Abstract
Despite the essential roles of sphingolipids as both structural components of membranes and critical signalling molecules, we have a limited understanding of how cells sense and regulate their levels. Here we reveal the function in sphingolipid metabolism of the ORM/ORMDL genes, a conserved gene family that includes ORMDL3, which has recently been identified as a potential risk factor for childhood asthma. Starting from an unbiased functional genomic approach, we identify Orm proteins as negative regulators of sphingolipid synthesis that form a conserved complex with serine palmitoyltransferase, the first and rate-limiting enzyme in sphingolipid production. We also define a regulatory pathway in which phosphorylation of Orm proteins relieves their inhibitory activity when sphingolipid production is disrupted. Changes in ORM gene expression or mutations to their phosphorylation sites cause dysregulation of sphingolipid metabolism. Our work identifies the Orm proteins as critical mediators of sphingolipid homeostasis and raises the possibility that sphingolipid misregulation contributes to the development of childhood asthma.
Introduction
As cells grow, divide and respond to their environment, they must synthesize lipids to meet metabolic demands while also ensuring the correct balance of a wide array of structurally and functionally diverse lipid species. At the same time, cells need to maintain the correct distribution of lipids within the membrane bilayers of different organelles and the plasma membrane. Regulatory mechanisms that ensure proper levels of some lipid species including sterols1–3 and various glycerolipids4–6 have been identified, with loss of such controls leading to a variety of disease states1, 7. While remarkable progress has been made in defining the protein machinery responsible for synthesizing8, 9 and transporting sphingolipids10–13, insights into how cells sense and maintain their levels are only now emerging14, 15. The need for precise regulation of sphingolipids may be particularly acute as, in addition to the structural roles of the terminal products, several biosynthetic intermediates—including sphingosine, ceramide, and their phosphorylated derivates—are signalling molecules that participate in key physiological and pathological processes16, 17.
Our investigations into how cells achieve sphingolipid homeostasis stem from analysis of the ORM/ORMDL genes, which have been implicated recently in asthma. Asthma has emerged as a major health problem, with rates of childhood disease increasing dramatically over the last three decades18. There is a strong heritable component that modulates asthma susceptibility, and many studies have been undertaken to identify genetic risk factors19. A breakthrough in these efforts came with the genome-wide association study of Moffatt et al.20, which identified single nucleotide polymorphisms (SNPs) at chromosome 17q21 near the ORMDL3 gene that confer an increased risk of childhood asthma. Subsequent studies have reproduced this finding in multiple ethnic groups21–23 . Additionally, the disease-associated polymorphisms have been shown to modulate expression of ORMDL3 and the adjacent gene GSDMB20, 24, suggesting that dysregulation of one or both of these genes may contribute to childhood asthma. Thus genetic variants near ORMDL3 are a widespread risk factor for developing childhood asthma that may raise the incidence of the disease by up to ~20%22, 25. However, translating these findings into an increased understanding of how ORMDL3 may contribute to asthma pathogenesis has been hampered by the lack of information on the function of ORM-family genes.
Members of the ORM gene family encode trans-membrane proteins localized in the Endoplasmic Reticulum (ER) and include two genes in the budding yeast S. cerevisiae (ORM1/2) and three genes in humans (ORMDL1/2/3)26. The Orm proteins have no known functional domains, and little is known about their cellular role; however, they are strongly conserved and have been proposed to share a common function26. Here we use a combination of global and targeted studies to identify Orm proteins as homeostatic regulators of the first and rate-limiting step in sphingolipid biosynthesis.
Results
Orm1/2 negatively regulate sphingolipid synthesis
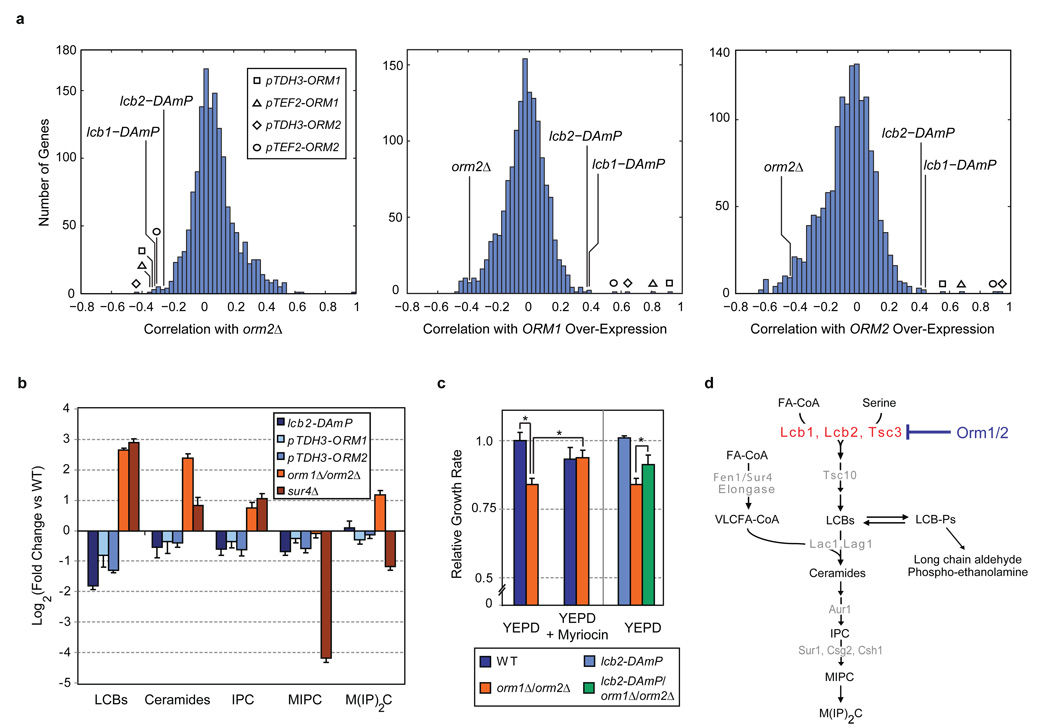
We began our investigation of S. cerevisiae ORM1 and ORM2 using a global approach to characterize gene function based on large-scale measurements of genetic interactions, termed epistatic mini-array profiles or E-MAPs27. In this strategy, the function of a given gene can be inferred without a priori hypotheses by comparing the spectrum of genetic interactions resulting from mutation of that gene with interaction patterns caused by mutations in other genes of defined function. When we compared the genetic interaction profile resulting from deletion of ORM2 to those obtained for a collection of >1400 yeast mutants focused on ER biology (manuscript in preparation and ref. 27), we observed a strong anti-correlation with the interaction patterns seen for hypomorphic alleles that reduce expression of LCB1 and LCB2 (lcb1-DAmP and lcb2-DAmP27), thus implying that ORM2 and LCB1/2 have opposing cellular roles (Fig. 1a and Supplementary Data). We also examined the genetic interactions that result from over-expression of ORM1 or ORM2 using the strong promoters pTDH3 and pTEF2, as it was shown that the asthma-associated polymorphisms near ORMDL3 are associated with increased expression of this gene20, 24. Over-expression of ORM1 or ORM2 produced a pattern of genetic interactions that was highly correlated with those seen for the lcb1-DAmP and lcb2-DAmP strains, indicating that increased Orm expression phenocopies reduced Lcb1/2 activity (Fig. 1a).
