Abstract
Antimicrobial peptides (AMPs) are an emerging class of antibiotics for controlling health effects of antibiotic-resistant microbial strains. Protegrin-1 (PG-1) is a model antibiotic among β-sheet AMPs. Antibiotic activity of AMPs involves cell membrane damage, yet their membrane interactions, their 3D membrane-associated structures and the mechanism underlying their ability to disrupt cell membrane are poorly understood. Using complementary approaches, including molecular dynamics simulations, atomic force microscopy (AFM) imaging, and planar lipid bilayer reconstitution, we provide computational and experimental evidence that PG-1, a β-hairpin peptide, forms ion channels. Simulations indicate that PG-1 forms channel-like structures with loosely attached subunits when reconstituted in anionic lipid bilayers. AFM images show the presence of channel-like structures when PG-1 is reconstituted in dioleoylphosphatidylserine/palmitoyloleoyl phosphatidylethanolamine bilayers or added to preformed bilayers. Planar lipid bilayer electrical recordings show multiple single channel conductances that are consistent with the heterogeneous oligomeric channel structures seen in AFM images. PG-1 channel formation seems to be lipid-dependent: PG-1 does not easily show ion channel electrical activity in phosphatidylcholine membranes, but readily shows channel activity in membranes rich in phosphatidylethanolamine or phosphatidylserine. The combined results support a model wherein the β-hairpin PG-1 peptide acts as an antibiotic by altering cell ionic homeostasis through ion channel formation in cell membranes.
Introduction
Antimicrobial peptides (AMPs) show a broad spectrum of cytotoxicity against bacteria, fungi, some enveloped viruses and even cancer cells (1,2). AMPs are often classified on the basis of biochemical feature (net charge) and/or structural characteristics (α-helices, β-sheets, extended linear, or disulfide S-S bonded) (3). Among the various native AMPs and their synthetic variants, cationic AMPs represent a predominant component of the innate immune system (4), suggesting that the mechanism for the cell membrane disruption involves strong electrostatic interactions with the negatively charged microbial membranes. Increasing evidence suggests that most AMPs possess a membrane lytic property (5–8). In this case, the cell membrane rupturing events can involve two steps. In the first step, initial depositions of monomeric or small oligomeric AMPs at the amphipathic interface of the microbial membrane, due to strong electrostatic interactions between peptides and lipids that leads to a significant thinning of the membrane. Most AMPs have a strong tendency to locate at the amphipathic interface of the bilayer (9–12). The second step is their diffusion into the membrane core in regions with low dielectric barrier and their clustering to form an ordered aggregate when the local peptide concentration reaches a critical value. Translocation of the peptides into the core of a target cell membrane can interfere with the cellular metabolism (1). It is generally accepted that most cationic AMPs mediate unregulated membrane permeation, leading to deregulation of ionic homeostasis and subsequent microbial cell death (13–15). However, the mechanism underlying AMP-induced permeation remains unclear (16).
Protegrin (PG) displays an antimicrobial activity with a great antibiotic potency (17). Native PGs were first isolated from porcine leukocyte cells (18). There are five known PG isomers, PG-1 to PG-5. These PGs share common features and conformations. Protegrin-1 (PG-1) is composed of 18 amino acids with a high content of positively charged arginine (Arg) and cysteine (Cys) residues. The structure of PG-1 in solution as determined by nuclear magnetic resonance (NMR) is a one-turn β-hairpin in which two antiparallel strands linked by a β-turn are stabilized by two disulphide bonds (19,20). The cationic nature of the peptide allows its interaction with the lipid matrix of bacterial membranes containing negatively charged lipids (21–23). Several studies have shown that PG-1 is able to alter the permeability of bilayers (13,24–27), monolayers (28) and modeled bilayers (29,30). In a previous modeling effort (29), the octameric PG-1 channels with different β-sheet arrangements were constructed in various lipid bilayer architectures that involve intrinsic barrel-stave and toroidal membrane pore topologies with both zwitterionic 1-palmitoyl 2-oleoyl phosphatidylcholine (POPC) and anionic POPC/1-palmitoyl 2-oleoyl phosphatidylglycerol (POPG) (mol ratio = 4:1). Remarkably, earlier molecular dynamic (MD) simulations using the U-shaped peptide with the β-strand-turn-β-strand motif (31–33) have predicted that the PG-1 channels exhibit β-sheet subunit morphology similar to the β-amyloid (Aβ) ion channels; the β-amyloid (Aβ) ion channels structure was confirmed by atomic force microscopy (AFM) imaging (34,35). Although computational studies at the atomic level predict a 3D structure of PG-1 channels, experimental verification of the channel conformations remains unavailable. Such information is needed to better understand the mechanism of PG-1 channel activity and the function of these channels in cell (patho-) physiology.
In this study, we present what we believe to be complementary lines of evidence that PG-1 forms ion channels in a lipid bilayer and exhibits channel-like activity. Using MD simulations, we modeled PG-1 channels in the anionic bilayer containing dioleoylphosphatidylserine/palmitoyloleoyl phosphatidylethanolamine (DOPS/POPE) (1:2). The PG-1 channels were composed of 10 β-hairpins. They were assembled initially into a β-sheet ring with either antiparallel (turn-next-to-tail) or parallel (turn-next-to-turn) β-sheet motifs in an NCCN packing mode (36). Consistent with previous observations (25,29,30), PG-1 channels consist of four subunits in the antiparallel arrangement and five subunits in the parallel β-sheet channels. AFM images of PG-1 reconstituted in anionic lipid bilayers show ion channel-like structures with predominantly three to five subunits. In planar lipid bilayers (PLBs), PG-1 shows ion channel activity that is concentration and lipid dependent. The combined results support a model where the β-hairpin antibiotic activity of PG-1 could be mediated by its channel forming properties and subsequent altering of cell ionic homeostasis.
Materials and Methods
MD simulations
The PG-1 channels were made with ten identical PG-1 β-hairpins, initially arranged to form a single layered β-sheet annular shape. The prerelaxed β-hairpin structure was obtained from preliminary simulations of the solution NMR structure of PG-1 monomer (20) in the lipid environment, as described in previous simulations (29). The minimized channel is embedded in the DOPS/POPE bilayer (mol ratio = 1:2) with the same packing described in Jang et al. (29) and Tang et al. (36). For the bilayer topology, the intrinsic barrel-stave membrane pore was initially prepared for the β-sheet channels (29). Because the simulation method follows closely the protocol described previously for PG-1 monomer (37), dimer (38), octamer (29), Aβ channels (31,32), and K3 channels (33), in this study we only describe briefly key parameters used for the decameric PG-1 channel simulations. The CHARMM program (39) was used to construct the set of starting points and to relax the systems to a production-ready stage. For production runs to 30 ns, the NAMD code (40) on a Biowulf cluster at the National Institutes of Health was used for the starting point. Averages were taken after 10 ns discarding the initial transient part of the trajectory. For details see the Materials and Methods section in the Supporting Material.
