1. INTRODUCTION
MicroRNAs (miRNAs) are 21 or 22 nucleotides noncoding RNAs known to possess important post-transcriptional regulatory functions [1]. Identifying targeting genes that miRNAs regulate is important for understanding their specific biological functions. Usually, miRNAs down-regulate target genes through binding to the complementary sites in the 3' untranslated region (UTR) of the targets. Since the binding of the miRNAs of animals is not a perfect one-to-one match with the complementary sites of their targets, it is difficult to find targets of animal miRNAs by accessing their alignment to the 3' UTRs of potential targets. More sophisticated computational approaches are desirable and have been proposed as a result. The most popular algorithms include TargetScan, miRanda, and PicTar. However, they share similar methodology and are restricted by the human observation of conserved nature of miRNAs and their targets.
In this article, we develop a statistical learning based approach that uses support vector machine (SVM) [2] sas a classifier to predict miRNA targets. SVM have been applied in many fields such as pattern recognition, computational biology, and medical image analysis [3]. With SVM, information is gained automatically from relevant data and therefore human bias can be removed in the decision process.
The design process can be summarized as follows: First, data set for the SVM algorithm was constructed, which consists of 3' UTR of targets and miRNAs sequences of 314 experimentally confirmed (positive) pairs and 186 negative target sequences, which were generated by knocking out the actual target sites of genes let-7 and lin-41. Secondly, 46 features were designed, based on data and existing knowledge of miRNA binding, for SVM implementation. Thirdly, the data set was equally divided into training and testing sets randomly. The SVM implemented by SVMlight [4] was optimized and trained on the training sets and then evaluated by test sets. The testing result shows an accuracy of 95.03% for the proposed algorithm, compared to miTarget's [5] 93.32%.
2. METHOD
2.1. Data set construction
Sequences of miRNA and target site pairs were downloaded from TarBase [6], which contains 314 positive pairs validated by experiments.
To generate sequences of negative targets, binding sites of the mRNAs of two miRNA:site pairs let-7:lin-41 and lin-14:lin-28a were masked first. Then, a sliding window of 30 nucleotides (nts) was used to search for sequences, which has more than 4 nts matches with the seed region of the corresponding miRNA. The seed region is defined as the 1st to the 8th nucleotide of a miRNA. 186 pseudo negative miRNA:site pairs were generated as a result. The rationale behind this practice is the fact that miRNA cannot repress its target expression if the binding site in mRNA is eliminated. To summarize, a data set was constructed including 314 positive pairs from TarBase and 186 negative pairs generated by masking binding sites.
2.2. Feature extraction
Extracting relevant features is a very important step that determines the efficacy and efficiency of the SVM algorithm. According to previous research [7], there are two obvious characters lie in miRNA:site pairs. First, miRNA and its site can bind together with a low free energy. Second, the seed region from the 1st to the 8th nucleotide of a miRNA always matches better than other regions. Based on these characters, two kinds of features were used in this algorithm: position specified features and regional features. There are totally 46 features extracted for the SVM classifier.
2.2.1. Position specified features
3' end of target site and 5' end of miRNA were first linked together with a sequence “LLLLLL”. Then, RNAfold [8] was used to generate the second structure of the sequence. In this step, both Watson-Crick pair and G–U wobble pair are allowed, as was shown in Figure 1. Next, going from the 5' end of miRNA, a sliding window of 2 nts was applied to obtain the match type of every 2 nts. Four types of match were recorded for a single nucleotide: GC match, AU match, GU match and mismatch and thus there are 16 possible types to every 2 nts combination. Then, the decimal numbers 1 to 16 were used to indicate each respective match type. Only first 20nts were counted, since the length of miRNA sequence varies around 20nt. Consequently, 19 features were extracted for 2-nt match type of miRNA:site pairs.20 features were extracted.
Figure 1.
