Abstract
The phenomenon of antibiotic resistance has created a need for the development of novel antibiotic classes with non-classical cellular targets. Unfortunately, target-based drug discovery against proteins considered essential for in vitro bacterial viability has yielded few new therapeutic classes of antibiotics. Targeting the large proportion of genes considered nonessential that have yet to be explored by HTS, e.g., RecA, can complement these efforts. Recent evidence suggests that RecA-controlled processes are responsible for tolerance to antibiotic chemotherapy and are involved in pathways that ultimately lead to full-fledged antibiotic resistance. Therefore inhibitors of RecA may serve as therapeutic adjuvants in combination chemotherapy of bacterial infectious diseases. Towards the goal of validating RecA as a novel target in the chemotherapy of bacterial infections, we have screened 35,780 small molecules against RecA. In total, 80 small molecules were identified as primary hits and could be clustered in six distinct chemotype clades. The most potent class of hits was further examined, and one member compound was found to inhibit RecA-mediated strand exchange and prevent ciprofloxacin-induced SOS expression in Escherichia coli. This compound represents the first small molecule demonstrating an ability to inhibit the bacterial SOS response in live bacterial cell cultures.
Keywords: RecA, antibiotic resistance, ATPase inhibition, ciprofloxacin, SOS response
INTRODUCTION
Antibiotic resistance is an escalating problem in the chemotherapy of bacterial infectious diseases1,2 that erodes the efficacy of current frontline therapeutic agents as well as undermining the profitability of developing new antibacterial agents.3 As a result, many pharmaceutical companies have curtailed their antibacterial drug discovery efforts,4 and since 1962 only three new classes of antibacterial agents have been introduced.5 Novel strategies will be required to overcome this looming crisis, and one potential solution to the problem involves screening targets considered non-essential for in vitro viability. Indeed, two-thirds of Escherichia coli genes have been characterized as non-essential,6 and exploring this previously ignored segment of the bacterial genome may offer possibilities for the discovery of non-traditional pharmaceutical targets and agents that attenuate pathogenicity or potentiate the pharmacologic effects of known antibacterial agents.7 In this context, we have focused on the bacterial RecA protein as a prospective target in the treatment of bacterial infectious diseases. RecA plays crucial roles in the repair of DNA damage and stalled replication, but also participates in processes that promote stress-induced mutation and horizontal gene transfer. We hypothesize that small molecule inhibitors of RecA may sensitize bacteria to established antibacterial agents and prevent the development and acquisition of genes conferring drug resistance.
E. coli RecA has been identified as a critical component of the response to antibacterial quinolones that interfere with topoisomerase II, leading to double-stranded DNA breaks and stalled replication forks, both of which are processed to ssDNA.8 In the event of such DNA damage, multiple ATP-bound RecA monomers coat the resulting ssDNA, forming a helical homopolymeric filament of RecA on DNA that has both enzymatic and signaling properties. This RecA-DNA filament (RDF) mediates recombinational DNA repair, an enzymatic function that exchanges strands between homologous DNA substrates, and activates autocleavage of the LexA repressor, a signaling activity that initiates the expression of SOS genes. The early SOS gene products, including RecA itself, help maintain genetic homeostasis by high-fidelity repair of the DNA damage, while the late SOS gene products introduce genome-wide mutations.
E. coli RecA's seemingly antagonistic activities are the basis for both short and long-term survival responses to antibiotic treatment. RecA-dependent DNA repair apparently leads to inherent ciprofloxacin tolerance, and ΔrecA strains are more susceptible to cell killing by ciprofloxacin.9 In addition, RecA-activated SOS mutagenesis results in genetic diversification that can lead to resistance-conferring mutations, and SOS-deficient strains adapt much more slowly to ciprofloxacin.10 RecA is a highly conserved bacterial protein and likely plays similar roles in other species,11 making it an attractive target for the development of new pharmaceutic adjuvants.12 Unfortunately, no cell-permeable natural product or synthetic small molecule inhibitors of RecA have been reported to date.
To advance our goal of obtaining cell-permeable small molecules that modulate RecA's biological activities in live bacterial cell cultures, we have screened a diverse collection of 35,780 drug-like small molecules for the inhibition of RecA. Because all of RecA's biological activities are expressed from an activated conformation that requires the binding ATP that is subsequently hydrolyzed,13 the compounds were screened using a colorimetric ATPase assay adapted for use with RecA. In all, we identified five distinct chemotype clades capable of abrogating the ATPase activity of RecA in vitro. One such class of molecules was found to significantly reduce the RecA-dependent activation of the SOS response in liquid cultures of E. coli resulting from exposure to ciprofloxacin.
MATERIALS AND METHODS
Materials
RecA was purified and stored as previously described.14 Poly(dT) single-stranded DNA (average length = 319 nucleotides) was purchased from Amersham Biosciences (Piscataway, NJ). Crystalline L-ascorbic acid and sulfuric acid were purchased from Fisher Scientific. ϕχ174 circular single-stranded DNA (cssDNA) and double stranded DNA (dsDNA) were purchased from New England Biolabs (Ipswich, MA). Unless otherwise stated all other reagents were purchased from Sigma-Aldrich (St. Louis, MO) at the highest level of purity possible. E. coli K-12 containing the sulA-gfp reporter gene fusion (SS996 strain)15 was generously provided by Dr. Steven Sandler at the University of Massachusetts at Amherst.
Compound libraries
The Challenge, Diversity and Natural Product libraries were obtained from the National Cancer Institute (NCI), Drug Synthesis and Chemistry Branch (Bethesda, MD) as 10 mM stocks in DMSO. The compounds were diluted to 1 mM stocks in sterile 96-well U-bottom microplates (Evergreen Scientific, Los Angeles, CA) in DMSO using a TOMTEC 96-channel pipet tower (TOMTEC, Hamden, CT).
The BRITE compound collection, consisting of a library of over 350,000 compounds generated by combinatorial chemistry synthetic routes, was a gift from Biogen Idec in 2006. Upon acquisition, compounds were placed into 100% DMSO at an initial concentration of 10 mM. All of the compound plates were stored in polypropylene deep-well blocks at 4 °C without humidity controls. Using a Biomek NX unit (Beckman-Coulter, Fullerton, CA), 0.5 μL of a 1 mM stock of library compounds in DMSO was pre-spotted onto Costar clear flat-bottom 384-well assay plates (Corning, Lowell, MA). In the left and right two columns of the assay plates 0.5 μL DMSO was spotted for the respective negative and positive control reactions.
