Abstract
PDZ proteins coordinate assembly of protein complexes that participate in diverse biological processes. GIPC is a multifunctional PDZ protein that interacts with several soluble and membrane proteins. Unlike most PDZ proteins, GIPC contains single PDZ domain and the mechanisms by which GIPC mediates its actions remain unclear. We investigated the possibility that in lieu of multiple PDZ domains, GIPC forms multimers. Here, we demonstrate that GIPC can bind to itself and that the PDZ domain is involved in GIPC–GIPC interaction. Gel filtration, sucrose gradient centrifugation and chemical cross-linking showed that whereas bulk of cytosolic GIPC was present as monomer, oligomers with an estimated molecular mass corresponding to GIPC homotrimer were readily detectable in the membrane fraction. Modeling of GIPC PDZ domain showed feasibility of trimerization. Immunogold electron microscopy showed that GIPC is present in clusters near vesicles. Our data suggest that oligomers of GIPC mediate its functions in melanocytes.
Keywords: PDZ proteins, GIPC, Protein folding, Oligomerization, Chemical cross-linking, Computational analysis
PDZ1 (Postsynaptic density-95/DISC large/ZO1) domain proteins, play important roles in assembling large signaling complexes involved in diverse biological processes such as phototransduction in Drosophila, synaptic transmission, trafficking of receptors and other membrane proteins, and maintenance of epithelial cell morphology and polarity [1–6]. PDZ protein GIPC was originally identified as a protein that interacts with GAIP (RGS 19), a regulator of G protein signaling and thought to play a role in trafficking of clathrin coated vesicles [7]. Subsequently, GIPC was shown to provide a link between signaling by NGF receptor Trk and MAP kinase pathways [8]. GIPC also binds a number of soluble and membrane bound proteins in a variety of cell types and is also known to bind certain viral oncoproteins [9–16]. We showed that in melanocytes GIPC binds C-terminus of newly synthesized melanosomal membrane protein TRP1 (tyrosinase related protein 1) in the Golgi region, and proposed a role for this interaction in trafficking of TRP1[17].
A common and striking feature of PDZ proteins, in general, is the presence of multiple PDZ domains and/or other protein–protein interacting and signaling domains within the same polypeptide. Moreover, PDZ proteins have also been shown to undergo self association to from multimers, thus generating complex signaling scaffolds [18–20]. GIPC, unlike most PDZ proteins, however, has a single PDZ domain and no other recognizable protein interacting domains. Other PDZ proteins that contain a single PDZ domain, PICK1 (protein interacting with C kinase 1) and ERBIN (ERBB2/HER2 receptor interacting protein) also have, respectively, a coiled–coiled domain and leucine-rich repeats, which are thought to mediate their functions [21,22]. The mechanisms that mediate the cell biological actions of GIPC however, remain to be understood. We hypothesized that, in addition to binding to its target protein TRP1 by PDZ domain, GIPC in melanocytes, also binds to other signaling proteins and to itself to form oligomers. In this study, we show that GIPC can bind to itself and occurs as trimers in vivo. GIPC–GIPC interaction occurs through surface interactions between PDZ domains. Immunogold electron microscopy showed clustering of GIPC molecules near intracellular membranes. Our data suggest a functional role for GIPC oligomers in trafficking of TRP1.
Materials and methods
GIPC expression plasmids
Cloning of full-length and ΔNH2-GIPC into pFLAG-CMV2 vector were described earlier [17]. N- and C-terminal deletion mutants ΔPDZ and ΔPDZ2 were generated using specific primers that incorporated a termination codon and an initiation codon at 126 and 225, respectively, and cloned into pFLAG-CMV2 vector (Sigma Aldrich Corp. St. Louis, MO). The C-terminal deletion mutant ΔACP-GIPC, lacking amino acid residues 248–333 was generated by digesting the pFLAG CMV2-GIPC plasmid with restriction enzyme Sma1 (at nucleotide 742 in the open reading frame of GIPC and the pFLAG-CMV2 vector at 1012) and the large plasmid fragment was religated generating a truncated GIPC protein with 1–247 amino acids. Expression plasmid for the fusion protein GIPC-EGFP was generated by cloning full-length GIPC into pEGFP-N3 vector (Clontech, Mountain View, CA). Mutations of cysteine residues at 100 and 189 positions to alanines were produced using QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using specific primers according to the manufacturer’s instructions.
Transfection, cell lysis and subcellular fractionation
Semi-confluent SK-MEL-23cl.22a (clone 22a) melanoma cells in 100 mm dishes were transfected with a total of 3–5µg of indicated plasmids using Lipofectamine Plus reagent (Invitrogen Life Technologies Inc., Carlsbad, CA) according to manufacturer’s instructions. Forty hours after transfection, cells were harvested, lysed in 50 mM phosphate buffer, pH 7.4, containing 1% Triton-X-100 and a mixture of protease inhibitors (Roche Diagnostics, Indianapolis, IN). Detergent lysates were cleared by centrifugation at 15,000g for 20 min. For preparation of cytosolic and membrane-bound proteins, clone 22a cells in semi-confluent 100 mm dishes were washed with ice-cold phosphate–buffered saline (PBS), harvested by scrapping, suspended in 50 mM phosphate buffer, pH 7.4 containing mixture of protease inhibitors and homogenized in Dounce homogenizer (20 strokes). Post nuclear supernatants (PNS) were centrifuged for 2h at 100,000g in a Beckman TLA-100.1 rotor at 4°C and supernatants were collected. The membrane pellet was solubilized in lysis buffer containing 1% Triton X-100 and cleared as described above. For sucrose gradient fractionation, the membrane fraction was washed with buffer containing 0.5 M NaCl for 1 h and clarified by centrifugation for 2 h at 100,000g [23]. The supernatant was collected and subjected to fractionation. The heavy membrane and light vesicle fractions were prepared by centrifuging the PNS at 10,000g for 30 min, and the supernatant (light vesicle fraction) was collected. The pellet (heavy membrane fraction) was then resuspended in SDS sample buffer. For cross-linking of GIPC with cupric orthophenanthroline (CuP), 48 h after transfection, cells were washed twice with PBS and once with 5 ml of lysis buffer (10 mM Tris–HCl, pH 7.4, and containing mixture of protease inhibitors). The cells were lysed with the lysis buffer and homogenized in Dounce homogenizer (20 strokes). The PNS was centrifuged at 100,000g for 2 h in a Beckman TLA-100.1 rotor. The resulting pellet was resuspended in buffer containing 20 mM Tris–HCl, pH 8.0, 1 mM MgCl2, 5 mM CaCl2 and 100 mM NaCl.
GST pull-down assay
GST and GST–GIPC fusion proteins were produced in Escherichia coli BL21 after induction with 0.1 mM isopropyl β-d-thiogalactopyranoside for 2 h. Cells were pelleted and resuspended in 300 µl B-PER, (Bacterial Protein Extraction Reagent, Pierce Biotechnology, Rockford, IL). Supernatants were incubated with glutathione (GSH)-Sepharose beads (Amersham Biosciences Corp., Piscataway, NJ) for 30 min and washed three times with 10 ml of PBS and resuspended in PBS. Lysates from clone 22a cells transfected with FLAG-GIPC and its deletion mutants, were prepared as described earlier. Five hundred microliter aliquots of cell lysates were incubated with 25 µg of GST protein immobilized on 50 µl of GSH–Sepharose beads for 1 h at 4°C followed by incubation with GST-fusion proteins immobilized on GSH–Sepharose beads. After extensive washing with lysis buffer and PBS, bound proteins were eluted by thrombin (Amersham) digestion for 16h at 22°C. The Sepharose beads were then centrifuged and the supernatants were resolved by 9% or 15% SDS–PAGE, transferred to PVDF membrane (PerkinElmer Life and Analytical Sciences, Boston, MA), and probed with anti-GIPC and/or anti FLAG mAb M2 (Sigma).