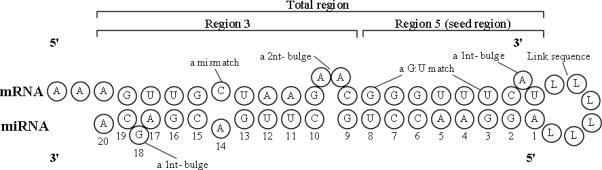
Secondary structure of miRNA and target site
To show the discriminative power of these features, the distributions of the 16 match types for both positive and negative miRNA:site pairs are plotted in Figure 2. As can be seen, the two distributions are quite different, indicating strong discriminative power of these features.
Figure 2.
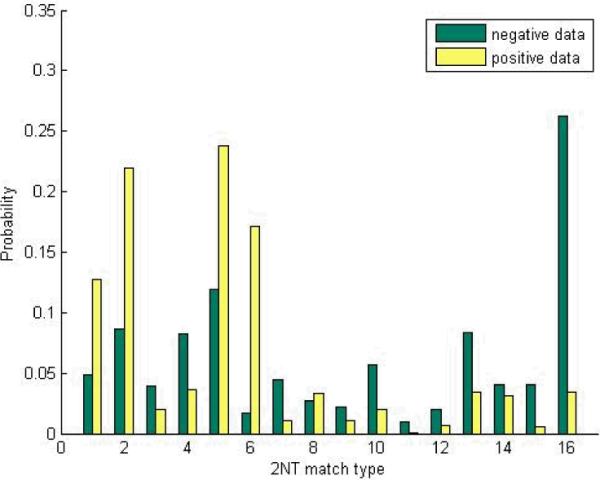
Distributions of 2-nt match type
2.2.2. Regional features
Since miRNA binding has different characteristics in different regions, an miRNA:site pair was divided into 3 parts (regions) and features are extracted to reflect the regional dependent characteristics. There are three regions. The total region refers to the whole miRNA:site pair; Region 5, which is also known as the seed region, refers to the 1st to 8th nucleotides; Region 3 refers to the rest of nucleotides from the 9th to 20th. For each one of the three region, 9 features were extracted that reflect the free energy of the region as well as the number of matches, mismatches, G:C matches, A:U matches, G:U matches, other mismatches, bulges in mRNA, and bulged nucleotides in mRNA.
2.3. Background of SVM
The machine learning method Support Vector Machine is used in two class classification based on the above extracted features. We used an SVM classifier to discriminate positive and negative miRNA:site pairs.
Suppose that there is a feature set x obtained from a data sample that belongs to a class y ε (−1,1), i.e.
| (1) |
where n is the size of the feature vector The objective of a classifier is to identify the correct class y based on the feature set x. In a rather simple case, a linear classifier such as LDA can be applied. However, in many other scenarios including the miRNA target identification, correct class cannot be assign linearly. Then, SVM can be applied to map the features into a high-dimensional space so that classification may be performed linearly by an implicit rule. The exact mapping is implemented by kernel functions. In our research, a radial basis function (RBF) kernel is used:
| (2) |
SVM always tries to find an optimal hyperplane to separate the positive and negative samples, however, the hyperplane cannot be found due to noise in data. So slack variables ξ are introduced to loosen the constraints:
| (3) |
A penalty constant C is also introduced to punish the noise:
| (4) |
An optimal hyperplane will be determined subject to (3) and (4). In our implementation, the SVM package SVMlight was used to construct miRNA target prediction models.
2.4. Parameter optimization and classifier evaluations
In order to optimize the parameters, C and γ, and evaluate the performance of the classifier, four steps were implemented repeatedly as follows. 1) The positive and negative data samples are equally divided into training and testing data sets by random sampling; 2) With C varying from 0.1 to 30 with a step size of 0.1 and γ from 0.01 to 3 with a step size of 0.01, five-fold cross validation based on training data was performed to evaluate the accuracy. Then an optimal parameter set, C and γ, can be found through maximizing the accuracy; 3) SVM was trained with the optimal C and γ based on the entire training data set. 4) Predictive power of the SVM obtained from step 3) is evaluated on the testing data set and the performance measures including sensitivity, specificity, and accuracy were calculated. These four steps were performed 100 times. The average sensitivity, specificity and accuracy among the 100 trial were reported as final performance measurement.