Phosphomolybdate blue ATPase assay
The phosphomolybdate blue dye was made as a 10X stock by adding 30 g ammonium molybdate to 87 mL concentrated sulfuric acid and adjusting the final volume to 250 mL with autoclaved MilliQ de-ionized water. On the day of use, fresh 1X phosphomolybdate blue dye was made by diluting the 10X stock in autoclaved MilliQ de-ionized water and ascorbic acid and SDS were added to final concentrations of 10% w/v and 1% w/v respectively.
The ATPase reactions were carried out in the 384-well plates that the compounds were spotted in, and the final volume in each well was 30.5 μL, giving final concentrations of 17 μM for the library compounds and 1.6 % for DMSO. A 2.25 mM stock of ATP was prepared in H2O and 10 μL of this was added to all wells of the assay plates using a Thermo Multidrop dispenser (Thermo Fisher Scientific, Waltham, MA), yielding a final ATP concentration of 0.75 mM. To columns 3–24 of the assay plates, using a Thermo Multidrop dispenser, was added 20 μL of a cocktail of containing RecA, poly(dT) ssDNA, MgOAc2 and Tris·Glycerol buffer (pH = 7.5) such that the final concentrations were 0.5 μM RecA, 5 μM-nts poly(dT), 10 mM MgOAc2, 25 mM Tris·HOAc and 5% v/v glycerol. For the negative control reactions, 20 μL of an identical solution containing no poly(dT) was added to columns 1 and 2 using a Thermo Multidrop dispenser. To ensure the uniformity of results, the assay plates were processed in batches of 10. Three Multidrop dispenser stations containing either ATP, the reaction cocktail or the negative control cocktail were placed in close proximity. The assay plates were serially moved between the Multidrop stations, each plate requiring 30 s to fill completely. An entire batch of 10 plates was processed in 5 min, and upon addition of all reagents the plates were transferred to a 37 °C air incubator and the ATPase reaction was allowed to proceed for 35 min. Subsequently, the plates were removed from the incubator and the reactions were stopped by the addition of 30 μL of the 1× phosphomolybdate blue dye reagent to all wells of the batch of 10 plates in the same order the reaction cocktail was added. The A650 signals of the stopped reactions were stable for at least 30 min, and all 10 assay plates could be easily measured in this window. Subsequent batches of 10 plates were staggered by 15 to 20 min, allowing the operator sufficient time to manage the manipulation of two batches of 10 plates simultaneously.
Typically, the plates were scanned for absorbance at 650 nm after standing 5 min at room temperature using a Spectramax Plus384 microplate reader (Molecular Devices, Sunnyvale, CA). Screening data informatics were processed using ActivityBase, XLfit (ID Business Solutions, Bridgewater, NJ) and Microsoft Excel 2003 (Microsoft, Redmond, WA). The percent-inhibition of RecA ATPase activity was analyzed on a plate-to-plate basis by comparing the A650 value per compound well with the plate-averaged control wells using the following relationship:
| Eq. (1) |
where A650 is the well-specific absorbance value, μmin is the plate-averaged minimum signal control value, and μmax is the plate-averaged maximum signal control value. No plate corrections were needed between run-sets.
To assess the quality of the HTS, determined the Z′ and Z-factor for each assay plate using the following formulas:16
| Eq. (2) |
| Eq. (3) |
where σmax and σmin are defined as the calculated standard deviations in the plate positive and negative controls, σCMPD is the standard deviation in the compound data, σPC is the standard deviation in the 100% inhibition control, μCMPD and μPC are the respective average values for the compound data and the 100% inhibition control.
Small molecule inhibition of RecA-mediated three-strand exchange reaction
The ϕχ174 RFI dsDNA was linearized to the RFIII form using XhoI endonuclease (New England Biolabs, Ipswich, MA) and subsequently gel-purified. Single-stranded DNA binding protein (SSB) was purchased from Promega (Madison, WI). Strand exchange promoted by RecA was monitored essentially as previously described.17 Briefly, 10 μM RecA was incubated at 37 °C for 10 min with 20 μM-nts ϕχ174 cssDNA in 1× Reaction Buffer (25 mM Tris·HOAc, 5% glycerol, pH = 7.5) with 10 mM Mg(OAc)2, 12 mM phosphocreatine, 10 U/mL creatine phosphokinase and in the presence or absence of 25 μM A1. After the initial incubation, 20 μM-nts ϕ χ174 linear dsDNA form III was added and the mixture was incubated for another 10 min at 37 °C. During this second incubation, 10 μL was removed as the 0 min aliquot and added to 3.3 μL stop dye (60 mM EDTA, 5% (w/v) SDS, 25% (w/v) glycerol, 0.2% bromophenol blue) to inactivate RecA and stop the strand exchange reaction. To initiate the strand exchange reaction 3 mM ATP and 2 μM SSB were added then placed in a Thermomixer R microplate shaker/incubator (Thermo Fisher Scientific, Waltham, MA) incubated at 37 °C. Aliquots (10 μL) were taken at 10, 30, 60 and 90 min and added to 3.3 μL of stop dye after the addition of ATP. All of the aliquots were run on a 0.8% agarose gel for 12 h at 30 V, then stained for 1.5 h with Sybr Gold (Invitrogen, Carlsbad, CA) for visualization.
Small molecule inhibition of RecA-dependent SOS activation
We assessed the ability of compound A1 to impact the RecA-dependent induction of the SOS response using E. coli K-12 having the sulA SOS promoter fused to the green fluorescent protein (gfp) reporter gene inserted at attλ on the chromosome (strain SS996).15 Fresh 2 mL LB cultures were inoculated with saturated overnight cultures of E. coli SS996 to OD600 = 0.3 in the presence and absence of 100 ng/μL ciprofloxacin and 100 μM A1. The highest final concentration of DMSO attained from the addition of these compounds was 0.15% and considered negligible. The cultures were grown in a shaking incubator at 37 °C for 90 min. At the end of this incubation, the cells were stained for 5 min at 25 °C directly in the LB medium with the addition of 3.7% formaldehyde and 10 μg/mL 4',6-diamidino-2-phenylindole (DAPI). Following staining, 1.6 mL of cells was centrifuged at 13,000 rpm (RPM) for 1 min and the supernatant was removed. The cells were washed three times with 10 mM MgSO4 (1 mL) to remove excess DAPI and finally resuspended in 10 mM MgSO4 (500 μL) for imaging.
