Abstract
TMPyP4 (Mesotetra(N-methyl-4-pyridyl)porphine) is known to have a high affinity for G-quadruplex DNA. However, there is still some controversy over the exact site(s) and mode(s) of TMPyP4 binding to G-quadruplex DNA. We examined TMPyP4 interactions with seven G-quadruplex forming oligonucleotides. The parent oligonucleotide is a 27-mer with a wild-type (WT) G-rich sequence of the Bcl-2 P1 promoter mid-region (5′-d(CGG GCG CGG GAG GAA GGG GGC GGG AGC-3′)). This sequence folds into at least three unique loop isomer quadruplexes. The two mutant oligonucleotides used in this study are shorter (23-mer) sequences in which nonquadruplex core bases were eliminated and two different (-G-G-) → (-T-T-) substitutions were made to restrict the folding complexity. The four additional mutant oligonucleotides were labeled by substituting a 2-aminopurine (2-AP) base for an A or G in either the first three-base lateral loop or the second five- or seven-base lateral loop (depending on the G→T mutation positions). Spectroscopic and microcalorimetric studies indicate that four molecules of TMPyP4 can be bound to a single G-quadruplex. Binding of the first two moles of TMPyP4 appears to occur by an end or exterior mode (K ≈ 1 × 107 M−1), whereas binding of the third and fourth moles of TMPyP4 appears to occur by a weaker, intercalative binding mode (K ≈ 1 × 105 M−1). As the mid-loop size decreases from seven to five bases, end binding occurs with significantly increased affinity. 2-AP-labeled Bcl-2 promoter region quadruplexes show increased fluorescence of the 2-AP base on addition of TMPyP4. The change in fluorescence for 2-AP bases in the second half of the TMPyP4 titration lends support to our previous speculation regarding the intercalative nature of the weaker binding mode.
Introduction
Short, single-stranded oligonucleotides with sequences taken from genomic regions such as telomeres, the regulatory region of insulin, and the promoter regions of the human c-MYC and Bcl-2 genes have also been reported to form G-quadruplex structures (1–4). Bcl-2 is an oncoprotein that is found in abundance in a wide range of human tumors, and has been reported to be high in breast, prostate, cervical, colorectal, and lung carcinoma cells (5). The human Bcl-2 gene has a GC-rich P1 promoter region located 1386–1423 basepairs upstream from the transcription start site. The P1 promoter, which is GC-rich and capable of forming G-quadruplex, has been identified as the principal transcriptional promoter for Bcl-2 (6).
G-quadruplexes and their formation, structure, stability, and interactions with small molecules are subjects of interest in a number of fields, including biology, molecular biology, genomics, biochemistry, and pharmaceutical science. Ligands exhibiting structural recognition and high affinity for G-quadruplex DNA have been characterized for their ability to stabilize quadruplex complexes (7,8). A number of porphyrin-based ligands have been the subject of cancer drug research because of their low toxicity and ability to inhibit telomerase (9,10). Even though it is well known that planar porphyrin ligands bind tightly to G-quadruplex structures, there is still some controversy regarding the specific site and/or molecular nature of their binding. Recent literature indicates that the interaction of drugs with 2-aminopurine (2-AP)-labeled DNA can be used to identify drug-binding sites and/or distances from the 2-AP label (11,12). It has been shown that 2-AP, a fluorescent analog of adenine, can be substituted for adenine in a DNA fragment without disrupting the structure of DNA (13). Investigators have followed interactions of TMPyP4 with a 2-AP-labeled human telomeric G-quadruplex by monitoring changes in the 2-AP fluorescence (14).
Haq et al. (15) previously proposed that TMPyP4 binds to G-quadruplex structures by intercalating between stacked G-tetrads. They reported that the binding stoichiometry at saturation was equal to (n − 1), where n is the number of stacked G-tetrads in the G-quadruplex motif. It has also been reported that for TMPyP4 binding to a variety of G-quadruplex structures, the saturation stoichiometry is consistent with both end-stacking and intercalation modes (16,17). Freyer et al. (17) reported that the thermodynamic signatures for the interactions of TMPyP4 with c-MYC P1 promoter sequence quadruplexes are consistent with two binding modes in which two moles of TMPyP4 bind by an end-stacking mechanism, and two moles of TMPyP4 bind by an intercalation mechanism.
