Abstract
Nucleophosmin (NPM), an oligomeric phosphoprotein and nucleolar target of the ARF tumor suppressor, contributes to several critical cellular processes. Previous studies have shown that the human NPM’s phosphorylation by cyclin E–cyclin-dependent kinase 2 (cdk2) on threonine (Thr) 199 regulates its translocation from the centrosome during cell cycle progression. Given our previous finding that ARF directly binds NPM, impeding its transit to the cytoplasm and arresting cells before S-phase entry, we hypothesized that ARF might also inhibit NPM phosphorylation. However, ARF induction did not impair phosphorylation of the cdk2 target residue in murine NPM, Thr198. Furthermore, phosphorylation of Thr198 occurred throughout the cell cycle and was concomitant with increases in overall NPM expression. To investigate the cell’s presumed requirement for NPM-Thr198 phosphorylation in promoting the processes of growth and proliferation, we examined the effects of a non-phosphorylatable NPM mutant, T198A, in a clean cell system in which endogenous NPM had been removed by RNA interference. Here, we show that the T198A mutant is fully capable of executing NPM’s described roles in nucleocytoplasmic shuttling, ribosome export and cell cycle progression. Moreover, the proliferative defects observed with stable NPM knockdown were restored by mutant NPM-T198A expression. Thus, we demonstrate that the reduction in NPM protein expression blocks cellular growth and proliferation, whereas phosphorylation of NPM-Thr198 is not essential for NPM’s capacity to drive cell cycle progression and proliferation.
Keywords: NPM, ribosome, p19ARF, centrosome
Introduction
A highly abundant and evolutionarily conserved nucleolar phosphoprotein, nucleophosmin/B23 (NPM), exhibits a dynamic subcellular localization throughout the cell cycle and has been reported to interact with RNA and a diverse suite of proteins, including p19/p14ARF, p53, nucleolin, ribosomal protein L5, GADD45a and a host of viral proteins (Li, 1997; Liu and Yung, 1999; Colombo et al., 2002; Brady et al., 2004; Gao et al., 2005; Yu et al., 2006). Consequently, NPM has been described as a key player in a number of cellular processes, such as the genotoxic stress response, ribosome biogenesis and centrosome duplication (Spector et al., 1984; Okuda, 2002; Yang et al., 2002; Maggi et al., 2008). Although a proteomic analysis of isolated centrosomes failed to corroborate previous reports of NPM’s direct association with the centrosome, several studies in cell culture systems and mouse models have indicated that NPM is a critical regulator of genomic stability and centrosome duplication, be it through a direct or indirect mechanism (Tokuyama et al., 2001; Grisendi et al., 2005).
To ensure the transmission of an intact, diploid genome from one generation to the next, mitotic cells must temporally coordinate the processes of centrosome duplication, DNA replication and cell cycle progression (Winey, 1999). Fibroblasts derived from Npm1−/− embryos rapidly display centrosomal amplification and chromosomal instability in the culture, leading to activation of p53, induction of p21-mediated growth arrest and premature expression of senescence markers (Grisendi et al., 2005). Previous studies have shown that human NPM was bound to single, unreplicated centrosomes in late G1 and underwent phosphorylation by cyclin E–cyclin-dependent kinase 2 (cdk2) at threonine 199 (Thr199; Thr198 in murine NPM), prompting NPM’s dissociation from the centrosome and its subsequent duplication (Okuda et al., 2000; Tokuyama et al., 2001). Other groups have observed NPM’s interaction with duplicated centrosomes in mitotic cells (Zatsepina et al., 1999), yet independent groups failed to detect NPM in preparations of purified centrosomes (Andersen et al., 2003; Cha et al., 2004). Consequently, NPM’s physical association with the centrosome and its purported role as a direct catalyst of centrosome duplication continue to be subjects of discussion and debate in the field.
In addition to NPM’s phosphorylation by cyclin E–cdk2, its nuclear export by the Ran–Crm1 complex has also been implicated in NPM’s induction of centrosome duplication. Overexpression of NPM nuclear export signal mutants or treatment with leptomycin B, an inhibitor of Crm1-mediated nuclear export, effectively impedes NPM export, resulting in NPM’s accumulation in the nucleus and its dissociation from the centrosome (Shinmura et al., 2005; Wang et al., 2005). In addition, human cells treated with leptomycin B or small interfering RNAs (siRNAs) targeting NPM display centrosome amplification, indicating that Crm1-mediated NPM nuclear export suppresses repeated centrosome duplication cycles, presumably through NPM’s observed localization to the centrosome (Shinmura et al., 2005; Wang et al., 2005). Using similar methods in primary mouse embryonic fibroblasts (MEFs), we have previously demonstrated that NPM expression and nucleocytoplasmic shuttling are required for cell cycle progression (Brady et al., 2004; Yu et al., 2006). The integration of our findings with previously published reports (Tokuyama et al., 2001; Shinmura et al., 2005; Wang et al., 2005) suggests that NPM may use its robust expression, nuclear export and phosphorylation at Thr198 to temporally coordinate the processes of centrosome duplication and cellular proliferation.
To date, phosphorylation of NPM-Thr198 has not definitively been shown to be essential for cell growth and proliferation. Nonetheless, centrosomes and their duplication are believed to play a crucial role in cell cycle progression, although recent studies have challenged this view (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001; Uetake et al., 2007). Recalling that an alanine substitution mutant (T199A) of human NPM failed to dissociate from the centrosome and initiate duplication (Tokuyama et al., 2001), we reasoned that parallel mutation of Thr198 in the murine NPM ortholog would severely compromise the proliferation of primary MEFs. Also, given our previous finding that the ARF tumor suppressor effectively blocked NPM nuclear export (Brady et al., 2004), a critical factor in NPM’s promotion of centrosome duplication and cellular proliferation, we hypothesized that ARF might also inhibit NPM-Thr198 phosphorylation. Here, we report that ARF cannot attenuate the phosphorylation of NPM. Moreover, we demonstrate that NPM expression levels, and not Thr198 phosphorylation, define the cell’s capacity to synthesize and export ribosomes, progress through the cell cycle and proliferate.
