The roles that RNA molecules play in the regulation of gene expression have only recently become apparent. Recent work in this area has uncovered several complex, RNA-mediated networks of gene regulation in eukaryotic systems. One newly discovered mechanism of RNA mediated gene regulation takes place at the level of transcription. In yeast, plant and mammalian systems, small RNAs targeted to gene promoters can result in a repression of transcription. Small RNA mediated transcriptional silencing has been shown to be operative by changes in chromatin structure at the targeted promoter. Specifically, silencing has been observed to correlate with decreases in certain active-state histone modifications, increases in various silent state histone methylation marks, and in some instances, DNA methylation at the targeted promoter. These epigenetic remodeling events represent a more stable, heritable form of gene regulation as opposed to the transitory post-transcriptional regulation observed in traditional RNAi mechanisms. Several recent findings have shed light on this newly discovered link between small RNA molecules and epigenetic regulatory machinery, notably in human cells.
RNA Interference Post-Transcriptional Gene Silencing
RNA interference (RNAi) is a process by which double-stranded RNA induces homology dependent inhibition of gene expression. 1-3 What is referred to as RNAi in animals is known as quelling in the filamentous fungus Neurospora crassa,4-6 and was initially described in plants where it was termed co-suppression.6 Upon further examination, it became apparent that RNAi is operative in targeted gene suppression by two distinct pathways involving small dsRNAs: transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS).7,8 The better characterized molecular pathway, PTGS, involves siRNAs targeting a gene transcript (mRNA), and in human cells has been shown to be operable in both the cytoplasm and nucleus.9,10 The endogenous equivalent to the PTGS based pathway utilizes micro-RNAs (miRNAs). The miRNA pathway involves the processing of the precursor RNA first by the RNAi related proteins Drosha and Dicer to small single stranded regulatory RNAs, ~21–22 nucleotides long. Once the small RNAs are processed they are then capable of interfering with mRNA expression through interactions with the 3′UTR or by directly targeting the homologous genes mRNA for degradation or translational inhibition,11 these silencing activities appear to take place in cytoplasmic P-bodies.12 Message targeted miRNAs may also function to repress translation by interactions of RISC with the ribosome. 13
Small RNAs and Transcriptional Regulation
The majority of work carried out with small RNAs in human cells has focused on siRNA and/or miRNA directed PTGS of cellular mRNAs, and the determination of complexes involved in this process. However, small RNAs can also be utilized to modulate gene expression by directly inducing epigenetic modifications at targeted gene promoters, resulting in transcriptional gene silencing.
Epigenetics is the study of meiotically and mitotically heritable changes in gene expression which are not coded for in the DNA. 14,15, Most current epigenetic studies focus specifically on chromatin structure and chemical DNA modifications. One common epigenetic modification is DNA methylation, specifically found on CpG dinucleotides. DNA methylation is a robust and long lasting epigenetic mark that is conserved and maintained by the activity of DNA methylatransferase 1 (DNMT1). DNMT1 functions to methylate hemi-methylated DNA during the action of DNA replication and cell division.16 De Novo DNA methylation is mediated by several proteins including DNMT3a and DNMT3b, and can be induced by a multitude of stimuli.17
Another common type of epigenetic regulation of gene expression involves chemical modifications of DNA-bound histones. Histone modifications can be both activating and deactivating, depending on the type of modification and the specific residue modified. The chemical modifications at various residues within histones and the affect these modifications have on gene expression is collectively referred to as the “histone code hypothesis”.18,19 Such modifications are most well characterized on histones 3 and 4 and include phosphorylation, ubiquitination, SUMOylation, acetylation and methylation. Importantly, histone 3 methylation on lysines 9 and 27 (K9 and K27, respectively) has been shown to direct the formation of repressive heterochromatin through the recruitment of heterochromatin protein 1 and polycomb group proteins.20,21