In this study, we used circular dichroism (CD), ultraviolet-visible (UV-Vis), fluorescence, differential scanning calorimetry (DSC), and isothermal titration calorimetry (ITC) techniques to probe the interactions between TMPyP4 and Bcl-2 P1 promoter quadruplexes. We employed mutant sequences to model two different loop isomer quadruplexes that may form in the Bcl-2 P1 promoter region. In addition, we modified the Bcl-2 mutant sequences to incorporate a single 2-AP base into the first or second lateral loop. Changes in the fluorescence of the loop 2-APs were used to identify the specific location of TMPyP4 binding.
Materials and Methods
The oligonucleotides used were purchased from Oligos Etc. (Wilsonville, OR). The wild-type (WT) 27-mer Bcl-2 mid-sequence and 23-mer mutant sequences are given in Scheme 1. X represents a 2-AP base substituted for a G or an A in the 23-mer G→T mutant 3:7:1 loop isomer sequence (G16,17-T16,17) or the 23-mer G→T mutant 3:5:3 loop isomer sequence (G19,20-T19,20). Bcl-2 model oligonucleotide stock solutions were prepared by dissolving weighed amounts of oligonucleotide into a BPES buffer solution (30 mM potassium phosphate, pH = 7.0, with 100 mM KCl). Oligonucleotide solutions were exhaustively dialyzed against the buffer using a 1000 molecular weight cutoff membrane, and the final concentration was determined by UV absorbance at 254 nm. Extinction coefficients were calculated by a nearest-neighbor method for single-stranded DNA (18). The extinction coefficients determined for the oligonucleotides in this study are as follows: ɛ254 = 2.08 × 105 M−1Cm−1 (WT 27-mer), ɛ254 = 2.27 × 105 M−1Cm−1 (3:7:1), ɛ254 = 2.19 × 105 M−1Cm−1 (3X:7:1), ɛ254 = 2.16 × 105 M−1Cm−1 (3:7X:1), ɛ254 = 2.29 × 105 M−1Cm−1 (3:5:3), ɛ254 = 2.21 × 105 M−1Cm−1 (3X:5:3), and ɛ254 = 2.18 × 105 M−1Cm−1 (3:5X:3), respectively. Oligonucleotide solutions were annealed by heating to 95°C followed by a slow cooling (≥2 h) back down to 25°C.
Scheme 1.
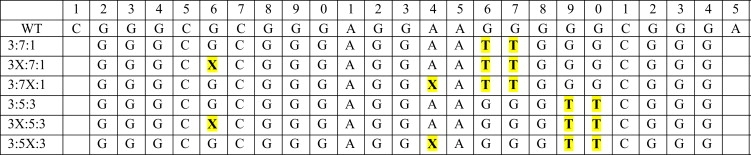
Bcl-2 model promoter oligonucleotide sequences
TMPyP4 (Mesotetra(N-methyl-4-pyridyl)porphine) was purchased from Frontier Scientific (Logan, UT). Stock solutions of TMPyP4 were prepared by dissolving weighed amounts of the drug into the final dialysate from the appropriate oligonucleotide solution, and the final TMPyP4 concentrations were determined by a UV/Vis spectrophotometer using the molar extinction coefficient ɛ424 = 2.26 × 105 M−1Cm−1 (19). All measurements were made at 25°C, and all solutions were made with BPES buffer (30 mM potassium phosphate, pH = 7.0, with 100 mM KCl) added as the supporting electrolyte.
The CD experiments employed a Jasco J-815 spectropolarimeter (Easton, MD) equipped with a temperature-controlled cuvette holder. All measurements were done at 25°C and CD spectra were recorded over a wavelength range of 200–600 nm. The nominal oligonucleotide concentration was 5 × 10−6 M. After annealing was completed, all seven oligonucleotides used in this study exhibited CD signatures characteristic of G-quadruplex formation.
The UV-Vis titration experiments employed an OLIS 8452A diode array spectrophotometer (Bogart, GA) equipped with a Quantum Northwest (Liberty Lake, WA) temperature-controlled cuvette holder. All measurements were made at 25°C. Absorption spectra were recorded over the range of 200 to 500 nm. In these experiments, the mole fraction of TMPyP4 varied from 0 to 1.0, and the total molar concentration, {[TMPyP4] + [oligonucleotide]}, was kept constant at 10 μM. Job plots were used to estimate the binding stoichiometry.
DSC experiments were performed using a VP-DSC calorimeter (Microcal, Northhampton, MA). A typical DSC experiment involved scanning the calorimeter temperature at a rate of 90°C/h over a temperature range of 10–95°C. The resulting thermogram was fit using Origin 7.0 software for the minimum number of two-state transitions needed to fit the data within expected experimental error.