The cells were diluted (1:10) in clear, flat-bottom 384-well imaging microplates (Greiner, Monroe, NC) in a volume of 50 μL, sealed and centrifuged for 15 min at 3000 rpm and imaged in a BD pathway 855 high-content epifluorescence confocal microscope, equipped with an Olympus 40X/0.90NA objective and a Hamamatsu Orca CCD camera. Multiplexed two-color images were acquired using the following filters; GFP: 488 nm/10 nm bandwidth excitation filter, FURA/FITC epifluorescence dichroic and 515 LP emission filter; DAPI: 380 nm/10 nm bandwidth excitation filter, 400 dclp dichroic and 435 nm long-pass emission filter. Bacterial cell binary masks were generated using the Attovision software (BD Biosciences, San Jose, CA) to delineate the cell boundaries using the DAPI channel. A normalized GFP intensity was calculated by integrating the GFP signal divided by the integrated DAPI intensity within the cell boundary. The average normalized GFP response per bacterium for a particular well was calculated and compared to other wells.
RESULTS
Assay optimization and validation
RecA is activated in the presence of ssDNA and ATP, forming the RDF that hydrolyzes ATP. Therefore, ATP hydrolysis serves as a useful indicator of RecA activation, and we chose to monitor the reduction of ATPase activity as a diagnostic for small-molecule mediated inhibition of RecA by in vitro screening. Although we successfully used a validated enzyme-linked fluorogenic ATPase assay18 to screen 2180 compounds from the National Cancer Institute against RecA (data not shown), we considered more cost-effective alternative assays. We have experience with several suitable assay technologies,18–20 including the well-known Malachite Green assay,19 but we elected adapt a phosphomolybdate blue (PMB) colorimetric phosphate detection assay21,22 to identify compounds that abrogated ATP hydrolysis by RecA. While the fluorogenic assay results can be observed in real-time, the PMB assay can be performed for roughly 2% of the cost per well and, in our hands, provides more reliable reaction endpoint data than the Malachite Green assay (Z' 0.73 vs. 0.68; see below).
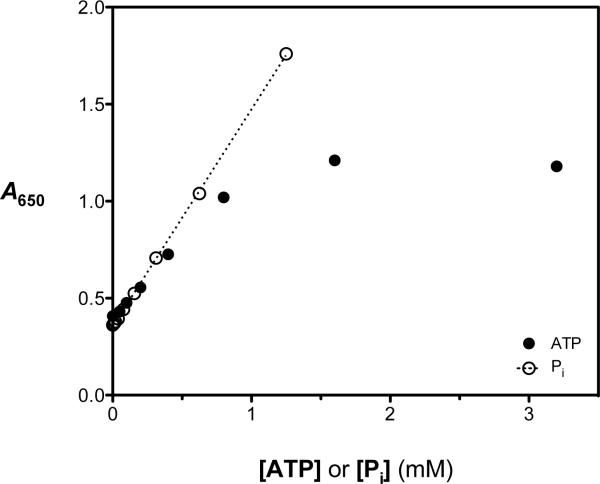
The PMB ATPase assay relies on the reaction of a molybdate-ascorbic acid complex with inorganic phosphate (Pi) to produce an aggregate phosphomolybdate complex having a strong absorbance in the 600 to 700 nm range. The A650 signal of the PMB phosphate detection assay remained linear to at least 1.25 mM Pi (Figure 1). Optimization of the assay protocol was undertaken to maximize the signal-to-noise ratio, which represents a challenge when using RecA because the equilibrium constant for its self-association during filament assembly and activation is modest. Thus, in spite of a relatively low turnover number for ATP hydrolysis (kcat ≤ 0.5 s−1), the absolute rate of ATP hydrolysis is high under conditions where RecA-DNA filament formation is favorable.
Figure 1.
Evaluation of the dependence of the phosphomolybdate blue (PMB) ATPase assay signal on free phosphate (open circles) and initial ATP (filled circles) concentrations under otherwise optimized conditions. In the presence of RecA (0.5 μM) and poly(dT) (5 μM-nts), the PMB assay reagent produced a linear colorimetric response (absorbance at 650 nm, A650) when free phosphate (Pi; open circles) was added up to 1.25 mM. In separate experiments, the RecA-DNA ATPase reaction was allowed to proceed with various starting ATP concentrations (0.01 – 3.5 mM). After 35 min, the reactions were stopped by addition of the PMB assay reagent and the A650 signal was measured. The A650 values were plotted as a function of initial [ATP] (filled circles), and the results demonstrate that the assay signal is linear with ATP concentration up to 0.8 mM but is substantially nonlinear between 0.8 and 3.2 mM ATP.
Guided by our experience with the ssDNA-dependent ATP hydrolysis reaction catalyzed by RecA,18,19 optimal conditions producing A650 signals in the assay's linear range in 384-well plates were identified when RecA, poly(dT) and ATP were present at 0.5 μM, 5 μM-nts and 0.75 mM, respectively (data not shown). Importantly, to keep the free phosphate concentration in the linear dynamic range of the assay reagent, the time of the reactions had to be limited to no more than 40 min; however, we elected to use 35 min as the longest allowable reaction time comfortably within the assay's linear dynamic range. Using the PMB assay, single time-points were taken in a 384-well plate by the addition of the PMB reagent solution to stop the ATPase reaction, and free phosphate was measured by comparison to a phosphate standard curve. Absorbance-versus-time plots were constructed for the hydrolysis reaction in the presence of various initial ATP concentrations and the data were analyzed as previously described to ensure that established kinetic parameters for steady-state ATP hydrolysis by RecA were reproduced.19 The RecA concentration (0.5 μM) was the minimal value necessary to achieve reproducible steady-state ATPase activity and the poly(dT) concentration (5 μM-nts) was just above that necessary to saturate the RecA under these reaction conditions.18,19 Above an initial ATP concentration of 0.75 mM, the A650 signal corresponding to generation of free phosphate increased non-linearly with increasing Pi concentration (Figure 1), due to the build-up of ADP, a natural feedback inhibitor of RecA-catalyzed ATP hydrolysis. Under these optimized conditions at 37 °C, neither the extent or kinetics of the reaction demonstrated sensitivity to the presence of DMSO, up to 10% (v/v).