Recently it has become clear that in many instances, epigenetic modifications can be induced and directed by small RNA molecules. Small RNA mediated transcriptional gene silencing (TGS) was first observed when doubly transformed tobacco plants exhibited a suppressed phenotype of a transgene that correlated with DNA methylation at the transgene promoter.22 TGS mediated by small RNAs was further substantiated in viriod-infected plants23 and was shown to be the result of the action of RNA-dependent DNA methylation (RdDM). RdDM requires a dsRNA to target the genomic loci.23,24 Recently, members of the Argonaute protein family in Arabidopsis have been shown to play an essential role in RdDM of gene promoter DNA and transposon silencing.25 Specifically, Ago4 is known to direct small RNA mediated silencing, and Ago4 mutants display reactivation of silent alleles, along with a corresponding decrease in both CpNpG DNA and H3K9 methylation. 26 Consequently, in plants siRNAs containing homologous sequences with gene promoter regions are capable of directing DNA and histone methylation of the homologous promoter and sequence specific transcriptional gene silencing. Importantly, RNA directed TGS is a longer-lasting if not permanent mode of silencing as epigenetic modifications are involved in the silencing whereas RNA directed PTGS is more transient as slicing of the target gene transcripts is involved. To date the ability of siRNAs to modulate TGS has been shown in plants (Arabidopsis), yeast (S. Pombe), flies (Drosophila), worms (C. elegans) and most recently in human cells.27
Comparative Analysis of RNA Mediated TGS in Various Organisms
Similarly to plants, the fission yeast S. Pombe also employs small RNA directed TGS. S. Pombe, however, lacks the epigenetic mechanism of DNA methylation. Instead S. Pombe utilizes Argonaute 1 (Ago1) to direct histone methylation and heterochromatin formation.25 RNAi mediated TGS in S. Pombe operates specifically through Histone 3 Lysine-9 methylation (H3K9).28 The result of targeted histone methylation is a compaction of the local DNA with it’s nucleosome and presumably a restriction in RNA polymerase II (RNAPII) activity at the targetted loci that results in transcriptional gene silencing. In S. Pombe mutants in dcr1 (Dicer homolog) and ago1 (Argonaute homolog) were shown to be reduced in centromeric repeat histone 3 lysine 9 di-methylation (H3K9me2), which is necessary for centromere function. 28 These data denote a link between small RNA targeting of specific genomic sequences and the recruitment of Swi6, which results in regulation of the heterochromatic state.28 Recently, RNA-mediated TGS in S. Pombe has been shown to also require some level of RNAPII activity,29 possibly for the expression of an RNA that overlaps the siRNA targeted genomic region. Indeed it has been shown that an RNA/Ago-1 complex can function to slice these gene associated RNAs resulting in TGS of the targeted gene in S. Pombe.30
RNAi mediated TGS appears to be conserved throughout many biological systems. Clearly, the majority of work and detailed characterization of the underlying mechanism involved in TGS has been carried out in S. Pombe and plants. However, other model organisms have demonstrated small dsRNA mediated TGS with slight variations in the underlying theme. In the fungi Neurospora crassa the silencing of homologous sequences has been termed quelling.31 Quelling tends to be used interchangeably in fungi to refer to both PTGS and TGS based mechanisms. Interestingly, in Neurospora both the PTGS and TGS pathways utilize histone 3 lysine 9 dimethylation and yet appear to be distinct from one another.32 In the fruit fly Drosophila, however, the two pathways of RNAi, PTGS and TGS, appear connected via piwi proteins.7 The piwi family of proteins is a composed of several homologous members (piwi/sign/elF2C/rde1/argonaute), which are conserved between many plant and animal species.33-35 Interestingly, in the nematode,C. Elegans, the PAZ-PIWI like protein Rde-1 plays an essential role in RNA mediated silencing in the soma. However, other RNA mediated silencing mechanisms, operative in the germline of C. Elegans, do not appear to require Rde-1.36,37 These data indicate a distinct mechanistic bifurcation in germline compared to somatic cell silencing and RNA mediated modes of gene silencing.
In C. elegans, RNA mediated transcriptional silencing of somatic transgenes has been shown to be the mediated by the ADAR-encoding genes, adr-1 and adr-2, and to be dependent on Rde-1.38 The observed silencing and requirements for adr-1, adr-2 and Rde-1 was characterized by investigation of siRNAs targeted to pre-mRNAs. The silencing corresponded with a decrease in both RNAPII localization and acetylated histones at the targeted genomic region.38 Interestingly, following an RNAi screen in C. elegans, genes encoding RNA-binding, polycomb, chromodomain proteins and histone methyltransferases were detected.38 These data were recapitulated in C. Elegans in an interesting set of experiments which demonstrated long-term transcriptional gene silencing by RNAi. In essence, one dose of siRNA was capable of modulating gene silencing that was inherited indefinitely in the absence of the original siRNA trigger.39 This observed long-term inheritance of siRNA mediated TGS appeared to require hda-4 (a class II histone deacetylase), K03D 10.3 (a histone acetyltransferase of the MYST family), isw1 (a homolog of the Yeast chromatin-remodeling ATPase ISW1), and mrg-1 (a chromodomain protein).39 Taken together these data strongly suggest that RNA mediated TGS in C. elegans contains a convergence of pathways that include epigenetic modifying factors and small RNAs. Overall, when comparisons are made between plants, yeast, worms and flies, one cannot help but notice that indeed RNA is more intricately involved in the regulation of gene expression than has previously been envisioned. This strongly suggests that other organisms might also utilize RNA directed mechanisms to regulate gene expression epigenetically, namely, humans.