ITC experiments were performed using a VP-ITC calorimeter (Microcal). A typical ITC experiment involved the addition of 25–50 (5.0 μL) injections of a dilute TMPyP4 solution into ∼1.5 mL of a dilute G-quadruplex DNA solution. Three replicate measurements were made. The integrated heat data obtained in an ITC titration were fit to a multiple-independent-site thermodynamic binding model using fitting algorithms from Mathematica 5.0 software.
Fluorescence titrations were performed using a FluoroMax-3 spectrofluorimeter (Jobin/Yvon, Edison, NJ) equipped with a Quantum Northwest temperature-controlled cuvette holder. All measurements were made at 25°C. The fluorescence of the 2-AP base in the 2-AP-labeled (3X:7:1), (3:7X:1), (3X:5:3), and (3:5X:3) oligonucleotides was followed as a function of added TMPyP4. Excitation of the 2-AP oligonucleotides was done at 310 nm and the emission spectra were recorded from 320 to 500 nm. All 2-AP-labeled oligonucleotides exhibited a fluorescence emission maximum at 370 nm.
Results and Discussion
As shown in Fig. 1, the CD spectrum is dependent on the sequence (and possibly the length) of the oligonucleotide. All three unlabeled oligonucleotides exhibit characteristic parallel G-quadruplex signatures with large positive peaks at 265 nm. The two mutant sequences show increased positive ellipticity in the 295 nm region in comparison to the WT sequence. Both the (G16,17 to T16,17) and (G19,20 to T19,20) mutations appear to drive the formation of G-quadruplex structures with mixed parallel/antiparallel structures, in contrast to the almost completely parallel structures that are predominant in the WT mixture of G-quadruplex motifs. The 3:5:3 (G19,20 to T19,20) mutant exhibits the highest fraction of antiparallel character. The binding of TMPyP4 does not significantly change any of the CD spectra (data not shown). In addition, there is no induced CD in the 440 nm region where TMPyP4 exhibits its absorbance maximum. The absence of the induced CD for bound TMPyP4 is consistent with previous observations made for TMPyP4/ c-MYC promoter quadruplex complexes with stoichiometries up to 4:1 (17). Apparently, both binding modes (end-stacking and intercalation) provide a symmetric environment for the bound porphyrin.
Figure 1.
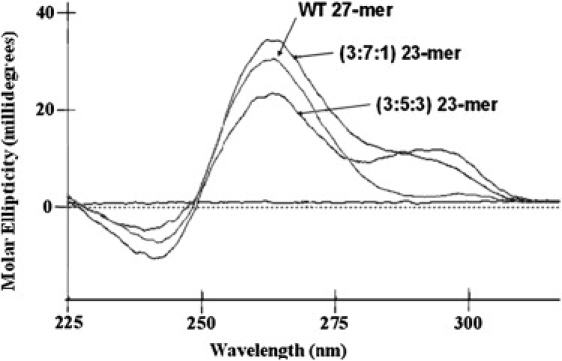
CD spectra for the annealed Bcl-2 WT 27-mer mid promoter sequence, the 3:7:1 23-mer loop isomer mutant sequence, and the 3:5:3 23-mer loop isomer sequence oligonucleotides. Molar ellipticity, θ, is plotted on the Y axis in millidegrees versus wavelength in nanometers.
The results of a typical UV-Vis titration in which a dilute solution of TMPyP4 was added to a dilute solution of the Bcl-2 3:7:1 loop isomer quadruplex are shown in Fig. 2. It is evident from these data that TMPyP4 forms a strong complex with this model Bcl-2 quadruplex construct. Similar data (not shown) were obtained for titration of the other oligonucleotides, i.e., the Bcl-2 WT 27-mer and the 3:5:3 23-mer loop isomer sequence quadruplexes. The data from these experiments are summarized in Table 1. The TMPyP4 Soret band exhibits a red shift of ∼18 nm (423–441 nm) upon TMPyP4 binding to the model quadruplexes. The isobestic point is located between 434 and 435.5 nm for the three different titrations. In addition, only the titration of the 3:7:1 quadruplex sequence exhibits a sharp isobestic point. The titrations for the WT and 3:5:3 quadruplexes exhibit isobestic points that are somewhat blurred. This is because the TMPyP4/oligonucleotide interaction occurs with different affinity for the different quadruplex isomers formed by these sequences, and only the 3:7:1 sequence appears to form a single quadruplex conformer (DSC data not shown).