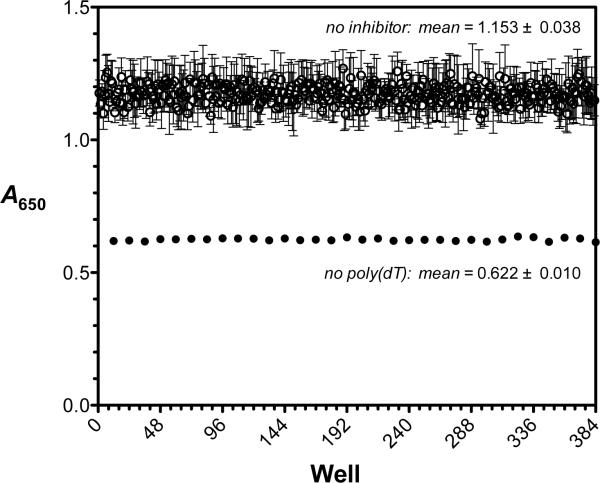
To evaluate the PMB ATPase assay for robust and reproducible behavior in a 384-well format, we compared the signal-to-background and signal-to-noise ratios as well as the Z' factors16 calculated across a 384-well microplate. Over a span of five days, three replicate assays were performed under the optimized conditions described above to test for interday, interplate and intercolumn variability. The A650 signal at 35 min was taken as the positive control value, the value at 35 min when poly(dT) was omitted from the reaction was taken as the negative control, and the mean positive control and background signals, as well as the coefficients of variation in each, were compiled for each of three assays (Figure 2). The overall quality of the PMB RecA HTS ATPase assay was assessed using the Z′ factor, which defines the difference between the positive and negative controls of the dynamic signal being measured and the data variation of that signal. Robust and reproducible assays have a Z′ factor ranging from 0.5 to 1. Consistent with a high-quality assay, the Z′ factor was 0.73 (Figure 2).
Figure 2.
Reproducibility of the phosphomolybdate blue (PMB) assay for RecA-DNA ATPase activity under optimal conditions. The absorbance at 650 nm (A650) was measured after 35 min in each well of a 384-well plate in the presence (open circles) or absence (filled circles) of 5 μM-nts poly(dT). The concentration of ATP was 0.75 mM in the experiments. Each data point indicates the A650 signal for each well from three different experiments on three different days; the error bars indicate the standard deviation of each mean value. The overall means for the positive signal control (no inhibitor added; open circles) and negative background control (no poly(dT) added; filled circles) were 1.153 ± 0.038 (CV = 3.3%) and 0.622 ± 0.010 (CV = 1.6%), respectively. The signal-to-background ratio was 1.8 and the signal-to-noise ratio was 14. The overall Z' factor was 0.73.
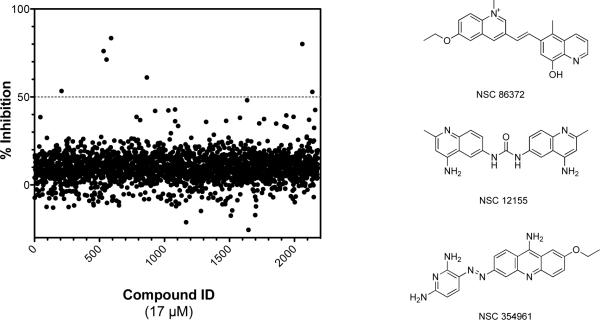
As further validation, the PMB assay was used to screen 2180 compounds combined from the NCI Challenge, Diversity and Natural Product library sets and the results were compared to those obtained using the previously validated fluorogenic ATPase assay.18 For this comparative analysis, we considered different cut-off values for identifying primary hits and selected 50% inhibition as the cut-off for both assays. This criterion provided maximum overlap among the bona fide hits from the two assays. Using this cut-off, the PMB ATPase assay resulted in a 0.32% hit rate, yielding seven primary hits above 50% inhibition (Figure 3), four of which were confirmed as bona fide hits in an IC50 study. Of the four confirmed hits, three were found in common between both the fluorogenic ATPase assay and the PMB ATPase assay. Remarkably, the three hits common to both assays shared a common chemotype (Figure 3). The ability of the PMB ATPase assay to mine the same hits from the NCI library as the fluorogenic ATPase assay18 confirmed that the former provides a reliable method for identifying inhibitors of RecA in multi-thousand compound libraries.
Figure 3.
Results from the screen of three National Cancer Institute libraries. A scatter plot of the relative inhibition (%) effected by 2180 compounds combined from three NCI libraries against RecA using the phosphomolybdate blue ATPase assay. Reactions proceeded for 35 min at 37 °C and contained RecA (0.5 μM), poly(dT) (5 μM-nts), ATP (0.75 mM), and assessed compounds (final concentration 17 μM). There were seven primary hits above a threshold of 50% inhibition. The structures of three confirmed hits sharing a similar scaffold from the National Cancer Institute screen are depicted at right. The IC50 values characterizing the three hits ranged from 4 to 6 μM.
Screening
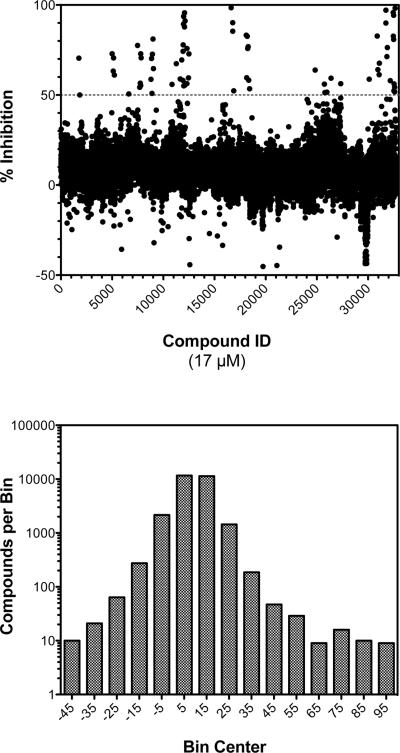
Although the NCI compounds used to validate the PMB ATPase assay inhibited RecA ATP hydrolysis in vitro with IC50 values between 4 to 6 μM, they failed to demonstrate an appreciable effect on the biological activities of RecA in liquid cultures of E. coli (data not shown). Nevertheless, the ability to efficiently identify these compounds indicated that the PMB ATPase assay was suitable for use with large chemical libraries. Therefore, we elected to screen a more diverse combinatorial library donated to the BRITE Center by Biogen Idec and comprising over 350,000 compounds. Using identical compound and reagent concentrations, we performed a screen of 33,600 representative compounds from this library. This “diversity” subset was selected to span the overall range of chemical diversity space represented in the larger collection (data not shown). The HTS assay of the compounds yielded a range of inhibitor activities, with > 91% of the compounds characterized by a relative inhibitory activity between −10% and 20% (Figure 4).