AFM sample preparation, imaging, and analysis
For AFM imaging, a 50-μL droplet of liposomes with and without reconstituted with the PG-1 peptide was allowed to adsorb for 5–15 min on freshly cleaved mica to create a supported lipid bilayer with preincorporated peptides. The sample was rinsed three times with phosphate buffer solution to eliminate all unincorporated peptides as well as liposomes that were not ruptured (34,35). Image analysis was carried out using protocols shown to be successful in our previous amyloid ion channels studies (34). Briefly, the sizes of the reconstituted channels in the membrane were obtained from the height images using cross-sectional analysis. Channel diameters were measured at two-thirds of the full height with respect to the lipid bilayer surface. Individual channel subunits were considered only if the neighboring features were separated from each other such that they were visible at the height 10% below their maximum height in the AFM image cross-section. Consistent with our previous AFM studies of other amyloid ion channels, as the PG-1 channels are present in noncrystalline and random clusters, no single particle averaging commonly used in electron microscopy studies were used. For details, see Materials and Methods in Supporting Material.
Formation of PLBs and data analysis
Vertical PLBs were generated by either Mueller-Rudin (41) or Montal-Mueller (42) methods, using concentrations (33,43) and lipid mixtures (44), as reported previously. Current versus time data acquisition and analysis were carried out as described in (45). For a complete description see Materials and Methods in the Supporting Material.
Results
MD Simulations
In previous MD simulations (29), we showed that PG-1 channels contained eight PG-1 β-hairpins that were initially assembled in an annular shape. During the relaxation process in the explicit lipid environment, the PG-1 channels divided into four or five subunits. We obtained three dimensional structures of PG-1 channels in the intrinsic barrel-stave and toroidal membrane pore settings of the lipid bilayers that consisted of both zwitterionic POPC and anionic POPC/1-palmitoyl 2-oleoyl phosphatidylglycerol (mol ratio = 4:1) lipids.
In this study, we further explored the PG-1 channel formation in the lipid bilayers. Here, PG-1 channels are prepared with 10 PG-1 β-hairpins that are assembled initially as an annular shape in the anionic bilayer DOPS/POPE (mol ratio = 1:2) (Fig. S1). The initial frustration in the annular conformation is gradually removed via relaxations of the lipid bilayer (Fig. S2). After the initial 5 ns of the simulation, the β-sheet channels of PG-1 retain the intermolecular H-bonds at localized β-sheets, with the discontinuities in the β-sheet network segregating the channel into subunits. The subunit appearance in the channel structure is caused by the β-strands optimization, which depends on the lipid dynamics during the simulation. The environmentally relaxed PG-1 channels in a fully hydrated lipid bilayer can be obtained after 30 ns of simulations with an all-atom representation. The PG-1 channels slightly increase both the outer diameter and inner pore size due to inhomogeneous channel shapes after relaxation (Fig. 1). We obtained the outer diameters, ∼5.5 and ∼5.3 nm for the antiparallel and parallel β-sheet channels of PG-1, respectively. The pore diameters are ∼1.4 and ∼1.3 nm for the antiparallel and parallel β-sheet channels, respectively. The PG-1 channels break into several subunits. The antiparallel β-sheet channel shows four subunits and the parallel β-sheet channel five subunits, consistent with our previous observation (29) and other β-sheet channels (31–33).
Figure 1.
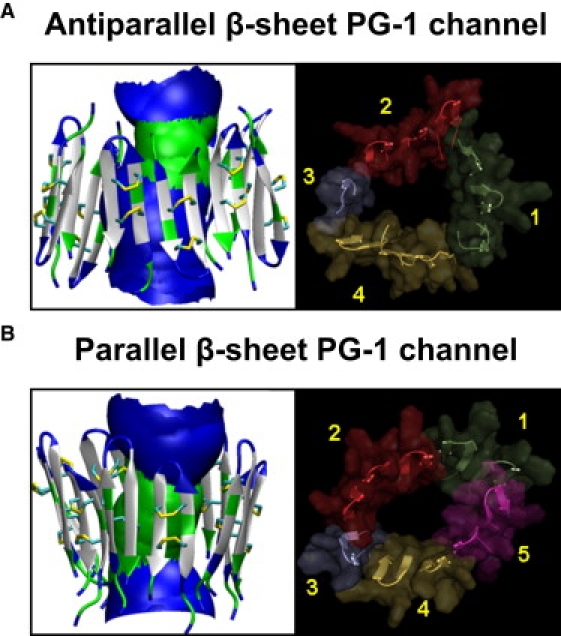
MD simulation of channel structure. Channel structures averaged over the simulation in a ribbon representation (left column) for the (A) antiparallel and (B) parallel β-sheet channels of PG-1 are shown in the lateral view from the lipid bilayer. In the peptides, hydrophobic residues are shown in white, polar and Gly residues are shown in green, and positively charged residues are shown in blue. In each channel, the averaged water pore structure calculated by the HOLE program (77) is embedded. For the pore structures in the surface representation, green denotes pore diameter in the range, 0.8 nm < d < 1.4 nm and blue denotes pore diameter of d > 1.4 nm. The simulated PG-1 channel structures with highlighted subunits (right column) are viewed from the top leaflet of the lipid bilayer for the antiparallel (top right) and parallel (bottom right) β-sheet channels of PG-1. The channels are depicted in a cartoon representation with a transparent surface. Each subunit in the channels is colored in a different color. The discontinuous β-sheet network determines the boundary between the subunits in the channels. Antiparallel β-sheet channels of PG-1 contain four subunits. Parallel β-sheet channels contain five subunits. The shapes of the subunits vary, with β-hairpin monomers, and β-sheet dimer or trimer.