Gel filtration
Gel filtration chromatography was performed with Sepharose 6B column (20 × 400 mm, 72 ml) (Amersham). The column was calibrated with ribonuclease A (13.7 kDa ± 15%), chymotrypsinogen A (25 kDa ± 25%), ovalbumin (43.0 kDa ± 15%) and albumin (67 kDa ± 10%) (Amersham). Each standard protein (2–5 mg) was dissolved in 1 ml of equilibration buffer (50 mM phosphate buffer, pH 7.4, containing 150 mM NaCl) and loaded on to the column in a 500 µl volume at a flow rate of 12 ml/h and eluted with the same buffer and flow rate and elution was monitored by measuring absorbance at 280nm. Soluble protein fraction (1.5 ml) obtained from clone 22a cells was subjected to gel filtration, 3 ml fractions were collected and analyzed for GIPC protein by SDS–PAGE followed by immunoblotting with anti-GIPC antibody.
Sucrose density gradient centrifugation
Clone 22a cells were harvested, lysed and cytosolic and membrane fractions were prepared as described earlier. Two hundred microliters of cytosolic and membrane fractions were layered on top of 4.5 ml of 8–40% (w/v) discontinuous sucrose gradient and centrifuged at 100,000 g for 24 h at 4°C in a Beckman SW 55Ti rotor for 24h [24]. After centrifugation, 500 µl fractions were collected from top of the gradient and analyzed by Western blotting with anti-GIPC antibody. The protein markers (200 µl of 5 mg/ml) described above in gel filtration section were loaded on to the gradient and fractionated in a parallel tube and the fractions were analyzed by spectrophotometry at 280 nm.
Chemical cross-linking
For chemical cross-linking with dithio-bis-maleimidoethane (DTME, Pierce) melanoma cells transfected with 3 µg of FLAG-GIPC plasmid, in culture, were resuspended at approximately 1 × 106/ml in cross-linking buffer (HBSS containing 10 mM HEPES, pH 7.4). Membrane permeable thiol cross-linker DTME (Pierce) was dissolved in DMSO and diluted 100 fold with the cross-linking buffer immediately prior to use. The cells were then incubated with varying concentrations of cross-linker for 1 h at 4°C with occasional gentle shaking. Reactions were stopped by two 5 min washes with HBSS containing 5 mM cysteine and lysing the cells in buffer containing 2 mM cysteine. The cells were centrifuged at 1200g for 10 min, washed in PBS and lysed in 50 mM phosphate buffer, pH 7.4, containing 150 mM NaCl, 1% Triton X-100, and a mixture of protease inhibitors. The cleavage of cross-linked proteins was done by boiling the cleared lysates with 1% 2-mercaptoethanol for 10 min. The cross-linked complexes were treated with SDS–PAGE sample buffer with or without reducing agent, and separated by SDS–PAGE (9%), analyzed by immunoblotting with anti-FLAG mAb M2. Chemical cross-linking of GIPC and cysteine mutants in membranes with CuP was performed essentially as described by Kota et al. [25]. The proteins were separated on 7.5% SDS–polyacryl-amide gel in the absence of any reducing agents. In control reactions, the membranes were first incubated for 10 min at room temperature with 20 mM NEM, 20 mM EDTA, and 0.5 mM PMSF and then were treated with CuP(400/1600 µM). Proteins were resolved by SDS–PAGE and visualized by immunoblotting with FLAG-mAb M2.
Immunoprecipitation and immunoblotting
Protein concentration in cell lysates was estimated using bicinchonnic acid (BCA) protein assay reagent kit (Pierce) and aliquots of lysates containing equal amount of protein were incubated overnight at 4°C either with anti-GFP mAb (3 µg), or a control IgG. The samples were then incubated with 50 µl of protein A–Sepharose beads for 1 h at 4°C. After extensive washing with lysis buffer, bound proteins were eluted by boiling the beads in SDS sample buffer, separated by SDS–PAGE. Protein from cell lysates, cell fractions, gel permeation chromatography, sucrose gradient fractions, immunoprecipitations and cross-linking studies were resolved by 7.5%–15% SDS–PAGE and transferred to PVDF. Blots were blocked with 3–5% milk in Tris-buffered saline–Tween 20 (TBST 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20) at room temperature, incubated with primary antibodies diluted in TBS [anti-FLAG mAb M2 (1:1000); anti-GFP mAb (1:1000, BD Sciences, Palo Alto, CA) anti-GIPC (1:2000)] overnight at 4°C, followed by horseradish peroxidase-conjugated donkey anti-mouse or rabbit antibodies (Amersham) for 1 h, and detected by chemiluminescence using ECL kit (Amersham).
Electron microscopy
Cultured normal human melanocytes derived from Caucasian foreskins were processed for routine EPON embedded electron microscopy as previously described [26]. Subsequently, ultrathin sections were collected on mesh nickel grids and prepared for immunolabeling as follows. Grids were washed 3 times with double distilled water, etched with 3% sodium metaperiodate (aqueous) for 30 min, and washed 3 times with carbonate buffer, pH 8.6, containing 1% bovine serum albumin (C/BSA buffer). The grids were then incubated overnight at 23°C in a humidified chamber with C/BSA buffer with GIPC antiserum or preimmune serum (1:100). Grids were then washed and incubated in 5 nm gold conjugated anti-rabbit immunoglobulin (1:100) prepared as described [27]. After washing the grids were stained with 2% uranyl acetate (aqueous) for 10 min at 23°C and viewed and photographed in a ZEOL 1230 transmission electron microscope. All tissue processing supplies were purchased from Ted Pella, Inc. Redding, CA.
Computational modeling
To create a homology-based fold model, GIPC PDZ domain sequence was threaded onto the backbone of GRIP1 PDZ domain using the SWISS-MODEL framework [28]. GIPC PDZ domain shares 33% identity (alignment score 70) with GRIP1 PDZ domain, which is known to dimerize and whose crystal structure has been solved [29]. To create the trimer model from the monomer, ZDOCK [30] was used to generate candidate docking interfaces between the monomers. The ClusPro server [31] was used to cluster the results and to look for solutions having a threefold symmetry. The quality and properties of the GIPC PDZ trimer model was assessed using additional tools. The FADE program was used to predict “hot spot” residues within the protein interfaces in the GRIP1 dimer and the GIPC trimer model [32,33]. The Swiss PdbViewer [28] was used to determine putative hydrogen bonds, and to analyze the biochemical properties (e.g., electrostatics, hydrophobicity) of the trimer structure. The software was also used to optimize slightly the structure, which was generated using rigid docking algorithms. Finally, we used the ConSurf Server [34] as an additional feasibility check for our model. ConSurf colors proteins according to the conservation level of each amino acid among sequences identified as being most homologous.