Results
NPM is pro-growth in the absence of Arf and a potent transforming oncogene in the absence of p53
To further investigate NPM’s contribution to cell proliferation and transformation, the impact of NPM overexpression in immortal Arf−/− or p53−/− MEFs was tested. Similar to transduction with oncogenic RasV12, exogenous expression of NPM induced a significant increase in Arf−/− cell size, as evidenced by flow cytometric measurements of forward and side scatter (Figure 1a). In agreement with previous findings in immortalized rodent cells (Kondo et al., 1997), overexpression of NPM significantly increased the size of p53−/−-transformed cell colonies that grew in soft agar, although not to the extent of RasV12 (Figure 1b).
Figure 1.
Nucleophosmin (NPM) drives oncogenic growth and proliferation. (a) Arf−/− mouse embryonic fibroblasts (MEFs) infected with retroviruses encoding the control vector, His-tagged NPM or H-RasV12, were fixed and subjected to forward and side scatter analysis by flow cytometry. The upper right quadrant represents the cell population showing increased size. (b) p53−/− MEFs infected with retroviruses encoding the control vectors, H-RasV12, His-NPM and His-NPM T198A, were seeded (3 × 103) in quadruplicate wells of a 24-well plate in media containing soft agar and were assessed for colony formation 14 days later. (c) Primary human breast, prostate and colon carcinoma tissue microarrays were obtained and immunohistochemically stained for NPM protein expression. Representative samples displaying negative staining for NPM are shown in the top panels and those exhibiting strong positive staining for NPM are shown in the bottom panels. The percentage of analyzed tumors showing positive NPM protein expression for each carcinoma type is indicated in the insets. (d) Arf−/− MEFs were infected with retroviruses encoding the control vector or His-tagged NPM for 72h, and were treated with colcemid, harvested and fixed for preparation and visualization of chromosomes with DAPI (4′,6-diamidino-2-phenylindole) (representative of 75 metaphase spreads counted, upper left). Cells infected in parallel were fixed and immunofluorescently stained with antibodies recognizing γ-tubulin to label centrosomes, and nuclei were demarcated with DAPI (upper right). For each condition, centrosomes from over 200 cells were counted in three separate experiments (n=3) and results graphed (plot, lower right). Exogenous His-tagged NPM protein expression was confirmed by western blot analysis (lower left).
To further address the putative role of NPM in promoting cell proliferation and transformation, 60 tissue samples from breast, prostate and colon carcinomas, were analyzed using NPM immunohistochemistry. Approximately 10–18% of Ki-67-positive tumor samples exhibited negative staining for NPM (Figure 1c, top panels), indicating that a subset of highly proliferative tumors does not upregulate NPM expression to drive proliferation. However, the remaining 82–90% of Ki-67-positive tumors did show positive staining for NPM, and nearly 50% of these samples displayed a strong nuclear/nucleolar NPM expression pattern, regardless of tumor type (Figure 1c, bottom panels).
Arf−/− MEFs, although immortal, remain diploid (Kamijo et al., 1997) and retain normal numbers of centrosomes when passaged in culture (Figure 1d, right panels). Genetic ablation of Npm1 results in centrosome amplification and genomic instability in MEFs (Grisendi et al., 2005), suggesting that NPM plays a critical regulatory role maintaining proper centrosome duplication. Given this and other corroborating reports (Okuda et al., 2000; Tokuyama et al., 2001; Wang et al., 2005), the influence of exogenous NPM expression on the ploidy and centrosome amplification in Arf−/− MEFs was examined. As shown in Figure 1d, NPM overexpression did not impact the overall chromosome number in Arf−/− MEFs, nor did it alter the number of centrosomes in these cells. Taken together, these findings demonstrate that the pro-growth and transforming properties of NPM are not coupled to the regulation of DNA ploidy changes or centrosome number.
Cell cycle position or ARF induction does not alter phosphorylation of NPM-Thr198
In response to hyper-proliferative cues, such as oncogenic signals emanating from Myc, E1A and Ras, ARF is induced, and antagonizes Mdm2, to promote p53-dependent pathways of growth arrest (Sherr and Weber, 2000). We have previously shown that ARF uses a common domain at its N terminus to bind both Mdm2 and NPM, resulting in the nucleolar sequestration of each protein independent of the other (Brady et al., 2004). ARF not only delocalizes Mdm2 to the nucleolus, away from active pools of nucleoplasmic p53, but also impairs Mdm2’s E3 ubiquitin ligase activity, thereby negatively regulating Mdm2 through two distinct mechanisms (Honda and Yasuda, 1999; Tao and Levine, 1999; Weber et al., 1999). Thus, ARF might employ a similar two-pronged approach attenuating NPM’s growth-promoting functions. As phosphorylation of human NPM by cyclin E–cdk2 was reported to be essential for the initiation of centrosome duplication in late G1 (Tokuyama et al., 2001), we considered that ARF might inhibit NPM phosphorylation in addition to retaining it in the nucleolus to arrest cell growth before S-phase entry (Weber et al., 2000).