Figure 1. Orm1/2 are negative regulators of sphingolipid synthesis.
a, Genetic interaction (E-MAP) profiles were generated for strains harbouring ORM2 deletion, ORM1 over-expression, and ORM2 over-expression alleles (orm2Δ, pTDH3-ORM1 & pTEF2-ORM1, and pTDH3-ORM2 & pTEF2-ORM2, respectively). Correlations between the E-MAP profiles of these strains and those of >1400 other yeast mutants are shown in histograms (see Methods and Supplementary Data). b, Lipids were extracted from the indicated strains and analyzed using a global mass spectrometry approach30. Amounts of the indicated metabolites are shown relative to wild-type (WT; average ± s.d., n = 4; see Methods and Supplementary Data). c, Logarithmic-phase growth rates for the indicated strains were measured in standard media or media supplemented with myriocin at 150 ng/ml. Growth rates (average ± s.d., n ≥ 3) are shown normalized to wild-type (WT); asterisks denote statistical significance (p < 0.05). d, Schematic of the sphingolipid biosynthesis pathway in S. cerevisiae, with selected metabolites and genes indicated. Abbreviations are as follows: FA-CoA (fatty acid co-enzyme A), VLCFA-CoA (very long chain fatty acid co-enzyme A), LCBs (long chain bases), LCB-Ps (long chain base phosphates), IPC (inositolphosphorylceramide), MIPC (mannosyl-inositolphosphorylceramide), and MIP2C (mannosyl-diinositolphosphorylceramide).
LCB1 and LCB2 encode serine palmitoyltransferase, an essential hetero-dimeric enzyme that catalyzes the first and rate-limiting step in sphingolipid biosynthesis9, 28. The synthesis of sphingolipids begins in the ER, where two classes of lipid metabolites, long chain bases (LCBs) and very long chain fatty acids (VLCFAs), are formed (Fig. 1d). LCBs are produced by the condensation of serine with fatty acids by serine palmitoyltransferase (Lcb1/2 in yeast and Sptlc1/2/3 in mammals), followed by Tsc10-mediated reduction.8, 9, 29. LCBs can then be phosphorylated or N-acylated with VLCFAs to form long chain base phosphates or ceramides, respectively, with the latter undergoing further modification to generate complex sphingolipids (Fig. 1d).
Thus our E-MAP data showing a correlation between increased ORM expression and decreased LCB1/2 function implicate the Orm proteins as negative regulators of sphingolipid synthesis and highlight the reaction carried out by Lcb1/2 as the possible step in the pathway at which Orm1/2 may act (Fig. 1a,d). We therefore examined the effects of altering Orm levels on cellular lipid composition using a mass spectrometry-based global lipidomic technique30. In agreement with the genetic interaction data, over-expression of ORM1 or ORM2 resulted in changes to the lipidome, including reduced LCB and ceramide levels, that closely resembled those seen for the lcb2-DAmP mutant (Fig. 1b and Supplementary Data). Conversely, cells deleted for ORM1/2 had highly elevated levels of LCBs and ceramides. In principle, this accumulation of LCBs caused by deletion of ORM1/2 could result from a failure to consume these species by reaction with VLCFAs to form ceramides, as is seen when SUR4 is deleted (Fig. 1b and ref. 30). However, in contrast to the sur4Δ strain, the orm1Δ/orm2Δ strain’s elevated LCB levels are also accompanied by increased amounts of the terminal sphingolipid mannosyl-diinositolphosphorylceramide (MIP2C). Thus, ORM1/2 deletion causes increased flux throughout the sphingolipid pathway. This increased flux is also the primary cause of the growth defect seen in orm1Δ/orm2Δ cells, as artificially decreasing Lcb1/2 activity, either through the use of myriocin, a drug that specifically inhibits Lcb1/231, or through the lcb2-DAmP allele, significantly suppresses the growth defect caused by loss of ORM1/2 (Fig. 1c).
A complex with Orm1/2, Lcb1/2, and Sac1
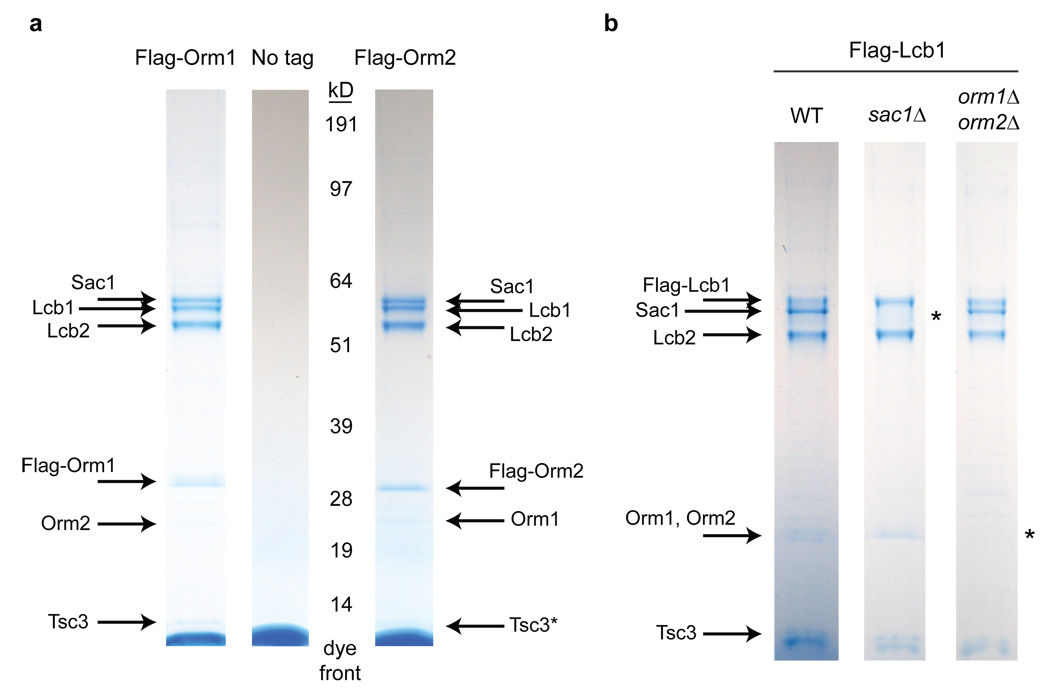
We next explored how Orm proteins modulate sphingolipid production by identifying proteins bound by Orm1 and Orm2 in vivo. Isolation of functional 3×Flag-tagged forms of Orm1 and Orm2 expressed from their endogenous loci revealed a previously uncharacterized, roughly stoichiometric complex (Fig. 2a). Mass spectrometry showed that this complex comprises the serine palmitoyltransferase proteins Lcb1, Lcb2 and Tsc3, in addition to the phosphoinositide phosphatase Sac132 (Supplementary Table 1). To further characterize physical interactions among these proteins, we utilized a 3×Flag-tagged form of Lcb1 that is able to rescue the inviability seen upon loss of LCB1 (although this allele does exhibit mild sensitivity to myriocin). Purification of 3×Flag-Lcb1 yielded the same components seen in the Orm1/2 pull-downs (Fig. 2b), indicating that the Orm-associated proteins are likely to exist in a single complex. Orm proteins and potentially their binding partners may also form higher-order oligomers, as we detected Orm2 co-immunoprecipitation with 3×Flag-Orm1 (and vice versa) and self-association of differently tagged copies of the same Orm protein (Fig. 2a and Fig. 4c).
Figure 2. Orm/Ormdl proteins form a complex with serine palmitoyltransferase.
a, Colloidal-stained gels are shown for proteins eluted after affinity purifications from strains expressing 3×Flag-Orm1 or 3×Flag-Orm2, or from an untagged control strain. Immunoprecipitated proteins were identified by mass spectrometry (see Supplementary Table 1; asterisk indicates the partially obscured protein Tsc3, whose presence was confirmed by mass spectrometry). b, Affinity purifications of 3×Flag-Lcb1 from wild-type (WT), sac1Δ, and orm1Δ/orm2Δ strains were performed as in a. Asterisks indicate bands that are lost in deletion backgrounds.