PG-1 and PG-3 are reported to form anion-selective channels in planar phospholipid bilayers (13). In our previous simulations (29), we also observed that the PG-1 channels yield selective anion conductance with low free energy profile for Cl− in the pore, whereas the channels prevent Na+ from crossing the pore by a high free energy barrier. In this study, we analyzed ion activity in the pore cavity. The system contains two cations, Mg2+ and Na+. These cations mostly interact with lipid headgroups, producing high probability distribution at z = ±2.4 nm, the lipid/water interfaces of both bilayer leaflets (Fig. 2). No deep penetration was observed for the cations. The Cl− anion enters through the water pore with relatively high probability distribution along the pore. Two peaks at both channel gates in the Cl− probability curves indicate that the Arg side-chains easily trap Cl− with the strong electrostatic interactions. PG-1 channels show anion permeability and are impermeable to cations. The charge states induced by ions (Mg2+, Na+, and Cl−) show total net charge in the pores of both channels during the simulations (Fig. S3).
Figure 2.
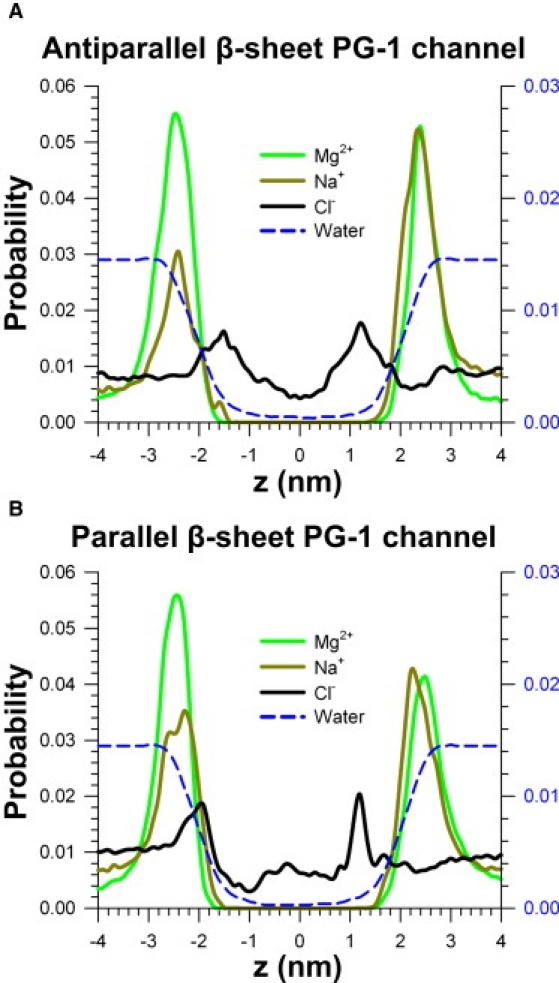
Probability distribution functions representing ionic permeation. Probability distribution functions for Mg2+ (green line), Na+ (orange line), Cl− (black line), and water (blue dashed line) as a function of the distance along the pore center axis for the (A) antiparallel and (B) parallel β-sheet channels of PG-1 are shown. The probability for water uses the scale on the right (blue labels). The high probability profile for Cl− in the pore indicates clearly that PG-1 forms an anionic conducting channel in the lipid bilayer.
AFM Imaging
Although other β–sheet rich peptides have been extensively studied in various physiological and nonphysiological conditions, not much is known about the PG-1 conformations under similar conditions. β-sheet rich peptides are known to present a multitude of architectures, from globular oligomers to fibrils (33,46–48). PG-1, when allowed to oligomerize without lipidic membrane, appear to form fibrillar structures (H. Jang, M. Mustata, R. Nussinov, and R. Lal, unpublished data) similar to other amyloid fibrils (49) and some antimicrobial peptides (50–55). In the presence of a lipid bilayer, PG-1 conformations change. When PG-1 is added to preformed anionic DOPS/POPE lipid bilayer, AFM images show globular PG-1 oligomers on the bilayer surface (Fig. 3 B). Higher resolution AFM shows large globular conglomerates on the bilayer surface (Fig. 3 C). On flattening and low pass filtering of a 100 nm × 100 nm image area, channel-like structures become quite visible (Fig. 3 C, inset).
Figure 3.
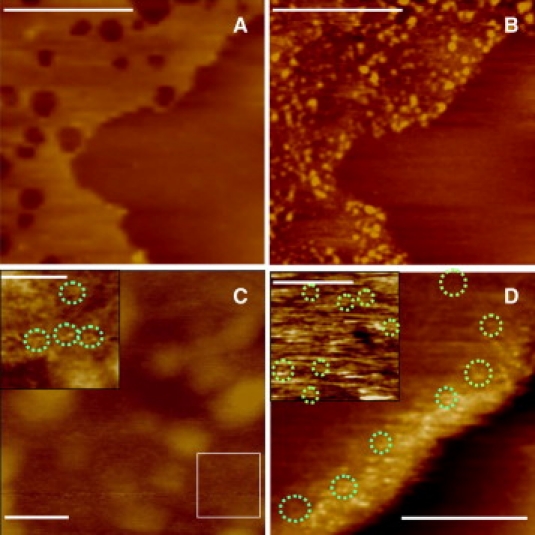
PG-1 forms channel-like structures in DOPS/POPE lipid bilayers. (A and B) AFM height images of DOPS/POPE bilayer before and after addition of PG-1 (conc 150 μg/mL, scale bar = 1 μm). When PG-1 is added to a preformed DOPS/POPE bilayer, large globular oligomers are observed preferentially on the bilayer surface.(C) AFM image of a membrane patch with PG-1 oligomer-like structures. Scale bar in C is 100 nm. The inset represents the highlighted square area after flattening and low pass filtering (inset scale bar = 50 nm). (D) AFM image of the edge of a lipid bilayer with incorporated PG-1 peptides (scan size: 100 nm × 100 nm) with the insert representing a 75 nm × 75 nm area in the central region of a bilayer (scale bars = 50 nm). In C and D, green circles highlight PG-1 channel-like structures.
AFM images of PG-1 peptides reconstituted in DOPS/POPE lipid bilayers (Fig. 3 D), significantly show no large conglomerates and only channel-like structures protruding out of the membrane plane are visible. Interestingly, the channel-like structures are visible both at the edges of the bilayer as well as closer to the center (Fig. 3 D, inset).