Results
GIPC–GIPC interaction in vivo
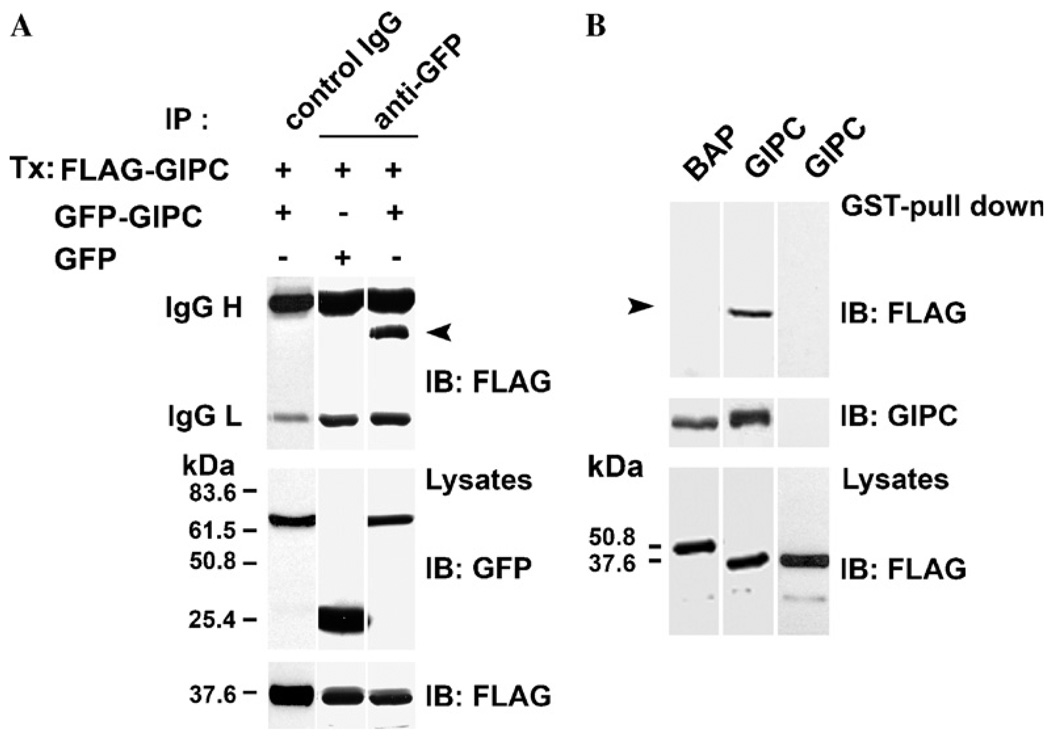
To investigate the possibility that GIPC binds to itself, we coexpressed GIPC proteins with two different epitope tags, FLAG–GIPC and GIPC–GFP, in clone 22a human melanoma cells and tested whether FLAG–GIPC co-precipitates with GIPC–GFP. Immunoprecipitation with anti-GFP mAb, followed by Western blotting with anti-FLAG mAb M2 showed that FLAG–GIPC readily co-precipitates with GIPC–GFP (Fig. 1A, top panel, lane 3) whereas no band corresponding to FLAG–GIPC could be seen when immunoprecipitated with a control IgG (Fig. 1A, top panel, lane 1) or in cells cotransfected with GFP and FLAG–GIPC and immunoprecipitated with anti-GFP mAb (Fig. 1A, top panel, lane 2). These data show that GIPC binds to itself in melanocytic cells.
Fig. 1.
GIPC–GIPC interaction in vivo and in vitro-(A) SK-MEL-23.cl.22a (clone 22a) cells were transfected with 5 µg of either GFP–GIPC or GFP constructs together with FLAG–GIPC full length plasmids. Lysates were immunoprecipitated (IP) with anti-GFP mAb (top lanes 2 and 3) or control IgG (top lane 1) and analyzed by immunoblotting (IB) using anti-FLAG mAb M2 (top panel). Total lysates were probed with anti-GFP mAb (middle panel) and anti-FLAG mAb (bottom panel). Immunoglobulin heavy and light chains of mAb GFP/or of control IgG are indicated on the left. The positions of molecular mass markers (kDa) are shown. Arrow indicates the M2 reactive band. (B) Lysates of clone 22a cells transfected with FLAG–GIPC, FLAG-BAP, were incubated with GST–GIPC or GST–thioredoxin fusion proteins immobilized on GSH–Sepharose beads for 1 h. The bound proteins were eluted with thrombin and analyzed by SDS–PAGE and immunoblotted with mAb M2. GST-pull down analysis of lysates of melanoma cells transfected with FLAG–GIPC or FLAG–BAP and incubated with GST–GIPC fusion protein (top panel, lanes 1 and 2), transfected with FLAG–GIPC and incubated with GST–thioredoxin fusion protein (top panel, lane 3). The blot was reprobed with anti-GIPC Ab (middle panel). Total cell lysates were probed with anti-FLAG mAb M2 (bottom panel). Arrow indicates the M2 reactive band.
GIPC–GIPC interaction in vitro
Analysis of GIPC amino acid sequence with protein domain search software (PROSITE and MOTIF) only showed the previously identified PDZ and the acyl carrier protein (ACP) domains and no other recognizable protein–protein interaction motifs. In order to identify the specific regions of GIPC involved in its oligomerization, we first tested the ability of FLAG–GIPC to bind to immobilized GST–GIPC fusion protein in vitro. As shown in Fig. 1B, thrombin digestion of GST–GIPC beads incubated with lysates of FLAG–GIPC transfected cells resulted in the release of mAb M2 reactive FLAG–GIPC band (Fig. 1B top panel, lane 2), while no M2 reactive bands could be seen by a similar treatment of GST–GIPC beads incubated with a control protein, FLAG-bacterial alkaline phosphatase (BAP) (Fig. 1B top panel, lane 1) or when the cell lysate transfected with FLAG–GIPC, was incubated with GST–thioredoxin fusion protein followed by digestion with thrombin (Fig. 1B top panel, lane 3). Probing the same blot with anti-GIPC antibody (Fig. 1B middle panel) showed that equal amount of GIPC was released (from GST–GIPC) by thrombin digestion from both FLAG–BAP and FLAG–GIPC incubated beads. These data showed that GIPC–GIPC interaction can be studied in vitro.
PDZ domain is required for GIPC–GIPC interaction
To determine the regions of GIPC that participate in this interaction, we constructed following FLAG epitope tagged N- and C-terminal deletion mutants lacking (i) the acyl carrier protein domain (ΔACP) between amino acid residues (aa) 248 to 333 and (ii) the PDZ domain (ΔPDZ) between aa 126 to 333 (iii) the N-terminal 90 aa (ΔNH2) and iv) the N-terminal 226 aa residues including most of the PDZ domain (ΔPDZ2) (Fig. 2B). Transient transfection and Western blot analysis with mAb M2 showed that these deletion constructs produced stable mutant GIPC polypeptides in vivo (Fig. 2A, lower panel), excepting for the 12 kDa ΔPDZ2, which could be stabilized by the addition of protease inhibitor leupeptin to the culture medium (Fig. 2A, lower panel, lane 5). GST-pull down analysis (Fig. 2A, top panel) of lysates containing the full-length and the mutant GIPC polypeptides showed that whereas the full-length, ΔACP and ΔNH2 GIPC proteins bound efficiently to GST–GIPC, the ΔPDZ and ΔPDZ2 mutants (lanes 4 and 5) failed to bind to the immobilized GST–GIPC. These data, summarized in Fig. 2B, suggest that the PDZ domain of GIPC is necessary for GIPC–GIPC interaction.