Figure 4.
A total of 33,600 compounds were selected from a donated Biogen Idec library to represent the overall chemical diversity of the library and were screened using the phosphomolybdate blue ATPase assay. Reactions proceeded for 35 min at 37 °C and contained RecA (0.5 μM), poly(dT) (5 μM-nts), ATP (0.75 mM), and assessed compounds (final concentration 17 μM). In the upper panel, the screening results are represented as a scatter plot of relative inhibition (%) effected by each compound. There were 73 primary hits above a threshold of 50% inhibition. In the lower panel, a frequency distribution of the assay results is depicted as a histogram, where the number of compounds in each bin (± 5% width) is plotted on a logarithmic scale.
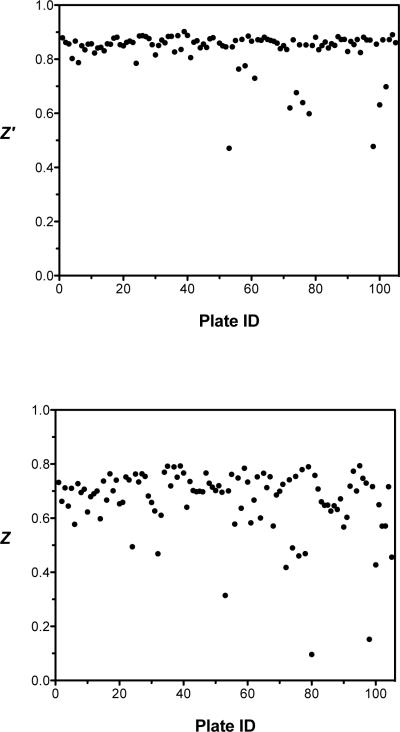
Overall, the HTS assay yielded an average Z′ factor of 0.83 and a Z factor of 0.67, demonstrating robust and reproducible behavior from plate to plate (Figure 5). Three plates had a Z factor below 0.4 and were rejected during data analysis. At a threshold of 50% inhibition, 73 hits (0.22% hit rate) were identified (Figure 4). Interestingly, 71 of the 73 primary hits could be categorized into one of five chemotype clades based on their molecular similarity. These hit groupings will be referred to as clades A to E, with 31, 4, 22, 9 and 5 compounds in clades A, B, C, D and E, respectively. The fact that multiple hits arise from the same chemotype was apparent in the raw data (Figure 4), in which groups of hits were observed on the same plate. This pattern likely results from the original grouping of similar compounds in the donated collection.
Figure 5.
Statistical analyses of each 384-well plate to determine the quality of the HTS assay performed on the Biogen Idec compounds. Z′ factor analysis was performed on each plate and the average Z′ factor value was 0.83 (upper panel). Z factor analysis was performed on each plate and the average Z factor value was 0.67 (lower panel). Three plates had a Z factor below 0.4 and were rejected during data analysis.
We independently synthesized nine compounds from clades A to C, and performed an IC50 study using the PMB ATPase assay to verify them as hits. IC50 values could be accurately measured for all compounds, with A1 – A4 yielding IC50 values of 8, 20, 22 and 24 μM, B1 – B4 yielding IC50 values of 21, 21, 71 and 104 μM, and C1 having an IC50 of 19 μM. Extrapolating these observations to the rest of the 71 primary hits belonging to one of the five chemotype clades, we conclude that the PMB ATPase effectively mined the 33,600 member library for structurally related hits, and the identification of false positives was minimized. The most potent hit, A1 (IC50 = 8 μM), was subjected to rigorous analysis of its activity against an in vitro RecA-mediated strand-exchange reaction and SOS induction in live bacterial cell cultures as described below.
Inhibition of RecA-catalyzed DNA three strand-exchange by clade A compounds
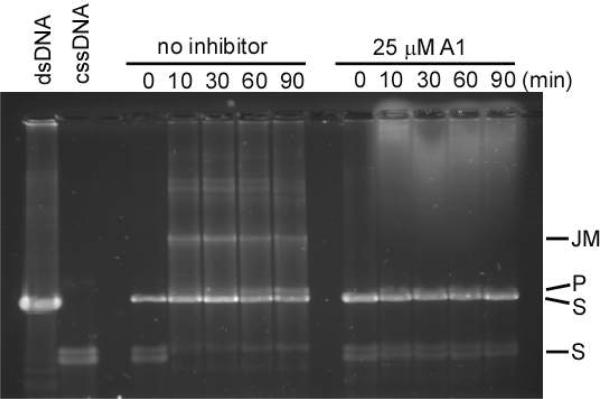
At a cellular level, RecA catalyzes a strand exchange reaction that is used to repair extensive DNA damage with homologous DNA. To determine if compound A1 could inhibit this biologically important reaction, it was assessed for inhibition of the RecA-mediated strand exchange reaction using an established in vitro assay that serves as a paradigm for the physiologic recombinational functions of RecA.17 In the assay, RecA is supplied with ϕχ174 cssDNA, linear dsDNA and ATP. Active RecA catalyzes the rapid exchange of a DNA strand, thereby converting the substrates into new linear ssDNA and nicked circular dsDNA products that are characterized by differences in their migration rates during gel electrophoresis. Because ATP hydrolysis by RecA is an absolute requirement for strand exchange,13 we examined the ability of A1 to interfere with this reaction. When A1 was present at 25 μM, the reaction did not proceed at all, indicated by the absence of both three DNA-strand intermediates bound to RecA as well as the newly resolved DNA products (Figure 6).
Figure 6.
The RecA mediated three-strand exchange reaction is inhibited by A1. In the absence of inhibitor, the formation of joint molecules intermediates (JM) and new nicked circular dsDNA product (P) from circular dsDNA and homologous cssDNA substrates (S) is observed. In the presence of A1 (25 μM) however, JM and P are not observed.