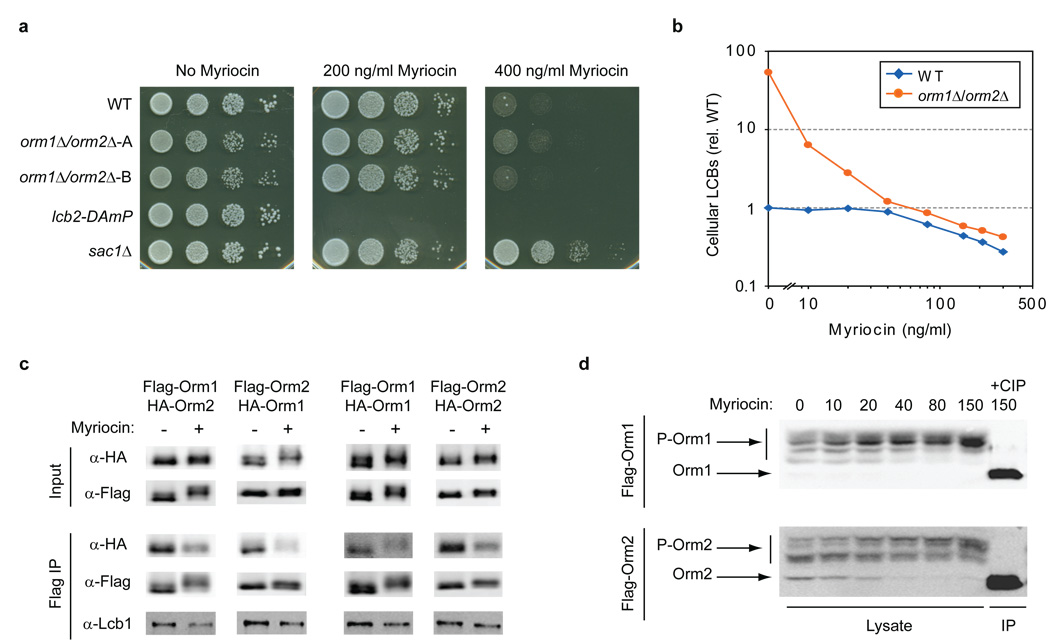
Figure 4. Orm1/2 are regulated in response to disruption of sphingolipid synthesis.
a, Serial dilutions of the indicated strains were spotted on plates with 0, 200, or 400 ng/ml myriocin and imaged after growth for 24–48 hr. b, LCBs were extracted and analyzed from wild-type (WT) and ORM1/2 deletion strains grown in media supplemented with the indicated concentrations of myriocin. The sums of C18 dihydrosphingosine and phytosphingosine peak intensities from n ≥ 2 experiments are shown (relative to wild-type LCB levels in the absence of myriocin). c, Native immunoprecipitations of 3×Flag-tagged Orm1 and Orm2 were performed from strains grown in standard media or media supplemented with 150 ng/ml myriocin and analyzed by Western blot. The indicated strains also expressed 3×HA-Orm1/2 from their endogenous loci (diploid strains were used to examine self-association of Orm1 and Orm2). d, Lysates were prepared from yeast expressing 3×Flag-Orm1/2 after growth in media containing the indicated concentrations of myriocin. Western blots show phosphorylated forms of 3×Flag-Orm1 (P-Orm1) and 3×Flag-Orm2 (P-Orm2) after separation on phosphate-affinity gels35. An immunoprecipitated (IP) sample treated with calf intestine phosphatase (CIP) shows the position of un-phosphorylated Orm1/2.
The presence of Sac1 in association with Lcb1/2 suggests a new connection between this phosphoinositide phosphatase and sphingolipid biosynthesis. Deletion of SAC1 leads to elevated LCB levels (Supplementary Fig. 1a and ref. 33) and resistance to the Lcb1/2 inhibitor myriocin (Fig. 4a). This resistance was also seen in cells expressing the sac1–834 catalytically inactive mutant of SAC1 (Supplementary Fig. 1b). Additionally, we found that Sac1 and Orm1/2 bind independently to Lcb1/2 (Fig. 2b) and that SAC1 and ORM1/2 deletions exhibit synthetic lethality (Supplementary Fig. 1c). These findings together suggest that Sac1 modulates serine palmitoyltransferase activity directly, but in a mode distinct from Orm1/2.
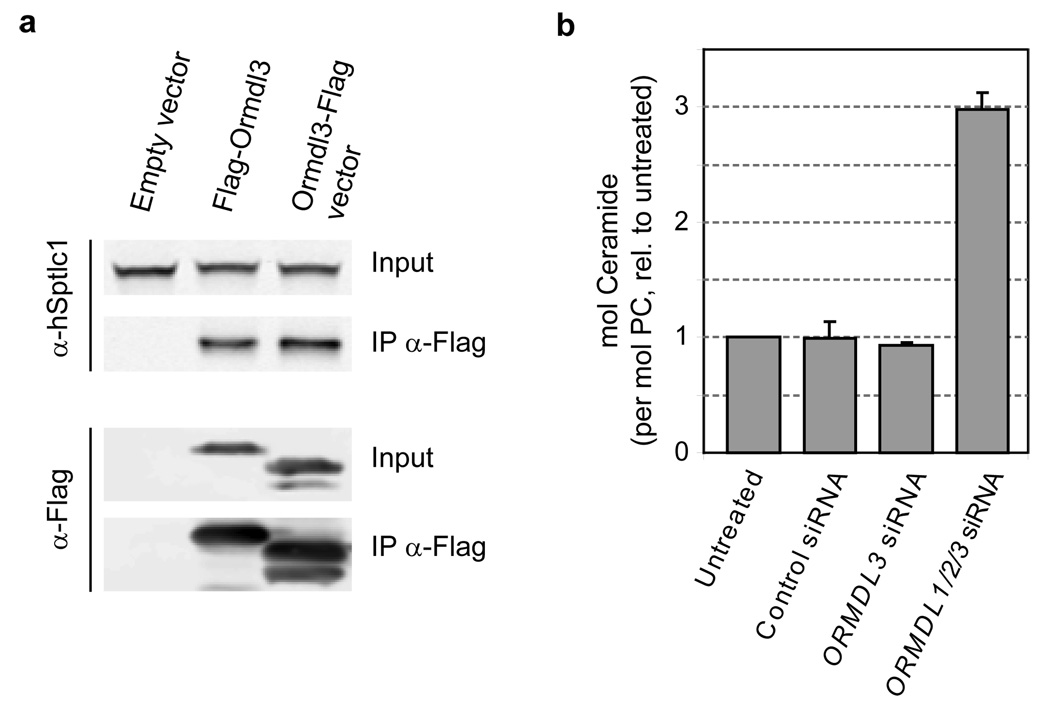
Finally, we showed that the functional complex between Orm proteins and serine palmitoyltransferase is conserved in human cells. Immunoprecipitations from HEK293T cells expressing 3×Flag-tagged Ormdl3 led to a prominent Sptlc1 band detected by Western blot (Fig. 3a). Additionally, we found that simultaneous knock-down by RNAi of ORMDL1/2/3 in Hela cells causes an approximately three-fold increase in ceramide levels (Fig. 3b, Supplementary Fig. 2 and Supplementary Data).