Fig. 2.
PDZ mediates GIPC–GIPC interaction. Lysates of clone 22a cells transfected with FLAG–GIPC full length and deletion mutants were incubated with GST–GIPC fusion proteins immobilized on GSH–Sepharose beads for 1 h. The bound proteins were eluted and analyzed as in Fig. 1B. (A) Interaction of FLAG–GIPC and deletion mutants with GST–GIPC immobilized on GSH-Sepharose beads (top panel). The bottom panel shows the expression levels of FLAG proteins. Arrows indicate M2 reactive bands. (B) A schematic of N-terminal and C-terminal deletion mutants of GIPC and their interaction with GST–GIPC immobilized on GST-Sepharose beads. (C) Role of csysteine mutations on binding of FLAG-GIPC to the GST–GIPC (top panel). The bottom panel shows the expression levels of FLAG–GIPC and cysteine mutants. Molecular markers (in kDa) are shown on the right for each panel. Arrow head indicates the mAb M2 reactive band and non specific bands are indicated by asterisks.
Role of cysteine residues in oligomerization of GIPC
There are two cysteine (C) residues within this region of interaction. To test whether these C residues participate in the oligomerization of GIPC, we first examined their bonding state and connectivity using a predictor program DISULFIND (www.cystein@cassandra.dsi.unifi.it). According to this program, both C residues, C100, and C189, appear to be free, i.e., not involved in intramolecular disulphide bonding; hence, may be available for intermolecular disulphide bonding with C residues on other GIPC molecules or other proteins. To test this, we created mutant GIPC proteins in which either one or both C residues were substituted with alanine(s) (C100A, C189A and C100,189A). We expressed these GIPC proteins in clone 22a melanoma cells and tested their interaction with wild type GIPC by GST pull-down assay. As shown in Fig. 2C, mutation of either one or both C residues had no effect on GIPC–GIPC interaction. In the same experiment, ΔPDZ–GIPC, as shown above (Fig. 2C, lane 5), did not bind to GST–GIPC.
Chemical cross-linking of GIPC
The above data showed that GIPC–GIPC interaction is likely mediated by the PDZ domain but not the C residues within this region. Therefore, we exploited the C residues for cross-linking studies. In cells treated with DTME, a prominent 115 kDa anti-GIPC reactive band appeared under non-reducing SDS–PAGE (Fig. 3A), in addition to the strong 37 kDa band corresponding to the monomeric form of GIPC. At higher concentrations of DTME additional high molecular weight bands could also be seen. When the lysates were electrophoresed under reducing conditions (Fig. 3B), only the monomeric 37 kDa form, but not the higher molecular weight bands, could be detected. Duplicate blots probed with mAb M2 (Fig. 3C) also revealed an M2 reactive band at 115 kDa in lysates of cells treated with DTME. This band was not observed upon reduction (Fig. 3D). These data show that GIPC polypeptides can be cross-linked in vivo by sulphhydryl cross linker. Migration of this cross-linked oligomer as a band of apparent molecular mass 115 kDa (approximately equal to three times the molecular weight of monomeric GIPC) under non-reducing SDS–PAGE, suggest that in vivo GIPC may exist as homotrimers.
Fig. 3.
Chemical cross-linking with DTME. Clone 22a cells transfected with 3 µg of FLAG–GIPC plasmids were incubated with 0.15 to 1.0 mg/ml of thiol-specific crosslinker DTME for 1 h at 4°C. Cells were lysed in PBS containing 1% Triton X-100 and analyzed by SDS–PAGE and immunoblotting with anti-GIPC antibody (A,B) anti-FLAG mAb M2 (C,D) under non-reducing and reducing conditions respectively. The arrow head indicates the immunoreactive band of approx. 115 kDa and of monomeric form of GIPC. The non-specific bands are indicated by the asterisks. The numbers on the right indicate the molecular mass (in kDa).
Subcellular distribution of GIPC oligomers
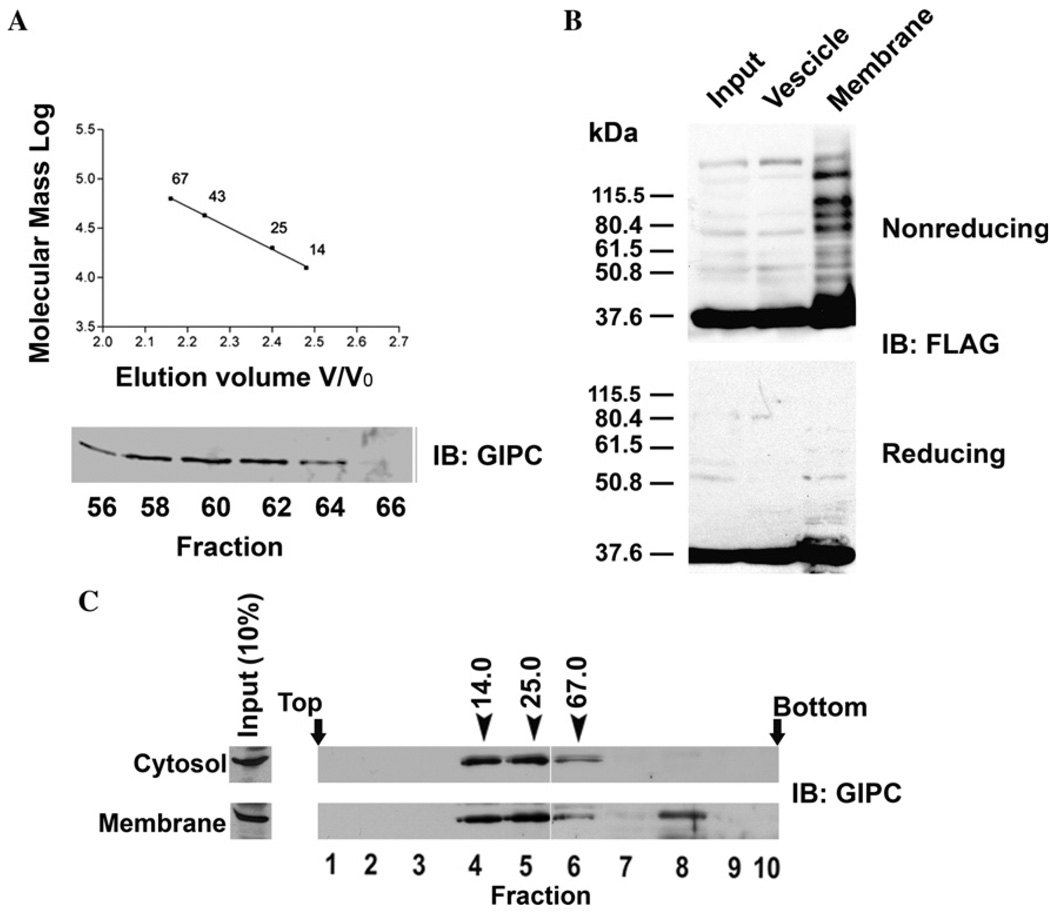
Earlier studies have shown that there are two intracellular pools of GIPC, cytosolic (50–70%) and membrane-associated (30–50%) [7,17]. Since we proposed a role for oligomerization of GIPC in its function, we reasoned that GIPC oligomers may be associated with or enriched on or near membranes where it interacts with integral transmembrane proteins. To test this, we first performed gel filtration chromatography of cytosolic soluble protein (100,000g supernatant) fraction of melanoma cells on a pre-calibrated Sepharose 6B column. Western blotting analysis of the fractions with anti-GIPC antibody showed that all immunoreactive GIPC protein eluted as a single peak corresponding to the monomeric GIPC at an estimated size of 36 kDa (Fig. 4A). Thus, bulk of soluble GIPC appears to exist in monomeric form. To test whether GIPC oligomers are associated with membranes, we fractionated the postnuclear supernatants of FLAG–GIPC transfected (as a trace to increase the sensitivity of detection) melanoma cells into heavy membrane (plasma membrane fragments, mitochondria, lysosomes, intact Golgi) and vesicle fractions (large and dense vesicles, all vesicles from plasma membrane, endoplasmic reticulum, Golgi-derived vesicles and endo-some) and analyzed GIPC oligomer distribution by Western blot analysis of SDS-solubilized (but not heat denatured) membrane proteins electrophoresed under reducing and non-reducing conditions and immunoblotted with mAb M2 (Fig. 4B). Under non-reducing conditions both membrane and light vesicular fractions showed a 37 kDa band corresponding to monomeric form of GIPC. In the heavy membrane fraction, in addition to the monomeric GIPC band, distinct bands corresponding to higher molecular forms of GIPC were also observed. These additional higher molecular weight bands were absent when the samples were heat denatured and reduced prior to SDS–PAGE and immunoblotting. The enrichment of oligomeric GIPC in the membrane fraction is consistent with its transient interaction with TRP1 and the proposed role of this interaction in intracellular protein transport [17].