Figure 2.
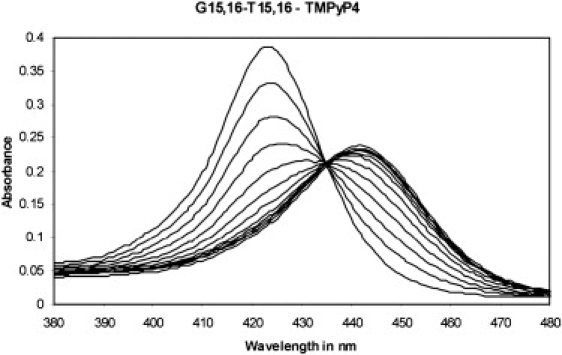
UV/Vis data for the TMPyP4 titration of the annealed Bcl-2 3:7:1 loop isomer mutant sequence. The data are indicative of a strong interaction between TMPyP4 and the Bcl-2 model quadruplex. The titration of the 3:7:1 loop isomer exhibits a sharp isobestic point at 434 nm. Titrations for the WT and the 3:5:3 loop isomer sequence quadruplexes exhibit blurred isobestic points ranging from 433 to 436 nm (see Table 1).
Table 1.
UV-Vis titration results for binding TMPyP4 to Bcl-2 promoter sequence G-quadruplexes
| Sequence | Soret band shift | Isobestic point | Hypochromicity |
|---|---|---|---|
| WT 27-mer | 423–441 (18 nm) | 433–436 nm | 59.7% |
| 3:5:3 mutant 23-mer | 422–439 (17 nm) | 433–436 nm | 43.3% |
| 3:7:1 mutant 23-mer | 422–441 (19 nm) | 434.0 nm | 40.2% |
The WT/TMPyP4 complex shows the largest decrease in absorbance (hypochromicity), with the 3:5:3 and 3:7:1 TMPyP4 complexes showing significantly less hypochromicity in the absorbance of the bound TMPyP4. This is an indication that the TMPyP4 is more effectively stacked on the G-tetrads in the WT quadruplex than in either of the mutant quadruplexes. It is interesting to note that the decrease in hypochromicity is correlated with the decreased average affinity as determined in the ITC experiments (see Table 1). Both the 3:5:3 and 3:7:1 mutant loop isomers exhibit similar hypochromicity, which must indicate that the TMPyP4 G-tetrad stacking interactions in these complexes are similar. Apparently, the antiparallel (or mixed parallel/antiparallel) conformations available to the loop isomer sequences result in both lower affinity for the TMPyP4 ligand and a less effective stacking of the TMPyP4 on (or between) the G- tetrads in the antiparallel quadruplex. Similar observations were reported by Arora and Maiti (20).
The composite Job plot (Fig. 3) shows data for TMPyP4 titrations of the parent WT Bcl-2 oligonucleotide and the two mutant 3:7:1 and 3:5:3 loop isomer sequences. All three quadruplexes exhibit the same binding stoichiometry, showing breaks in the Job plot at ∼2:1 and at ∼4:1. The uncertainty in the saturation stoichiometry, anywhere from 3.5:1 to 4.5:1 as determined in the Job plots, results from small errors in the long extrapolation of the absorbance change data in the Job plot. Larger stoichiometries are necessarily more uncertain: the difference in mole fraction at the break point in the Job plot is only 0.05 in comparing stoichiometries of 3:1 and 4:1, whereas the mole fraction difference in the Job plot is 0.166 in comparing stoichiometries of 2:1 and 3:1, or 0.25 in comparing stoichiometries of 1:1 and 2:1. Nevertheless, the UV-Vis titration endpoints are in reasonable agreement with fluorescence and ITC data, which obviously exhibit two different binding modes (with each mode comprised of two binding sites).
Figure 3.