Figure 3. ORM gene function is conserved in human cells.
a, HEK293T cells were transfected with either an empty vector or a vector encoding Ormdl3 fused to the 3×Flag epitope (N- or C-terminal fusion). Immunoprecipitations with anti-Flag agarose were analyzed by Western blot against human serine palmitoyltransferase (α-hSptlc1) and against the Flag epitope (α-Flag). b, Hela cells were transfected with siRNAs directed against the indicated genes. After 72 hr, cells were harvested and their lipids were analyzed by mass spectrometry. Ceramide levels normalized to phosphatidylcholine (PC) are shown (relative to untreated cells; average ± s.d., n = 3).
Together, these results establish that Orm proteins are components of a novel and conserved protein complex, which we term the SPOTS complex (Serine Palmitoyltransferase, Orm1/2, Tsc3, and Sac1). Additionally, our data suggest that Orm1/2 negatively regulate sphingolipid synthesis by acting directly on Lcb1/2 (for example by altering Lcb1/2 catalytic activity, accessibility of substrates to the enzyme, or its sub-cellular localization).
Homeostatic regulation of Orm1/2 activity
Why would cells include negative regulators of serine palmitoyltransferase, Orm1/2, as core components of this essential enzyme complex? This seemingly paradoxical arrangement prompted us to investigate whether Orm-mediated inhibition of LCB production might be regulated in response to changes in the cellular need for sphingolipid synthesis. We therefore examined how ORM1/2 deletion affects sensitivity to the Lcb1/2 inhibitor myriocin. Growth in the presence of myriocin is strongly dependent on the cellular capacity for LCB production, as the lcb2-DAmP strain is highly sensitive to myriocin (Fig. 4a), and SAC1 deletion, which leads to increased LCB levels, confers myriocin resistance (Fig. 4a). Thus, naively we might expect deletion of ORM1/2 to also confer pronounced resistance to this drug. However, we observed that wild-type and orm1Δ/orm2Δ strains exhibit comparable growth in the presence of myriocin (Fig. 4a). Thus, loss of ORM1/2 strongly impacts cell growth under standard conditions but has little apparent effect when sphingolipid synthesis is disrupted, suggesting that Orm1/2 may be inactivated in response to myriocin treatment.
To further investigate this possibility, we compared the effects of myriocin on the LCB levels of wild-type and orm1Δ/orm2Δ strains. As described above, the ORM1/2 deletion strain exhibits highly elevated LCB levels in the absence of myriocin. However, with increasing doses of myriocin, LCB levels in the orm1Δ/orm2Δ strain decrease strongly and approximately converge with those of wild-type (Fig. 4b), supporting our growth data indicating that Orm1/2 may be inactivated in response to myriocin treatment (Fig. 4a). Interestingly, we also found that wild-type cells maintain roughly constant LCB levels even at intermediate myriocin concentrations (up to ~40 ng/ml). This robustness is not due to a failure of myriocin to affect Lcb1/2 at these concentrations, as LCB levels in the orm1Δ/orm2Δ strain are reduced strongly by the same drug concentrations (Fig. 4b; for further evidence see Fig. 4d). Rather, these results suggest that progressive inactivation of Orm1/2 may provide cells with a homeostatic mechanism to maintain nearly constant sphingolipid output in the face of increasing Lcb1/2 inhibition. Consistent with this hypothesis, LCB levels in the wild-type strain begin to decline only at myriocin concentrations where Orm-mediated inhibition of LCB synthesis appears to be fully relieved, as evidenced by convergence of wild-type and orm1Δ/orm2Δ LCB levels (Fig. 4b).
Orm1/2 regulation by phosphorylation
We next investigated the mechanism by which Orm1/2 activity might be decreased in response to myriocin treatment. Growth in myriocin did not alter Orm1/2 expression levels (Fig. 4c and Supplementary Fig. 7) or abolish their ability to interact with Lcb1/2 and Sac1 (Supplementary Fig. 3). However, we observed a notable reduction in localization of GFP-Orm2 to the cortical but not peri-nuclear ER after myriocin treatment (Supplementary Fig. 4), raising the possibility that changes in sub-cellular localization contribute to regulation of SPOTS complex activity. We also found a strong reduction in Orm protein co-immunoprecipitation in cells treated with myriocin (Fig. 4c), both for Orm1–Orm2 interaction and for self-association of Orm1 and of Orm2. A weak reduction in Orm-Lcb1 binding was also observed in response to myriocin (Fig. 4c). These findings suggest a model in which Orm-Lcb1/2 sub-complexes can self-associate into higher-order oligomers in a manner that is inhibited in response to disruptions to sphingolipid synthesis.
We also detected in our Orm1/2 co-immunoprecipations a slight shift in the electrophoretic mobility of Orm1 caused by myriocin treatment, indicating that Orm proteins may be post-translationally modified (see Fig. 4c, Flag-Orm1 and HA-Orm1 inputs). Analysis of 3×Flag-Orm1 and 3×Flag-Orm2 in SDS/PAGE gels that incorporate a phosphate-binding agent that aids in the separation of phosphorylated species35 resolved multiple phosphorylated forms for both Orm1 and Orm2 that collapsed to faster-migrating species upon phosphatase treatment (Fig 4d). Importantly, growth in myriocin induced a dose-dependent shift to more highly phosphorylated forms, and this response occurred over the same concentrations of myriocin that led to homeostatic inactivation of Orm1/2, as determined by LCB measurements (compare Fig. 4b and Fig. 4d). We also observed increased Orm1/2 phosphorylation after shutting off expression of downstream sphingolipid synthetic enzymes such as Tsc10 or Lag1 (in a lac1Δ background; Supplementary Fig. 5), further indicating that phosphorylation of Orm proteins is a homeostatic response to disruption of sphingolipid production (see Fig. 5c). Finally, we did not observe substantial changes in Orm1/2 phosphorylation upon shutting off expression of AUR1 or CSG2 (Supplementary Fig. 5), thus providing preliminary evidence that a metabolite upstream of these enzymes, such as ceramide, is the sphingolipid intermediate whose levels are sensed to regulate Orm1/2 activity.
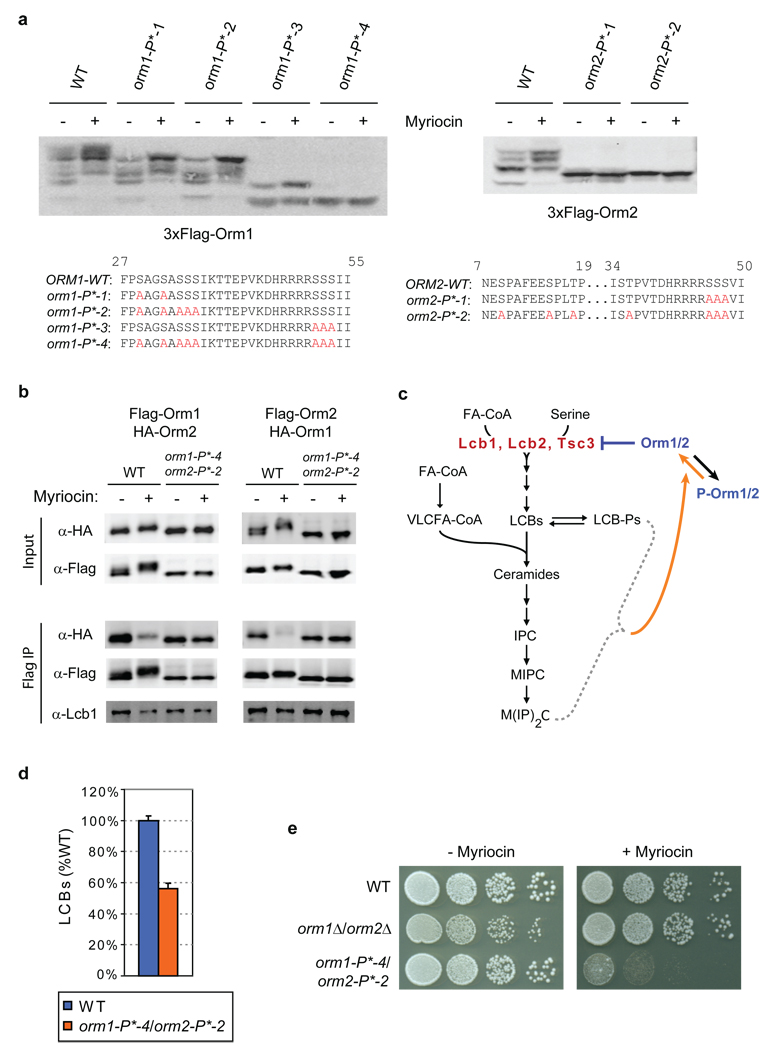
Figure 5. Mutation of phosphorylated Orm1/2 residues impairs sphingolipid homeostasis.