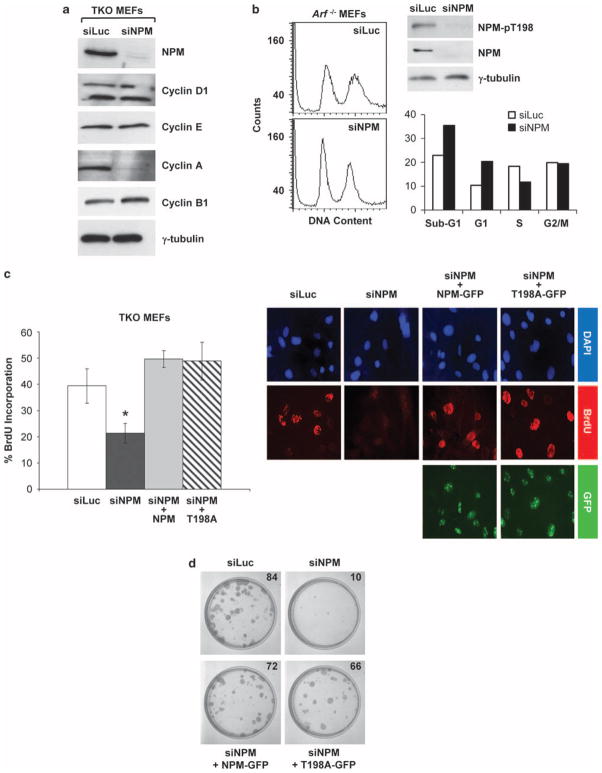
Alignment of human and mouse NPM amino acid sequences revealed 94% identity and 97% similarity. As shown in Figure 2a (lower panel), Thr199 in human NPM corresponds to Thr198 in murine NPM. A polyclonal antibody raised against a phosphopeptide surrounding Thr198 in murine NPM was generated to specifically detect phospohoThr198 in NPM (Figure 2a, underlined sequence). The phosphospecific NPM-Thr198 antibody (NPM-pT198) reacted with a protein band migrating at approximately 38 kDa in whole cell lysates from asynchronously growing triple knockout (TKO) MEFs (Arf−/− p53−/− Mdm2−/−) (Figure 2a, lane 1), but failed to detect the corresponding band in lysates from contact-inhibited TKO MEFs (Figure 2a, lane 2) or in purified recombinant NPM proteins expressed in Escherichia coli (Figure 2a, lane 3). Re-probing of this membrane with a monoclonal antibody recognizing NPM showed that a 38 kDa protein band was present in all three lanes, indicating that the polyclonal antibody reacts specifically with NPM phospho-Thr198 proteins, but does not cross-react with non-phosphorylated NPM. In addition, TKO MEFs infected with siRNAs targeting the 3′-UTR of endogenous NPM were used to show specificity of the antibody to Thr198. Phosphorylation of Thr198 was reduced at a level consistent with reduction in total NPM protein after siNPM infection (Figure 2a, right panel). Rescue of NPM knockdown with an ectopic RNA interference-resistant NPM-GFP (green fluorescent protein) protein resulted in a restoration of NPM phosphorylation at Thr198 (Figure 2a, right panel, lane 3 arrow), whereas rescue with an NPM T198A-GFP mutant resulted in a non-observable phosphorylation with the phospho-T198 antibody (Figure 2a, right panel, lane 4). This demonstrates that our NPM phospho-T198 antibody is specific for Thr198.
Figure 2.
Characterization of murine nucleophosmin-threonine 198 (NPM-Thr198) phosphorylation. (a) Whole cell lysates from actively cycling (lane 1) and contact-inhibited (lane 2) triple knockout mouse embryonic fibroblasts (TKO MEFs), and purified recombinant murine NPM proteins (lane 3) were equally loaded and separated on denaturing polyacrylamide gels, followed by western blot analysis with antibodies recognizing total NPM or phospho-Thr198 NPM (NPM-pT198). Shown is the amino acid alignment for the cyclin-dependent kinase (cdk) target region of human (upper sequence) and murine (lower sequence) NPM, with a line denoting the phosphopeptide that was used to generate the custom phosphospecific NPM-Thr198 antibody. TKO MEFs were infected with lentiviruses encoding siLuc, siNPM or siNPM + NPM-GFP (green fluorescent protein), or siNPM + NPM T198A-GFP and harvested 48-h post-infection for western blot analysis using antibodies recognizing phospho-NPM T198, NPM or GFP. The arrow points to the shifted form of NPM-pT198-GFP. (b) Low-passage (p4) TKO MEFs were synchronized into quiescence by culturing in medium containing 0.1% serum for 48 h. Cells were released into medium containing 10% serum and harvested at the indicated time points. Whole cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), and protein expression levels for cyclin D1, cyclin B1, NPM and NPM-pT198 were determined by immunoblotting with the appropriate antibodies; γ-tubulin was included as a control for equal protein loading. Levels of NPM and NPM-pT198 were quantified by densitometry and graphed as percent of asynchronous levels. Data are representative of three independent experiments (n = 3). (c) TKO MEFs were treated for 24 h with aphidicolin (1 μg/ml), nocodazole (1 μg/mol/l) or roscovitine (10 μg/mol/l), and harvested for western blot analysis using antibodies recognizing NPM, cyclin B1, γ-tubulin and phospho-NPM T198. Fold change indicates levels of phospho-NPM to total NPM after normalization to γ-tubulin. (d) TKO MEFs were infected with retroviruses encoding the SRα-MSV-tkneo vector, full-length p19ARF or a p19ARFΔ1 – 14 mutant, which lacks the NPM-binding domain. Cells were harvested and lysed at 4 days post-viral transduction, and protein lysates were separated by SDS-PAGE. Expression levels of ectopic ARF and endogenous NPM proteins were assessed by western blot analysis with the indicated antibodies, and equal protein loading was confirmed by immunoblotting for γ-tubulin.
To determine whether or not phosphorylation of murine NPM-Thr198 is a cyclin E–cdk2-specific event within the context of cell cycle progression, TKO MEFs were serum-starved and synchronized in G0, evidenced by the cells’ low expression levels of cyclin D1 protein (Figure 2b, lane 2). After release into serum, phospho-Thr198 NPM expression increased, achieving maximal levels at 24-h post-serum addition (Figure 2b). Notably, the observed increase in phospho-Thr198 NPM levels coincided with the increased expression of total NPM protein (Figure 2b). Quantitative comparison of protein band intensities confirmed that phospho-Thr198 NPM protein levels increased in parallel with total NPM protein expression. Given that cyclin D1 protein expression levels were maximal at approximately 8 h after the cells’ release into serum, yet abundant levels of phospho-T198 NPM were already evident by 4-h post-stimulation, this result suggests that cyclin E–cdk2 is not the sole kinase which phosphorylates NPM-Thr198 within the cell (Figure 2b). These data instead indicate that NPM-Thr198 seems to be constitutively phosphorylated throughout the cell cycle rising only when overall protein levels of NPM increase, and likely undergoes phosphorylation at Thr198 by one or more kinases, with overall NPM abundance being the limiting substrate. To further explore this possibility, cells were growth arrested at various points of the cell cycle. Aphidicolin-induced G1/S-phase arrest did not alter phospho-T198 compared with dimethyl sulfoxide controls (Figure 2c, lane 2). We did observe a modest increase in Thr198 phosphorylation (1.4-fold) with nocodazole treatment, consistent with an overall increase in NPM abundance (Figure 2b). Inhibition of cdk2 with roscovitine resulted in no change in Thr198 phosphorylation (Figure 2c, lane 4), suggesting that kinases other than cdk2 are quite capable of phosphorylating this residue throughout the cell cycle.