Clade A compounds attenuate ciprofloxacin-induced SOS gene expression
After demonstrating that clade A compounds were potent inhibitors of RecA's in vitro ATPase and recombinase activities, we examined their effect on RecA-dependent induction of the SOS response in liquid cultures of E. coli. The Sandler group recently reported the ability to measure SOS gene expression in E. coli K-12 having a sulA-gfp fusion reporter gene inserted into the chromosomal DNA (strain SS996).15 The sulA promoter is LexA-regulated, and is induced 100-fold during the SOS response.15 We used SS996 cells to determine the differential SOS expression effected by A1 following treatment with ciprofloxacin, a topoisomerase II specific DNA-damaging antibiotic known to stimulate de-repression of the SOS regulon.23 The RecA-dependent expression of GFP (corresponding to SOS induction) could be measured amongst a population of bacteria exposed to ciprofloxacin and comparatively analyzed to bacterial cultures given a combination of ciprofloxacin and A1.
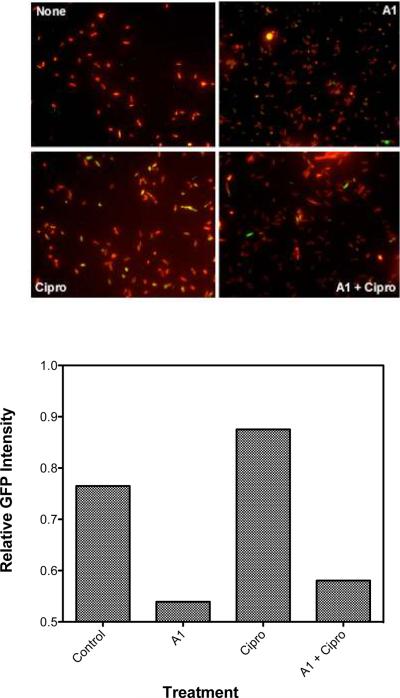
Cultures of SS996 were given either no compound, A1 only, ciprofloxacin only or a combination of A1 and ciprofloxacin and allowed to incubate for 90 min at 37 °C. The bacteria were then fixed and stained with formaldehyde and DAPI, then examined using a 40X objective and filters selected for DAPI and GFP. The fluorescence intensity of the DAPI and GFP channels was analyzed and a ratio of GFP/DAPI intensity was calculated to determine the relative GFP expression per cell (Figure 7). There was a noticeable decrease in SOS expression in the presence of A1. Untreated bacteria had a relative GFP expression of 0.76, while those treated with ciprofloxacin saw this value increase to 0.87. In the presence of ciprofloxacin and A1, the relative GFP expression was observed to drop to 0.58. Accordingly, at a value of 0.53, the bacteria treated with only A1 exhibited the lowest relative GFP expression. The apparent effect of A1 alone was unexpected. We tentatively attribute the observation that SOS was induced to a modest degree in the absence of ciprofloxacin to one or a combination of the following facts: (1) many replication forks are inactivated in aerobically growing bacteria even in the absence of SOS-inducing conditions;24 and (2) any one of a number of environmental stresses, including oxidative stress, osmotic stress, acoustic cavitations, pH changes, and high pressure, can induce the SOS response.25
Figure 7.
Inhibition of SOS induction by A1. Cultures of E. coli K-12 (2 mL, OD600 = 0.3) containing a RecA/LexA-controlled sulA-gfp fluorescent reporter gene were given no compound, A1 (100 μM) only, ciprofloxacin (100 ng/mL) only, or a combination of the two and incubated for 90 min at 37 °C. Then, the bacteria were briefly stained with DAPI and immediately examined under a high-content epifluorescence microscope in 384-well microplates using filters selected for DAPI and GFP (left). The intensity of the GFP signal was normalized to the DAPI signal for all bacteria in each image and a ratio of GFP/DAPI intensity was calculated to yield the total GFP response (right). A1 inhibited both wild type and ciprofloxacin-induced SOS expression to similar levels.
DISCUSSION
The development of drug-resistant bacteria is an unavoidable outcome of antibacterial chemotherapy of infectious diseases. Reducing the rate and extent of development of resistance could have profound influence on human health. Although the mechanisms that facilitate the de novo development, clonal spread, and horizontal transfer of resistance factors are not fully understood, the rapid rate at which antibiotic-resistant pathogens arise is likely due to a combination of genetic mutation introduced by stress-induced non-replicative polymerases and gene transfer from commensal organisms. Recently, RecA has emerged as a crucial player in these phenomena as well as in the modulation of cell killing by bactericidal antibiotics.
One of our labs has undertaken a systematic approach towards developing inhibitors of RecA that could attenuate bacterial DNA repair and stress-induced SOS mutagenesis, thereby diminishing the organism's inherent antibiotic tolerance and its ability to evolve resistance. Transition metal complexes,26 nucleotide analogs,12,19,20 structured peptides,27 and polysulfated naphthyl compounds18 have previously been identified as inhibitors of the in vitro activities of RecA. Unfortunately, these compounds were not cell-permeable, or were determined to have a pleiotropic effect on live bacteria, and the discovery of more drug-like small molecule inhibitors of RecA was desirable. Towards this end, a well-known phosphomolybdate blue phosphate detection assay21,22 was adapted to screen a collection of 35,780 small molecules for inhibition of ATP hydrolysis by RecA, resulting in the discovery of six classes of small molecules that inhibited RecA in the range of 10 to 30 μM. The most potent class of inhibitors, clade A compounds, were examined more closely to determine their effect on the biological activities of RecA that lead to antibiotic tolerance and resistance.
The first such activity of RecA associated with antibiotic survival is its ability to catalyze a DNA strand exchange reaction that is employed in the recombinational repair of extensive chromosomal damage.11 We examined the inhibitory activity of A1 using an in vitro assay that provides a paradigm for the in vivo recombinational activity of RecA.17 The presence of A1 at 25 μM completely inhibited the RecA-catalyzed strand exchange reaction.
At a physiologic level, RecA-catalyzed strand exchange can lead to two outcomes depending on the source of the homologous dsDNA: (1) the DNA may originate from a sister chromatid present during replication, replacing the damaged DNA with an exact copy or (2) the DNA may originate from a foreign source taken up through transformation, transduction or conjugation, and introduce genetic variation. The former possibility restores DNA homeostasis with wild-type genes and allows bacteria to tolerate higher doses of antibiotics that precipitate DNA damage,11 while the latter possibility leads to horizontal transfer of genes that may confer antibiotic resistance.28 Therefore, we are currently investigating the ability of A1 to interfere with these processes that are fundamental to the survival of bacteria in the face of antibacterial exposure.