Fig. 4.
Subcellular distribution of GIPC oligomers. (A) Gel filtration chromatography was performed as detailed in the materials and methods with Sepharose 6B column (20 × 400 mm, 72 ml) which was calibrated with ribonuclease A (14 kDa), chymotrypsinogen A (25 kDa), ovalbumin (43 kDa) and albumin (67 kDa). Soluble protein fraction (1.5 ml) obtained from clone 22a cells was subjected to gel filtration, and the fractions collected were analyzed by SDS–PAGE followed by immunoblotting with anti-GIPC antibody. (B) Clone 22a cells transfected with FLAG-GIPC were harvested 40 h after transfection. Heavy membrane and light vesicle fractions were prepared as described in the methods, and were resolved by SDS–PAGE and probed with anti FLAG-mAb M2. (C) Clone 22a cells grown to 80% confluence were fractionated in to soluble protein and membrane fractions (see Materials and methods). The membrane associated proteins were eluted and soluble and membrane associated proteins were loaded on to a 8 to 40% (wt/vol) stepwise sucrose density gradient. After centrifugation for 24 h at 100,000g, 0.5 ml fractions were collected from the top and were analysed by SDS–PAGE and western blotting with anti-GIPC antibody. Fraction numbers are indicated below the immunoblot. Arrows indicate the positions of the marker proteins ribonuclease (14 kDa), chymotrypsinogen A (25 kDa) and albumin (67 kDa) processed on a gradient identically.
Additionally, we performed sucrose density gradient (8–40% sucrose) fractionation of endogenous soluble proteins and proteins eluted from particulate fraction by 0.5 M NaCl and analyzed by Western blotting with anti-GIPC antibody. Cytosolic GIPC appeared in fractions 4–7 [(between the peaks of chymotrypsinogen A (25 kDa) and albumin (67 kDa)], whereas GIPC eluted from membranes was distributed in two regions of the gradient, one corresponding to the monomeric form and the other corresponding to the sedimentation of protein of molecular weight >67 kDa (Fig. 4C). Thus, only membrane associated GIPC appears to be present in oligomeric state.
Chemical cross-linking with CuP
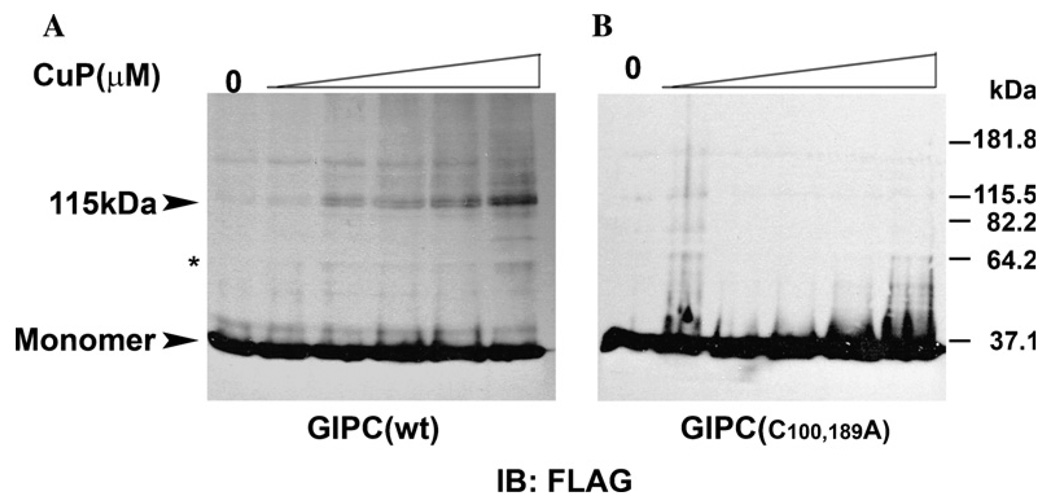
To investigate this further we performed disulfide cross-linking with CuP. Membranes from cells expressing FLAG-tagged wild type GIPC or the C100,189A mutants were prepared and treated with different concentrations of CuP [25]. As shown in Fig. 5A, in the absence of CuP, GIPC migrated predominantly as a 37 kDa species, corresponding to monomer with a very faint immunoreactive band at 115 k Da approximately three times the size of the monomer. In the presence of CuP, intensity of the band at 115kDa increased with increasing concentration of CuP with concomitant appearance of other bands corresponding to the higher oligomeric forms GIPC. A weak band of a putative dimeric GIPC could also be observed at higher concentrations of the cross-linker (Fig. 5A). In control cells treated with NEM prior to the addition of cross-linker (see Materials and methods), a faint band corresponding to trimeric form of GIPC was evident which is consistent with our earlier result which showed that lack of involvement of cysteine residues in GIPC–GIPC interaction. However this oligomerization did not result in marked decrease in the monomeric forms of GIPC suggesting that even in the membranes only a small portion of GIPC may exist as oligomers. Similar CuP treatment of membranes of cells expressing cysteine mutants, however, did not produce any bands corresponding to the oligomeric forms of GIPC, due to the lack of cysteines for cross-linking of GIPC oligomers by CuP (Fig. 5B).
Fig. 5.
Chemical cross-linking of membrane GIPC by CuP. Clone 22a cells were transfected with (A) FLAG-tagged wild type GIPC and (B) cysteine mutant plasmids. Membranes and cytosolic fractions were prepared as detailed in the Materials and methods. The membranes were treated with CuP and then with 20 mM NEM containing 20 mM EDTA and 0.5 mM PMSF. Cross linked proteins were separated by SDS–PAGE and visualized by immunoblotting with anti-FLAG mAb M2. The arrow head indicates the immunoreactive band of approx.115 kDa. A band at 64.2 kDa at higher concentrations of CuP is indicated by the asterisk.