Higher resolution AFM image of PG-1 channels show subunit arrangements that are in good agreement with the above described MD simulation models. Fig. 4 shows channel-like structures when PG-1 is incorporated into DOPS/POPE bilayers. Representative structures with identifiable subunits (highlighted in Fig. 3, C and D), are further magnified to show the distinctive subunit organizations (Fig. 4, A–C). Forty-nine pores were analyzed using the section feature of the Veeco software. In the cross-section of the diameter of pore structure (Fig. 4 D) the outer diameter was measured to be at two-thirds level of the full height with respect to the lipid bilayer surface, as marked by the red arrows in Fig. 4 E. The green arrows mark the position where the inner pore diameter was measured. The range of the inner and outer pore diameters are plotted in Fig. 4, F and G. The size distribution was fitted to a Gaussian distribution with the inner diameters centered at 2.07 nm, and the outer diameters centered at 5.2 nm. The last graph shows a frequency count of the subunit organization (Fig. 4 H). The number of subunits ranged from three to five, though the majority is three or four (43% and 48% occurrences, respectively). Statistical analysis shows that the dimensions of the channels' inner and outer diameters are strongly dependent on the number of subunits: pores with three subunits had on average an inner diameter of 1.96 nm and an outer diameter of 5.13 nm (n = 21); pores with four subunits, had inner and outer diameters 2.09 nm and 5.30 nm (n = 24), respectively. The few pores comprised of five subunits had an average 2.24 nm and 5.76 nm inner and outer diameters (n = 4), respectively. The multiple subunit stoichiometry observed here is consistent with previous observations that several β-sheet pore formers tend to display multiple-conductances when reconstituted in PLBs (13,15,33,34,56–58).
Figure 4.
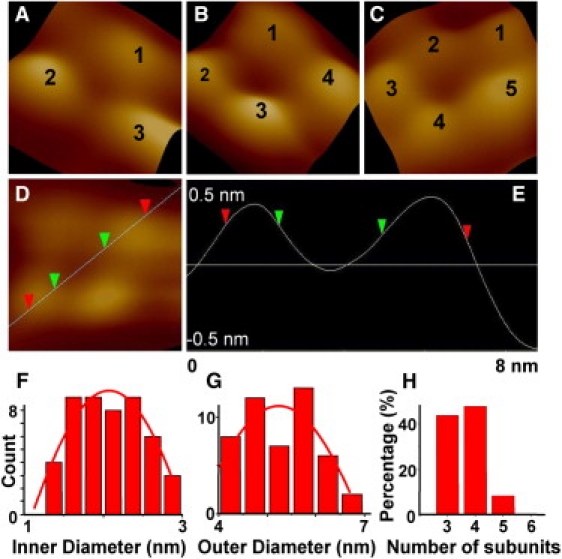
The structure of PG-1 ion channels incorporated in DOPS/POPE lipid bilayers from high resolution AFM imaging. (A–C) Examples of channel-like structures with different subunit organizations. (D and E) Inner and outer diameters of PG-1 channels that were measured from the cross-section profile of the channel height. The red arrows mark the outer diameter, and the green arrows mark the regions where the inner diameter was measured. The graphs shown in F–H represent pore (F) inner and (G) outer diameter distributions and (H) channel subunits distribution.
Electrical conductance of PG-1 reconstituted in PLBs
PG-1 activity in various membrane compositions using folded or painted bilayers were examined (Fig. 5). In membranes made with anionic lipids DOPS/DOPE and DOPS/POPE, ionic permeation was observed for sub and low micromolar (0.2–1.5 μM) PG-1 concentrations (Fig. 5 A). This finding is in agreement with previous works using anionic lipids (13,24) and supports the notion that PG-1 binds more avidly to anionic lipids (13,23,28). The activity presented in Fig. 5 A also shows multiple conductance and could be defined as spiky or bursting (15).
Figure 5.
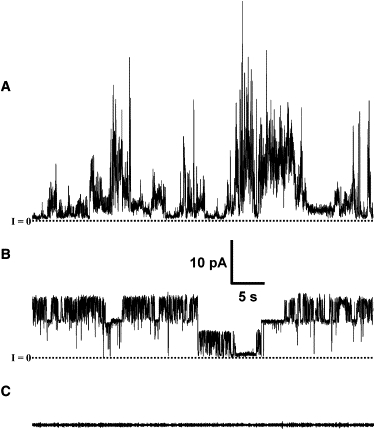
PG-1 channel activity is dependent on bilayer composition. (A) PG-1 activity in the anionic DOPS/DOPE 1:1 bilayer. Note the short lived and spiky behavior. (B) Membrane activity of PG-1 in DOPC/DOPE 1:1 bilayer. The activity shown here is well sustained with fast openings and closings. (C) Typical PG-1 activity in 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine or DOPC bilayers. Occasionally PC bilayers present short lived, low amplitude very sparse step like current jumps. Membranes were formed by the folded technique. Traces shown were recorded at 40 mV of applied potential. The electrolyte solution used was 100 mM KCl, 10 mM Hepes, and 1 mM MgCl2. PG-1 was added directly into one side of the recording chamber.
PG-1 also shows channel activity in membranes made with the zwitterionic lipid mixture DOPC/DOPE, although requiring somewhat higher concentration (Fig. 5 B and Fig. 6). Channel activity can be well defined and occasionally very stable. However, activity can also appear as in Fig. 5 A. Fig. 6 shows a 14-min trace with what seems to be two very stable channels, or one with two stable conductances of ∼190 pS and ∼100 pS. The activity presented in Fig. 6 showed voltage independence, although it should be noted that the voltage dependency data were collected after the membrane was held at 40 mV for >10 min (Fig. 6 A, top two current versus time traces).
Figure 6.
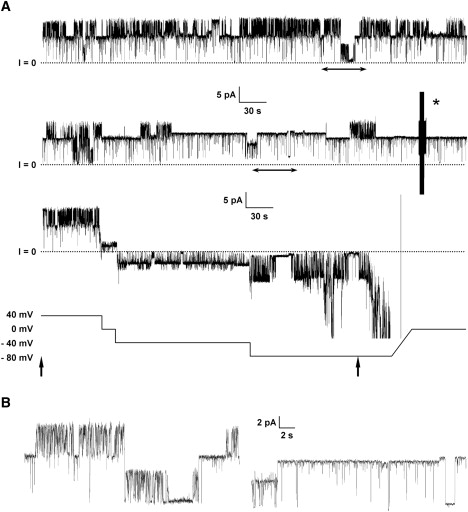
PG-1 membrane activities on zwitterionic DOPC/DOPE bilayers. (A) A 14-min trace presented in 5-min intervals. The top two 5-min fragments are at 40 mV. The calculated conductances for both steps shown are 100 pS and 190 pS. Using the lower current versus time trace shown in A, the conductance calculated as the slope of the linear fit for the current versus voltages 40, 0, −40, and −80 mV yields a conductance of ∼100 pS. The double arrow head under the traces show the current versus time fragments used to make B. The asterisk (∗) symbol marks a capacitance measurement to check membrane integrity during recording. The vertical arrow below the voltage versus time trace in A shows the trace fragment used to calculate the IV plot.