The ROC was further plotted to show the performance. To this end, a threshold varying between maximal score and minimal score was used to get correspond sensitivity and specificity. A score, which stands for the distance between sample and hyperplane, can be calculated when classify the sample. The ROC was plotted with the scores generated by 100 repeated evaluations.
3. RESULTS AND FUTURE WORK
3.1. Results
The testing results were obtained. As a comparison, an existing algorithm miTarget was also implemented on the same dataset. The performance was shown in Table 1 and the ROC is plotted in Figure 3. It can be seen that the proposed algorithm outperforms miTarget.
Table 1.
Performance of proposed algorithm and miTarget
| Algorithm | Sensitivity | Specificity | Accuracy | ROC |
|---|---|---|---|---|
| miTarget | 94.5031 | 94.9013 | 93.32 | 0.9881 |
| Proposed | 95.5606 | 96.5449 | 95.028 | 0.9886 |
Figure 3.
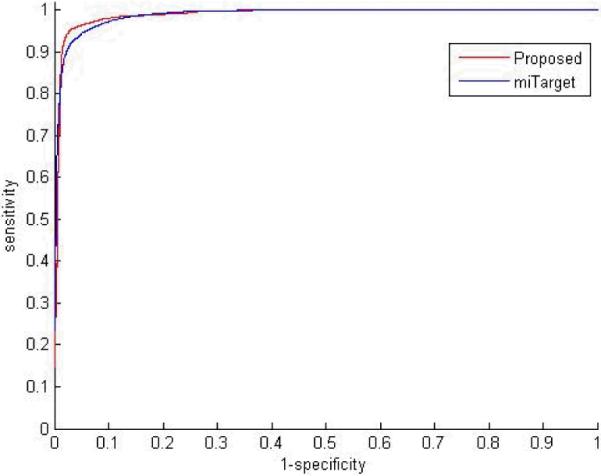
ROC of proposed algorithm and miTarget
3.2. Future work
Based on this work, additional improvements can be carried out. First, since the binding structure of miRNA:site pair generated by RNAfold is not consistent with that supplied by TarBase. Therefore, an algorithm that can better predict the actual binding structure of miRNA:site pairs is highly desirable.
Secondly, approximate site information must be provided and only in-site features was considered in this paper, which restricts its practical use. It is shown in [7] that additional statistical characteristics of miRNA targets can be found not only inside sites but also in whole 3'UTR. Therefore how to design useful features for multiple sites and out-site information will be pursued next.
It is also of great interest to integrate additional data sources such as microarray data into target prediction. These data can provide additional information that cannot be captured by sequence data and thus further improve the prediction accuracy.
4. ACKNOWLEDGMENTS
Hui Liu and Lin Zhang are supported by ?????
Yufei Huang is supported by an NSF Grant CCF-0546345.
5. REFERENCES
- [1].Lai E. Runts of the genome assert themselves. Current Biology. 2003;13:R925–36. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- [2].Boser BE, Guyon IM, Vapnik V. A training algorithm for optimal margin classifiers. Pittsburgh; 1992. [Google Scholar]
- [3].Wang X, Naqa I. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics Advance Access. 2007 Nov 29; doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- [4].Joachims T. In Advances in Kernel Methods: Support Vector Machines. MIT Press; Cambridge, MA: 1998. Making large-scale support vector machine learning practical; pp. 169–184. [Google Scholar]
- [5].Kim S, Nan J, Rhee J, Lee W, Zhang B. miTarget: microRNA target gene prediction using a support vector machine. BMC Bioinformatics. 2006;7:411. doi: 10.1186/1471-2105-7-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sethupathy P, Corda B, Hatziegeorgiou TarBase: A comprehensive database of experimentally supported animal microRNA targets. A.G. 2006. RNA. 12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Grimson Andrew, Farh Kyle Kai-How, Johnston Wendy K, Garrett-Engele Philip, Lim Lee P, Bartel David P. MicroRNA Targeting Specificity in Mammals: Determinants beyond Seed Pairing. Molecular Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P. Fast Folding and Comparison of RNA Secondary Structures. Monatshefte f. Chemie. 1994;125:167–188. [Google Scholar]