RNA Mediated TGS in Human Cells
Observations of RNA mediated TGS in human cells has lagged behind work done in Arabidopsis, S. Pombe and C. Elegans. However, recent studies have revealed that RNA mediated TGS is operative in human cells and that the observed silencing is the result of small RNA directed methylation of histone 3 lysine’s 9 and 27 (H3K9 and H3K27, respectively), and DNA methylation at the targeted promoter.40-45 However, DNA methylation may not be a conserved feature in all cases of small RNA-mediate TGS in human cells46-48 or may be a function of the duration of targeted small RNA exposure to the target loci in the cell.
More recently it has become clear that only the 21 bp antisense (‘guide’) strand of the promoter targeted small RNA is required to modulate TGS of the targeted promoter.43,45 While the role of histone methylation in small RNA mediated TGS is well established in human cells, only recently have components of the RNAi pathway been shown to be implicated in directing the small RNAs to the targeted promoter regions and modulating the silent state chromatin modifications. To date the components of the RNAi pathway shown to be involved in small RNA mediated TGS are the Argonautes (Ago-1 and Ago-2).43,49 Interestingly, time-course experiments suggest that the argonautes are functional in TGS prior to the appearance of histone methylation at the targeted promoter as silent state histone modifications increase at the targeted loci concomitantly will the enrichment of Ago-1 diminishes.43
In addition to Ago 1, the DNA methyltransferase DNMT3a has been implicated in the silencing activity of promoter targeted small RNAs. DMNT3a has been show to elute specifically with small RNAs at the targeted gene promoter regions of some genes.45 One implication of the epigenetic mechanism of transcriptional regulation induced by promoter targeted small dsRNAs is the possibility for long-term heritable changes in expression levels. Recent work from our lab indicates that long-term gene suppression by promoter targeting may in fact be possible. In addition, long term silencing may be achieved after relatively short periods of exposure to the effector small RNA molecule. Interestingly, the time scale for establishment of long-term silencing correlates with the appearance of DNA methylation. This would be predicted as DNA methylation of promoters is a more heritable, long-term form of epigenetic gene silencing than histone modifications alone. Long-term silencing is not, however, associated with a loss of RNA polymerase II (RNAPII) at the targeted promoter, although it appears to remain in an inactive state.
Promoter associated RNAs and TGS
As discussed previously, active RNAPII has been implicated in small RNA-mediated TGS in human cells and shown to co-immunopreciptate with Ago-1 at siRNA targeted gene promoter loci.43 A role for RNAPII in this process is not necessarily intuitive and was initially explained by two competing models. The first was the RNA-DNA model, in which transcription by active RNAPII results in a melting of the DNA duplex at the promoter, making one strand available for hybridization with the small RNA (Fig. 1A). This small RNA would either already be in complex with epigenetic remodeling proteins, or would recruit them to the site upon hybridizing. Supporting this model was the observation that RNAPII has been shown to associate with complete unfolding of 1.85 out of 3 nucleosomes upstream of the transcription start site for the PH05 promoter,50 and the siRNA EF52 target site, shown to initiate TGS in human cells, is ~1 nucleosome upstream of the TATAA transcriptional start site.41,44 Moreover, RNAPII is associated with nearly 60 subunits and a mass in excess of 3 million Da, including the crescent-shaped Mediator, which essentially envelopes the RNA Pol-II core subunits.50 This model seems most plausible when small RNAs are targeted to RNAPII start sites, or TATAA, subsequently resulting in a small RNA/DNA interaction and robust transcriptional silencing.42 However, silencing of regions upstream of the transcriptional start site or TATAA also result in gene silencing but perhaps this silencing is operative via a different mechanism involving an RNA-RNA mediated interaction (Fig. 1B).