Bilayer membranes made with only phosphatidylcholine (PC) lipids required higher PG-1 concentrations and elicited only infrequent electrical activity. More generally PC membranes simply broke, and could not be refolded, suggesting that PG-1 interacts differently with PC bilayers. Two types of PC phospholipids, DOPC and 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine, were used in this study. In folded membranes, the PG-1 concentration needed to elicit membrane activity in PC+ phosphatidylethanolamine (PE) bilayers is slightly higher (1–3 μM) than the concentration needed for the phosphatidylserine (PS) + PE bilayers (0.2–1.5 μM) and is even higher for PC only bilayers (2–10 μM). Even in these conditions, on occasion, no ionic permeation was seen in PC membranes (Fig. 5 C).
Because the bilayers made by different techniques differ slightly in thickness that may affect pore ionic conduction (59–61), PG-1 channel behavior in both folded and painted membranes were examined. The PG-1 monomer has 18 amino acids and is folded to a β-hairpin that precludes it spanning through the whole bilayer thickness. Thicker membranes (painted bilayers) are expected to conduct less efficiently. A direct comparison of PG-1 channel activity in PS/PE bilayers created by the two methods is shown in Fig. S4. In general, PS/PE painted bilayers, seem to allow for somewhat longer duration of channel openings, although current amplitudes seem to remain somewhat comparable. As PG-1 shows some commonality with amyloids (in their aggregation and channel-like features (34,62–64)), we tested whether or not PG-1 activity would be inhibited by Zn2+, an amyloid ion channel blocker. Unlike amyloid β channels (56), and as expected (29), PG-1 channel conductance was not inhibited by Zn2+.
Discussion
MD simulation and AFM results show that PG-1 forms channel structures in anionic membranes and electrical conductance measurements show channel behavior. PG-1 channels seem to be lipid dependent in AFM and PLB studies. AFM images show pores only in membranes with anionic lipids (Fig. 3, B and D). No channel-like structures were observed in DOPC bilayers. In DOPC membrane, PG-1 incorporation at the same concentrations show only membrane disruption without channel like structures, consistent with the work of Lam et al. (16). The outer diameters of the antiparallel and parallel β-sheet PG-1 channels in the MD simulations are ∼5.5 and ∼5.3 nm, respectively. This is consistent with the outer diameter of 5.3 nm measured from AFM images. However, the pore diameters, ∼1.4 and ∼1.3 nm for the antiparallel and parallel β-sheet channels, respectively, are slightly smaller than the averaged pore diameter of 2.04 nm measured from AFM images. The disparity could be due to the experimental designs: in AFM images, the diameter is measured at 2/3 of the full height of the pore compared to the lipid bilayer, whereas MD calculates the pore diameter at the pore center. Due to the torus shape, the center of the pore would be narrower than the gate (Fig. 1). AFM images of PG-1 reconstituted in DOPS/POPE show channel diameter larger than those simulated earlier (25,29,30), which were modeled in other lipid compositions, suggesting that the PG-1 channels measured by AFM contain more than eight PG-1 β-hairpins. MD simulations with 10 PG-1 β-hairpins show stability and the predicted outer and inner diameters are in better agreement with the AFM values.
The decameric PG-1 channel modeling suggests subunit morphology that is shared by other β-sheet channels (29,31–33). The antiparallel and parallel β-sheet channels of PG-1 obtain four and five subunits, respectively, consistent with AFM images presented in this study (Fig. 4) as well as previous observations (25,29,30). In the simulation, subunit formation varies even for the same channel in the lipid bilayer, strongly depending on the fluidic bilayer dynamics. No PG-1 channels with three subunits were observed, perhaps due to limited simulation trajectories. The pores imaged by AFM in DOPS/POPE show mainly trimeric (43%) and tetrameric (48%) (with infrequent pentameric (9%)) subunits, with inner diameters for trimeric, tetrameric, and pentameric subunits of 1.96 nm (n = 21), 2.09 nm (n = 24), and 2.24 nm (n = 4), respectively (Fig. 4). Unlike in amyloid pores (33–35), we did not observe any hexameric channels from a pool of 49 pores analyzed in this study.
PG-1 channels exhibit anionic selectivity (13,29). In the MD simulations, the pore of the PG-1 channel exhibits relatively high Cl− probability distribution across the lipid bilayer. This is to be expected for an oligomeric pore where each monomer contains six positive Arg residues. The results presented in this study together with previous evidence by Sokolov et al. (13) showing that PG-1 channels in bilayers made with lipid A or LPS become cation-selective, all point toward channel structures that are influenced strongly by the lipid headgroup composition. It is likely that lipids headgroups are very close to the entrance of the PG-1 pore mouth or form part of the pore lining itself.
Functional PG-1 channel activity as seen in PLB experiments shows dynamic multiconductance and differs significantly depending on the headgroups of the phospholipids used to form the bilayers. The findings presented in this study confirm and expand on the previous work by Sokolov et al. (13), and Mangoni et al. (24). We believe the novel finding that PG-1 can form channels in PE rich zwitterionic membranes might help explain the toxicity of PG-1 in mammalian models (65,66). This observation is consistent with the recently proposed dynamic nature of β-sheet amyloid pores (31–33,67), where loosely-interacting 3-6 dynamic units associate and dissociate leading to complex ionic flux.
In terms of the mechanism of action and the underlying structure, the β-hairpin antibiotic PG-1, and the U-shaped amyloids with β-strand-turn-β-strand motif, share several theoretical and experimental features, including positive charge(s) that facilitate interaction with negatively charged membranes, the ability to form pores and amyloid-like fibrils (H. Jang, M. Mustata, R. Nussinov, and R. Lal, unpublished data), the lack of preference for incorporation, and permeation into preformed PC bilayers. Several similarities between cationic AMPs and certain amyloids' effects on membranes were observed in electrophysiological (15), structural imaging (34,35), and in MD simulations (29,31–33). An obvious difference is the two disulfide bonds (Cys6-Cys15 and Cys8-Cys13) that add stability to the PG-1 β-hairpin when compared to the Cys bridgeless β amyloids. Nonetheless mutated sequences unable to form cysteine-bonds (68) or PG-1 with reduced cysteines are still bactericidal (24).