Given that ARF’s interaction with NPM represents one of its p53-independent functions, ARF’s impact on NPM-Thr198 phosphorylation in TKO MEFs was examined. Retroviral-mediated transduction of p19ARF into TKO MEFs failed to produce an appreciable change in phospho-Thr198 NPM protein levels (Figure 2d, lanes 1 and 3). TKO MEFs that were transduced to express p19ARFΔ1 – 14, a mutant lacking the NPM-binding domain (Brady et al., 2004), showed a very subtle increase in phospho-Thr198 NPM levels (Figure 2d, lane 2). In combination with the earlier result showing that NPM-Thr198 is constitutively phosphorylated, these data indicate that this particular NPM phosphorylation site is not subject to either positive (that is, cdk-mediated) or negative (that is, ARF-directed) regulation throughout the cell cycle, but is instead constantly being phosphorylated as total levels of NPM rise in the cell.
Mutation of NPM-Thr198 does not impair its oligomerization or nucleocytoplasmic shuttling
Although this current study demonstrates that ARF induction does not influence NPM-Thr198 phosphorylation (Figure 2c), our previously published findings have shown that ARF effectively blocks NPM nucleocytoplasmic shuttling, a critical function of NPM that is essential for cellular growth and proliferation (Brady et al., 2004; Yu et al., 2006). Thus, the requirement of phosphorylation of Thr198 for NPM’s nucleocytoplasmic shuttling was examined using a non-phosphorylatable alanine substitution mutant, T198A.
Given NPM’s well-documented capacity to form homo-oligomers (Liu and Chan, 1991; Yung and Chan, 1987; Namboodiri et al., 2004), the ability of ectopically-expressed T198A to hetero-oligomerize with endogenous NPM was examined. We have previously shown that NPM functional mutants often form hetero-oligomers with wild-type NPM and act as dominant-negative NPM molecules, inhibiting the function of wild-type NPM (Yu et al., 2006). Immunoprecipitation of retrovirally transduced His-tagged wild-type NPM or mutant T198A proteins from TKO MEFs, followed by NPM western blot analysis revealed that T198A formed complexes with endogenous NPM proteins, similar to ectopic wild-type NPM (Figure 3a). If NPM-T198A mutants were non-functional, we expected that they would act as dominant-negative mutants, preventing the function of endogenous wild-type NPM. Heterokaryon shuttling assays using constructs encoding either wild-type NPM or mutant T198A were then performed to answer this biological question. This experimental system assesses NPM’s capacity to shuttle between the nucleus and cytoplasm, a property that defines NPM’s role in promoting cell growth and proliferation (Yu et al., 2006; Maggi et al., 2008), demonstrated by the transit of the visibly-tagged protein of interest from a transfected human donor cell into an untransfected murine recipient cell (Tao and Levine, 1999; Yu et al., 2006). Similar to wild-type NPM (24/24, 100% shuttling), mutant NPM-T198A shuttled from the nuclei/nucleoli of transiently transfected human HeLa cells into the nuclei/nucleoli of fused, untransfected mouse NIH3T3 cells, demonstrating that NPM’s nucleocytoplasmic shuttling is not dependent on its phosphorylation at Thr198 and is not inhibited by mutant NPM-T198A molecules (Figure 3b).
Figure 3.
A non-phosphorylatable T198A nucleophosmin (NPM) mutant is not a dominant-negative mutant and displays normal nucleocytoplasmic shuttling. (a) Triple knockout mouse embryonic fibroblasts (TKO MEFs) were transduced with SRα-MSV-tkneo retroviruses encoding the control vector or His-tagged NPM proteins (wild type or T198A mutant, as indicated). Cells were harvested at 4 days post-infection, and protein complexes were immunoprecipitated on agarose beads using non-immune mouse serum (NMS) or an antibody recognizing the His tag. Washed beads were boiled in sample buffer, and proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to polyvinylidene fluoride membranes. Expression of endogenous NPM (lower band), and ectopic His-tagged wild type or T198A-mutant NPM (upper band) proteins was visualized using an antibody against NPM. (b) NIH 3T3 cells were seeded onto HeLa cells that had been cotransfected with a plasmid encoding a Myc-tagged NPC-M9-positive shuttling control and a His-tagged plasmid encoding either wild type or T198A-mutant NPM. Heterokaryon assays were carried out as described in the Materials and methods, and expression of NPC-M9, and either wild type or T198A-mutant NPM was visualized using antibodies recognizing the Myc epitope (green) and His tag (red), respectively; nuclei were demarcated with DAPI (4′,6-diamidino-2-phenylindole). Human and mouse nuclei are labeled h and m, respectively, and mouse cells are circled in white. Shuttling efficiency numbers are provided for a total of three independent experiments.
NPM knockdown impairs ribosome biogenesis and centrosome duplication, but is rescued by mutant NPM-T198A
The T198A mutant’s ability to hetero-oligomerize with endogenous NPM and shuttle to the cytoplasm could potentially mask this mutant’s true phenotype. More specifically, the T198A mutant does not display dominant-negative behavior within the cell, unlike our previously described NPMdL mutant, which blocks NPMdL-NPM hetero-oligomers from shuttling (Yu et al., 2006). To address this possibility, an NPM knockdown-rescue lentiviral construct was engineered encoding both a short hairpin RNA targeting the 3′-UTR of murine NPM (siNPM) and an siRNA-resistant cDNA corresponding to either wild type (siNPM + NPM-GFP) or mutant (siNPM + T198A-GFP) murine NPM. This strategy allowed the simultaneous reduction of endogenous NPM protein levels and ectopic expression of GFP-tagged NPM rescue proteins with high efficiency in TKO MEFs, as confirmed by NPM western blot analysis (Figure 4a).