a, Lysates were prepared from strains expressing wild-type (WT) or phospho-site mutant 3×Flag-Orm1 or 3×Flag-Orm2 after growth in standard media or media supplemented with 150 ng/ml myriocin. Western blots are shown after protein separation on phosphate-affinity gels. Residues mutated to alanine in the indicated phospho-mutants are highlighted in red below. b, Native immunoprecipitations of 3×Flag-tagged wild-type or phospho-mutant Orm1 and Orm2 were performed from strains expressing wild-type or phospho-mutant 3×HA-Orm1/2 after growth in standard media or media supplemented with 100 ng/ml myriocin. Western blots were analyzed as in Fig. 4c. c, Model for homeostatic regulation of Orm1/2, in which Orm proteins act as negative regulators of serine palmitoyltransferase (Lcb1/Lcb2/Tsc3), whose inhibitory activity is dependent on levels of downstream sphingolipids. P-Orm1/2 denotes phosphorylated Orm1/2; see Fig. 1d for abbreviations used. d, LCBs were extracted and analyzed from wild-type (WT) and 3×Flag-tagged ORM1/2 double phospho-mutant strains grown in standard media. Data were analyzed as in Fig. 4b (average ± s.d., n = 3). e, Serial dilutions of wild-type (WT), ORM1/2 deletion, and 3×Flag-tagged ORM1/2 double-phospho-mutant strains were spotted on plates with 0 or 280 ng/ml myriocin and imaged after growth for 24–36 hr.
To test our phosphorylation-mediated feedback model directly, we identified residues of Orm1 and Orm2 that are phosphorylated. Through a combination of targeted mutagenesis and mass spectrometry, we found several sites of Orm1 and Orm2 phosphorylation in the N-terminal regions of these proteins (Fig. 5a and Supplementary Fig. 6). Importantly, mutation of these sites to alanine blocks formation of the slower-migrating phosphorylated species seen in phosphate-affinity gels (Fig. 5a). This loss of phosphorylation is unlikely to be due to these mutations resulting in non-functional Orm proteins, as the phospho-mutants are expressed at levels comparable to wild-type and rescue the slow growth of the orm1Δ/orm2Δ strain (Supplementary Fig. 7 and data not shown). Furthermore, immunoprecipitations of phospho-mutant Orm1/2 revealed that these proteins not only maintain the ability to interact with Lcb1 and each other, but that blocking phosphorylation prevents the change in higher-order assembly of the SPOTS complex normally seen in response to myriocin (Fig. 5b). Thus Orm1/2 phosphorylation appears to be the event that triggers dynamic regulation of SPOTS complex oligomerization.
We next investigated the in vivo role of Orm1/2 phosphorylation. Consistent with a basal level of Orm1/2 phosphorylation that is important for steady-state LCB production, we observed a substantial (two-fold) reduction in LCB levels in the ORM1/2 phospho-mutant strain under standard growth conditions (Fig. 5d). Furthermore, we found that this strain is highly sensitive to myriocin treatment (Fig. 5e). This result thus confirms a key prediction of our homeostatic model and indicates that although cells may possess additional mechanisms to regulate sphingolipid biosynthesis, such pathways cannot compensate for disruption of the critical feedback control provided by Orm1/2. Thus, Orm proteins are dynamic negative regulators of serine palmitoyltransferase whose inhibitory activity is dependent on adequate levels of downstream sphingolipids (Fig. 5c).
Discussion
Our studies identify Orm proteins as homeostatic regulators of sphingolipid metabolism and demonstrate that alteration of these controls leads to misregulation of sphingolipid production (Fig. 5c). Specifically, loss of the Orm1/2-mediated brake on serine palmitoyltransferase activity results in toxic accumulation of sphingolipids, while mutations that prevent this brake from being removed render cells unable to survive disruptions to sphingolipid synthesis.
A phosphorylation-based feedback loop provides a mechanistic basis for homeostatic regulation of sphingolipid production and exhibits a number of features that make it well-suited to precisely control sphingolipid levels. First, phosphorylation of a direct regulator of serine palmitoyltransferase may enable cells to respond rapidly to changing environments. Second, Orm1/2 exhibit partial phosphorylation under standard growth conditions, which may position the Orm1/2-based sphingolipid rheostat for maximal sensitivity to changes in lipid metabolism. Additionally, multi-site phosphorylation of Orm1 and Orm2 has the potential to generate several distinct and stable phospho-states, which may provide cells with a graded mechanism to finely adjust lipid production to match cellular needs. Finally, multiple phosphorylation sites may also render Orm1/2 responsive to multiple physiologic or metabolic cues.
Our identification of the SPOTS complex reveals a novel mechanism by which cells may be able to coordinate a variety of important secretory pathway processes. While a link between sphingolipids and phosphoinositides has been appreciated for some time29, 33, 36, we provide evidence that the phosphoinositide phosphatase Sac1 directly modulates sphingolipid metabolism, as it is associated physically with serine palmitoyltransferase. These results are of particular interest given the role of phosphoinositides, including the Sac1 substrate phosphatidylinositol-4-phosphate (PI4P), in the trafficking of both proteins and sphingolipids from the ER and Golgi apparatus10, 11, 32. Additionally, changes in Sac1 localization from the ER to the Golgi have been shown in both yeast and mammals to regulate protein flux through the secretory pathway in response to nutrient levels37. We therefore suggest that the SPOTS complex may represent a dynamic coordinating center that couples changes in sphingolipid and nutrient levels to the activity and localization of key enzymes of lipid metabolism and trafficking. This model is consistent with our finding that Orm proteins self-associate in a regulated fashion and with evidence that mammalian serine palmitoyltransferase is part of a high-molecular-mass complex38.
Finally, our characterization of ORM gene function provides a framework for investigating whether alterations in ORMDL3 activity and sphingolipid metabolism play a role in asthma pathogenesis. A number of studies support a role for sphingolipids such as ceramide and sphingosine-1-phosphate in asthma-associated inflammatory processes, including mast cell degranulation, airway hyper-responsiveness, and immune cell trafficking39–43. Our findings now link this growing body of evidence to the identification of SNPs near ORMDL3 that are associated with childhood asthma20. Specifically, our observation that altered Orm levels strongly impact sphingolipid metabolism raises the testable hypothesis that misregulation of sphingolipids may have a direct and causative role in the development of asthma. A broader potential role for sphingolipids in inflammatory disease is also suggested by reports that SNPs near ORMDL3 are associated with Crohn’s disease44, type I diabetes45 and primary biliary cirrhosis46. Finally, the existence of a rich set of pharmacologic tools targeting the sphingolipid pathway31 raises the possibility of therapeutic intervention to counteract the potential effects of changes in ORMDL3 activity.