In conclusion, we provide what we believe to be theoretical and experimental evidence that PG-1, a β-hairpin AMP, forms lipid dependent ion channels similar to amyloid channels and other β-sheet pore formers, including bacterial toxins and other AMPs (29,31,32,34,35,64). The structural and functional similarities between amyloids and β-hairpin-AMPs suggests a possible shared mechanism of action (15,69,70); namely channel-mediated dysregulation of cellular ionic homeostasis. Dysregulation of cell ionic homeostasis by amyloid ion channels play a key role in neurodegenerative diseases (34,35,57,63,64,71–76). Membrane coated microbes are rarely resistant to PG-1, but therapeutic dosages of PG-1 are toxic to humans. This study provides what we believe to be a new understanding of the biophysical and physiological nature of β-hairpin PG-1 mediated microbial toxicity and would be useful for the development of newer, safer, and more effective PG-1 based antibiotics.
Supporting Material
Detailed methods and four figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00275-4.
Supporting Material
Acknowledgments
We thank S. Ramachandran at the University of Chicago for critical reading of the manuscript. R.C. acknowledges custom-made software used for current recordings developed by D. J. Estes, PhD, and J. D. Uram, PhD.
This work has been supported by National Institutes of Health (National Institute on Aging) extramural program (R.L.). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. All simulations had been performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Footnotes
R. Capone's, F. Teran Arce's, and R. Lal's present address is Bioengineering and Mechanical Aerospace Engineering, University of California San Diego, La Jolla, California.
Contributor Information
Ruth Nussinov, Email: ruthnu@helix.nih.gov.
Ratnesh Lal, Email: rlal@uchicago.edu.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Hoskin D.W., Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sitaram N., Nagaraj R. Host-defense antimicrobial peptides: importance of structure for activity. Curr. Pharm. Des. 2002;8:727–742. doi: 10.2174/1381612023395358. [DOI] [PubMed] [Google Scholar]
- 4.Hancock R.E., Diamond G. The role of cationic antimicrobial peptides in innate host defenses. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 5.Gottler L.M., Ramamoorthy A. Structure, membrane orientation, mechanism, and function of pexiganan—a highly potent antimicrobial peptide designed from magainin. Biochim. Biophys. Acta. 2009;1788:1680–1686. doi: 10.1016/j.bbamem.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papo N., Shahar M., Shai Y. A novel lytic peptide composed of DL-amino acids selectively kills cancer cells in culture and in mice. J. Biol. Chem. 2003;278:21018–21023. doi: 10.1074/jbc.M211204200. [DOI] [PubMed] [Google Scholar]
- 7.Mecke A., Lee D.K., Banaszak Holl M.M. Membrane thinning due to antimicrobial peptide binding: an atomic force microscopy study of MSI-78 in lipid bilayers. Biophys. J. 2005;89:4043–4050. doi: 10.1529/biophysj.105.062596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heller W.T., Waring A.J., Huang H.W. Membrane thinning effect of the beta-sheet antimicrobial protegrin. Biochemistry. 2000;39:139–145. doi: 10.1021/bi991892m. [DOI] [PubMed] [Google Scholar]
- 9.Sayyed-Ahmad A., Kaznessis Y.N. Determining the orientation of protegrin-1 in DLPC bilayers using an implicit solvent-membrane model. PLoS One. 2009;4:e4799. doi: 10.1371/journal.pone.0004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marassi F.M., Opella S.J., Merrifield R.B. Orientation of cecropin A helices in phospholipid bilayers determined by solid-state NMR spectroscopy. Biophys. J. 1999;77:3152–3155. doi: 10.1016/S0006-3495(99)77145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi S., Huster D., Hong M. Orientation and dynamics of an antimicrobial peptide in the lipid bilayer by solid-state NMR spectroscopy. Biophys. J. 2001;81:2203–2214. doi: 10.1016/S0006-3495(01)75868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaser R.W., Sachse C., Ulrich A.S. Concentration-dependent realignment of the antimicrobial peptide PGLa in lipid membranes observed by solid-state 19F-NMR. Biophys. J. 2005;88:3392–3397. doi: 10.1529/biophysj.104.056424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokolov Y., Mirzabekov T., Kagan B.L. Membrane channel formation by antimicrobial protegrins. Biochim. Biophys. Acta. 1999;1420:23–29. doi: 10.1016/s0005-2736(99)00086-3. [DOI] [PubMed] [Google Scholar]
- 14.Yang L., Weiss T.M., Huang H.W. Crystallization of antimicrobial pores in membranes: magainin and protegrin. Biophys. J. 2000;79:2002–2009. doi: 10.1016/S0006-3495(00)76448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kourie J.I., Shorthouse A.A. Properties of cytotoxic peptide-formed ion channels. Am. J. Physiol. Cell Physiol. 2000;278:C1063–C1087. doi: 10.1152/ajpcell.2000.278.6.C1063. [DOI] [PubMed] [Google Scholar]
- 16.Lam K.L.H., Ishitsuka Y., Lee K.Y. Mechanism of supported membrane disruption by antimicrobial peptide protegrin-1. J. Phys. Chem. B. 2006;110:21282–21286. doi: 10.1021/jp0630065. [DOI] [PubMed] [Google Scholar]
- 17.Miyasaki K.T., Lehrer R.I. Beta-sheet antibiotic peptides as potential dental therapeutics. Int. J. Antimicrob. Agents. 1998;9:269–280. doi: 10.1016/s0924-8579(98)00006-5. [DOI] [PubMed] [Google Scholar]
- 18.Kokryakov V.N., Harwig S.S., Lehrer R.I. Protegrins: leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 19.Roumestand C., Louis V., Chavanieu A. Oligomerization of protegrin-1 in the presence of DPC micelles. A proton high-resolution NMR study. FEBS Lett. 1998;421:263–267. doi: 10.1016/s0014-5793(97)01579-2. [DOI] [PubMed] [Google Scholar]
- 20.Fahrner R.L., Dieckmann T., Feigon J. Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. Chem. Biol. 1996;3:543–550. doi: 10.1016/s1074-5521(96)90145-3. [DOI] [PubMed] [Google Scholar]
- 21.Gidalevitz D., Ishitsuka Y., Lee K.Y. Interaction of antimicrobial peptide protegrin with biomembranes. Proc. Natl. Acad. Sci. USA. 2003;100:6302–6307. doi: 10.1073/pnas.0934731100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heller W.T., Waring A.J., Huang H.W. Multiple states of β-sheet peptide protegrin in lipid bilayers. Biochemistry. 1998;37:17331–17338. doi: 10.1021/bi981314q. [DOI] [PubMed] [Google Scholar]