Figure 4.
Nucleophosmin-threonine 198 (NPM-Thr198) phosphorylation is dispensable for centrosome duplication, rRNA synthesis and ribosome export. (a) Triple knockout mouse embryonic fibroblasts (TKO MEFs) were infected with green fluorescent protein (GFP)-tagged lentiviruses encoding short hairpin RNAs directed against luciferase (siLuc) or NPM (siNPM), as well siNPM lentiviruses encoding siRNA-resistant NPM wild type or T198A-mutant cDNAs (siNPM + 6NPM and siNPM + T198A, respectively). Cells were selected in puromycin for 2 days, and at 48-h post-selection, whole cell lysates were harvested, separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with an antibody recognizing NPM to verify protein knockdown and ectopic expression of wild type and T198A-mutant NPM-GFP fusion proteins (38 kDa endogenous protein and ~64 kDa NPM-GFP fusion protein); equal protein loading was confirmed by western blot analysis for γ-tubulin. (b) TKO MEFs infected with the indicated lentiviruses were re-plated onto glass coverslips, and at 96-h post-selection, were fixed and stained with an antibody recognizing γ-tubulin to permit visualization and quantitation of centrosome number per cell. Shown is the overlay of pFLRu-siNPM-NPM-GFP or pFLRu-siNPM-T198A-GFP expression (green), γ-tubulin-marked centrosomes (red) and nuclei marked with DAPI (4′,6-diamidino-2-phenylindole) (blue). For each condition, 200 cells were counted in three separate experiments, and the results from a representative experiment are shown in the bar graph. (c) At 48-h post-selection for expression of the indicated lentiviruses, cytosolic extracts from 3 × 106 TKO MEFs per condition were isolated and separated over a 7–47% sucrose gradient. Gradients were fractionated and ribosomal subunits were detected by measuring RNA absorbance at 254 nm.
Knockdown of endogenous NPM in TKO MEFs resulted in an increase in the number of cells containing a single centrosome and a concomitant decrease in the number of cells exhibiting two centrosomes (Figure 4b, black bars). A slight, but reproducible, increase in the number of cells displaying more than two centrosomes was observed, which is consistent with another group’s findings in Npm1−/− MEFs (Figure 4b, black bars) (Grisendi et al., 2005). Ectopic expression of wild-type NPM and T198A reversed some of the centrosome defects observed upon NPM loss (cells with two centrosomes), but neither was capable of limiting cells with centrosome numbers greater than two (Figure 4b, gray and hatched bars). In addition, colocalization of ectopic wild type or T198A NPM with centrosomes was not observed, although cells displaying NPM-GFP-positive nucleoli adjacent to tubulin-positive centrosomes were observed (Figure 4b, arrows).
We have previously shown that NPM nucleocytoplasmic shuttling is essential for nuclear export and for the formation of cytosolic ribosomes (Yu et al., 2006; Maggi et al., 2008). Having confirmed that the T198A mutant efficiently shuttles from the nucleolus/nucleus to the cytoplasm (Figure 3b), we next aimed to determine whether NPM-Thr198 phosphorylation is necessary for NPM’s established role in the assembly and transport of translationally competent ribosomes. We observed that knockdown of NPM in TKO MEFs produced a striking reduction in the populations of 40S, 60S and 80S cytosolic ribosomal subunits, as well as a significant attenuation in the levels of actively translating polysomes (Figure 4c). Importantly, expression of either wild-type NPM or the T198A mutant was sufficient to rescue the siNPM-induced ribosomal defect, restoring all cytosolic ribosomal populations to levels present in control siLuc-infected cells (Figure 4c). Consistent with our findings from nuclear export assays, this result demonstrates that NPM plays a critical role in ribosome biogenesis that is not dependent on its phosphorylation at Thr198.
Cell proliferation is dependent on NPM expression levels, but not its phosphorylation at Thr198
A previous study has suggested that phosphorylation of human NPM at Thr199 is necessary for proper S-phase entry and cellular proliferation (Tokuyama et al., 2001). Given that the T198A mutant was fully competent in executing NPM’s described roles in shuttling, centrosome duplication and ribosome biogenesis (Figures 3 and 4), the influence of the T198A mutant on cellular proliferation was examined. Stable knockdown of endogenous NPM in TKO MEFs severely compromised the cells’ ability to enter S-phase, as evidenced by decreased cyclin A expression (Figure 5a) and bromodeoxyuridine (BrdU) incorporation into replicating DNA (Figure 5c). Ectopic expression of either wild-type NPM or T198A-mutant siRNA-resistant proteins was sufficient to fully rescue incorporation of BrdU into the DNA of NPM knockdown cells (Figure 5c). In addition, knockdown of NPM in diploid Arf−/− MEFs resulted in a substantial increase in G1 cells (Figure 5b), suggesting that loss of NPM imposes a block before S-phase entry. To further investigate the potential long-term effects of NPM loss on cell proliferation, foci formation assays were conducted in parallel. Stable knockdown of NPM significantly inhibited foci formation by TKO MEFs, a proliferative defect that was fully reversed upon rescue with either wild-type NPM or T198A-mutant siRNA-resistant proteins (Figure 5d). Thus, these data demonstrate that phosphorylation of NPM on Thr198 is dispensable for cell cycle progression and cellular proliferation, whereas adequate NPM protein expression is essential.
Figure 5.