Methods Summary
Phosphorylation analysis by phosphate-affinity SDS/PAGE
Lysates for phosphorylation analysis were prepared by pelleting and immediately resuspending logarithmically growing yeast in SDS lysis buffer (50mM Tris-Cl, pH 6.8, 2% SDS, 10% glycerol) with protease inhibitors (Complete EDTA-free protease inhibitor tablets; Roche) at 65°C. SDS/PAGE gels with phosphate affinity reagent35 (synthesized according to standard synthetic procedures and kindly provided by D. Fiedler) were prepared with a 7.5% acrylamide resolving gel with 50µM MnCl2 and 50µM phosphate affinity reagent (375mM Tris, pH 8.8, 0.1% SDS). Gels were run at 70V for 2.5 hr, rinsed twice for 5 min in transfer buffer with 1mM EDTA, and rinsed twice more for 5 min in transfer buffer without EDTA prior to transfer to nitrocellulose membranes. Western blots were performed as described below.
Western blots
Proteins were transferred from SDS/PAGE gels to nitrocellulose membranes and probed for proteins of interest using standard Western blot procedures. Antibodies used were as follows: rabbit anti-Flag (Sigma Aldrich), mouse anti-Flag (M2; Sigma Aldrich), mouse anti-HA antibodies 12CA5 (Roche) and HA.11 (Covance Inc.), mouse anti-yPgk1 (Molecular Probes), rabbit anti-yLcb1 (raised against peptide antigen SYIKKSHHDDPYRTTC; Abgent, Inc), mouse anti-hSptlc1 (Becton Dickinson), and rabbit anti-hOrmdl1/2/3 (raised against peptide antigen KGTPFETPDQGKARLC; Abgent, Inc.). Western blots were scanned using an Odyssey fluorescent scanner (Li-Cor Biosciences).
Methods
Growth media, plasmids, and strain construction
All yeast experiments were conducted in strain BY4741 (S288c), a related strain suitable for E-MAP experiments27, and derivatives thereof. Strains were grown in YEPD media or media supplemented with myriocin (Sigma Aldrich; ≥12 hr growth in myriocin in all cases), with the exception of samples prepared for lipidome analysis, which were grown in synthetic defined media.
Deletion, DAmP, over-expression (pTEF2 and pTHD3), mCherry fusion, and tetracycline-repressible promoter strains were constructed by standard PCR-based methods (see Supplementary Information). Strains expressing N-terminally epitope-tagged or GFP-tagged forms of wild-type and mutant Orm1, Orm2, Lcb1, and Sac1 were made by integration of DNA fragments encoding these gene variants into strains in which these genes had been deleted with the URA3 counter-selectable marker. For the essential gene LCB1, a diploid strain was used for integration of the tagged allele containing the 3×Flag epitope inserted between codons 9 and 10, as previously reported47. ORM1 and ORM2 mutants, as well as the previously described sac1–8 mutation34, were created by standard PCR mutagenesis.
HEK293T cells (ATCC) and Hela-Kyoto cells (gift from A. Hyman) were grown in DMEM high-glucose media supplemented with penicillin, streptomycin, glutamine, and 10% heat-inactivated fetal calf serum. ORMDL3 was cloned from HEK293T cDNA into expression plasmids containing N- or C-terminal 3×Flag epitopes (p3×FLAG-CMV-10 and p3×FLAG-CMV-14; Sigma Aldrich).
E-MAP data collection and analysis
E-MAP data were collected and analyzed as described previously27 (manuscript in preparation). Genetic interaction patterns were compared for pairs of mutants by first identifying those genetic backgrounds for which E-MAP data was available for both mutants of interest. The similarity of the genetic interaction scores for the two mutants was then calculated by computing the cosine correlation between their interaction scores across these shared mutant backgrounds. For a given mutant of interest such as orm2Δ, these pair-wise comparisons were carried out with all other mutant strains having at least 200 shared genetic interaction data points. For the ORM1 and ORM2 over-expression mutants, an average of the pTDH3 and pTEF2 interaction profiles was used as the query profile for correlation calculations.
Lipidome analysis by mass spectrometry
Lipidome analyses were conducted as described previously30 with minor modifications based on the protocol of Lester and Dickson48 to improve analysis of LCBs. Samples were harvested from 20ml cultures of yeast cells growing logarithmically in synthetic defined media or from ~4–6 ×106 Hela cells grown as described above. Cells were washed in water or 155mM ammonium bicarbonate at 4°C, and cell pellets were then frozen immediately in liquid nitrogen. Yeast data are from two independent samples each analyzed twice; Hela cell data are from samples analyzed in triplicate.
Growth rate measurements
Logarithmic growth rates were measured by OD600 from n ≥ 3 replicate samples. Saturated cultures of the relevant strains were diluted into fresh media and grown for >7 hr prior to growth analysis. Myriocin-treated cultures were grown for 3 hr in the presence of drug prior to growth rate analysis.
Native affinity purifications of solubilized membrane proteins (large-scale, yeast)
ER-derived microsomes were prepared from yeast cells grown in YEPD (~4000 OD600 units harvested at OD600 ~1.4). Cells were harvested, washed in water at 4°C, resuspended in lysis buffer (50mM Hepes-KOH, pH 6.8, 150mM KOAc, 2mM MgOAc, 1mM CaCl2, 200mM sorbitol) supplemented with protease inhibitors (Complete EDTA-free cocktail; Roche) and frozen in a drop-by-drop fashion in liquid nitrogen. Frozen cells were then pulverized in a ball mill (6× 3 min at 30Hz; Retsch), thawed in lysis buffer at 4°C, and dounced 10 times to homogeneity. After clarification by two consecutive centrifugations at 1000 × g, microsomes were pelleted at 44000 × g, resuspended in 0.5ml lysis buffer, and then diluted with 14ml IP buffer (50mM Hepes-KOH, pH 6.8, 150mM KOAc, 2mM MgOAc, 1mM CaCl2, 15% glycerol) with 2% digitonin (Calbiochem) supplemented with protease inhibitors. After nutating for 1.5 hr at 4°C, un-solubilized material was removed by centrifugation at 44000 × g, and the remaining supernatant was added to anti-Flag resin (150µl bed volume; Sigma Aldrich). Immunoprecipitations were nutated for 3 hr at 4°C and then washed 4×10ml with IP buffer with 0.1% digitonin. Bound proteins were eluted twice with 150µl IP buffer with 0.25% digitonin and 2mg/ml 3×Flag peptide (Sigma Aldrich). Eluates were combined, separated on SDS/PAGE gels, and stained with a colloidal blue staining kit (Invitrogen). Samples were prepared for phospho-site mapping using the same protocol, except that phosphatase inhibitors (Halt phosphatase inhibitor cocktail; Pierce) were included in the buffers used.
Native affinity purifications of solubilized membrane proteins (small-scale, yeast)
Lysates for small-scale immunoprecipitations were prepared from logarithmically growing yeast cells (~25 OD600 units) grown in YEPD. Cultures were harvested, washed in water at 4°C, and resuspended in 500µl IP buffer (see above) with 0.1% digitonin and supplemented with protease and phosphatase inhibitors. Cell lysates were prepared by bead-beating at 4°C, followed by addition of 500µl IP buffer with 1.9% digitonin to raise the final digitonin concentration to 1%. Membrane proteins were solubilized by nutating lysates at 4°C for 40 min. Unsolubilized material was next removed by centrifugation at 100,000 × g, and clarified lysates were incubated with anti-Flag resin (25µl bed volume) for 2.5 hr. The anti-Flag resin was then washed four times with 1ml IP Buffer containing 0.1% digitonin, and bound proteins were eluted by addition of 30µl IP Buffer with 0.25% digitonin and 2mg/ml 3×Flag peptide.