Computational modeling
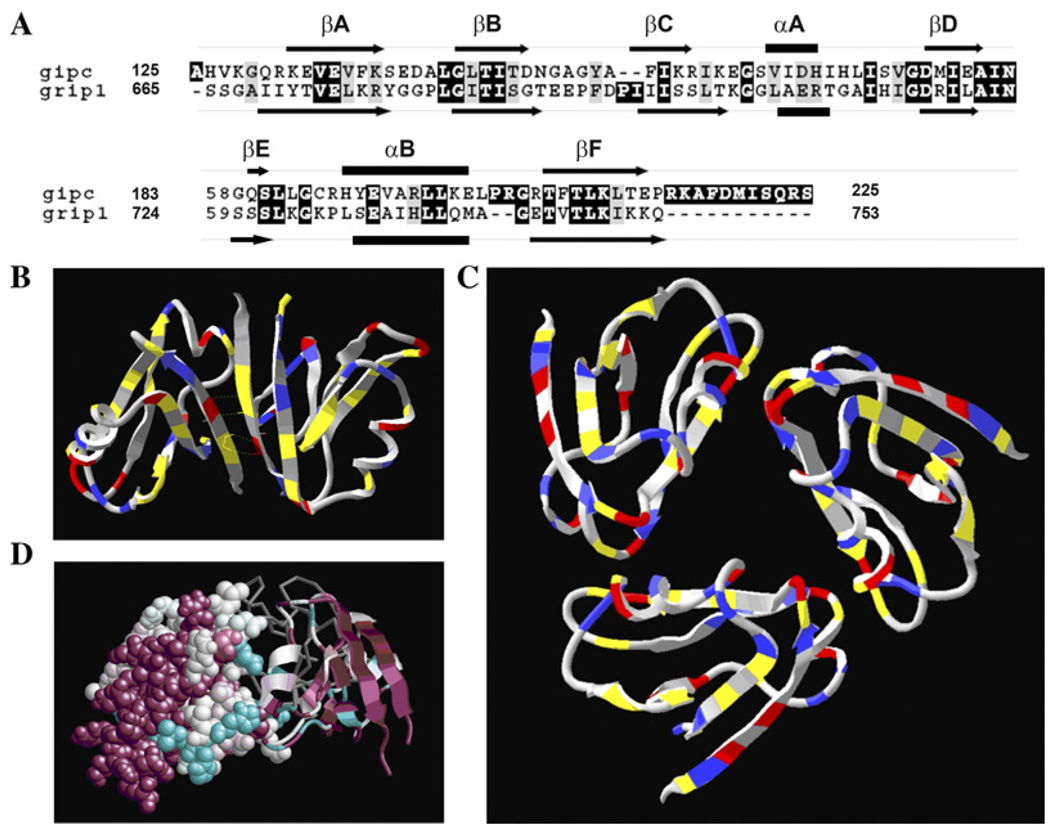
In order to test the feasibility of trimer formation, we generated a model for the GIPC PDZ domain folding and oligomerization using GRIP1 PDZ6 domain backbone as template. GIPC PDZ domain shares 33% identity (alignment score 70) (Fig. 6A) with GRIP1 PDZ6 domain, which is known to dimerize and whose crystal structure has been solved [29]. Through trial and error, it was found that the majority of the GIPC PDZ domain would fold onto the backbone of the GRIP1 PDZ6 domain, PDB 1N7F. The dimeric structure of GRIP1 PDZ domain is shown in Fig. 6B, with its key interface contacts highlighted. As expected the FADE program predicted the residues shown (Fig. 6B) as hotspots residues within the protein interfaces of GRIP dimer. However, since our biochemical data suggested formation of a trimer, we tested the feasibility of trimerization of GIPC PDZ domain by constructing a model by homology-based protein folding and rigid body molecular docking. The ClusPro server [31] was used to cluster the results and to look for solutions having a three-fold symmetry. The model generated as a result of this process is shown in Fig. 6C. In the GIPC PDZ trimer, the FADE program identified several residues that may be involved in hydrogen bonds or salt bridges. A comparison of GRIP PDZ6 dimer and our putative GIPC PDZ trimer model with residues colored according to their biochemical properties reveals why GIPC is unlikely to dimerize according to the motif adopted by GRIP1. In particular, Fig. 6B shows four hydrogen bonds between sequences TVE that are reflected across the interface. In GIPC, this sequence is EVE, which has two positive charges rather than a charged/polar mix. Reflecting this sequence leads to unfavorable electrostatic interactions and the loss of hydrogen bonds. This provides added evidence that GIPC PDZ domain adopts a different multimeric structure than the known GRIP1 dimer.
Fig. 6.
Computational analysis of GIPC trimer. (A) PDZ domain sequence (aa125-aa225) of GIPC is aligned with that of GRIP 1 PDZ6 using the program ClustalW [45]. Highly conserved residues are shaded in black and gray. The predicted secondary structure elements of GIPC PDZ [46] and GRIP1 PDZ6 [29] are shown as arrows (β-sheet), bars (α-helix) and lines (connecting loops). (B) The GRIP dimer is colored according to amino acid type-charged (red/blue), hydrophobic (gray) or neutral polar (yellow). The sequence TVE is found symmetrically across the interface, resulting in hydrogen bonds between a pair of polar and charged residues. In the GIPC PDZ domain, which is believed to form a trimer, this sequence is replaced by EVE. (C) A feasible model for a trimer of PDZ domains. The model has pairs of putative hydrogen bonds at each interface. In addition, the termini are highly accessible. (D) Conservation data for the trimer model, as generated by ConSurf. One monomer is shown with full atomic resolution, a second as a ribbon, and the third as a gray backbone. Highly conserved regions are displayed in magenta and unconserved regions in cyan. Conserved regions, and in particular the peptide-binding region, are exposed in the model trimer.
Immunogold staining
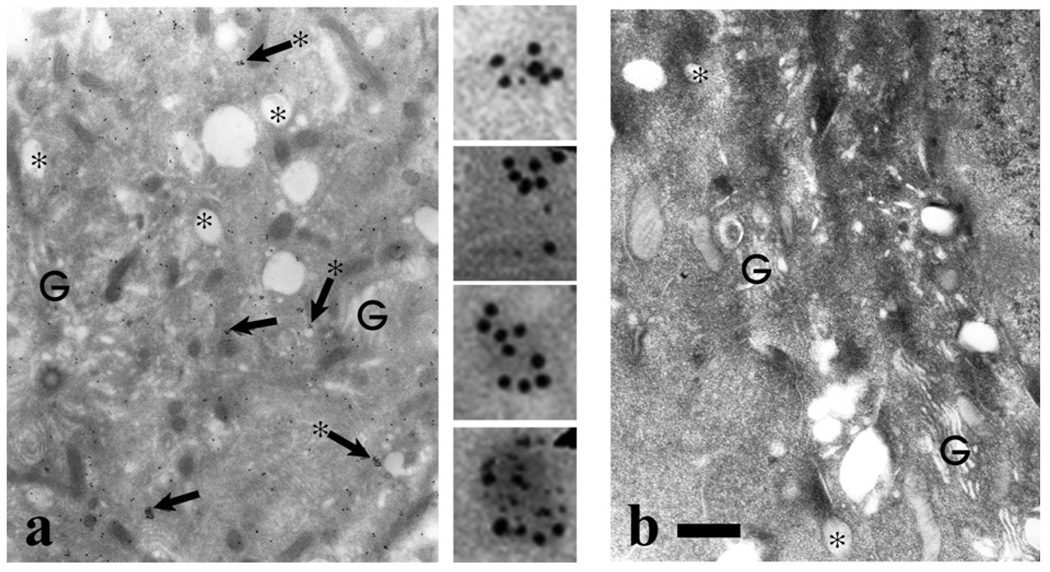
To examine the intracellular distribution of GIPC, we performed immunogold electron microscopic studies using cultured human neonatal foresk in melanocytes and affinity-purified anti-GIPC antibody. As shown in Fig. 7a, gold particles were localized throughout the cytoplasm and could be frequently found as clusters near/on membrane vesicles around the Golgi region. These observations support our biochemical results and implicate oligomeric GIPC in intracellular membrane events.