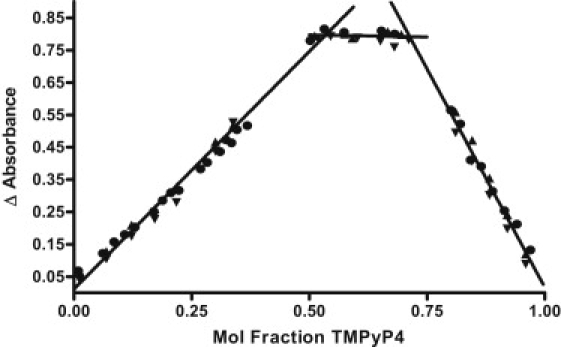
Composite Job plot generated from the UV/Vis titration data obtained for all three Bcl-2 model sequences. The solid circles (●) correspond to the data for the TMPyP4 titration of the Bcl-2 WT 27-mer sequence, the solid triangles (▴) are the data for the titration of the 3:7:1 23-mer loop isomer, and the inverted solid triangles (▾) represent the data for the 3:5:3 23-mer loop isomer.
The DSC thermogram for the unfolding of the annealed Bcl-2 WT 27-mer G-quadruplex (or mixture of quadruplex conformers) is shown in Fig. 4. The WT thermogram indicates the presence of at least three folded forms (e.g., loop or chain isomers). The Tm values observed for three independent WT melting transitions are ∼68°C, 78°C, and 89°C. The melting curves for both the 3:5:3 and 3:7:1 mutant sequences are simpler. The melting curve for the 3:7:1 mutant exhibits a single two-state transition with Tm = 68°C. The melting curve for the 3:5:3 mutant exhibits an asymmetric or broadened transition that cannot be fit for a single two-state transition, with the predominant (>> 95%) species having Tm = 78°C. The 3:7:1 mutant quadruplex is similar in stability to the lowest melting conformer in the WT mixture, with both having Tm values of ∼68°C. The Tm for the predominant species in the 3:5:3 mutant quadruplex mixture, Tm = 78°C, is similar to the Tm for the predominant species in the WT melting curve.
Figure 4.
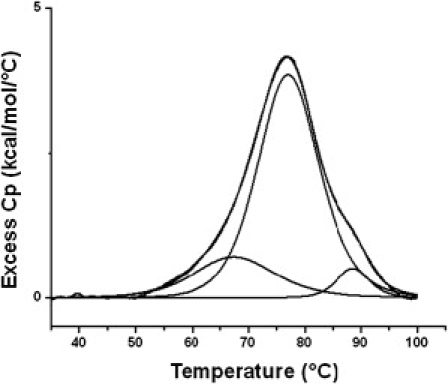
Excess heat capacity data are shown for the thermal denaturation of the WT 27-mer Bcl-2 mid promoter sequence. The DSC thermogram has been fit by deconvolution of the raw data into three independent overlapping two-state transitions, with approximate Tm values of 68°C, 78°C, and 89°C.
Using the similarities in the Tm values, we might speculate that the mixture of G-quadruplex conformers formed by the Bcl-2 WT sequence includes 3:5:3 and 3:7:1 loop conformers. The caveat is that neither mutant quadruplex is a completely parallel structure, whereas the WT sequence forms predominantly parallel motifs. Our best guess is that the WT sequence forms a mixture of three different G-quadruplex conformers, with a 3:5:3 mid-loop parallel conformer being predominant (comprising at least 70% of the mixture) and a 3:7:1 parallel conformer representing <20% of the mixture. At this point, we are not prepared to speculate on the structure of the highest melting conformer in the WT quadruplex mixture.
Typical ITC enthalpograms for the titration of the Bcl-2 WT and the two loop isomer quadruplex constructs with TMPyP4 are shown in Fig. 5. The ITC data and best-fit thermodynamic parameters (see Fig. 5 and Table 2) illustrate four significant points:
-
1)
Changes to the oligonucleotide sequence have a profound effect on TMPyP4 affinity, at least for the high-affinity site. The WT sequence exhibits the highest affinity followed by the 3:5:3 loop isomer, with the 3:7:1 loop isomer showing a 40-fold reduction in TMPyP4 affinity when compared to the WT sequence. The reduced affinity exhibited by the two loop isomers must reflect differences between the energetics for TMPyP4 stacking on G-tetrads in the antiparallel conformation versus the parallel conformation.
-
2)
The influence of end bases that have been removed from the mutant sequences is largely insignificant (see for comparison the differences in the K1 values reported for the two mutants versus the differences in the K1 values for the WT and the 3:5:3 mutant sequences).
-
3)
All three sequences show similar energy change profiles for mode 1 versus mode 2 binding. In all cases, mode 1 binding has the more favorable entropy change and mode 2 binding has the more favorable enthalpy change. It should be noted, however, that mode 1 binding for both mutant loop isomers is accompanied by a more favorable enthalpy change than is seen for the WT sequence, which again is probably indicative of the differences in stacking TMPyP4 on antiparallel versus parallel G-quadruplex structures.