Figure 1.
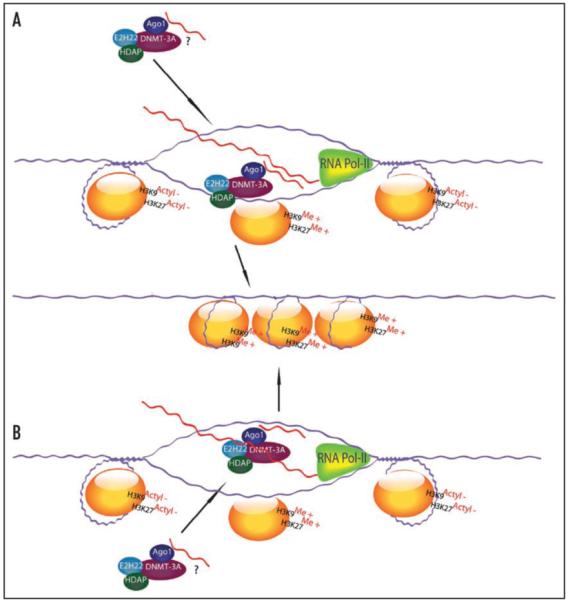
Putative models for small RNA directed TGS in human cells. (A) The RNA/RNA model of transcriptional gene silencing is shown where the antisense strand of the small RNA either binds or associates with Ago-1 and possibly Ezh2, DNMT3a or HDAC-1 and then localizes to the elongating RNAPII expressed transcript that spans the targeted promoter loci. This model is supported by data showing that RNAPII activity and a low copy promoter associated RNA is involved in small RNA directed TGS in human cells.41,45 Alternatively, or more common at TATAA loci and transcriptional start sites the RNA/DNA model may be operative. (B) In the RNA/DNA model the antisense strand of the small RNA may localize with the transcriptional silencing complex containing Ago-1, Ezh2, DNMT3a and HDAC-1 to the genomic DNA. The localization of these components to the transcriptional start site might alter later RNAPII binding and activity. This model is supported by data showing transcriptional start sites are susceptible to small antigene RNA targeting.42
In the RNA/RNA model RNAPII synthesizes a low-copy number transcript that corresponds with the targeted promoter to which the small RNA binds (Fig. 1B). Initial support for this model came from the observations that in S. pombe T7-mediated transcribed genomic regions were not susceptible to siRNA-mediated TGS while RNAPII regions were, and that heterochromatin formation in mouse cells involves HP1 proteins and treatment with RNase causes a dispersion of heterochromatin-associated protein (HP1) from pericentromeric foci.51 Recent findings support the RNA-RNA model and incorporate a role for RNAPII, which has been shown to be required for TGS,44 in small RNA mediated TGS (Fig. 1B). This conclusion was reached by the identification of low-copy promoter transcripts spanning several human gene promoters.41 Blocking of these low copy promoter-associated transcripts using complimentary phosphorothioate oligonucleotides (ODNs) results in abrogation of silencing at the small RNA targeted promoter. The characterization of this novel transcript, in all genes that were assessed, and of its role in transcriptional regulation represents a significant step towards understanding the complex and varied roles that RNA molecules play in regulating cellular activities.
Many questions remain regarding small RNA mediated TGS in human cells. Perhaps the most pressing issue is the identification of an endogenous trigger for this process in human cells. To date all examples of small RNA directed TGS in human cells has been the result of transfecting small RNAs designed a priori to target a particular gene promoter loci. Thus, essentially two scenarios are plausible, either (1) the mechanism which is being employed to transcriptionally silence gene promoter loci is a vestigial mechanism, or (2) the mechanism is endogenously active at some stage in cell development. In either instance, directed control of gene transcription is feasible with small antisense RNAs targeted to particular regions of RNAPII gene promoters. If this particular mechanism proves to be vestigial, then there is little doubt this pathway should be capitalized on for therapeutic benefit as off-target effects, which often result from targeting physiologically relevant pathways, would be limited. Clearly, the identification of an endogenously expressed, promoter targeted small RNA which mediates TGS in human cells would solidify this phenomenon as a physiologically relevant process.