The second activity of RecA implicated in the antibiotic survival response is its ability to induce the SOS regulon by stimulating the autoproteolysis of the LexA repressor.10,23,29–31 In E. coli, the SOS response is characterized by the temporal activation of up to 40 genes that gradually increase the severity of the response taken towards DNA damage.32 At first, the early SOS gene products are involved in the repair of moderate DNA damage by nucleotide excision (e.g., UvrABCD) and recombination (e.g., RecA),33 but damage that is irreparable by these means or that persists for an extended period causes the late SOS genes to be expressed. Late SOS gene products include the error-prone DNA polymerases, PolIV and PolV, which promote global mutagenesis in an apparent last-ditch effort to resume normal cellular function.33 The induction of the SOS response is quickly amplified by severe DNA damage, as RecA is over-expressed in the intermediary stages of the pathway. Hence, it is likely that the RecA-activated SOS response initially allows for higher tolerance to antibacterials by up-regulating DNA repair pathways, but can ultimately lead to antibacterial resistance by means of up-regulating genomic mutation and horizontal gene transfer if its expression is sustained.
To assess whether inhibitor A1 could attenuate SOS stimulated by the bactericidal agent ciprofloxacin, RecA-dependent SOS gene expression was quantified in live cultures of E. coli K-12 cells having a sulA-gfp fusion.15 We found that A1 was capable of substantially reducing the induction of SOS under conditions simulating natural environmental stress and also under conditions of genomic stress stimulated by the DNA damaging antibiotic ciprofloxacin.
In summary, we have used a high-throughput target-based screening approach to identify a class of compounds, best represented by its highest-affinity member A1, that has the following activities: inhibition of RecA's ssDNA-dependent ATPase activity in vitro, inhibition of RecA's DNA strand exchange activity in vitro, and attenuation of ciprofloxacin-induced SOS gene expression in live E. coli cells. Overall, these results represent the first proof-of-concept that a small-molecule inhibitor targeting RecA can attenuate the SOS response in living bacteria. Although, the experiments described herein have focused on the effect of ciprofloxacin on E. coli, there is compelling evidence suggesting that this strategy may be generalized to other antibacterials and other organisms.
While the mechanism by which quinolone antibacterials such as ciprofloxacin result in RecA-activating persistent ssDNA is implied by their cellular target, recent data from the Collins laboratory demonstrated that, in E. coli, members of three major classes of bactericidal agents result in the production of DNA-damaging hydroxyl radicals.9 The same study also showed that E. coli with loss-of-function recA mutations are sensitized to cell killing by all three classes of bactericidal agents. This suggests that a cell-permeable RecA inhibitor may enhance the potency of a broad spectrum of antibacterial agents.
In a formal sense, RecA is not essential for bacterial growth on supplemented laboratory media. Nevertheless, bacteria having loss-of-function mutations in the recA gene have yet to isolated from clinical sources and in vitro studies indicate that such RecA-deficient bacteria are sensitized to antibiotic treatment9 and develop resistance much more slowly or not at all (Singleton et al., unpublished results). Indeed, RecA is nearly ubiquitous among bacteria and has one of the slowest evolutionary rates.34 RecA's high degree of conservation suggests that it likely plays roles in other species similar to those in E. coli.11 In the rare bacterial species lacking a defined SOS response, RecA is heavily relied upon for DNA repair and other biological processes that may promote survival and even pathogenesis in a human host.35,36 Taken together, these data suggest that RecA may provide an ideal candidate for target-based drug discovery of inhibitors that potentiate cell killing by frontline bactericidal agents and attenuate the development of resistance in a broad spectrum of bacterial pathogens.
Acknowledgments
The authors wish to thank Professor Steven Sandler (University of Massachusetts at Amherst) for the gracious gift of E. coli K-12 with sulA-gfp reporter fusions. This work was supported by a grant to S.F.S. from the National Institutes of Health (GH5B114).
REFERENCES
- 1.Diekema DJ, BootsMiller BJ, Vaughn TE, Woolson RF, Yankey JW, Ernst EJ, Flach SD, Ward MM, Franciscus CL, Pfaller MA, Doebbeling BN. Antimicrobial Resistance Trends and Outbreak Frequency in United States Hospitals. Clin. Infect. Dis. 2004;38(1):78–85. doi: 10.1086/380457. [DOI] [PubMed] [Google Scholar]
- 2.Talbot GH, Bradley J, Edwards JE, Jr., Gilbert D, Scheld M, Bartlett JG. Bad Bugs Need Drugs: An Update on the Development Pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 2006;42(5):657–668. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 3.Power E. Impact of Antibiotic Restrictions: The Pharmaceutical Perspective. Clin. Microbiol. Infect. 2006;12(Suppl 5):25–34. doi: 10.1111/j.1469-0691.2006.01528.x. [DOI] [PubMed] [Google Scholar]
- 4.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for Bad Bugs: Confronting the Challenges of Antibacterial Discovery. Nat. Rev. Drug Discov. 2007;6(1):29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 5.Lock RL, Harry EJ. Cell-Division Inhibitors: New Insights for Future Antibiotics. Nat. Rev. Drug Discov. 2008;7(4):324–338. doi: 10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- 6.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia Coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottarel G, Wierzbowski J. Combination Drugs, an Emerging Option for Antibacterial Therapy. Trends Biotechnol. 2007;25(12):547–555. doi: 10.1016/j.tibtech.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Power EG, Phillips I. Induction of the Sos Gene (Umuc) by 4-Quinolone Antibacterial Drugs. J. Med. Microbiol. 1992;36(2):78–82. doi: 10.1099/00222615-36-2-78. [DOI] [PubMed] [Google Scholar]