Nucleophosmin (NPM) expression, but not threonine 198 (Thr198) phosphorylation, is essential for cell cycle progression and cell proliferation. (a) Triple knockout mouse embryonic fibroblasts (TKO MEFs) infected with lentiviruses encoding siLuc or siNPM expression constructs were harvested 48-h post-infection for western blot analysis using antibodies recognizing NPM, cyclin D1, cyclin E, cyclin A, cyclin B1 and γ-tubulin. (b) Arf−/− MEFs infected with lentiviruses encoding siLuc or siNPM expression constructs were harvested 48-h post-infection, fixed, stained with propidium iodide and analyzed by flow cytometry. Cell cycle analysis was performed using FACSCalibur software and plotted (lower right panel). Cell lysates were harvested from duplicate plates for western blot analysis using antibodies recognizing NPM, phospho-NPM T198 and γ-tubulin. (c) TKO MEFs were infected with siNPM lentiviruses encoding siRNA-resistant NPM wild type or T198A-mutant cDNAs and at 96-h post-selection, were re-plated onto glass coverslips, allowed to adhere and pulsed with 10 μmol/l bromodeoxyuridine (BrdU). At 20 h after BrdU addition, cells were fixed, stained and quantitated for incorporation of BrdU. For each condition, 200 cells were counted in three separate experiments, and results from a representative experiment were plotted (left panel). Shown are the relative patterns for BrdU uptake (red) and NPM-GFP or T198A-GFP rescue expression (green) for a given field of cells for each condition; cell nuclei are demarcated by DAPI (4′,6-diamidino-2-phenylindole) (right panel). (d) TKO MEFs infected and selected as in (a) were re-plated in duplicates at a density of 3 × 102 per 100 mm dish. Fresh media was replenished every fourth day for a period of 12 days, at which time cells were fixed in methanol, stained with Giemsa and counted.
Discussion
A multifunctional and dynamic nucleolar phosphoprotein, NPM, has been described as a critical mediator and regulator of numerous processes within the cell, including protein chaperoning, ribosome biogenesis, centrosome duplication and genomic stability (Okuwaki et al., 2001; Okuda, 2002; Okuwaki et al., 2002; Colombo et al., 2005; Maggi et al., 2008). Given this list of disparate, but basic, cellular functions that require NPM, it is not surprising that NPM also plays essential roles in embryonic development (Grisendi et al., 2005) and cell cycle progression (Brady et al., 2004).
In support of this hypothesis, ectopic expression of NPM in immortalized fibroblasts not only increased cell size but also supplied the cell with signals that are necessary for enhanced proliferation and anchorage-independent growth. On the basis of our data and that of other groups, we propose that upregulation of NPM can promote transformation. In agreement with this idea, a subset of adult leukemias carries an NPM mutation, which encodes a second nuclear export signal at NPM’s extreme carboxy terminus (Falini et al., 2005). Further study of this mutant revealed that it dictates increased nucleocytoplasmic shuttling of NPM (Colombo et al., 2006), and our laboratory has previously shown that proper cell cycle progression requires NPM nuclear export (Brady et al., 2004). In addition, numerous laboratories (Itahana et al., 2003; Bertwistle et al., 2004; Brady et al., 2004) have demonstrated that NPM is a functional target of the nucleolar ARF tumor suppressor, implying that the transformation properties of NPM can be antagonized by the ARF tumor suppressor. The fact that we have shown NPM to be oncogenic in the absence of p53 and Arf suggests that NPM’s role in promoting transformation is not to simply antagonize these two tumor suppressors.
Previous studies have demonstrated that human NPM undergoes phosphorylation at Thr199 (Thr198 in mouse), and that cyclin E–cdk2 targets this Thr residue to relieve NPM-mediated repression of centrosome duplication and cell cycle progression (Okuda et al., 2000; Tokuyama et al., 2001). In considering this argument, one would predict that centrosome duplication would be repressed under conditions of increased NPM expression or nuclear export. However, this has not been observed in acute myelogenous leukemia patients who carry NPMc+ mutants (Falini et al., 2005) or in our current study of the cellular effects of NPM overexpression. Although intriguing, the existing model concerning the role of NPM and its phosphorylation at Thr199 in the process of centrosome duplication does not account for the mounting evidence which links NPM overexpression and nuclear export to increased cell growth and proliferation. We have provided evidence that induction of NPM protein expression is the critical limiting factor in NPM’s ability to promote cell growth and proliferation.
Our studies have revealed that ARF’s binding to NPM cannot block phosphorylation of NPM at Thr198. In addition, a non-phosphorylatable mutant of NPM, T198A, does not block cell cycle progression, centrosome duplication, nuclear export or cytosolic ribosome accumulation in the absence of endogenous wild-type NPM. Moreover, we observed that NPM-Thr198 is constitutively phosphorylated throughout the cell cycle, and any increase in Thr198 phosphorylation parallels the increase in total NPM protein expression. Although our data indicates that phosphorylation of NPM-Thr198 does not influence NPM function, we do not discount the importance of NPM in centrosome duplication. In agreement with others’ published findings from NPM knockout mice (Grisendi et al., 2005) and cell lines (Okuda et al., 2000; Tokuyama et al., 2001), we have shown that loss of NPM deregulates centrosome duplication. However, we propose that this might be a downstream effect, which may not be directly mediated by NPM. In cells undergoing acute NPM loss, we observed a decrease in the number of actively translating ribosomes at time points (48 h) preceding the observed defects in centrosome duplication and S-phase entry (96–120 h). Therefore, our data supports a model in which NPM’s direct command over ribosome biogenesis and protein translation could result in indirect changes in a downstream target that plays a critical role in the process of centrosome duplication. Thus, translational targets of the ribosome might in turn also promote cellular proliferation and transformation.
Materials and methods
Cell culture
The Arf−/− MEFs, Arf−/−/p53−/−/Mdm2−/− MEFs (TKO MEFs, provided by Gerard Zambetti, St Jude Children’s Research Hospital), NIH3T3 and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 0.1 mM non-essential amino acids and 100 U each of penicillin and streptomycin. TKO MEFs were synchronized into quiescence by culturing at sub-confluency in medium supplemented with 0.1% fetal bovine serum for 48 h.