Fig. 7.
Immunogold localization of GIPC in human melanocytes- Normal human melanocytes were processed for immunoelectron microscopy using (a) GIPC antiserum or (b) preimmune antiserum. In panel (a) regions indicated by the arrows, are shown digitally enlarged. In the Golgi (G) area prevalent in stage I melanosomes (asterisks), GIPC localization throughout the cytosol with frequent clustering (arrows) that in many cases was localized on the cytoplasmic side of vesicles (arrows with asterisks). Bar = 1.0 µm.
Discussion
In this paper we show that the small (333 amino acids-long, ~36 kDa), ubiquitously expressed PDZ protein, GIPC, binds to itself both in vitro and in vivo and this interaction requires the PDZ domain. Chemical crosslinking and sedimentation studies also showed that endogenous GIPC in melanoma cells can form oligomers with a molecular mass that corresponds to trimeric GIPC. Self association of PDZ proteins, through both PDZ-independent and PDZ dependent mechanism, to form higher molecular complexes has been documented [19,35]. These PDZ–PDZ interactions are distinct from canonical PDZ–peptide interactions in that they do not involve recognition of a COOH-terminal sequence and require the full tertiary structures of both PDZ proteins [36]. The ability of the PDZ domains to form both homo-and heteromeric interactions has been proposed as a mechanism by which PDZ proteins form higher-order scaffold for the components of the signal transduction pathways [37].
Proteins containing single PDZ domain have also been shown to dimerize. PICK1 (protein interacting with C kinase 1) contains single PDZ domain and a coiled-coiled domain which is thought to mediate its dimerization [38]. Our data also shows that GIPC–GIPC interaction is PDZ dependent. Other investigators have employed yeast two-hybrid analysis to investigate GIPC–GIPC interaction. These studies have yielded contradictory observations. While Bunn et al., showed that GIPC can bind to itself through non-PDZ interaction, Wang et al., did not find evidence for interactions between SEMCAP-1(GIPC) molecules [10,16]. Jeanneteau et al., on the other hand, suggested that GIPC dimerizes through its N-terminal region [14]. The discordance between our finding and that of Jeanneteau et al., may be due to the limitations of the methods employed. The N-terminal deletion mutant construct used by Jeanneteau et al., lacked the N-terminal 118 amino acids compared to our mutant which lacked only N-terminal 91 amino acids. Moreover, in our study a truncated GIPC containing the N-terminal region, but not the PDZ domain, did not bind to GIPC (Fig. 2A). Interestingly, Jeanneteau et al., showed that mutant GIPC containing only the PDZ domain also did not bind to GIPC. These observations suggest that intact PDZ domain in the context of its flanking regions may be required for GIPC–GIPC binding.
Crystal structure of the GRIP1 PDZ6-ligand complex revealed that GRIP1 PDZ6 domains form dimers. The dimeric interface between the two PDZ domains involves a βA strand and an αA–βD loop from each monomer [29]. The dimeric interaction was supported by six hydrogen bonds between two anti-parallel βA strands and hydrophobic forces between non-polar atoms in the interface. In Shank 1 PDZ dimer, the monomers are held tightly by six hydrogen bonds, four water-bridged hydrogen bonds, and numerous van der Waals interactions [39]. The interface between dimeric PDZ domains involves a βA strand and a βB/βc loop from each monomer. In Shank 1 and GRIP PDZ dimers, the peptide binding pockets are located at the distal sides of the complex and oriented in antiparlallel fashion. Such independent target biniding by PDZ multimers was also detected with InaD and NHERF proteins [20,40]. Moreover, crystal structure of the Shank1 PDZ domain suggests that dimeric configuration of the PDZ domain may facilitate multimeric organization [41,42]. Our biochemical data and computer modeling indicate that the trimerization of GIPC, rather than dimerization is favoured. A second feasible trimer can be created by structural alignment of the folded monomer model to be part of a hexameric assembly found in the Protein Data Bank file1PDR. At this time, we do not claim either model to be precisely correct, but describe them here to establish the structural and biophysical feasibility of a GIPC PDZ trimer. The model has pairs of putative hydrogen bonds at each interface. In addition, the termini are highly accessible. This is essential as the PDZ domains are embedded within a larger protein. Thus, the above model is structurally feasible in the context of the entire protein. Multimerization of PDZ protein has also been shown to involve covalent linkages such as disulfide bridges or require covalent modification through palmitoylation of cysteines [43,44]. Although GIPC contains two cysteine residues, one within (C100) and another outside (C189) the PDZ domain, we show that mutations of these cysteine residues did not effect GIPC–GIPC interaction. Thus, similar to binding of ligands to PDZ domain, it appears that GIPC-GIPC binding is also a weak interaction involving only non-covalent linkages.
Based on these data, we propose that GIPC binds simultaneously to its target protein and to itself. Simultaneous homophilic and target binding has been proposed for InaD, where two types of interaction occurred through different interface [20]. The Shank1 PDZ domain interact with C-terminal domain of βPIX, and the leucine zipper domain mediates the homodimerization of βPIX. It has been suggested that such interactions are advantageous, because it would enable dimeric PDZ domains to efficiently colocalize with dimeric target proteins [41].
Based on its ability to bind to and its role in trafficking of membrane bound TRP1, we surmised that the multimeric GIPC may be localized to the membranes. In support of this we found that multimeric complexes of GIPC could be isolated from membranes by salt wash. Taken together our biochemical and electron microscopic data suggest that oligomerization of GIPC allows clustering of TRP1 at the ER and Golgi membranes for vesicular transport, and regulation of vesicular transport of TRP1 by GIPC trimers provides a potential regulatory step in melanosome biogenesis.
Acknowledgments
We thank Dr. Michael F. Hoffman, University of Wisconsin, for providing GST-thioredoxin fusion protein construct and Dr. Raymond Boissy, University of Cincinnati, for help with electron microscopy. This work was supported by NIH Grant R01AR048913.
Footnotes
Abbreviations used: GFP, green fluorescent protein; GIPC, GAIP-interacting protein C-terminus; GST, glutathione S-transferase; PDZ domain, postsynaptic density 95/disc large/ZO-1 domain; PAGE, polyacrylamide gel electrophoresis; DTME, dithio-bis-maleimidoethane; CuP, cupric orthophenanthroline; GRIP, glutamate receptor interacting protein.