- 9.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell. 2007;130(5):797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 10.Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of Mutation and Combating the Evolution of Antibiotic Resistance. PLoS Biol. 2005;3(6):e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roca AI, Cox MM. Reca Protein: Structure, Function, and Role in Recombinational DNA Repair. Prog. Nucleic Acid Res. Mol. Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 12.Lee AM, Ross CT, Zeng BB, Singleton SF. A Molecular Target for Suppression of the Evolution of Antibiotic Resistance: Inhibition of the Escherichia Coli Reca Protein by N(6)-(1-Naphthyl)-Adp. J. Med. Chem. 2005;48(17):5408–5411. doi: 10.1021/jm050113z. [DOI] [PubMed] [Google Scholar]
- 13.Gruenig MC, Renzette N, Long E, Chitteni-Pattu S, Inman RB, Cox MM, Sandler SJ. Reca-Mediated Sos Induction Requires an Extended Filament Conformation but No Atp Hydrolysis. Mol. Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singleton SF, Simonette RA, Sharma NC, Roca AI. Intein-Mediated Affinity-Fusion Purification of the Escherichia Coli Reca Protein. Protein Expr. Purif. 2002;26(3):476–488. doi: 10.1016/s1046-5928(02)00571-5. [DOI] [PubMed] [Google Scholar]
- 15.McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. Measurement of Sos Expression in Individual Escherichia Coli K-12 Cells Using Fluorescence Microscopy. Mol. Microbiol. 2004;53(5):1343–1357. doi: 10.1111/j.1365-2958.2004.04225.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 17.Roman LJ, Kowalczykowski SC. Relationship of the Physical and Enzymatic Properties of Escherichia Coli Reca Protein to Its Strand Exchange Activity. Biochemistry. 1986;25(23):7375–7385. doi: 10.1021/bi00371a020. [DOI] [PubMed] [Google Scholar]
- 18.Wigle TJ, Singleton SF. Directed Molecular Screening for Reca Atpase Inhibitors. Bioorg. Med. Chem. Lett. 2007;17(12):3249–3253. doi: 10.1016/j.bmcl.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee AM, Wigle TJ, Singleton SF. A Complementary Pair of Rapid Molecular Screening Assays for Reca Activities. Anal. Biochem. 2007;367(2):247–258. doi: 10.1016/j.ab.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wigle TJ, Lee AM, Singleton SF. Conformationally Selective Binding of Nucleotide Analogues to Escherichia Coli Reca: A Ligand-Based Analysis of the Reca Atp Binding Site. Biochemistry. 2006;45(14):4502–4513. doi: 10.1021/bi052298h. [DOI] [PubMed] [Google Scholar]
- 21.Lin TI, Morales MF. Application of a One-Step Procedure for Measuring Inorganic Phosphate in the Presence of Proteins: The Actomyosin Atpase System. Anal. Biochem. 1977;77(1):10–17. doi: 10.1016/0003-2697(77)90284-6. [DOI] [PubMed] [Google Scholar]
- 22.Hergenrother PJ, Haas MK, Martin SF. Chromogenic Assay for Phospholipase D from Streptomyces Chromofuscus: Application to the Evaluation of Substrate Analogs. Lipids. 1997;32(7):783–788. doi: 10.1007/s11745-997-0101-5. [DOI] [PubMed] [Google Scholar]
- 23.Cirz RT, Jones MB, Gingles NA, Minogue TD, Jarrahi B, Peterson SN, Romesberg FE. Complete and Sos-Mediated Response of Staphylococcus Aureus to the Antibiotic Ciprofloxacin. J. Bacteriol. 2007;189(2):531–539. doi: 10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The Importance of Repairing Stalled Replication Forks. Nature. 2000;404(6773):37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 25.Erill I, Campoy S, Barbe J. Aeons of Distress: An Evolutionary Perspective on the Bacterial Sos Response. FEMS Microbiol. Rev. 2007;31(6):637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee AM, Singleton SF. Inhibition of the Escherichia Coli Reca Protein: Zinc(Ii), Copper(Ii) and Mercury(Ii) Trap Reca as Inactive Aggregates. J. Inorg. Biochem. 2004;98(11):1981–1986. doi: 10.1016/j.jinorgbio.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 27.Cline DJ, Holt SL, Singleton SF. Inhibition of Escherichia Coli Reca by Rationally Redesigned N-Terminal Helix. Org. Biomol. Chem. 2007;5(10):1525–1528. doi: 10.1039/b703159a. [DOI] [PubMed] [Google Scholar]
- 28.Hastings PJ, Rosenberg SM, Slack A. Antibiotic-Induced Lateral Transfer of Antibiotic Resistance. Trends Microbiol. 2004;12(9):401–404. doi: 10.1016/j.tim.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Cirz RT, Gingles N, Romesberg FE. Side Effects May Include Evolution. Nat. Med. 2006;12(8):890–891. doi: 10.1038/nm0806-890. [DOI] [PubMed] [Google Scholar]
- 30.Cirz RT, O'Neill BM, Hammond JA, Head SR, Romesberg FE. Defining the Pseudomonas Aeruginosa Sos Response and Its Role in the Global Response to the Antibiotic Ciprofloxacin. J. Bacteriol. 2006;188(20):7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cirz RT, Romesberg FE. Controlling Mutation: Intervening in Evolution as a Therapeutic Strategy. Crit. Rev. Biochem. Mol. Biol. 2007;42(5):341–354. doi: 10.1080/10409230701597741. [DOI] [PubMed] [Google Scholar]
- 32.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative Gene Expression Profiles Following Uv Exposure in Wild-Type and Sos-Deficient Escherichia Coli. Genetics. 2001;158(1):41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley WL. Lex Marks the Spot: The Virulent Side of Sos and a Closer Look at the Lexa Regulon. Mol. Microbiol. 2006;62(5):1228–1238. doi: 10.1111/j.1365-2958.2006.05444.x. [DOI] [PubMed] [Google Scholar]
- 34.Rocha EP, Cornet E, Michel B. Comparative and Evolutionary Analysis of the Bacterial Homologous Recombination Systems. PLoS Genet. 2005;1(2):e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kline KA, Sechman EV, Skaar EP, Seifert HS. Recombination, Repair and Replication in the Pathogenic Neisseriae: The 3 R's of Molecular Genetics of Two Human-Specific Bacterial Pathogens. Mol. Microbiol. 2003;50(1):3–13. doi: 10.1046/j.1365-2958.2003.03679.x. [DOI] [PubMed] [Google Scholar]
- 36.Kramer N, Hahn J, Dubnau D. Multiple Interactions among the Competence Proteins of Bacillus Subtilis. Mol. Microbiol. 2007;65(2):454–464. doi: 10.1111/j.1365-2958.2007.05799.x. [DOI] [PubMed] [Google Scholar]