Plasmid constructs
The pSRα-MSV-tkneo retroviral expression vectors encoding p19ARF, p19ARFΔ1 14 and full-length murine NPM were used as described previously (Brady et al., 2004). The His-T198A NPM mutant was amplified from pET28a-NPM using the following mutagenic primers: 5′-ATCTGTACGAGATGCA CCAGCCAAAAATGC-3′ (sense) and 5′-GTGCATTTTTGG CTGGTGCATCTCGTACAG-3′ (antisense). The resultant His-T198A cDNA was sub-cloned into pcDNA3.1 using EcoRI and BamHI, and into pSRα-MSV-tkneo using EcoRI; pcDNA3.1-Myc-NPC-M9 was gift from J Alan Diehl (University of Pennsylvania, USA). The pFLRu-GFP-siLuc and pFLRu-GFP-siNPM vectors were provided by Gregory Longmore (Washington University, USA) (Pelletier et al., 2007). To generate the pFLRu-siNPM-NPM-GFP and pFLRu-siNPM-T198A-GFP rescue constructs, murine cDNAs encoding wild type or T198A-mutant NPM were sub-cloned into the EcoRI and BamHI sites of the pFLRu-GFP-siNPM vector. The lentiviral envelope and packaging vectors, pHCMV.G and CMVΔR8.2, were gifts from Sheila Stewart (Washington University).
Virus production and infection
Retroviral production and infection using pBabe-H-RasV12 and SRα-MSV-tkneo vectors were carried out according to methods described previously (Brady et al., 2004; Roussel et al., 1995). Lentiviruses encoded by the pFLRu-GFP vectors were packaged in 293T cells after cotransfection of the pHCMV.G, CMVΔR8.2 and pFLRu-GFP lentiviral vectors using Fugene 6 (Roche, Indianapolis, IN, USA). Primary MEFs were infected for 4 h with freshly harvested lentiviral supernatants in the presence of 8 μg/ml protamine sulfate, and at 24-h post-infection, puromycin (2 μg/ml) was added to the cells for a selection period of 48 h where appropriate.
Flow cytometry
The Arf−/− MEFs were infected with retroviruses encoding the control vector, His-NPM or RasV12, and were harvested at 72 h. Cells were fixed and resuspended in 1X phosphate-buffered saline/1% fetal bovine serum with or without propidium iodide before analysis using a FACSCalibur (Becton Dickson, Rockville, MD, USA).
Foci formation
Mouse embryonic fibroblasts were infected with lentiviral expression supernatants and were seeded (2 × 103) onto 100 mm dishes. Cells were grown for 14 days in complete medium, fixed in 100% methanol and stained for 30 min with 50% Giemsa.
Soft agar colony formation
The p53−/− MEFs were infected with control vector, His-NPM, His-NPM-T198A or RasV12 retroviruses, and were seeded (1 × 103) in triplicates onto 60 mm dishes. Colonies were allowed to grow for 14 days in complete medium supplemented with fetal bovine serum and Noble Agar.
Immunohistochemistry using the common cancer tissue array
The TARP4 tissue array was purchased from NCI Tissue Array Research Project. The tissues used to construct arrays were obtained from the Cooperative Human Tissue Network (CHTN). Each tissue array slide contained 600 samples. De-paraffinized tissue sections were first treated with 3% H2O2 for 30 min followed by antigen retrieval by heating in citra plus solution (BioGenex, San Ramon, CA, USA). After subjecting to avidin block, biotin block and power block for 15 min, the sections were incubated with mouse anti-NPM antibody (Zymed, San Francisco, CA, USA) for 1 h. After further incubation with biotinylated multi-link antibody for 45 min and peroxidase-labeled streptavidin for 30 min, the staining was developed by reaction with 3,3′-diaminobenzidine tetrahydrochloride substrate–chromogen solution.
Karyotyping analysis
The Arf−/− MEFs were infected with control vector or His-NPM retroviruses, and at 72-h post-infection were treated with colcemid (10 μg/ml) for 16 h. Cells were harvested in 75 mM KCl for 6 min at 37 °C. Cells were fixed in methanol:acetic acid (3:1) and washed. The cells were resuspended in 2 ml fixative and one drop was allowed to fall onto frosted glass slide. DNA was stained with DAPI (4′,6-diamidino-2-phenylindole) and fluorescent signals were detected.
Immunoprecipitation and western blot analysis
Immunoprecipitation of cell lysates was performed as previously described (Brady et al., 2004). Antibodies recognizing γ-tubulin, cyclin D1, His (Santa Cruz, Santa Cruz, CA, USA), p19ARF (Abcam, Cambridge, MA, USA), NPM (Zymed) and NPM (custom rabbit polyclonal, Sigma, St Louis, MO, USA) were used in western blot analyses. The custom phosphospecific polyclonal antibody recognizing phospho-NPM (Thr198) was generated commercially (Zymed) and raised against the following phosphopeptide: CSVRDpTPAKN (Tufts University Peptide Core).
Heterokaryon assay
The HeLa cells (2 × 105) were seeded onto glass cover slips in six-well dishes and transfected with constructs encoding either His-tagged wild type or T198A-mutant NPM in combination with a Myc-tagged NPC-M9 plasmid (a gift from J Alan Diehl, University of Pennsylvania). Heterokaryon assays were performed as previously described (Yu et al., 2006).
Indirect immunoflourescence
The Arf−/− or TKO MEFs were infected with SRα-MSV-tkneo retroviruses or pFLRu-GFP lentiviruses as indicated, and seeded onto glass cover slips. Cells were washed with phosphate-buffered saline, fixed at room temperature using 10% formalin/10% methanol, followed by 1% NP-40 in phosphate-buffered saline for 5 min at room temperature. Cells were stained with an antibody recognizing γ-tubulin (Sigma), followed by FITC or rhodamine X-conjugated immunoglobulins. Nuclei were counterstained with DAPI.
BrdU incorporation
The Arf−/− or TKO MEFs were infected with SRα-MSV-tkneo retroviruses or pFLRu-GFP lentiviruses as indicated. Cells were seeded onto glass cover slips and subjected to BrdU incorporation analysis (Brady et al., 2004).