Gene Activation
In addition to the suppressive activities of promoter targeted small RNAs, evidence is mounting which suggests that small RNAs are involved in many additional aspects of transcriptional regulation. One such finding is the observation that small RNAs targeted to promoter regions low in GC content and high in complexity can activate certain genes.52,43 Activation in these cases seemed to be mediated by histone modifications, and in one instance53 was shown to confer long-term activation. Importantly, this work demonstrated that promoter targeted small RNA molecules do not inherently function in only an inhibitory manner. Another interesting recent finding is that a liver specific miRNA is involved in the positive regulation of hepatitis C replication in human liver.54
However, in other instances gene activation by small RNAs was shown to be the result of off-target effects.55 In this example, a small RNA was found to activate HIV-1 LTR transcription. Upon examination it became apparent that the small RNA was targeting a non-coding RNA, C10orf76, which appeared to exhibit a cell wide regulatory effect. When C10orf76 was suppressed by RNAi, increased cell-wide gene expression was observed.55 While the mechanism of small RNA directed transcriptional activation are not completely understood, these observations provide yet another example of RNA mediated gene regulation. Clearly, these recent findings point to the existence of a complex genetic regulatory network mediated by RNA.
Non Coding RNAs
With the discovery of RNAi came an explosion of new small RNA species such as microRNAs (miRNAs) and piwi associated RNAs (piRNAs).56 The observation and characterization of these small RNA species has significantly changed the perception of how gene expression is controlled.57 It has also become apparent that several non-coding RNAs (ncRNA)58-60 are present within the cell and that the majority of the entire human genome appears to be transcribed.61,62 It is also very intriguing that ncRNA expression profiles do not appear to be conserved between various organisms,63 suggesting a role for ncRNAs in the differential aspects of various species. NcRNAs have been shown to be involved in X inactivation, dosage compensation, imprinting and polycomb mediated silencing.62 Interestingly, many of the ncRNAs that have been detected have been in the antisense orientation relative to gene transcription58,64 contain some homology to gene promoter loci.65 These observations support earlier notions that sequence specific ncRNAs might be involved in the regulation of gene promoter activity. Indeed, such an eventuality has been observed in S. cerevisiae where the ncRNA, SRG1 has been observed to interfere with SER3 gene expression.66,67
Evolutionary and Therapeutic Implications
There are many implications from this work in the fields of evolutionary biology and medicine. One prominent evolutionary theory states that at some point in Earth’s evolutionary history, organisms may have existed which relied entirely on RNA for the storage of information as well as the completion of enzymatic cellular processes. RNA is well suited for these functions as the molecule is able to encode information as well as act enzymatically, as a ribozyme, to catalyze necessary chemical reactions.68 As our understanding of the various roles that RNA plays in gene regulation expands, the prospect that all cellular processes once relied on RNA molecules seems more and more possible.
As aberrant gene expression is the cause of many human diseases, the ability to specifically modulate human gene expression holds exciting potential for the development of novel therapeutics. In fact, adaptations of the PTGS and TGS pathways of RNAi are currently being investigated as potential treatments for diseases ranging from AIDS to cancer. In addition to gene therapy applications, it is also possible that previously uncharacterized factors involved in these forms of genetic regulation may prove to be feasible targets for the development of new therapeutics.