- 23.Jing W., Prenner E.J., Lohner K. Headgroup structure and fatty acid chain length of the acidic phospholipids modulate the interaction of membrane mimetic vesicles with the antimicrobial peptide protegrin-1. J. Pept. Sci. 2005;11:735–743. doi: 10.1002/psc.702. [DOI] [PubMed] [Google Scholar]
- 24.Mangoni M.E., Aumelas A., Chavanieu A. Change in membrane permeability induced by protegrin 1: implication of disulphide bridges for pore formation. FEBS Lett. 1996;383:93–98. doi: 10.1016/0014-5793(96)00236-0. [DOI] [PubMed] [Google Scholar]
- 25.Mani R., Cady S.D., Hong M. Membrane-dependent oligomeric structure and pore formation of a beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. USA. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang M., Waring A.J., Hong M. Phosphate-mediated arginine insertion into lipid membranes and pore formation by a cationic membrane peptide from solid-state NMR. J. Am. Chem. Soc. 2007;129:11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- 27.Gottler L.M., de la Salud Bea R., Marsh E.N. Using fluorous amino acids to probe the effects of changing hydrophobicity on the physical and biological properties of the β-hairpin antimicrobial peptide protegrin-1. Biochemistry. 2008;47:9243–9250. doi: 10.1021/bi801045n. [DOI] [PubMed] [Google Scholar]
- 28.Ishitsuka Y., Pham D.S., Lee K.Y. Insertion selectivity of antimicrobial peptide protegrin-1 into lipid monolayers: effect of headgroup electrostatics and tail group packing. Biochim. Biophys. Acta. 2006;1758:1450–1460. doi: 10.1016/j.bbamem.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Jang H., Ma B., Nussinov R. Models of toxic beta-sheet channels of protegrin-1 suggest a common subunit organization motif shared with toxic Alzheimer beta-amyloid ion channels. Biophys. J. 2008;95:4631–4642. doi: 10.1529/biophysj.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langham A.A., Ahmad A.S., Kaznessis Y.N. On the nature of antimicrobial activity: a model for protegrin-1 pores. J. Am. Chem. Soc. 2008;130:4338–4346. doi: 10.1021/ja0780380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang H., Zheng J., Nussinov R. Models of beta-amyloid ion channels in the membrane suggest that channel formation in the bilayer is a dynamic process. Biophys. J. 2007;93:1938–1949. doi: 10.1529/biophysj.107.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang H., Zheng J., Nussinov R. New structures help the modeling of toxic amyloidbeta ion channels. Trends Biochem. Sci. 2008;33:91–100. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Mustata M., Capone R., Nussinov R. K3 fragment of amyloidogenic beta(2)-microglobulin forms ion channels: implication for dialysis related amyloidosis. J. Am. Chem. Soc. 2009;131:14938–14945. doi: 10.1021/ja9049299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quist A., Doudevski I., Lal R. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin H., Bhatia R., Lal R. Amyloid beta protein forms ion channels: implications for Alzheimer's disease pathophysiology. FASEB J. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 36.Tang M., Waring A.J., Hong M. Intermolecular packing and alignment in an ordered β-hairpin antimicrobial peptide aggregate from 2D solid-state NMR. J. Am. Chem. Soc. 2005;127:13919–13927. doi: 10.1021/ja0526665. [DOI] [PubMed] [Google Scholar]
- 37.Jang H., Ma B., Nussinov R. Interaction of protegrin-1 with lipid bilayers: membrane thinning effect. Biophys. J. 2006;91:2848–2859. doi: 10.1529/biophysj.106.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang H., Ma B., Nussinov R. Conformational study of the protegrin-1 (PG-1) dimer interaction with lipid bilayers and its effect. BMC Struct. Biol. 2007;7:21. doi: 10.1186/1472-6807-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks B.R., Bruccoleri R.E., Karplus M. CHARMM—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 40.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller P., Rudin D.O., Wescott W.C. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature. 1962;194:979–980. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
- 42.Montal M., Mueller P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. USA. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capone R., Quiroz F.G., Mayer M. Amyloid-beta-induced ion flux in artificial lipid bilayers and neuronal cells: resolving a controversy. Neurotox. Res. 2009;16:1–13. doi: 10.1007/s12640-009-9033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capone R., Blake S., Mayer M. Designing nanosensors based on charged derivatives of gramicidin A. J. Am. Chem. Soc. 2007;129:9737–9745. doi: 10.1021/ja0711819. [DOI] [PubMed] [Google Scholar]
- 45.Macrae, M. X., S. Blake, …, J. Yang. 2009. A semi-synthetic ion channel platform for detection of phosphatase and protease activity. ACS Nano. 3:3567–3580. [DOI] [PMC free article] [PubMed]
- 46.Ban T., Yamaguchi K., Goto Y. Direct observation of amyloid fibril growth, propagation, and adaptation. Acc. Chem. Res. 2006;39:663–670. doi: 10.1021/ar050074l. [DOI] [PubMed] [Google Scholar]
- 47.Ozawa D., Yagi H., Goto Y. Destruction of amyloid fibrils of a beta2-microglobulin fragment by laser beam irradiation. J. Biol. Chem. 2009;284:1009–1017. doi: 10.1074/jbc.M805118200. [DOI] [PubMed] [Google Scholar]
- 48.Kozhukh G.V., Hagihara Y., Goto Y. Investigation of a peptide responsible for amyloid fibril formation of beta 2-microglobulin by Achromobacter protease I. J. Biol. Chem. 2002;277:1310–1315. doi: 10.1074/jbc.M108753200. [DOI] [PubMed] [Google Scholar]
- 49.Khurana R., Ionescu-Zanetti C., Carter S.A. A general model for amyloid fibril assembly based on morphological studies using atomic force microscopy. Biophys. J. 2003;85:1135–1144. doi: 10.1016/S0006-3495(03)74550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Auvynet C., El Amri C., Rosenstein Y. Structural requirements for antimicrobial versus chemoattractant activities for dermaseptin S9. FEBS J. 2008;275:4134–4151. doi: 10.1111/j.1742-4658.2008.06554.x. [DOI] [PubMed] [Google Scholar]