-
4)
All three quadruplex-forming sequences bind four moles of TMPyP4/moleof oligonucleotide at saturation.
Figure 5.
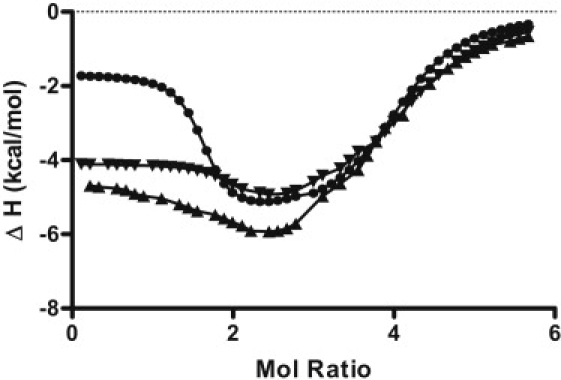
Typical ITC titration data (points) and nonlinear regression fits (solid lines) are shown for three titration experiments in which a dilute solution of TMPyP4 was added to a dilute solution of the annealed oligonucleotide in the calorimeter cell. The solid circles (●) correspond to the data for the TMPyP4 titration of the Bcl-2 WT 27-mer sequence, the solid triangles (▴) are the data for the titration of the 3:7:1 23-mer loop isomer, and the inverted solid triangles (▾) represent the data for the 3:5:3 23-mer loop isomer. The best-fit parameters for the nonlinear regression analysis of these ITC data are given in Table 2.
Table 2.
ITC-derived thermodynamic parameters for binding TMPyP4 to Bcl-2 promoter sequence G-quadruplexes
| Thermodynamic parameter | WT | 3:7:1 | 3:5:3 |
|---|---|---|---|
| K1 × 10−7 | 4.2 (± 0.4) | 0.10 (± 0.01) | 1.1 (± 0.1) |
| ΔG1 (kcal/mol) | −10.4 | −8.2 | −9.6 |
| ΔH1 (kcal/mol) | −1.8 (± 0.1) | −4.6 (± 0.1) | −4.5 (± 0.1) |
| −TΔS1 (kcal/mol) | −8.6 | −3.6 | −5.2 |
| K2 × 10−5 | 3.6 (± 0.5) | 1.3 (± 0.2) | 1.7 (± 0.2) |
| ΔG2 (kcal/mol) | −7.6 | −7.0 | −7.1 |
| ΔH2 (kcal/mol) | −6.1 (± 0.1) | −12.3 (± 0.1) | −6.7 (± 0.1) |
| −TΔS2 (kcal/mol) | −1.5 | 5.3 | −0.4 |
Parameters are for a two-independent-sites binding model. The uncertainties listed for the two fitting parameters, Ki and ΔHi, were determined from a Monte Carlo analysis.
The 2-AP fluorescence data are summarized in Fig. 6. The 2-AP fluorescence is shown to increase as TMPyP4 is added to all four of the loop-labeled quadruplexes. Quenching of the 2-AP fluorescence by neighbor quadruplex bases is reduced in all four titrations. The fluorescence titration curve is comprised of three linear regions with different slopes: the first (steepest) for end binding, the second (slightly less steep) for intercalation, and the third (essentially flat) after binding-site saturation. The fluorescence intensity of the 2-AP incorporated into the first loop is increased by approximately the same amount on TMPyP4 binding for both the 3X:5:3 or the 3X:7:1 loop isomers. The fluorescence intensity of the 2-AP incorporated into the second loop is increased by TMPyP4 binding to a greater degree in the 3:7X:1 versus the 3:5X:3 loop isomer.
Figure 6.
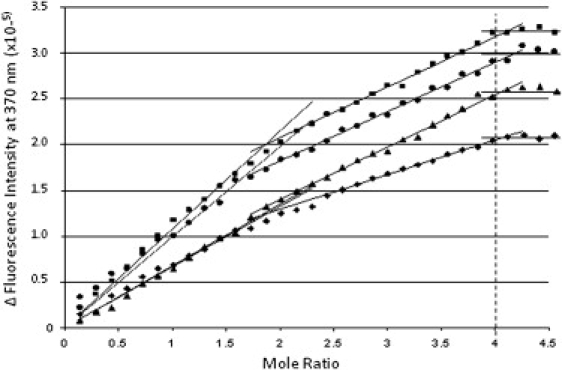
2-AP fluorescence emission intensity plotted as a function of the mol ratio of added TMPyP4 (TMPyP4/DNA). Data shown as solid squares (■) are for the TMPyP4 titration of the 3X:7:1 loop isomer with a 2-AP base substitution at position 6 in the first three-base loop, and those indicated by solid triangles (▴) are for the titration of the 3X:5:3 loop isomer also with a 2-AP base substitution at position 6. Data shown as (●) are for the TMPyP4 titration of the 3:7X:1 loop isomer with a 2-AP base substitution at position 14, and data shown as solid diamonds (♦) are for the titration of the 3:5X:3 loop isomer, again with a 2-AP base substitution at position 14.