Ribosome fractionation
At 4 days post-infection with pFLRu-GFP lentiviruses, TKO MEFs were subjected to ribosome fractionation analysis (Maggi et al., 2008).
Acknowledgments
We are indebted to Sheila Stewart, Gregory Longmore, J Alan Diehl, Martine Roussel, Charles Sherr and Gerard Zambetti for gifts of plasmid constructs, antibodies and primary TKO MEFs. In addition, we would like to thank Sheila Stewart, Helen Piwnica-Worms, Michael Tomasson and John Majors for insightful discussions throughout the course of this study. SNB was supported by the Cancer Biology Pathway. CLP was a trainee in the Lucille P Markey Special Emphasis Pathway in Human Pathobiology. JDW was funded through the National Institutes of Health and Department of Defense Era of Hope Scholar Award in Breast Cancer Research.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SN, Yu Y, Maggi LB, Jr, Weber JD. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha H, Hancock C, Dangi S, Maiguel D, Carrier F, Shapiro P. Phosphorylation regulates nucleophosmin targeting to the centrosome during mitosis as detected by cross-reactive phosphorylation-specific MKK1/MKK2 antibodies. Biochem J. 2004;378:857–865. doi: 10.1042/BJ20031173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, Zamponi R, Marine JC, et al. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol. 2005;25:8874–8886. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L, et al. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 2006;66:3044–3050. doi: 10.1158/0008-5472.CAN-05-2378. [DOI] [PubMed] [Google Scholar]
- Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- Gao H, Jin S, Song Y, Fu M, Wang M, Liu Z, et al. B23 regulates GADD45a nuclear translocation and contributes to GADD45a-induced cell cycle G2-M arrest. J Biol Chem. 2005;280:10988–10996. doi: 10.1074/jbc.M412720200. [DOI] [PubMed] [Google Scholar]
- Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Minamino N, Nagamura-Inoue T, Matsumoto M, Taniguchi T, Tanaka N. Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene. 1997;15:1275–1281. doi: 10.1038/sj.onc.1201286. [DOI] [PubMed] [Google Scholar]
- Li YP. Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J Virol. 1997;71:4098–4102. doi: 10.1128/jvi.71.5.4098-4102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HT, Yung BY. in vivo interaction of nucleophosmin/B23 and protein C23 during cell cycle progression in HeLa cells. Cancer Lett. 1999;144:45–54. doi: 10.1016/s0304-3835(99)00184-6. [DOI] [PubMed] [Google Scholar]
- Liu QR, Chan PK. Formation of nucleophosmin/B23 oligomers requires both the amino- and the carboxyl-terminal domains of the protein. Eur J Biochem. 1991;200:715–721. doi: 10.1111/j.1432-1033.1991.tb16236.x. [DOI] [PubMed] [Google Scholar]
- Maggi LB, Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, et al. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol Cell Biol. 2008;28:7050–7065. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namboodiri VM, Schmidt-Zachmann MS, Head JF, Akey CW. Purification, crystallization and preliminary X-ray analysis of the N-terminal domain of NO38, a nucleolar protein from Xenopus laevis. Acta Crystallogr D Biol Crystallogr. 2004;60:2325–2327. doi: 10.1107/S0907444904023959. [DOI] [PubMed] [Google Scholar]
- Okuda M. The role of nucleophosmin in centrosome duplication. Oncogene. 2002;21:6170–6174. doi: 10.1038/sj.onc.1205708. [DOI] [PubMed] [Google Scholar]
- Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001;506:272–276. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- Okuwaki M, Tsujimoto M, Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol Biol Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier CL, Maggi LB, Jr, Brady SN, Scheidenhelm DK, Gutmann DH, Weber JD. TSC1 Sets the rate of ribosome export and protein synthesis through nucleophosmin translation. Cancer Res. 2007;67:1609–1617. doi: 10.1158/0008-5472.CAN-06-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel MF, Theodoras AM, Pagano M, Sherr CJ. Rescue of defective mitogenic signaling by D-type cyclins. Proc Natl Acad Sci USA. 1995;92:6837–6841. doi: 10.1073/pnas.92.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Tarapore P, Tokuyama Y, George KR, Fukasawa K. Characterization of centrosomal association of nucleophosmin/B23 linked to Crm1 activity. FEBS Lett. 2005;579:6621–6634. doi: 10.1016/j.febslet.2005.10.057. [DOI] [PubMed] [Google Scholar]
- Spector DL, Ochs RL, Busch H. Silver staining, immunofluorescence, and immunoelectron microscopic localization of nucleolar phosphoproteins B23 and C23. Chromosoma. 1984;90:139–148. doi: 10.1007/BF00292451. [DOI] [PubMed] [Google Scholar]
- Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, et al. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Budhu A, Forgues M, Wang XW. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat Cell Biol. 2005;7:823–830. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- Weber JD, Jeffers JR, Rehg JE, Randle DH, Lozano G, Roussel MF, et al. p53-independent functions of the p19 (ARF) tumor suppressor. Genes Dev. 2000;14:2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Winey M. Cell cycle: driving the centrosome cycle. Curr Biol. 1999;9:R449–R452. doi: 10.1016/s0960-9822(99)80279-6. [DOI] [PubMed] [Google Scholar]
- Yang C, Maiguel DA, Carrier F. Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 2002;30:2251–2260. doi: 10.1093/nar/30.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maggi LB, Jr, Brady SN, Apicelli AJ, Dai MS, Lu H, et al. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol Cell Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung BY, Chan PK. Identification and characterization of a hexameric form of nucleolar phosphoprotein B23. Biochim Biophys Acta. 1987;925:74–82. doi: 10.1016/0304-4165(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Zatsepina OV, Rousselet A, Chan PK, Olson MO, Jordan EG, Bornens M. The nucleolar phosphoprotein B23 redistributes in part to the spindle poles during mitosis. J Cell Sci. 1999;112:455–466. doi: 10.1242/jcs.112.4.455. [DOI] [PubMed] [Google Scholar]