Native affinity purifications of solubilized membrane proteins (small-scale, human cells)
HEK293T cells grown in DMEM high-glucose media supplemented with penicillin, streptomycin, glutamine, and 10% heat-inactivated fetal calf serum were transfected using Fugene6 transfection reagent (Roche) according to the manufacturer’s instructions. After ~48 hr growth, transfected cells were harvested by trypsinization and washed with PBS. Cell pellets were then resuspended at 4°C in 1ml IP buffer (see above) with 2% digitonin and protease inhibitors and nutated for 45 min to solubilize membrane proteins. Insoluble material was removed by centrifugation at 100,000 × g, and the clarified supernatant was incubated with anti-Flag resin (25µl bed volume) for 2.5 hr. The remainder of the immunoprecipitation was done in the same fashion as the yeast small-scale affinity purifications (see above).
Denaturing immunoprecipitations and phosphatase treatment
A 50ml yeast culture in logarithmic phase growth was harvested and resuspended in 10ml 5% tricholoroacetic acid and incubated for 10 min at 4°C. Cell pellets were washed once in 1ml 5% TCA, washed twice in 1ml acetone, and then air-dried. 200µl acid-washed glass beads and 150µl SDS lysis buffer (50mM Tris-Cl, pH 7.8, 1mM EDTA, 1% SDS, 2M Urea) supplemented with protease inhibitors were added to the dried pellets, which were then lysed by bead-beating (3× 60s at 4°C). Lysates were diluted by addition of 1350µl Tween IP buffer (100mM Tris-Cl, pH 7.8, 200mM NaCl, 0.5% Tween-20, also supplemented with protease inhibitors) and clarified at 20000 × g. Supernatants were then added to anti-Flag agarose (25µl bed volume) and immunoprecipitated for 2.5 hr at 4°C. The anti-Flag resin was then washed 2× 1ml with Tween IP buffer, 1× 1ml with Tween/Urea wash buffer (100mM Tris-Cl, pH 7.8, 100mM NaCl, 0.5% Tween-20, 2M Urea), and 2× 1ml in phosphatase buffer (NEBuffer 3; New England Biolabs), with 0.1% Tween-20). The washed resin was then resuspended in 54µl phosphatase buffer + 6µl calf intestine phosphatase (New England Biolabs) and incubated for 1 hr at 37°C. After phosphatase treatment, the resin was sequentially washed with 1ml Tween IP buffer and 1ml in SDS/DOC wash buffer (50mM Tris-Cl, pH 7.8, 0.1% SDS, 0.1% Na-deoxycholate). Bound proteins were then eluted by addition of 60µl 2× SDS buffer (100mM Tris-Cl, pH 6.8, 4% SDS, 20% glycerol) and incubation for 10 min at 65°C.
RNAi and plasmid transfection of Hela cells
Hela cells grown in DMEM high-glucose media supplemented with penicillin, streptomycin, glutamine, and 10% heat-inactivated fetal calf serum were transfected using HiPerFect transfection reagent (Qiagen) according to the manufacturer’s instructions. After ~72 hr growth, transfected cells were harvested by trypsinization for lipidomic and gene expression analysis. For each targeted gene, a pool of four siRNAs was used (ON-TARGETplus SMARTpool; Dharmacon), with siRNA sequences as follows: ORMDL1 – UGGCAAGUUUCUAUACGAA, GGAGUUGGCUUGCUUCAUA, GCUUAUAAGUUAAAGGGCA, GGACCAGCUUAACUUAUAA; ORMDL2 – UGGUAGGAUUGCUGCAUAU, AGUUUAAUCAGGAUGGGUA, GUGUACUGCUGCCGAAGUU, AGUAUGAUGCUGCGCACUU; ORMDL3 – GAACAUGGACCACGCAGUU, CACUAAGUACGACCAGAU, CGGUACGGCUUCUGGAUUG, UGGGUAGGGAGCUGUCUAA.
RT-PCR analysis of gene expression in Hela cells
RNA was extracted from Hela cells using the RNeasy kit (Qiagen) after cell homogenization (QIAshredder; Qiagen). Extracted RNA was reverse-transcribed using AMV RT (Promega) and amplified with Taq polymerase (GoTaq; Promega). Real-time SYBR Green fluorescence was monitored using a DNA Engine Opticon System (MJ Research), and the following primers were used: RPL19 – ATGTATCACAGCCTGTACCTG, TTCTTGGTCTCTTCCTCCTTG; ORMDL1 – CTGACCAGGGTAAAGCAAGG, TCCAAAGATCCGAACACCAT; ORMDL2 – CATACGGTGAAAGGGACACC, CGGCAGCAGTACACTTAGCA; ORMDL3 –AACCCCTGCTCACTTGTTTG, CAAAGACCCATCCCACACTT.
Live-cell yeast fluorescence microscopy
Yeast cells expressing Sec63-mCherry and GFP-Orm1 or GFP-Orm2 were grown in rich media with or without myriocin and spotted on a concanavalin A-coated 25 × 50 mm coverslip covered with a 22 × 22 mm coverslip. Cells were imaged through the 25 × 50 mm coverslip using a Zeiss Axiovert 200M microscope (Carl Zeiss Microimaging Inc.) equipped with a spinning disk confocal scanhead and laser system (Yokogawa CS10, Yokogawa Electronics; Solamere). Images were collected as Z-stacks with 0.5 µm spacing between frames using µManager software and an ImagEM C9100-13 EM-CCD camera (Hamamatsu Photonics).
Serial-dilution cell spotting assays
Logarithmically growing yeast were spotted onto agar plates in 10-fold serial dilutions from a starting OD600 density of 0.3 to a final OD600 density of 3E-4. Plates were imaged after growth for 24–48 hr. Plates with myriocin are standard YEPD supplemented with 0.1% tergitol and 2mM citric acid.
LCB extraction and analysis by HPLC
LCB extraction and analysis was carried out as described by Lester and Dickson48. Myriocin-treated samples were grown for 14–16 hr in the presence of drug prior to extractions. For extractions, 50ml of a logarithmically growing culture was harvested and re-suspended in 10ml 5% TCA at 4°C. Following a 10 min incubation, cells were washed once in 5% TCA and three times in H2O at 4°C. Cell pellets were then re-suspended in 75µl H2O followed by addition of 425µl 70.6mM TEA/EtOH. Lipids were extracted by bath sonication (3× 99s, 30W) and incubation at 65°C for 30 min. 150µl of this extract was derivatized by reaction for 40 min at room temperature with 30µl of AccQ Fluor reagent (Waters Corp.). Ester-linked lipids were de-acylated by addition of 22.5µl of 1.5M KOH/MeOH. Following 30 min incubation at 37°C, the reaction was neutralized by addition of 22.5µl 1.74M AcOH/MeOH. Samples were analyzed on an Agilent Chemstation HPLC using a reverse-phase C18 column (ZORBAX Eclipse XDB-C18; Agilent).
Protein identification by mass spectrometry
Gel slices were trypsinized and mass spectra were acquired on a Bruker Reflex III MALDI-TOF mass spectrometer. Proteins were identified by searching NCBInr database using MS-Fit program on Protein Prospector (UCSF, http://prospector.ucsf.edu)49.