Although they are not conclusive, the fluorescence changes for the 2-AP bases incorporated into the first and second lateral loops seem to support intercalation as the mechanism for mode 2 binding. This interpretation is based on the similarities in the two binding modes insofar as both show dramatic increases in the fluorescence of the 2-AP loop bases on TMPyP4 binding up to the point where both of the specific binding sites are saturated. The addition of excess TMPyP4 has little or no effect on the fluorescence of the labeled loop bases. Our interpretation of this result is that nonspecific exterior interactions, e.g., aggregation of the ligand on the quadruplex surface or in quadruplex grooves, do not change the environment of the labeled loop bases. In effect, end binding and intercalation are expected to be similar from the standpoint that both result in part from the stacking of TMPyP4 on (or between) a G-tetrad(s).
Conclusions
We have shown that Bcl-2 promoter region mid-G-rich sequences fold into at least three unique G-quadruplexes. This is consistent at least in part with previously reported structures for Bcl-2 promoter region sequences in solution (21,22). We propose that the 3:7:1 and 3:5:3 loop isomer quadruplexes interact with TMPyP4 in a similar manner, at least as far as binding site(s) and binding mode(s) are concerned. Both mutant quadruplexes appear to bind a total of four moles of TMPyP4 per mole of quadruplex.
Two moles of TMPyP4 bind to the higher-affinity end sites, and two moles of TMPyP4 bind to the lower-affinity intercalation sites. CD signatures and DSC data are consistent with the Bcl-2 WT-27-mer forming a mixture of three (or more) parallel quadruplex conformers, whereas both the (3:7:1) and the (3:5:3) mutants fold into single folded conformers with mixed parallel/antiparallel structure. The mutation of a G to a T at positions 16 and 17, or positions 19 and 20, appears to have a significant effect on backbone orientation. Spectroscopic and microcalorimetric data indicate that the 3:5:3 loop isomer is most similar to the WT quadruplex, at least in terms of the thermal stability and the thermodynamics for binding TMPyP4. Changes in the fluorescence of the 2-AP bases inserted into the first and second lateral loops are again consistent with two binding modes: end binding and a second mode that may be intercalation. ITC results indicate that TMPyP4 affinity is strongly dependent on the size of the second lateral loop in the Bcl-2 promoter sequence quadruplex. As the second lateral loop size is increased from five to seven bases, TMPyP4 binding decreases ∼10-fold (or 40-fold in comparison to the WT sequence). There also appears to be a correlation between the thermal stability of a quadruplex and the number of unstructured bases in the quadruplex loop regions. As the second lateral loop size is increased from five to seven bases, the melting temperature is decreased by 10°C.
In summary, TMPyP4 binding to Bcl-2 promoter sequence quadruplexes is very similar to TMPyP4 interactions with c-MYC promoter sequence quadruplexes (17), at least in terms of affinity and mechanism. The Bcl-2 WT 27mer sequence forms a mixture of at least three predominantly parallel quadruplex conformers. Perhaps the most unexpected and maybe even significant result is that selective G→T mutations in the third G run yield sequences that fold into quadruplex structures with a significant antiparallel character.
Stoichiometry at saturation is once again given by (n + 1), where n is the number of stacked G-tetrads in the G-quadruplex structure. New information from this study includes insight into lateral loop involvement in TMPyP4 binding, and the 2-AP fluorescence data lend support to our previous speculation about intercalation (17).
Acknowledgments
N. Nagesh thanks Dr. Lalji Singh, director of the Centre for Cellular and Molecular Biology (Hyderabad, India), for allowing him to pursue this work with Dr. Edwin Lewis at the Northern Arizona University Molecular Biophysics Laboratory (Flagstaff, AZ).
Contributor Information
Narayana Nagesh, Email: nagesh@ccmb.res.in.