References
- 1.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 2.Montell C. Curr. Opin. Neurobiol. 1998;8:389–397. doi: 10.1016/s0959-4388(98)80066-4. [DOI] [PubMed] [Google Scholar]
- 3.Short DB, Trotter KW, Reczek D, Kreda SM, Bretscher A, Boucher RC, Stutts MJ, Milgram SL. J. Biol. Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- 4.Simske JS, Kaech SM, Harp SA, Kim SK. Cell. 1996;85:195–204. doi: 10.1016/s0092-8674(00)81096-x. [DOI] [PubMed] [Google Scholar]
- 5.Hall RA, Premont RT, Chow CW, Blitzer JT, Pitcher JA, Claing A, Stoffel RH, Barak LS, Shenolikar S, Weinman EJ, Grin-stein S, Lefkowitz RJ. Nature. 1998;392:626–630. doi: 10.1038/33458. [DOI] [PubMed] [Google Scholar]
- 6.Snow BE, Hall RA, Krumins AM, Brothers GM, Bouchard D, Brothers CA, Chung S, Mangion J, Gilman AG, Lefkowitz RJ, Siderovski DP. J. Biol. Chem. 1998;273:17749–17755. doi: 10.1074/jbc.273.28.17749. [DOI] [PubMed] [Google Scholar]
- 7.De Vries L, Lou X, Zhao G, Zheng B, Farquhar MG. Proc. Natl. Acad. Sci. USA. 1998;95:12340–12345. doi: 10.1073/pnas.95.21.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lou X, Yano H, Lee F, Chao MV, Farquhar MG. Mol. Biol. Cell. 2001;12:615–627. doi: 10.1091/mbc.12.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotthardt M, Trommsdorff M, Nevitt MF, Shelton J, Richardson JA, Stockinger W, Nimpf J, Herz J. J. Biol. Chem. 2000;275:25616–25624. doi: 10.1074/jbc.M000955200. [DOI] [PubMed] [Google Scholar]
- 10.Bunn RC, Jensen MA, Reed BC. Mol. Biol. Cell. 1999;10:819–832. doi: 10.1091/mbc.10.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ligensa T, Krauss S, Demuth D, Schumacher R, Camonis J, Jaques G, Weidner KM. J. Biol. Chem. 2001;276:33419–33427. doi: 10.1074/jbc.M104509200. [DOI] [PubMed] [Google Scholar]
- 12.Blobe GC, Liu X, Fang SJ, How T, Lodish HF. J. Biol. Chem. 2001;276:39608–39617. doi: 10.1074/jbc.M106831200. [DOI] [PubMed] [Google Scholar]
- 13.Jeanneteau F, Guillin O, Diaz J, Griffon N, Sokoloff P. Mol. Biol. Cell. 2004;15:4926–4937. doi: 10.1091/mbc.E04-04-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeanneteau F, Diaz J, Sokoloff P, Griffon N. Mol. Biol. Cell. 2004;15:696–705. doi: 10.1091/mbc.E03-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu LA, Chen W, Martin NP, Whalen EJ, Premont RT, Lefkowitz RJ. J. Biol. Chem. 2003;278:26295–26301. doi: 10.1074/jbc.M212352200. [DOI] [PubMed] [Google Scholar]
- 16.Wang LH, Kalb RG, Strittmatter SM. J. Biol. Chem. 1999;274:14137–14146. doi: 10.1074/jbc.274.20.14137. [DOI] [PubMed] [Google Scholar]
- 17.Liu TF, Kandala G, Setaluri V. J. Biol. Chem. 2001;276:35768–35777. doi: 10.1074/jbc.M103585200. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 19.Dong H, Zhang P, Song I, Petralia RS, Liao D, Huganir RL. J. Neurosci. 1999;19:6930–6941. doi: 10.1523/JNEUROSCI.19-16-06930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu XZ, Choudhury A, Li X, Montell C. J. Cell Biol. 1998;142:545–555. doi: 10.1083/jcb.142.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staudinger J, Lu J, Olson EN. J Biol Chem. 1997;272:32019–32024. doi: 10.1074/jbc.272.51.32019. [DOI] [PubMed] [Google Scholar]
- 22.Jaulin-Bastard F, Saito H, Le Bivic A, Ollendorff V, Marchetto S, Birnbaum D, Borg JP. J. Biol. Chem. 2001;276:15256–15263. doi: 10.1074/jbc.M010032200. [DOI] [PubMed] [Google Scholar]
- 23.Christgau S, Schierbeck H, Aanstoot H, Aagaard L, Begley K, Kofod H, Hejnaes K, Baekkeskov S. J. Biol. Chem. 1991;266:21257–21264. [PubMed] [Google Scholar]
- 24.Radke K, Carter V, Moss P, Dehazya P, Schliwa M, Martin G. J. Cell Biol. 1983;97:1601–1611. doi: 10.1083/jcb.97.5.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kota P, Reeves PJ, RajBhandary UL, Khorana HG. Proc. Natl. Acad. Sci. U S A. 2006;103:3054–3059. doi: 10.1073/pnas.0510982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huizing M, Sarangarajan R, Strovel E, Zhao Y, Gahl WA, Boissy RE. Mol. Biol. Cell. 2001;12:2075–2085. doi: 10.1091/mbc.12.7.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charara A, Pare JF, Levey AI, Smith Y. Neuroscience. 2005;131:917–933. doi: 10.1016/j.neuroscience.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 29.Im YJ, Park SH, Rho SH, Lee JH, Kang GB, Sheng M, Kim E, Eom SH. J. Biol. Chem. 2003;278:8501–8507. doi: 10.1074/jbc.M212263200. [DOI] [PubMed] [Google Scholar]
- 30.Chen R, Li L, Weng Z. Proteins: Structure, Function, and Genetics. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 31.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell JC, Kerr R, Ten Eyck LF. J. Mol. Graph. Model. 2001;19:324–329. doi: 10.1016/s1093-3263(00)00079-6. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell JC, Shahbaz S, Ten Eyck LF. J. Mol. Sim. 2004;30:97–106. [Google Scholar]
- 34.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 35.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 36.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 37.Fanning AS, Anderson JM. J. Clin. Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. J. Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Im YJ, Lee JH, Park SH, Park SJ, Rho SH, Kang GB, Kim E, Eom SH. J. Biol. Chem. 2003;278:48099–48104. doi: 10.1074/jbc.M306919200. [DOI] [PubMed] [Google Scholar]
- 40.Lau AG, Hall RA. Biochemistry. 2001;40:8572–8580. doi: 10.1021/bi0103516. [DOI] [PubMed] [Google Scholar]
- 41.Im YJ, Lee JH, Park SH, Park SJ, Rho SH, Kang GB, Kim E, Eom SH. J. Biol. Chem. 2003;278:48099–48104. doi: 10.1074/jbc.M306919200. [DOI] [PubMed] [Google Scholar]
- 42.Im YJ, Park SH, Rho SH, Lee JH, Kang GB, Sheng M, Kim E, Eom SH. J. Biol. Chem. 2003;278:8501–8507. doi: 10.1074/jbc.M212263200. [DOI] [PubMed] [Google Scholar]
- 43.Hsueh YP, Kim E, Sheng M. Neuron. 1997;18:803–814. doi: 10.1016/s0896-6273(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 44.Christopherson KS, Sweeney NT, Craven SE, Kang R, El-Husseini AE-D, Bredt DS. J. Cell Sci. 2003;116:3213–3219. doi: 10.1242/jcs.00617. [DOI] [PubMed] [Google Scholar]
- 45.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGuffn LJ, Bryson K, DT J. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]