Identification of phosphorylation sites by mass spectrometry
Proteins eluted from native affinity purifications of 3×Flag-Orm1 or 3×Flag-Orm2 (see above) were precipitated using ice-cold acetone. After incubation overnight at −20°C, proteins were pelleted at 4°C by centrifugation. Protein pellets were then processed, and the resulting peptide mixtures were analyzed on a hybrid LTQ-Orbitrap mass spectrometer, as described previously50. Database searches were performed using the SGD non-redundant database, as described50.
Supplementary Material
Acknowledgments
The authors acknowledge A. Falick, S. Zhou and D. King at the Howard Hughes Medical Institute Mass Spectrometry Laboratory, University of California, Berkeley for identification of proteins by mass spectrometry; M. Schuldiner and S. Wang for E-MAP data collection; D. Fiedler and K. Shokat for kindly providing the phosphate-binding acrylamide reagent; E. Griffis and M. D’Ambrosio for assistance with fluorescence microscopy; N. Ingolia for a codon-optimized mCherry-tagging plasmid; I. Poser and A. Hyman for helpful advice and providing the Hela-Kyoto cell line; and M. Bassik, G. Brar, V. Denic, A. Frost, N. Ingolia, E. Quan, B. Toyama, and other members of the Weissman lab for insightful discussions. This work was supported by funding from Deutsche Forschungsgemeinschaft SFB/TR 13 projects A1 (K.S.) and D1 (A.S.), EUFP6 PRISM (K.S.), ETH Zurich (R.A.), the National Heart, Lung, and Blood Institute, National Institute of Health (N01-HV-28179) (R.A.), SystemsX.ch, the Swiss initiative for systems biology (R.A.), the Boehringer Ingelheim Fonds (B.B.), the Howard Hughes Medical Institute (J.S.W), the University of California, San Francisco Strategic Asthma Basic Research Center (J.S.W.), the National Science Foundation Graduate Research Fellowship Program (D.K.B), and the Fannie and John Hertz Foundation (D.K.B).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
D.K.B. designed, carried out, and interpreted experiments. C.S.E performed and analyzed lipidomic measurements with the support of A.S. and K.S. S.R.C. oversaw E-MAP data collection and analysis. B.B. carried out and analyzed protein mass spectrometry experiments to identify sites of phosphorylation under the supervision of R.A. J.S.W designed and interpreted experiments. D.K.B. and J.S.W. prepared the manuscript.
Author Information
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J Lipid Res. 2009;(50 Suppl):S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev. 2009;109:1561–1574. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burg JS, et al. Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab. 2008;8:522–531. doi: 10.1016/j.cmet.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loewen CJ, et al. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 5.Martin CE, Oh CS, Jiang Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim Biophys Acta. 2007;1771:271–285. doi: 10.1016/j.bbalip.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Kurat CF, et al. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Fischer J, et al. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 8.Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Cowart LA, Obeid LM. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim Biophys Acta. 2007;1771:421–431. doi: 10.1016/j.bbalip.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanada K, et al. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 11.D'Angelo G, et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- 12.Kumagai K, Kawano M, Shinkai-Ouchi F, Nishijima M, Hanada K. Interorganelle trafficking of ceramide is regulated by phosphorylation-dependent cooperativity between the PH and START domains of CERT. J Biol Chem. 2007;282:17758–17766. doi: 10.1074/jbc.M702291200. [DOI] [PubMed] [Google Scholar]
- 13.Klemm RW, et al. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185:601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aronova S, et al. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 2008;7:148–158. doi: 10.1016/j.cmet.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vacaru AM, et al. Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J Cell Biol. 2009;185:1013–1027. doi: 10.1083/jcb.200903152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 17.Lebman DA, Spiegel S. Cross-talk at the crossroads of sphingosine-1-phosphate, growth factors, and cytokine signaling. J Lipid Res. 2008;49:1388–1394. doi: 10.1194/jlr.R800008-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akinbami LJ, Moorman JE, Garbe PL, Sondik EJ. Status of childhood asthma in the United States, 1980–2007. Pediatrics. 2009;123(Suppl 3):S131–S145. doi: 10.1542/peds.2008-2233C. [DOI] [PubMed] [Google Scholar]
- 19.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 20.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 21.Galanter J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, et al. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64:629–635. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouzigon E, et al. Effect of 17q21 Variants and Smoking Exposure in Early-Onset Asthma. N Engl J Med. 2008 doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 24.Verlaan DJ, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavendale R, Macgregor DF, Mukhopadhyay S, Palmer CN. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J Allergy Clin Immunol. 2008;121:860–863. doi: 10.1016/j.jaci.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Hjelmqvist L, et al. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-6-research0027. RESEARCH0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 28.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta. 2003;1632:16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 29.Beeler T, et al. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+sensitive csg2Delta mutant. J Biol Chem. 1998;273:30688–30694. doi: 10.1074/jbc.273.46.30688. [DOI] [PubMed] [Google Scholar]
- 30.Ejsing CS, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delgado A, Casas J, Llebaria A, Abad JL, Fabrias G. Inhibitors of sphingolipid metabolism enzymes. Biochim Biophys Acta. 2006;1758:1957–1977. doi: 10.1016/j.bbamem.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 32.Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brice SE, Alford CW, Cowart LA. Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem. 2009;284:7588–7596. doi: 10.1074/jbc.M808325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kearns BG, et al. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blagoveshchenskaya A, Mayinger P. SAC1 lipid phosphatase and growth control of the secretory pathway. Mol Biosyst. 2009;5:36–42. doi: 10.1039/b810979f. [DOI] [PubMed] [Google Scholar]
- 38.Hornemann T, Wei Y, von Eckardstein A. Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochem J. 2007;405:157–164. doi: 10.1042/BJ20070025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera J, Proia RL, Olivera A. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat Rev Immunol. 2008;8:753–763. doi: 10.1038/nri2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ammit AJ, et al. Sphingosine 1-phosphate modulates human airway smooth muscle cell functions that promote inflammation and airway remodeling in asthma. Faseb J. 2001;15:1212–1214. doi: 10.1096/fj.00-0742fje. [DOI] [PubMed] [Google Scholar]
- 41.Masini E, et al. Ceramide: a key signaling molecule in a Guinea pig model of allergic asthmatic response and airway inflammation. J Pharmacol Exp Ther. 2008;324:548–557. doi: 10.1124/jpet.107.131565. [DOI] [PubMed] [Google Scholar]
- 42.Ryan JJ, Spiegel S. The role of sphingosine-1-phosphate and its receptors in asthma. Drug News Perspect. 2008;21:89–96. doi: 10.1358/dnp.2008.21.2.1188195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Idzko M, et al. Local application of FTY720 to the lung abrogates experimental asthma by altering dendritic cell function. J Clin Invest. 2006;116:2935–2944. doi: 10.1172/JCI28295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009 doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirschfield GM, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gable K, Slife H, Bacikova D, Monaghan E, Dunn TM. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J Biol Chem. 2000;275:7597–7603. doi: 10.1074/jbc.275.11.7597. [DOI] [PubMed] [Google Scholar]
- 48.Lester RL, Dickson RC. High-performance liquid chromatography analysis of molecular species of sphingolipid-related long chain bases and long chain base phosphates in Saccharomyces cerevisiae after derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Anal Biochem. 2001;298:283–292. doi: 10.1006/abio.2001.5368. [DOI] [PubMed] [Google Scholar]
- 49.Jiménez CR, L H, Y Q, Burlingame AL. In: In-Gel Digestion of Proteins for MALDI-MS Fingerprint Mapping. Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT, editors. New York: John Wiley & Sons, Inc.; 1998. [DOI] [PubMed] [Google Scholar]
- 50.Huber A, et al. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.