Edwin A. Lewis, Email: elewis@chemistry.msstate.edu.
References
- 1.Simonsson T. G-quadruplex DNA structures—variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 2.Hammond-Kosack M.C., Kilpatrick M.W., Docherty K. Analysis of DNA structure in the human insulin gene-linked polymorphic region in vivo. J. Mol. Endocrinol. 1992;9:221–225. doi: 10.1677/jme.0.0090221. [DOI] [PubMed] [Google Scholar]
- 3.Simonsson T., Pecinka P., Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baretton G.B., Diebold J., Löhrs U. Apoptosis and immunohistochemical bcl-2 expression in colorectal adenomas and carcinomas. Aspects of carcinogenesis and prognostic significance. Cancer. 1996;77:255–264. doi: 10.1002/(SICI)1097-0142(19960115)77:2<255::AID-CNCR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.Heckman C., Mochon E., Boxer L.M. The WT1 protein is a negative regulator of the normal bcl-2 allele in t(14;18) lymphomas. J. Biol. Chem. 1997;272:19609–19614. doi: 10.1074/jbc.272.31.19609. [DOI] [PubMed] [Google Scholar]
- 7.Gavathiostis E., Heald R.A., Searle M.S. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d (TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003;334:24–36. doi: 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Haider S.M., Parkinson G.N., Neidle S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 9.Han H., Langley D.R., Hurley L.H. Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 10.Dixon I.M., Lopez F., Meunier B. Porphyrin derivatives for telomere binding and telomerase inhibition. ChemBioChem. 2005;6:123–132. doi: 10.1002/cbic.200400113. [DOI] [PubMed] [Google Scholar]
- 11.Bailly C., Waring M.J. The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA. Nucleic Acids Res. 1998;26:4309–4314. doi: 10.1093/nar/26.19.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbieri C.M., Kaul M., Pilch D.S. Use of 2-aminopurine as a fluorescent tool for characterizing antibiotic recognition of the bacterial rRNA A-site. Tetrahedron. 2007;63:3567–6574. doi: 10.1016/j.tet.2006.08.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen O.F.A., van Stokkum I.H.M., van Amerongen H. Probing the structure and dynamics of a DNA hairpin by ultrafast quenching and fluorescence depolarization. Biophys. J. 2001;81:1115–1126. doi: 10.1016/S0006-3495(01)75768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura T., Kawai K., Fujitsuka M., Majima T. Detection of the G-quadruplex-TMPyP4 complex by 2-aminopurine modified human telomeric DNA. Chem. Commun. (Camb.) 2006;(4):401–402. doi: 10.1039/b514526k. [DOI] [PubMed] [Google Scholar]
- 15.Haq I., Trent J.O., Chowdhry B.Z., Jenkins T.C. Intercalative G-tetraplex stabilization of telomeric DNA by a cationic porphyrin. J. Am. Chem. Soc. 1999;121:1768–1779. [Google Scholar]
- 16.Wei C., Jia G., Li C. A spectroscopic study on the interactions of porphyrin with G-quadruplex DNAs. Biochemistry. 2006;45:6681–6691. doi: 10.1021/bi052356z. [DOI] [PubMed] [Google Scholar]
- 17.Freyer M.W., Buscaglia R., Lewis E.A. Biophysical studies of the c-MYC NHE III1 promoter: model quadruplex interactions with a cationic porphyrin. Biophys. J. 2007;92:2007–2015. doi: 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plum G.E. Determination of oligonucleotide molar extinction coefficients. In: Beaucage S.L., Bergstrom D.E., Herdewijn P., Matsuda A., editors. Current Protocols in Nucleic Acid Chemistry. Vol. 7.3. John Wiley and Sons; New York: 2000. pp. 1–17. [Google Scholar]
- 19.Pasternack R.F., Huber P.R., Hinds L.de C. On the aggregation of meso-substituted water-soluble porphyrins. J. Am. Chem. Soc. 1972;94:4511–4517. doi: 10.1021/ja00768a016. [DOI] [PubMed] [Google Scholar]
- 20.Arora A., Maiti S. Effect of loop orientation on quadruplex-TMPyP4 interaction. J. Phys. Chem. 2008;112:8151–8159. doi: 10.1021/jp711608y. [DOI] [PubMed] [Google Scholar]
- 21.Dai J., Dexheimer T.S., Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dexheimer T.S., Sun D., Hurley L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]