References
- 1.Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:15502–7. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishikura K. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell. 2001;107:415–8. doi: 10.1016/s0092-8674(01)00581-5. [DOI] [PubMed] [Google Scholar]
- 3.Sharp PA. RNA interference. Genes and Development. 2001;15:485–90. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 4.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Hutvagner G, Zamore PD. RNAi: nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–32. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 6.Tijsterman M, Ketting RF, Plasterk RH. The genetics of RNA silencing. Annu Rev Genet. 2002;36:489–519. doi: 10.1146/annurev.genet.36.043002.091619. [DOI] [PubMed] [Google Scholar]
- 7.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in drosophila. Mol Cell. 2002;9:315–27. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 8.Sijen T, Vign I, Rebocho A, Blokland R, Roelofs D, Mol J, Kooter J. Transcriptional and posttranscriptional gene silencing are mechansitically related. Curr Biol. 2001;11:436–40. doi: 10.1016/s0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- 9.Langlois MA, Boniface C, Wang G, Alluin J, Salvaterra PM, Puymirat J, Rossi JJ, Lee NS. Cytoplasmic and nuclear retained DMPK mRNAs are targets for RNA interference in myotonic dystrophy cells. J Biol Chem. 2005;280:16949–54. doi: 10.1074/jbc.M501591200. [DOI] [PubMed] [Google Scholar]
- 10.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol. 2005;12:133–7. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 11.Leung AK, Sharp PA. Function and localization of microRNAs in mammalian cells. Cold Spring Harb Symp Quant Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 15.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 16.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 17.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 18.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 19.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 20.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 21.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–64. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Matzke MA, Primig M, Trnovsky J, Matzke AJM. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. The EMBO Journal. 1989;8:643–9. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wassenegger M, Graham MW, Wang MD. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–76. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 24.Mette MF, Aufsatz W, Van der Winden J, Matzke AJM, Matzke MA. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. The EMBO Journal. 2000;19:5194–201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct Mechanisms Determine Transposon Inheritance and Methylation via Small Interfering RNA and Histone Modification. PLoS Biol. 2003;1:67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–9. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 27.Morris KV. siRNA-mediated transcriptional gene silencing: the potential mechanism and a possible role in the histone code. Cell Mol Life Sci. 2005;62:3057–66. doi: 10.1007/s00018-005-5182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of Heterchromatic Silencing and Histone H3 Lysine-9 Methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 29.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–9. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 30.Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–7. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 31.Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–53. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 32.Chicas A, Forrest EC, Sepich S, Cogoni C, Macino G. Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation necessary for transgenic tandem repeat stabilization in Neurospora crassa. Mol Cell Biol. 2005;25:3793–801. doi: 10.1128/MCB.25.9.3793-3801.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA. 2000;97:11650–4. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 35.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–32. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 36.Dernburg AF, Zalevsky J, Colaiacovo MP, Villeneuve AM. Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 2000;14:1578–83. [PMC free article] [PubMed] [Google Scholar]
- 37.Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–8. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- 38.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005 doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- 40.Castanotto D, Tommasi S, Li M, Li H, Yanow S, Pfeifer GP, Rossi JJ. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol Ther. 2005;12:179–83. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422–7. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol. 2005;1:210–5. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 43.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006 doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 44.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 45.Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen Z, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2005:12. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park CW, Chen Z, Kren BT, Steer CJ. Double-stranded siRNA targeted to the huntingtin gene does not induce DNA methylation. Biochem Biophys Res Commun. 2004;323:275–80. doi: 10.1016/j.bbrc.2004.08.096. [DOI] [PubMed] [Google Scholar]
- 47.Svoboda P, Stein P, Filipowicz W, Schultz RM. Lack of homologous sequence-specific DNA methylation in response to stable dsRNA expression in mouse oocytes. Nucleic Acids Res. 2004;32:3601–6. doi: 10.1093/nar/gkh697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 49.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;9:787–92. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 50.Boeger H, Bushnell DA, Davis R, Griesenbeck J, Lorch Y, Strattan JS, Westover KD, Kornberg RD. Structural basis of eukaryotic gene transcription. FEBS Lett. 2005;579:899–903. doi: 10.1016/j.febslet.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 51.Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–81. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 53.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 55.Weinberg MS, Barichievy S, Schaffer L, Han J, Morris KV. An RNA targeted to the HIV-1 LTR promoter modulates indiscriminate off-target gene activation. Nucleic Acids Res. 2007;35:7303–12. doi: 10.1093/nar/gkm847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartig JV, Tomari Y, Forstemann K. piRNAs—the ancient hunters of genome invaders. Genes Dev. 2007;21:1707–13. doi: 10.1101/gad.1567007. [DOI] [PubMed] [Google Scholar]
- 57.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–8. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Mattick JS, Suzuki H. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mattick JS. A new paradigm for developmental biology. The Journal of experimental biology. 2007;210:1526–47. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 60.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 61.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 62.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;1128:763–76. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 63.Babak T, Blencowe BJ, Hughes TR. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genomics. 2005;6:104. doi: 10.1186/1471-2164-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Timmons JA, Good L. Does everything now make (anti)sense? Biochem Soc Trans. 2006;34:1148–50. doi: 10.1042/BST0341148. [DOI] [PubMed] [Google Scholar]
- 65.Finocchiaro G, Carro MS, Francois S, Parise P, DiNinni V, Muller H. Localizing hotspots of antisense transcription. Nucleic Acids Res. 2007;35:1488–500. doi: 10.1093/nar/gkm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–4. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 67.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cech TR. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987;236:1532–9. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]