- 51.Code C., Domanov Y., Kinnunen P.K. Amyloid-type fiber formation in control of enzyme action: interfacial activation of phospholipase A2. Biophys. J. 2008;95:215–224. doi: 10.1529/biophysj.108.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Code C., Domanov Y.A., Kinnunen P.K. Activation of phospholipase A2 by temporin B: formation of antimicrobial peptide-enzyme amyloid-type cofibrils. Biochim. Biophys. Acta. 2009;1788:1064–1072. doi: 10.1016/j.bbamem.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Gössler-Schöfberger R., Hesser G., Jilek A. An orphan dermaseptin from frog skin reversibly assembles to amyloid-like aggregates in a pH-dependent fashion. FEBS J. 2009;276:5849–5859. doi: 10.1111/j.1742-4658.2009.07266.x. [DOI] [PubMed] [Google Scholar]
- 54.Sood R., Domanov Y., Kinnunen P.K. Binding of LL-37 to model biomembranes: insight into target vs. host cell recognition. Biochim. Biophys. Acta. 2008;1778:983–996. doi: 10.1016/j.bbamem.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 55.Zhao H., Sood R., Kinnunen P.K. Interaction of the antimicrobial peptide pheromone Plantaricin A with model membranes: implications for a novel mechanism of action. Biochim. Biophys. Acta. 2006;1758:1461–1474. doi: 10.1016/j.bbamem.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 56.Arispe N., Pollard H.B., Rojas E. Zn2+ interaction with Alzheimer amyloid beta protein calcium channels. Proc. Natl. Acad. Sci. USA. 1996;93:1710–1715. doi: 10.1073/pnas.93.4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirakura Y., Kagan B.L. Pore formation by beta-2-microglobulin: a mechanism for the pathogenesis of dialysis associated amyloidosis. Amyloid. 2001;8:94–100. doi: 10.3109/13506120109007350. [DOI] [PubMed] [Google Scholar]
- 58.Kourie J.I., Hanna E.A., Henry C.L. Properties and modulation of alpha human atrial natriuretic peptide (alpha-hANP)-formed ion channels. Can. J. Physiol. Pharmacol. 2001;79:654–664. [PubMed] [Google Scholar]
- 59.Lundbaek J.A., Andersen O.S. Spring constants for channel-induced lipid bilayer deformations. Estimates using gramicidin channels. Biophys. J. 1999;76:889–895. doi: 10.1016/S0006-3495(99)77252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinac B., Hamill O.P. Gramicidin A channels switch between stretch activation and stretch inactivation depending on bilayer thickness. Proc. Natl. Acad. Sci. USA. 2002;99:4308–4312. doi: 10.1073/pnas.072632899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mobashery N., Nielsen C., Andersen O.S. The conformational preference of gramicidin channels is a function of lipid bilayer thickness. FEBS Lett. 1997;412:15–20. doi: 10.1016/s0014-5793(97)00709-6. [DOI] [PubMed] [Google Scholar]
- 62.Arispe N., Rojas E., Pollard H.B. Alzheimer disease amyloid beta protein forms calcium channels in bilayer membranes: blockade by tromethamine and aluminum. Proc. Natl. Acad. Sci. USA. 1993;90:567–571. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kagan B.L., Hirakura Y., Lin M.C. The channel hypothesis of Alzheimer's disease: current status. Peptides. 2002;23:1311–1315. doi: 10.1016/s0196-9781(02)00067-0. [DOI] [PubMed] [Google Scholar]
- 64.Lin M.C., Mirzabekov T., Kagan B.L. Channel formation by a neurotoxic prion protein fragment. J. Biol. Chem. 1997;272:44–47. doi: 10.1074/jbc.272.1.44. [DOI] [PubMed] [Google Scholar]
- 65.Frecer V. QSAR analysis of antimicrobial and hemolytic effects of cyclic cationic antimicrobial peptides derived from protegrin-1. Bioorg. Med. Chem. 2006;14:6065–6074. doi: 10.1016/j.bmc.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 66.Langham A.A., Khandelia H., Kaznessis Y.N. Correlation between simulated physicochemical properties and hemolycity of protegrin-like antimicrobial peptides: predicting experimental toxicity. Peptides. 2008;29:1085–1093. doi: 10.1016/j.peptides.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jang H., Arce F.T., Nussinov R. Misfolded amyloid ion channels present mobile β-sheet subunits in contrast to conventional ion channels. Biophys. J. 2009;97:3029–3037. doi: 10.1016/j.bpj.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai J.R., Huck B.R., Gellman S.H. Design of non-cysteine-containing antimicrobial β-hairpins: structure-activity relationship studies with linear protegrin-1 analogues. Biochemistry. 2002;41:12835–12842. doi: 10.1021/bi026127d. [DOI] [PubMed] [Google Scholar]
- 69.Kourie J.I., Henry C.L. Protein aggregation and deposition: implications for ion channel formation and membrane damage. Croat. Med. J. 2001;42:359–374. [PubMed] [Google Scholar]
- 70.Mahalka A.K., Kinnunen P.K. Binding of amphipathic alpha-helical antimicrobial peptides to lipid membranes: lessons from temporins B and L. Biochim. Biophys. Acta. 2009;1788:1600–1609. doi: 10.1016/j.bbamem.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 71.Hirakura Y., Azimov R., Kagan B.L. Polyglutamine-induced ion channels: a possible mechanism for the neurotoxicity of Huntington and other CAG repeat diseases. J. Neurosci. Res. 2000;60:490–494. doi: 10.1002/(sici)1097-4547(20000515)60:4<490::aid-jnr7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 72.Arispe N., Pollard H.B., Rojas E. beta-Amyloid Ca(2+)-channel hypothesis for neuronal death in Alzheimer disease. Mol. Cell. Biochem. 1994;140:119–125. doi: 10.1007/BF00926750. [DOI] [PubMed] [Google Scholar]
- 73.Kagan B.L. Amyloidosis and protein folding. Science. 2005;307 doi: 10.1126/science.307.5706.42b. 42–43, author reply 42–43. [DOI] [PubMed] [Google Scholar]
- 74.Kagan B.L., Azimov R., Azimova R. Amyloid peptide channels. J. Membr. Biol. 2004;202:1–10. doi: 10.1007/s00232-004-0709-4. [DOI] [PubMed] [Google Scholar]
- 75.Pollard H.B., Arispe N., Rojas E. Ion channel hypothesis for Alzheimer amyloid peptide neurotoxicity. Cell. Mol. Neurobiol. 1995;15:513–526. doi: 10.1007/BF02071314. [DOI] [PubMed] [Google Scholar]
- 76.Pollard H.B., Rojas E., Arispe N. A new hypothesis for the mechanism of amyloid toxicity, based on the calcium channel activity of amyloid beta protein (A beta P) in phospholipid bilayer membranes. Ann. N. Y. Acad. Sci. 1993;695:165–168. doi: 10.1111/j.1749-6632.1993.tb23046.x. [DOI] [PubMed] [Google Scholar]
- 77.Smart O.S., Goodfellow J.M., Wallace B.A. The pore dimensions of gramicidin A. Biophys. J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


