of ligand-receptor complexes using magnetic tweezers. Anal. Chem. 2005;77:3023–3028. doi: 10.1021/ac050057+. [DOI] [PubMed] [Google Scholar]
- 7.Ribeck N., Saleh O.A. Multiplexed single-molecule measurements with magnetic tweezers. Rev. Sci. Instrum. 2008;79:094301. doi: 10.1063/1.2981687. [DOI] [PubMed] [Google Scholar]
- 8.Smith S.P., Bhalotra S.R., Prentiss M. Inexpensive optical tweezers for undergraduate laboratories. Am. J. Phys. 1999;67:26–35. [Google Scholar]
- 9.Harvey E.N., Loomis A.L. A microscope centrifuge. Science. 1930;72:42–44. doi: 10.1126/science.72.1854.42. [DOI] [PubMed] [Google Scholar]
- 10.Hiramoto Y., Kamitsubo E. Centrifuge microscope as a tool in the study of cell motility. Int. Rev. Cytol. 1995;157:99–128. doi: 10.1016/s0074-7696(08)62157-9. [DOI] [PubMed] [Google Scholar]
- 11.Cheng D., Halvorsen K., Wong W.P. Note: High-precision microsphere sorting using velocity sedimentation. Rev. Sci. Instrum. 2010;81:026106. doi: 10.1063/1.3302828. [DOI] [PubMed] [Google Scholar]
- 12.Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 13.Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Halvorsen K., Springer T.A. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuert G., Albrecht C., Gaub H.E. Dynamic force spectroscopy of the digoxigenin-antibody complex. FEBS Lett. 2006;580:505–509. doi: 10.1016/j.febslet.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 16.Greenleaf W.J., Woodside M.T., Block S.M. Passive all-optical force clamp for high-resolution laser trapping. Phys. Rev. Lett. 2005;95:208102. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinrich V., Wong W.P., Evans E. Imaging biomolecular interactions by fast three-dimensional tracking of laser-confined carrier particles. Langmuir. 2008;24:1194–1203. doi: 10.1021/la7027059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


